Abstract
Objective
Although counting of circulating tumour cells (CTC) has attracted a broad interest as potential markers of tumour progression and treatment response, the lack of functional characterisation of these cells had become a bottleneck in taking these observations to the clinic. Our objective was to culture these cells in order to understand them and exploit their therapeutic potential to the full.
Design
Here, hypothesising that some CTC potentially have cancer stem cell (CSC) phenotype, we generated several CTC lines from the blood of patients with advanced metastatic colorectal cancer (CRC) based on their self-renewal abilities. Multiple standard tests were then employed to characterise these cells.
Results
Our CTC lines self-renew, express CSC markers and have multilineage differentiation ability, both in vitro and in vivo. Patient-derived CTC lines are tumorigenic in subcutaneous xenografts and are also able to colonise the liver after intrasplenic injection. RNA sequencing analyses strikingly demonstrate that drug metabolising pathways represent the most upregulated feature among CTC lines in comparison with primary CRC cells grown under similar conditions. This result is corroborated by the high resistance of the CTC lines to conventional cytotoxic compounds.
Conclusions
Taken together, our results directly demonstrate the existence of patient-derived colorectal CTCs that bear all the functional attributes of CSCs. The CTC culture model described here is simple and takes <1 month from blood collection to drug testing, therefore, routine clinical application could facilitate access to personalised medicine.
Clinical Trial Registration
ClinicalTrial.gov NCT01577511.
Keywords: COLORECTAL CANCER, LIVER METASTASES, DRUG TOXICITY
Significance of this study.
What is already known on this subject?
Circulating tumour cells (CTCs) contain key prognostic markers for patients with metastatic colorectal cancer (CRC).
CTCs are scarce among blood cells and they are also heterogeneous.
Functional characterisation of CTCs is thus needed.
In vitro CTC models are lacking in the CRC field.
What are the new findings?
CTC lines contain functional cancer stem cells.
CTC lines are genetically and phenotypically heterogeneous.
Identification of gene subset commonly enriched in cultured CTC of the present study and previously published CTCs from colon and other cancers.
CTC lines express high levels of drug metabolism genes and are resistant to conventional therapies.
How might it impact on clinical practice in the foreseeable future?
This study is the first experimental demonstration that CTCs isolated from patients with CRC express cancer stem cell phenotype and can be used to determine drug sensitivity thus, culturing CTCs could drive a personalised approach to patients with metastatic CRC.
Introduction
Circulating tumour cells (CTCs) are commonly present in the blood of solid cancer patients,1 transit through the bloodstream and constitute seeds for subsequent metastasis development in distant organs.2 This process is responsible for the vast majority of deaths from colorectal cancer (CRC),3 making it the third leading cause of cancer death in the developed world. In recent years, CTCs have attracted interest as a precious tool to better understand mechanisms underlying metastatic progression and also as clinically relevant prognostic markers, since the number of CTCs has been correlated with poor prognosis notably in patients with CRC.4
Two important obstacles currently hamper our ability to gain deeper understanding of CTCs: their heterogeneity and scarcity. These problems have recently been partially overcome by single cell analyses such as RNA or exon sequencing.5 6 While these studies did not address the functional aspects of CTC biology, they did identify different CTC subpopulations within a single blood sample.7 Heterogeneity of CTCs has been demonstrated at the phenotypic level in breast cancer.8 In CRC, potential CTC markers such as plastin 3 have been proposed but are yet to be validated,9 and aneuploidy has been used to detect CTCs that undergo epithelial to mesenchymal transition.10 Although the scarcity of CTCs has restricted the number of functional studies, subpopulations of metastasis-initiating breast cancer CTCs11 and tumorigenic lung cancer CTCs12 have been described in vivo, and molecular characterisation studies have suggested that CTC-driven metastatic progression might rely upon their cancer stem cell (CSC) properties.13 Culturing human CTCs would overcome the difficulty of characterising these rare cells and allow both researchers and clinicians to study them. Accordingly, recent publications in several cancers14–17 have described in vitro CTC culture models. However, for CRC research, thorough general functional characterisation of CTCs still represents a major challenge as systemic CTC number is particularly low compared with other solid cancers.18
In order to functionally characterise colorectal CTCs, we developed CTC lines from several patients with metastatic CRC, by growing them under conditions that promote the survival of self-renewing cells. Our CTC lines were compared with some of the established patient-derived cells isolated from primary tumours and liver metastases in our team; and grown under the same conditions. We demonstrate that CTC lines contain cells that have the functional characteristics of CSCs as they have maintained their self-renewal and multilineage differentiation properties. These cells robustly express CSC markers and were able to initiate metastasis development in vivo. Strikingly, we found clear overexpression of genes involved in xenobiotic resistance in our CTC lines and furthermore our cytotoxicity assay corroborates the potential usefulness of this model to predict patient drug response for individual patients.
Results
Establishment of tumorigenic colorectal CTC lines
Three CTC lines from four attempts were established (CTC41, CTC44 and CTC45) from chemotherapy-naïve patients with metastatic CRC (stage IV), while attempts from patients with lower stage CRC or chemotherapy treated were unsuccessful. Details about efficiency rates of CTC culture are provided in online supplementary table S1. All described experiments were performed on cells grown as spheres in suspension to promote the survival of CSCs (figure 1A). Indeed, suspension cultures include absence of serum in order to decrease cell differentiation and maintenance of isolated cells, which is restricted to cancer cells with an undifferentiated phenotype.
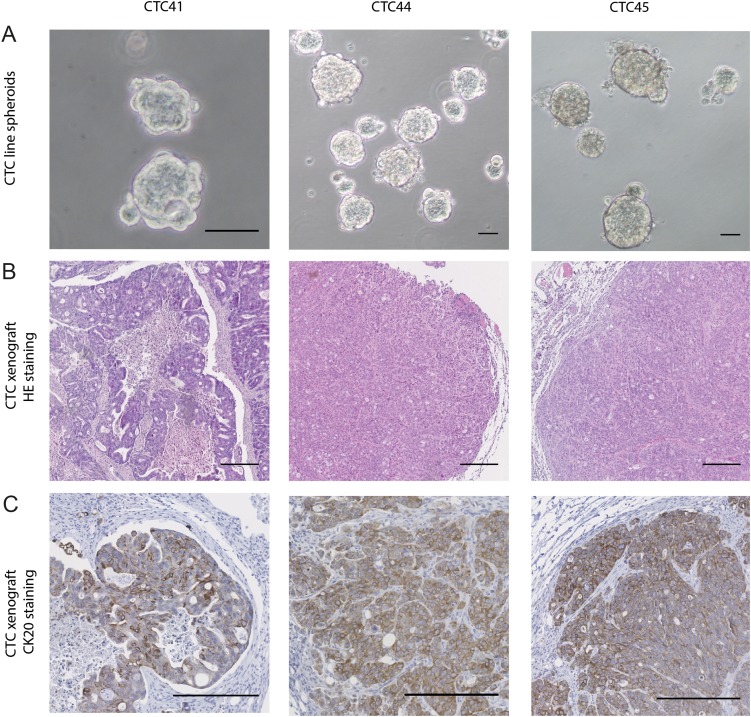
Figure 1.
(A) Images of spheroids formed by circulating tumour cell (CTC)41, CTC44 and CTC45 lines (scale bar 50 μm). (B) H&E staining on tumours following subcutaneous injections of CTC lines into nude mice (scale bar 250 µm). (C) CK20 staining on tumours following subcutaneous injections of CTC lines.
gutjnl-2016-311447supp001.pdf (56KB, pdf)
To ascertain the tumorigenicity of CTC lines and their origin, we injected them subcutaneously in the flank of nude mice and we showed that all CTC lines were able to initiate tumours. Histological examination of dissected tumour xenografts was performed by a clinical pathologist and showed the characteristics of typical invasive colorectal adenocarcinoma with both proliferative and necrotic areas (figure 1B), which was validated with CK20 staining (figure 1C). H&E staining on primary and metastasis biopsies from patients 44 and 45 are shown in online supplementary figure S1.
gutjnl-2016-311447supp007.pdf (1.1MB, pdf)
CTC cell lines contain multipotent cells responsible for phenotypic heterogeneity
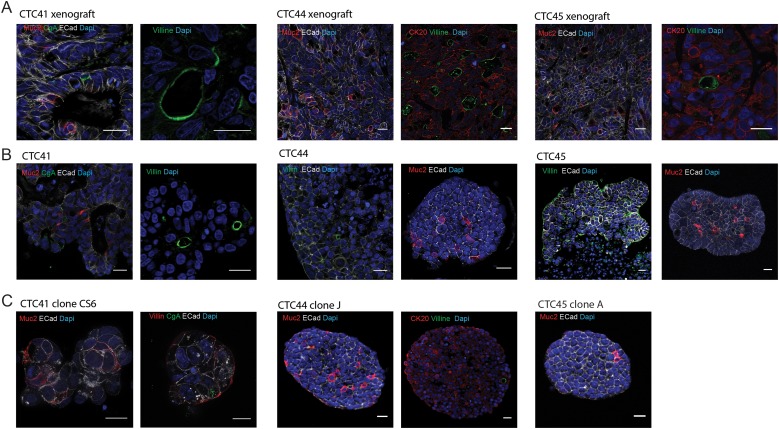
We then observed that the three CTC lines were able to differentiate towards all three main intestinal lineages in vivo (figure 2A) and in vitro within spheres (figure 2B). Indeed, terminally differentiated cells expressing markers of enteroendocrine-like cells (chromogranin-A), goblet cells (mucin-2) and enterocyte cells (villin) were represented within CTC spheres and CTC-derived xenografts. To determine whether the presence of cells with multiple different phenotypes emerged from the presence of cells with multipotent ability within these cell lines, we amplified several clones established from single cells. Multiple lineages were also represented in several of these single cell-derived clones (figure 2C), demonstrating that phenotypic heterogeneity in these patient-derived CTC populations emerges from the presence of multipotent cells, which strongly suggests that CSCs are present in these cell populations.
Figure 2.
(A) Immunofluorescent staining of tumour xenografts obtained after subcutaneous injection of circulating tumour cell (CTC) lines into the flank of nude mice (scale bar 20 μm). (B) Immunofluorescent staining of tumour spheres formed in vitro from CTC lines (scale bar 20 μm). (C) Immunofluorescent staining of representative tumour spheres derived from single-cell clones of CTC lines (scale bar 20 μm). Names of stained intestinal and epithelial markers are specified within each photograph in the corresponding colour. E-cadherin (ECad) and cytokeratin 20 (CK20) are epithelial markers. Mucin 2 (Muc2) stains goblet cells, villin stains enterocytes and chromogranin A (CgA) stains enteroendocrine cells.
CTC lines display hallmarks of CSCs
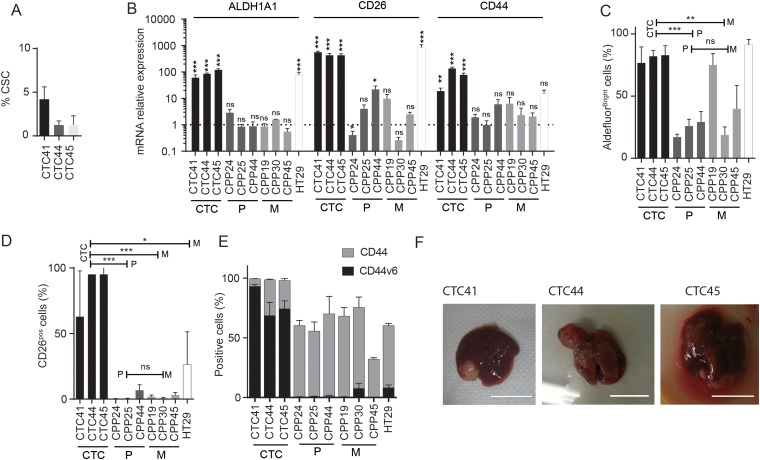
We then determined that the CTC lines had the ability to self-renew over long periods (20 passages) when grown as spheroids in serum-free medium at very low density. Using extreme limiting dilution analysis19 on spheres that were passaged at least 3 times, we quantified CSC frequency and found that CTC41, CTC44 and CTC45, respectively contained 4.2, 1.3 and 1.2% self-renewing cells (figure 3A). In following experiments, we compared CTC lines with cell lines freshly established from primary tumours (CPP24, CPP25 and CPP44) or liver metastasis biopsies (CPP19, CPP30 and CPP45) and grown in suspension under similar conditions (table 1). Importantly, the CTC44 and CTC45 lines were isolated from the same patients, respectively, as CPP44 and CPP45. Clinical data for each of these patients are detailed in the online supplementary table 2. We also used HT29, a cell line known to have a strong CSC phenotype.20
Figure 3.
(A). Cancer stem cell (CSC) frequency quantified in circulating tumour cell (CTC) lines after more than seven passages as tumour spheres using the extreme dilution assay (ELDA). Presence or absence of spheres is quantified as a binary outcome and stem cell frequency is calculated,19 and expressed as percentage of total cell number. (B) Expression of mRNAs encoding CSC markers such as ALDH1A1, CD26 and CD44, measured using reverse transcription-quantitative PCR analysis in CTC lines and cells derived from primary colon tumours (P) or liver metastases (M) of patients with CRC. Expression of mRNAs is expressed relative to the mean expression level across all primary and metastatic tumour-derived cell lines (which was set to 1). Results are expressed as mean±SEM, n>3, statistical analyses were performed using a Mann–Whitney U test. (C) Percentage of cells with high ALDH-activity in CTC lines, HT29 and tumour-derived cell lines, quantified using the Aldefluor assay kit (STEMCELL Technologies) and measured by flow cytometry. (D) Percentage of CD26-positive cells in CTC lines, HT29 and tumour-derived cell lines quantified by flow cytometry. (C and D) Results are expressed as mean±SEM, n>3, statistical analysis: Mann–Whitney U test comparing the mean value of each group of cells lines (CTC, P and M). (E) Percentage of CD44-positive (grey bars) and CD44 v6-positive (black bars) cells in CTC, HT29 and tumour-derived cell lines analysed by flow cytometry. (F) Photographs of liver metastases formed after intrasplenic injection of CTC lines in NOD/SCID mice (scale bar 1 cm).
Table 1.
Origin of different tumour patient-derived cell lines and the potential treatment given to patients before sampling
| Patient number | Patient-derived cell line name | Origin | Treatment before sampling |
|---|---|---|---|
| 41 | CTC41 | Blood | None |
| 44 | CTC44 | Blood | None |
| CPP44 | Primary tumour | None | |
| 45 | CTC45 | Blood | None |
| CPP45 | Liver metastasis | None | |
| 24 | CPP24 | Primary tumour | Bevacizumab Cetuximab FOLFIRI |
| 25 | CPP25 | Primary tumour | Capecitabine |
| 19 | CPP19 | Liver metastasis | Bevacizumab FOLFIRI |
| 30 | CPP30 | Liver metastasis | Bevacizumab FOLFOX4 FOLFIRI Xelox |
CTC, circulating tumour cell.
gutjnl-2016-311447supp002.pdf (67.2KB, pdf)
To validate the strong CSC phenotype of our CTC lines, we focused on aldehyde dehydrogenase (ALDH1A1) expression and enzymatic activity, which has been suggested as a specific marker for the CSC/progenitor population in CRC.21 Our results showed that ALDH1A1 mRNA was highly expressed in CTC lines compared with other tumour patient-derived cell lines (figure 3B, left) and the majority of CTCs within the lines had strong ALDH activity (figure 3C). We also found strong expression of putative CSC markers within our CTC lines such as CD133 and EpCAM (data not shown).
Together with the multipotent differentiation ability demonstrated above, these results represent the first functional demonstration in cancer that CTCs do contain CSCs.
CTC lines are endowed with strong metastatic potential
We then quantified the expression of two markers specifically associated with CSCs with metastatic potential in CRC: CD26 and CD44v6.22 23 CD26 was highly expressed in CTC lines both at the mRNA and protein levels (figure 3B,D) and CD44v6 expression was strongly enriched in CTC lines, while barely any expression was detected in other patient-derived tumour cell lines (figure 3E). In contrast, the overall protein expression of CD44 (all isoforms) was not higher in CTC cell lines, although RNA was enriched (figure 3B). In addition, immunostaining for CD44V6 (as well as for the CSC marker ALDH) (figure 3E) was strong in tumours grown after subcutaneous CTC injection in immunocompromised mice (see online supplementary figure S2). To functionally validate the metastatic potential in vivo, we injected CTC lines independently in the spleen of nude mice. Within 4 weeks, all CTC lines injected led to the formation of metastasis in the liver and in the lung also for CTC45 (figure 3F).
CTC lines are genetically heterogeneous
In line with the study by Yu et al describing mutation differences between primary tumour and CTC in breast cancer,14 next-generation sequencing analysis performed on four frequently mutated genes demonstrated that some cancer-associated mutations or variants were different between CTC lines and the respective primary tumour and metastasis (table 2). Strikingly, CTC44 and CTC45 lines were carrying the BRAF V600E mutation, whereas primary tumours and metastasis were diagnosed as KRAS G12V-mutated for patient 44 and KRAS G12D-mutated for patient 45, which means that the BRAF V600E mutation was absent from these samples. The presence of the BRAF V600E mutation in the CTC lines was confirmed by pyrosequencing. Additional staining of tissue sampled from areas of the primary and liver metastasis (different from those used for sequencing) revealed the presence of small BRAF V600E-positive areas (see online supplementary figure S3).
Table 2.
Variants detected in patient tumour and metastasis and CTC lines
| Patient | Variant | Tumour | CTC line | Metastasis |
|---|---|---|---|---|
| 44 | EGFR Q787Q | Yes | Yes | Yes |
| KRAS G12V | Yes | No | Yes | |
| PIK3CA E545K | Yes | No | Yes | |
| BRAF V600E | No | Yes | No | |
| 45 | EGFR Q787Q | Yes | Yes | Yes |
| KRAS G12D | Yes | No | Yes | |
| BRAF V600E | No | Yes | No |
CTC, circulating tumour cell.
To further analyse the intra cell line heterogeneity, we used the CTC41 cell line, which homogeneously carries the heterozygous BRAF V600E mutation (see online supplementary figure S4), identical to that detected in the patient tumour (not shown). We generated 10 CTC41 subclones from single cells and analysed their genomic DNA by next-generation sequencing using the pan-cancer integrated DNA technologies (IDT) panel, which includes 2290 target regions covering 0.8 MB of primarily exonic regions. Five hundred six high-confidence variants (342 heterozygous, 164 homozygous) spanning 95 genes were identified in all CTC41 subclones, including multiple well-recognised colon cancer-associated genes (see online supplementary table S3), consistent with the CRC origin of these cells. In addition, we detected a hemizygous androgen receptor variant (NM_000044.3:c.1617-7T>G) in one of the ten subclones (see online supplementary figure S5). This result indicates that genetic heterogeneity is present within the CTC41 cell line, and is likely to underestimate the actual heterogeneity level as the IDT panel only covers less than 0.03% of the human genome.
gutjnl-2016-311447supp003.pdf (167.2KB, pdf)
The mRNA expression profile of human colon CTC lines reveals similarities with CTCs from other cancers and is enriched for xenobiotics metabolism genes
We then performed RNA sequencing on biological triplicates of our three CTC lines as well as of three primary colorectal tumour-derived cell lines grown using a similar approach, two derived in our laboratory (CPP1 and CPP44) and one commercially available (DLD-1). Using unsupervised clustering we found that the three CTC lines clustered together, away from the primary tumour cells (see online supplementary figure S6). A total of 6096 genes were differentially expressed in CTCs compared with primary CRC-derived cells using two differential expression-testing packages (DESeq2 and Voom, see the Materials and methods section,24–26). This included 2791 (45.8%) upregulated genes and 3305 downregulated genes (54.2%) (see online supplementary figure S6).
Further analysis of RNASeq results for the CTC44 cell line and cells from the matching primary tumour (CPP44) corroborated the common origin of these samples while suggesting that CTCs circulating in this patient only represented a fraction of cells from the primary tumour (see online supplementary results and discussion ).
gutjnl-2016-311447supp008.pdf (259.8KB, pdf)
gutjnl-2016-311447supp009.pdf (271.7KB, pdf)
This differentially expressed gene list was then compared with four previously published studies that identified ‘CTC-specific’ gene sets in melanoma, breast, prostate and CRCs,27–30 in order to determine the potential level of overlap between our and their list of differentially expressed genes gene. As shown in online supplementary table S4, differentially upregulated genes from the present study were enriched in the gene sets from three of these four studies, excluding that from melanoma, suggesting that CTCs emerging from very different cancers may share some key characteristics and that some of these characteristics are still detectable in our cultured CTCs. Six genes were detected as differentially expressed in all four studies (including the present one): AGR2, CEACAM5, CLDN3, CK18, EpCAM and FGFR3.
gutjnl-2016-311447supp004.pdf (169.5KB, pdf)
gutjnl-2016-311447supp005.pdf (811.3KB, pdf)
gutjnl-2016-311447supp006.pdf (85.9KB, pdf)
In addition, the list of genes differentially expressed in CTCs versus primary tumour-derived cells was used to perform a generally applicable gene set enrichment for pathway analysis.31 The most striking upregulated feature distinguishing CTC lines from primary tumour-derived cell related to their metabolic activity, highlighting their enhanced drug/xenobiotics metabolising activity, in particular via cytochrome P450 pathway (p=0.0109, see online supplementary figure S7), suggesting that these cells may display enhanced resistance to conventional cytotoxic compounds.
Drug sensitivity on CTC lines as a potential prediction tool for personalised medicine
As demonstrated above, xenobiotic resistance was the most represented common pathway within CTC lines and isolated CTCs have been suggested in breast cancer as a good predictive model to screen potential alternative drug treatments in order to select those that are the most likely to be effective.14 However, in order to inform therapeutic decisions by clinical oncologists, such screening approaches must be conducted within a short timeframe after blood sample collection. Using our approach we were able to generate sufficient cellular material (5 million cells) within 3 weeks of sample collection, allowing us to perform cytotoxicity assays. As a proof of concept for CTCs from patients with CRC, we quantified the sensitivity of our CTC lines to an in vitro cytotoxic regimen inspired by standard-of-care chemotherapy combinations for patients with CRC (FIRI: 5-fluorouracil (5-FU) and SN-38, the active metabolite of irinotecan). Overall, we found that CTC lines were significantly more resistant to FIRI than primary and metastatic tumour-derived cells grown under the same conditions (figure 4A).
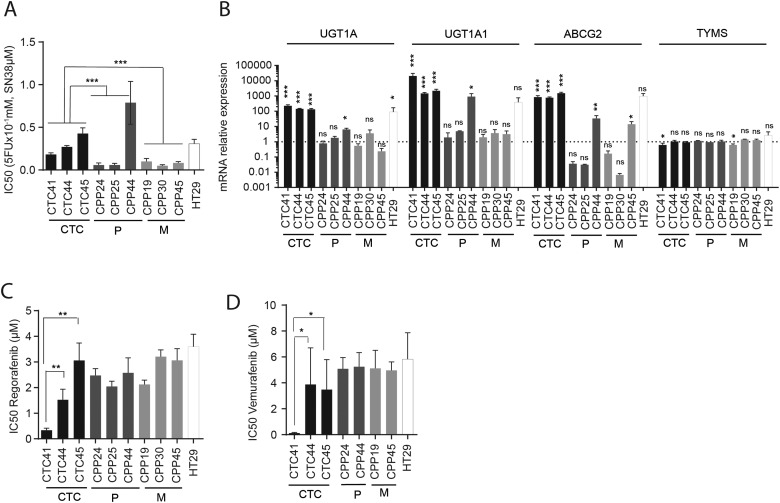
Figure 4.
(A) IC50 of 5-FU + SN-38 (active metabolite of irinotecan), a common combination of chemotherapies, on the cell viability of circulating tumour cell (CTC) lines, primary (P) or metastatic (M) tumour-derived cell lines and HT29, quantified using the Cell Titer Glow assay. Results are expressed as mean±SEM, n>3, statistical analysis: Mann–Whitney U test comparing the mean value of each group of cells lines (CTC, P and M). (B) Relative expression of mRNAs encoding proteins involved in chemotherapy resistance (UGT1A, UGT1A1, MDR1, ABCG2 and TYMS) quantified by qPCR on CTC lines, P or M tumour-derived cell lines. Expression of mRNAs is expressed relative to the mean expression level across all primary and metastatic tumour-derived cell lines, which was set to 1. Results are expressed as mean±SEM, n>3, statistical analysis was performed by Mann–Whitney U test. (C) IC50 of regorafenib (multikinase inhibitor) on the cell viability of CTC lines, P or M tumour-derived cell lines and HT29, quantified using the Cell Titer Glow assay. (D) IC50 of vemurafenib (BRAF inhibitor) on the cell viability of CTC lines, P or M tumour-derived cell lines and HT29. Results are expressed as mean±SEM with n>3. Statistical analysis: Mann–Whitney U test comparing mean of each cell lines subgroup (A) or each cell line (B and C).
We then confirmed by reverse transcription-quantitative PCR that CTC lines displayed a high expression of genes associated with irinotecan resistance, including UGT1A isoforms and ABCG2, in comparison with other patient-derived cell lines. In contrast, no difference was observed for the expression of TYMS (involved in 5-FU metabolism) (figure 4B).
Finally, we tested the sensitivity of our CTC lines to the multikinase inhibitor regorafenib and the BRAF V600 inhibitor vemurafenib (figure 4C,D). For regorafenib, a multikinase inhibitor approved by the US Food and Drug Administration and European Medicines Evaluation Agency for treatment-refractory patients with CRC, we found that sensitivity varied greatly in all samples tested, with CTC41 the most sensitive to this compound (figure 4C). Since each of the CTC lines carried the V600E BRAF mutation, we also tested the toxicity of the V600E BRAF inhibitor vemurafenib, on these cells. Despite the fact that vemurafenib is demonstrated to be poorly efficient on BRAF-mutated CRCs,32 CTC41 was found to be also very sensitive to this inhibitor (figure 4D).
Discussion
Here, we established three CTC lines displaying CSC phenotype, metastatic potential and phenotypic and genetic heterogeneity. These CTC lines have specific drug metabolising abilities compared with patient-derived CRC cells as proved with RNA sequencing analysis.
The CTC lines display phenotypic heterogeneity which is shown here by the presence of different intestinal lineages in vitro and in vivo, validating both the origin of these lines and the robustness of the described model. Overall, our data provide the first experimental demonstration that cells with multilineage differentiation potential circulate in the blood of patients with cancer and suggest that they are the likely source of CSCs in CRC metastases.33
Using single cell clone experiments, we have demonstrated that this phenotypic heterogeneity came from the ability of some multipotent cells to differentiate towards different intestinal lineages. Together with the ability of these cells to self-renew and to highly express CSC markers, this latter result functionally demonstrates, for the first time, that CSCs are present within CTCs. The theory that CTC populations include cells that display CSC characteristics has been subject to debate in the literature since molecular characterisation reported both CSC marker expression within CTC34–37 and the absence of a CSC signature after RNA sequencing analysis.14
Our CTC lines express high levels of metastatic CSC markers, CD26 and CD44v6.22 23 We show here for the first time that CTC lines from patients with CRC are able to induce liver metastases following intrasplenic injection. Our results demonstrate the ability of CTC to generate tumours in distant organs, which has already been shown in another model for another cancer.11
Unexpectedly, we describe the growth of predominantly BRAF-mutated cells in our CTCs despite the fact that BRAF V600E mutation appears to be detected minority in other samples from the same patient. Potential reasons for this finding could be the selective enrichment of BRAF V600E cells under our culture conditions, or alternatively the fact that BRAF-mutated CTCs were predominant at the time of sampling. Although, in the similar conditions BRAF wild-type cell lines from primary and/or metastasis biopsies were established, which suggest that culture condition selection is unlikely.
This result highlights the intratumoural heterogeneity and suggests that some circulating clones may carry bad prognosis mutations.38
Tumour heterogeneity has clinical implications in patient-specific responses to therapy and the rapid emergence of resistance to targeted therapies. Oncologists increasingly use molecular characterisation of a sample of primary or metastatic tumour to guide their selection of treatments for individual patients. Yet, intertumour and intratumour heterogeneities pose a challenge to personalised cancer medicine because a single biopsy cannot always accurately capture the complete genomic landscape of a patient’s cancer. For example, heterogeneity of the KRAS mutational profile between primary and matched metastatic samples is detected in 10%–23% of patients carrying KRAS-mutated colorectal tumours.39 40 Recently, a complete genetic analysis of cancer evolution in patients with prostate cancer proposed a complex model41 42 involving polyclonal seeding in multiple waves and transfer of diverse tumour clones between metastatic sites. In our study, the differential presence of genetic variants in individual CTC line subclones (eg, androgen receptor variant) for CTC41 and the presence of BRAF V600E mutated cells within CTC44 and CTC45 lines suggest that tumour cells with different genetic/phenotypic make up were circulating in the blood of patients at the time of sample collection. Consequently, CTC may be a ‘liquid biopsy’ of the primary tumour and may also provide a snapshot of an otherwise undetected heterogeneous tumour state at primary and/or secondary sites.
Despite the fact that our CTC lines were maintained in culture for several months, the analysis of RNA sequencing results indicated that differentially expressed genes in these cells shared similarities with those identified in three prior studies conducted on freshly isolated CTCs.30 43 44 In particular, six genes were detected as differentially expressed in all four studies (including the present one): AGR2, CEACAM5, CLDN3, KRT18 (encoding cytokeratin 18), EpCAM (TACSTD1) and FGFR3, suggesting that these genes could form part of a core CTC signature across multiple cancers.
Corroborating the robust CSC phenotype that we described in the present study, at least four of these proteins (AGR2, KRT18, EpCAM and FGFR3) were previously proposed as CSC markers in several cancers.45–48 Thus, the expression of AGR2 was found to correlate with that of the CSC marker LGR5 in patients with CRC, leading to the suggestion that detection of AGR2/LGR5 levels may reflect the presence of CTCs with CSC properties in CRC.49 Cytokeratin 18 was identified as one of a small number of CSC markers using an unbiased proteomics approach in gastric cancer46 and was enriched in self-renewing prostate CSCs in vitro.50 High EpCAM expression has been shown to be a feature of CSCs in several carcinomas including liver and colorectal.47 51 FGFR3 was shown to mediate the paracrine effects of FGF9 in the oestrogen-driven expansion of breast CSCs.52
Strikingly also, the six genes identified here as characterising CTCs across several studies encode proteins that localise at the extracellular membrane and/or are secreted in the extracellular space. This characteristic could be highly valuable to propose alternative markers to identify CTCs and to purify them directly from the blood of patients. Most of these proteins were proposed to actively participate in the metastatic process of various cancers, notably by interacting with surrounding normal cells.53–56
From a clinical point of view, it is noteworthy that most of these proteins have been suggested as putative biomarkers either for the presence of metastasis or poor outcome for the patient.57–61
CTCs have also been suggested as a potential useful tool to derive predictive information and thereby inform therapeutic decisions.62 Extensive drug testing was recently performed on CTC cell lines carrying various mutational profiles established from patients with breast cancer, highlighting the interest of such approaches to personalise the identification of sensitivity to cytotoxic or targeted anticancer compounds.14 Interestingly, CTCs were previously shown to be more resistant than primary tumour cells from matching patients due to an enhanced DNA damage response ability,63 and the pathway analysis of differentially expressed genes in our CTC lines pointed towards a robust enrichment of drug-metabolising networks in CTCs.
In addition, CTC-derived xenografts were shown to mirror the donor patient's response to platinum and etoposide chemotherapy.12 The time necessary for xenografts to develop may reduce the applicability of this latter approach to derive predictive information for patients with metastatic CRC, for whom survival times are unfortunately very short. In contrast, the CTC culture model described here is simple and rapid (<1 month from blood collection to drug testing), implying that using this approach to test available treatments could be particularly useful for patients with treatment-refractory tumours such as BRAF-mutated CRC, pancreas cancer or melanoma as well as for patients with CTCs that reflect the presence of minor/undetected clones within metastatic samples. Our preliminary clinical data on a small number of cell lines potentially suggest that toxicity assays on CTC might predict patient response to drugs. For instance a patient from whom the CTC41 line was established rapidly died after being treated with FOLFIRI and the CTC41 line was shown to be resistant to this combination of chemotherapies in vitro (figure 4A). Furthermore, since kinase inhibitors regorafenib and vemurafenib induce many side effects, drug sensitivity assays could be proposed on CTC lines to potentially predict patient response for these drugs and spare patients, by diminishing the risk of leading to severe side effects without any impact on tumour cells.
In conclusion, as suggested by the differential responses to chemotherapeutic cocktail and targeted inhibitor recorded between CTC lines and tumour-derived cell lines, we speculate that generating CTC cell lines using the approach described in this study will provide an invaluable tool to rapidly test and potentially predict treatment response for individual patients, thus facilitating the access of patients to personalised medicine in the future.
Materials and methods
Establishment of circulating tumour patient-derived cell lines
On the day of surgery, hospital shipped two EDTA tubes of blood for each patient through a taxi which was dedicated to this project and during the whole project, blood had never waited more than 4 hours following the sampling to be processed, the average time being 2 hours. Blood samples were then pooled to reach a total volume of 8–10 mL, they were then incubated at room temperature for 20 min with 50 µL of Rosette Sep Human Circulating Epithelial Tumor Cell Enrichment Cocktail (STEMCELL Technologies) per mL of blood diluted in phosphate buffered saline (PBS) containing 2% of fetal bovine serum (FBS) v/v. After 20 min, the mix was gently put on 15 mL of lymphocyte separation medium (LSM, Eurobio) and centrifuged 20 min at 1200g without brake. Cells located at the interface between serum and LSM were delicately harvested, washed twice in PBS containing 2% of FBS and resuspended in M12 medium (1 mL/well) in ultralow attachment 24-well plates (Corning). M12 medium contains advanced DMEM-F12 (Gibco), 2 mmol/L of l-glutamine, 100 Unit/mL of penicillin and streptomycin, N2 supplement (Gibco), 20 ng/mL of epidermal growth factor (R&D) and 10 ng/mL of fibroblast growth factor-basic (R&D). CTC41 has been partially described previously.64
Statistical analysis
For each experiment, data are shown as mean SEM of at least three independent experiments. GraphPad Prism 6 software was used for data analysis. The Mann–Whitney U test was used to analyse the difference between two groups of quantitative variables; α value was set at 5%.
Acknowledgments
Philippe Crespy was helpful for stainings and in vivo experiments. Julian Venable, Philippe Jay and Riccardo Fodde for their help in writing the manuscript. The authors wish to acknowledge the Centre Hospitalier Regional Universitaire (CHRU) of Montpellier for making the CTC41 sample available.
Footnotes
Contributors: FH and JP designed the research; FG and JMP designed part of the research; FG, EB, OV, ELL, KP, CM, LC-T and JMP performed the experiments; FG, JP and FH analysed the data; EC-J, NR, JFB, MP, CP, SB and JCB contributed to clinical part; LZ, SL, GRT and AH contributed to analytic tools, JP and FH wrote the manuscript. JMP and FG discussed results and commented on the manuscript.
Funding: Université Montpellier I (BQR to FH), La ligue contre le cancer (to JP and FG), FH is supported by The University of Melbourne Department of Pathology and by the National Health and Medical Research Council (NHMRC) of Australia (grants #1049561, 1064987, 1069024). SIRIC: Grant “INCa-DGOS-Inserm 6045”.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Patient CTC and tumor-derived cell lines (CTC 44, 45 and CPP1, 19, 24, 25, 30, 44 and 45) of colon cancer cells were obtained from CRC patient blood and biopsies provided by CHU-Carémeau (Nîmes, France, ClinicalTrial.gov Identifier#NCT01577511).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Dobrila-Dintinjana R, Vanis N, Dintinjana M, et al. Etiology and oncogenesis of pancreatic carcinoma. Coll Antropol 2012;36:1063–7. [PubMed] [Google Scholar]
- 2. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559–64. 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 3. Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127:679–95. 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 4. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213–21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 5. Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777–82. 10.1038/nbt.2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479–84. 10.1038/nbt.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Powell AA, Talasaz AH, Zhang H, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 2012;7:e33788 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580–4. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yokobori T, Iinuma H, Shimamura T, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res 2013;73:2059–69. 10.1158/0008-5472.CAN-12-0326 [DOI] [PubMed] [Google Scholar]
- 10. Pecot CV, Bischoff FZ, Mayer JA, et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov 2011;1:580–6. 10.1158/2159-8290.CD-11-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539–44. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- 12. Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897–903. 10.1038/nm.3600 [DOI] [PubMed] [Google Scholar]
- 13. Tinhofer I, Saki M, Niehr F, et al. Cancer stem cell characteristics of circulating tumor cells. Int J Radiat Biol 2014;90:622–7. 10.3109/09553002.2014.886798 [DOI] [PubMed] [Google Scholar]
- 14. Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014;345:216–20. 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176–87. 10.1016/j.cell.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bobek V, Gurlich R, Eliasova P, et al. Circulating tumor cells in pancreatic cancer patients: enrichment and cultivation. World J Gastroenterol 2014;20:17163–70. 10.3748/wjg.v20.i45.17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolostova K, Matkowski R, Gürlich R, et al. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology Published Online First: 11 Apr 2015. 10.1007/s10616-015-9866-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bidard FC, Ferrand FR, Huguet F, et al. Disseminated and circulating tumor cells in gastrointestinal oncology. Crit Rev Oncol Hematol 2012;82:103–15. 10.1016/j.critrevonc.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 19. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 20. Touil Y, Igoudjil W, Corvaisier M, et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin Cancer Res 2014;20:837–46. 10.1158/1078-0432.CCR-13-1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 2009;69:3382–9. 10.1158/0008-5472.CAN-08-4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pang R, Law WL, Chu AC, et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell 2010;6:603–15. 10.1016/j.stem.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 23. Todaro M, Gaggianesi M, Catalano V, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014;14:342–56. 10.1016/j.stem.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Law CW, Chen Y, Shi W, et al. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Onstenk W, Sieuwerts AM, Weekhout M, et al. Gene expression profiles of circulating tumor cells versus primary tumors in metastatic breast cancer. Cancer Lett 2015;362:36–44. 10.1016/j.canlet.2015.03.020 [DOI] [PubMed] [Google Scholar]
- 28. Mostert B, Sieuwerts AM, Kraan J, et al. Gene expression profiles in circulating tumor cells to predict prognosis in metastatic breast cancer patients. Ann Oncol 2015;26:510–16. 10.1093/annonc/mdu557 [DOI] [PubMed] [Google Scholar]
- 29. Luo X, Mitra D, Sullivan RJ, et al. Isolation and molecular characterization of circulating melanoma cells. Cell Rep 2014;7:645–53. 10.1016/j.celrep.2014.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smirnov DA, Zweitzig DR, Foulk BW, et al. Global gene expression profiling of circulating tumor cells. Cancer Res 2005;65:4993–7. 10.1158/0008-5472.CAN-04-4330 [DOI] [PubMed] [Google Scholar]
- 31. Luo W, Friedman MS, Shedden K, et al. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 2009;10:161 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227–35. 10.1158/2159-8290.CD-11-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brabletz T, Jung A, Spaderna S, et al. Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer 2005;5:744–9. 10.1038/nrc1694 [DOI] [PubMed] [Google Scholar]
- 34. Kasimir-Bauer S, Hoffmann O, Wallwiener D, et al. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res 2012;14:R15 10.1186/bcr3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Markiewicz A, Książkiewicz M, Wełnicka-Jaśkiewicz M, et al. Mesenchymal phenotype of CTC-enriched blood fraction and lymph node metastasis formation potential. PLoS ONE 2014;9:e93901 10.1371/journal.pone.0093901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katoh S, Goi T, Naruse T, et al. Cancer stem cell marker in circulating tumor cells: expression of CD44 variant exon 9 is strongly correlated to treatment refractoriness, recurrence and prognosis of human colorectal cancer. Anticancer Res 2015;35:239–44. [PubMed] [Google Scholar]
- 37. Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol 2011;29:1547–55. 10.1200/JCO.2010.30.5151 [DOI] [PubMed] [Google Scholar]
- 38. Pakneshan S, Salajegheh A, Smith RA, et al. Clinicopathological relevance of BRAF mutations in human cancer. Pathology 2013;45:346–56. 10.1097/PAT.0b013e328360b61d [DOI] [PubMed] [Google Scholar]
- 39. Artale S, Sartore-Bianchi A, Veronese SM, et al. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J Clin Oncol 2008;26:4217–19. 10.1200/JCO.2008.18.7286 [DOI] [PubMed] [Google Scholar]
- 40. Paliogiannis P, Cossu A, Tanda F, et al. Mutational concordance between primary and metastatic colorectal adenocarcinoma. Oncol Lett 2014;8:1422–6. 10.3892/ol.2014.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353–7. 10.1038/nature14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong MK, Macintyre G, Wedge DC, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 2015;6:6605 10.1038/ncomms7605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Onstenk W, Kraan J, Mostert B, et al. Improved Circulating Tumor Cell Detection by a Combined EpCAM and MCAM CellSearch Enrichment Approach in Patients with Breast Cancer Undergoing Neoadjuvant Chemotherapy. Mol Cancer Ther 2015;14:821–7. 10.1158/1535-7163.MCT-14-0653 [DOI] [PubMed] [Google Scholar]
- 44. Mostert B, Sieuwerts AM, Bolt-de Vries J, et al. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol 2015;9:920–32. 10.1016/j.molonc.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma SR, Wang WM, Huang CF, et al. Anterior gradient protein 2 expression in high grade head and neck squamous cell carcinoma correlated with cancer stem cell and epithelial mesenchymal transition. Oncotarget 2015;6:8807–21. 10.18632/oncotarget.3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morisaki T, Yashiro M, Kakehashi A, et al. Comparative proteomics analysis of gastric cancer stem cells. PLoS ONE 2014;9:e110736 10.1371/journal.pone.0110736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA 2007;104:10158–63. 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dvorak P, Dvorakova D, Hampl A. Fibroblast growth factor signaling in embryonic and cancer stem cells. FEBS Lett 2006;580:2869–74. 10.1016/j.febslet.2006.01.095 [DOI] [PubMed] [Google Scholar]
- 49. Valladares-Ayerbes M, Blanco-Calvo M, Reboredo M, et al. Evaluation of the adenocarcinoma-associated gene AGR2 and the intestinal stem cell marker LGR5 as biomarkers in colorectal cancer. Int J Mol Sci 2012;13:4367–87. 10.3390/ijms13044367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castillo V, Valenzuela R, Huidobro C, et al. Functional characteristics of cancer stem cells and their role in drug resistance of prostate cancer. Int J Oncol 2014;45:985–94. 10.3892/ijo.2014.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guan DX, Shi J, Zhang Y, et al. Sorafenib enriches epithelial cell adhesion molecule-positive tumor initiating cells and exacerbates a subtype of hepatocellular carcinoma through TSC2-AKT cascade. Hepatology 2015;62:1791–803. 10.1002/hep.28117 [DOI] [PubMed] [Google Scholar]
- 52. Fillmore CM, Gupta PB, Rudnick JA, et al. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA 2010;107:21737–42. 10.1073/pnas.1007863107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu H, Zhao J, Lin L, et al. Proteomic study explores AGR2 as pro-metastatic protein in HCC. Mol Biosyst 2012;8:2710–18. 10.1039/c2mb25160d [DOI] [PubMed] [Google Scholar]
- 54. Bramswig KH, Poettler M, Unseld M, et al. Soluble carcinoembryonic antigen activates endothelial cells and tumor angiogenesis. Cancer Res 2013;73:6584–96. 10.1158/0008-5472.CAN-13-0123 [DOI] [PubMed] [Google Scholar]
- 55. Ni J, Cozzi PJ, Duan W, et al. Role of the EpCAM (CD326) in prostate cancer metastasis and progression. Cancer Metastasis Rev 2012;31:779–91. 10.1007/s10555-012-9389-1 [DOI] [PubMed] [Google Scholar]
- 56. Rangel LB, Agarwal R, D'Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res 2003;9:2567–75. [PubMed] [Google Scholar]
- 57. Alavi M, Mah V, Maresh EL, et al. High expression of AGR2 in lung cancer is predictive of poor survival. BMC Cancer 2015;15:655 10.1186/s12885-015-1658-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gebauer F, Wicklein D, Horst J, et al. Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PLoS ONE 2014;9:e113023 10.1371/journal.pone.0113023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morris KL, Tugwood JD, Khoja L, et al. Circulating biomarkers in hepatocellular carcinoma. Cancer Chemother Pharmacol 2014;74:323–32. 10.1007/s00280-014-2508-7 [DOI] [PubMed] [Google Scholar]
- 60. Hiraga T, Ito S, Nakamura H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int J Cancer 2016;138:1698–708. 10.1002/ijc.29921 [DOI] [PubMed] [Google Scholar]
- 61. Ach T, Schwarz-Furlan S, Ach S, et al. Genomic aberrations of MDM2, MDM4, FGFR1 and FGFR3 are associated with poor outcome in patients with salivary gland cancer. J Oral Pathol Med Published Online First: 14 Dec 2015. 10.1111/jop.12394 [DOI] [PubMed] [Google Scholar]
- 62. Toss A, Mu Z, Fernandez S, et al. CTC enumeration and characterization: moving toward personalized medicine. Ann Transl Med 2014;2:108 10.3978/j.issn.2305-5839.2014.09.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gong C, Liu B, Yao Y, et al. Potentiated DNA Damage Response in Circulating Breast Tumor Cells Confers Resistance to Chemotherapy. J Biol Chem 2015;290:14811–25. 10.1074/jbc.M115.652628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cayrefourcq L, Mazard T, Joosse S, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res 2015;75:892–901. 10.1158/0008-5472.CAN-14-2613 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-311447supp001.pdf (56KB, pdf)
gutjnl-2016-311447supp007.pdf (1.1MB, pdf)
gutjnl-2016-311447supp002.pdf (67.2KB, pdf)
gutjnl-2016-311447supp003.pdf (167.2KB, pdf)
gutjnl-2016-311447supp008.pdf (259.8KB, pdf)
gutjnl-2016-311447supp009.pdf (271.7KB, pdf)
gutjnl-2016-311447supp004.pdf (169.5KB, pdf)
gutjnl-2016-311447supp005.pdf (811.3KB, pdf)
gutjnl-2016-311447supp006.pdf (85.9KB, pdf)