Abstract
We present a clinical device for simple, rapid, and viable isolation of circulating tumor cells (CTCs) from cancer patient bloods. In spite of the clinical importance of CTCs, the lack of easy and non-biased isolation methods is a big hurdle for implementing CTC into clinical use. The present device made of photosensitive polymer was designed to attach to conventional syringe to isolate the CTCs at minimal resources. Its unique tapered-slits on the filter are capable not only to isolate the cell based on their size and deformability, but also to increase sample flow rate, thus achieving label-free rapid viable CTC isolation. We verified our device performance using 9 different types of cancer cells at the cell concentration from 5 to 100cells/ml, showing that the device capture 77.7% of the CTCs while maintaining their viability of 80.6%. We extended our study using the 18 blood samples from lung, colorectal, pancreatic and renal cancer patients and captured 1-172 CTCs or clustered CTCs by immunofluorescent or immunohistochemical staining. The captured CTCs were also molecularly assayed by RT-PCR with three cancer-associated genes (CK19, EpCAM, and MUC1). Those comprehensive studies proved to use our device for cancer study, thereby inaugurating further in-depth CTC-based clinical researches.
Keywords: Circulating tumor cells, tapered-slit filter, viable rare cell isolation, photosensitive polymer, clinical cancer study.
1. Introduction
Circulating tumor cells (CTCs) are the tumor cells in blood, originated from primary tumor site and responsible for cancer metastasis. After pre-clinical studies revealed their presence in cancer patient blood, subsequent clinical studies have been conducted and showed that their counts have close relavance to overall survival and metastatic potential. 1,2 Those studies elucidated the potential role of CTC in tumor progression and metastasis, however, still have been limited to study their heterogeniety and the difference from primary tumor. In addition, in order to clarify their ambiguous and heterogeneous properties, label-free separation method and their molecular profiling are demanding. To date, the only FDA-approved CTC detection technique, CellSearch® and most afterward techniques rely on surface affinity between CTC and epithelial cell adhesion molecule (EpCAM), in spite of several design alteration and variation.3, 4 Although the EpCAM-based isolation methods can capture CTC in specific manner, however, they have difficulty in capturing EpCAM weak or negative CTC which comes from epithelial mesenchymal transition (EMT) or non-epithelial tumor types such as melanoma. Moreover, due to their irreversible antibody interacion, those methods need additional chemical treatment or cleavable linker chemistry for releasing the captured cells for downstream analysis.5 Their low repeatability and needs of controlled experiment setup are also the obstacles for simple clinical applications. Alternatively, the physical property-based CTC isolation methods have been prepared and proposed for solving those issues with the merit of rapid and simple CTC isolations. 6-9 Among them, size-based CTC isolation have been widely studied and remarkable microfluidics-based devices utilizing size of the cell for CTC isolation have been suggested recently. Those isolated the CTCs based on different motion trend in specially designed channels and in order to enhance the purity and throughput, various design such as multiorifice channel 10, spiral channel 11, 12, contraction-expansion arrayed channel 13 have been proposed. Recent advance in this field achieved over 85% target cell recovery from the heterogeneous cell mixture, and succefully captured the CTCs from the patient sample with breast and lung cancer. 14 However, those devices commonly need pre-processing, such as red blood cell lysis and buffy coat isolation, and steady sample control and optimized condition are crucial for the best result, which make it hard to isolate and examine the CTCs in limited resource condition.
The filtration is one of the simplest and most widely studied method for capturing the bigger cells from the others. Since after the vast interest toward the circulating tumor cells for liquid biopsy, considerable number of filters have been developed for CTC isolation and have showed the possibilities of those device for CTC-based liquid biopsy. 6, 7, 15, 16
Recent studies of microfilter have showed comparable results with FDA approved technique 17-19 and the overall CTC count was even much higher than that of CellSearch® method. Because this method is applicable to variable cancer types regarldess of their EpCAM expression, it is proper to use this device for studying cancer heterogeneity without biased view. In spite of those significant merits of filtration method, however, the previous CTC filters designing in straight holes are limited to increase the throughput due to concentrated cell stress on edge, resulting in the captured cell damages or lysis at high throughput condition. 20, 21 In addition, most of previous microfiltration studies have been verified their CTC isolation performance by immunofluorescent staining only 16, 19, which is not enough to show them as CTCs. Therefore, comprehensive performance verification including downstream analysis of captured CTCs are urgently needed for the microfiltration method to prove their clinical usefulness.
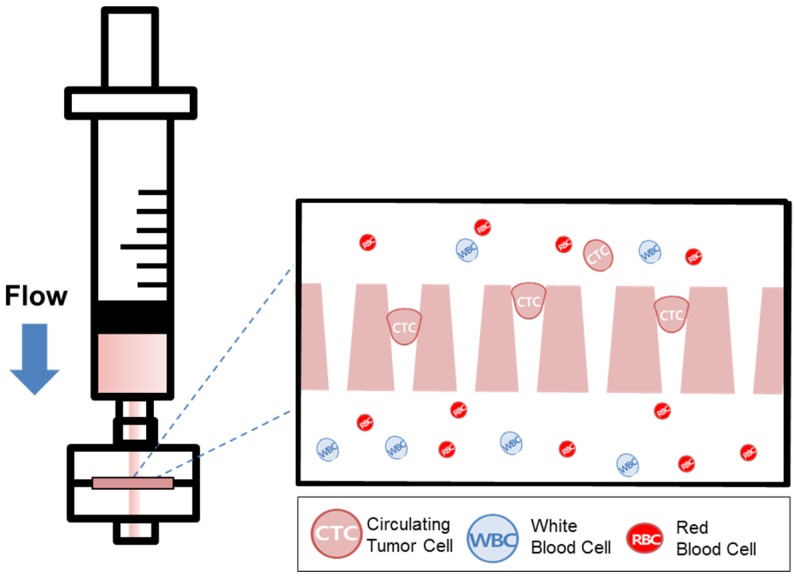
Recently, our group introduced the uniquely designed membrane filter, tapered-slit filter (TSF), having wider cell entrance and gradually narrower exit in order to both reduce the captured cell stress and capture the CTCs specifically taking advantage of both size and deformability. 22 The previous microfilter showed the meaningful progress on viable CTC isolation compared to previous CTC filters. However, its total sample processing capacity and their operational method were still low and difficult for applying it to clinical sample and setup, respectively. Here, we present the photosensitive polymer-based circular-shaped microfilter having optimized tapered-slits for clinical use. This microfabricated filter is inserted in a filter cassette and simply connected to commercial syringe to isolate the CTCs from unprocessed clinical samples from cancer patients (Fig.1). Therefore, the device that is simple, rapid, and label-free is prepared for finding further functional and molecular inherence of CTCs.
Figure 1.
Schematic diagram of the photosensitive polymer-based microfilter device for the label-free rapid viable circulating tumor cell isolation.
We tested our device's performance varying 4 types of cancer cell lines of lung (A549 and NCI-H23) and colorectal cancers (DLD-1 and SW620) at the 5 different cell concentrations from up to 100cells/ml to low to 5cells/ml, which is sufficiently low enough to mimick CTCs in the blood. In addition, 7 different types of cancer are also tested for verifying wide clinical application of the present device independent to types of cancer. Finally, we extended our studies using 18 blood samples collected from lung (n=6), colorectal (n=6), pancreatic (n=4), renal cancer (n=2) patients, and 2 samples from healthy donors, and examined with at least two confirmation methods among immunofluorescent staining, immunohistochemical staining, and RT-PCR. Those comprehensive pathological and molecular examiniation of captued CTCs by the present device aim to fulfill the practical clinical use of our device for not only CTC enumeration but further downstream molecular analysis of CTC.
2. Result and Discussion
2.1. Development of Photosensitive polymer-based Microfilter
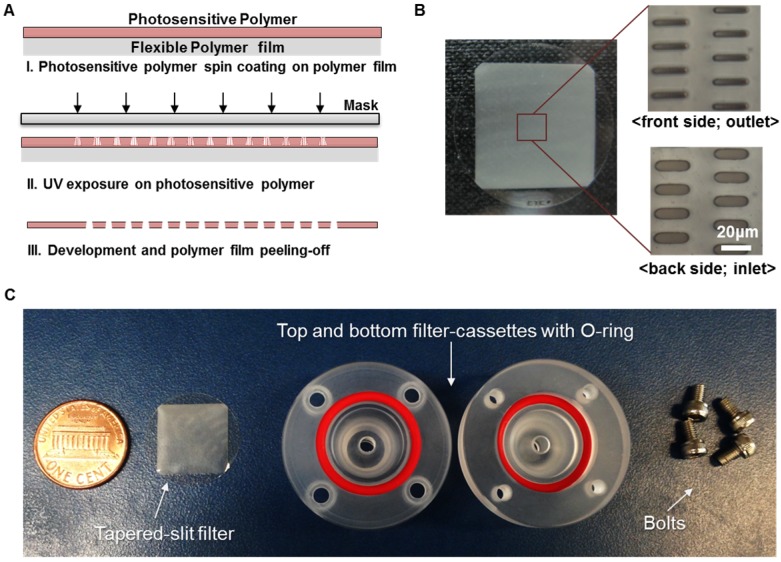
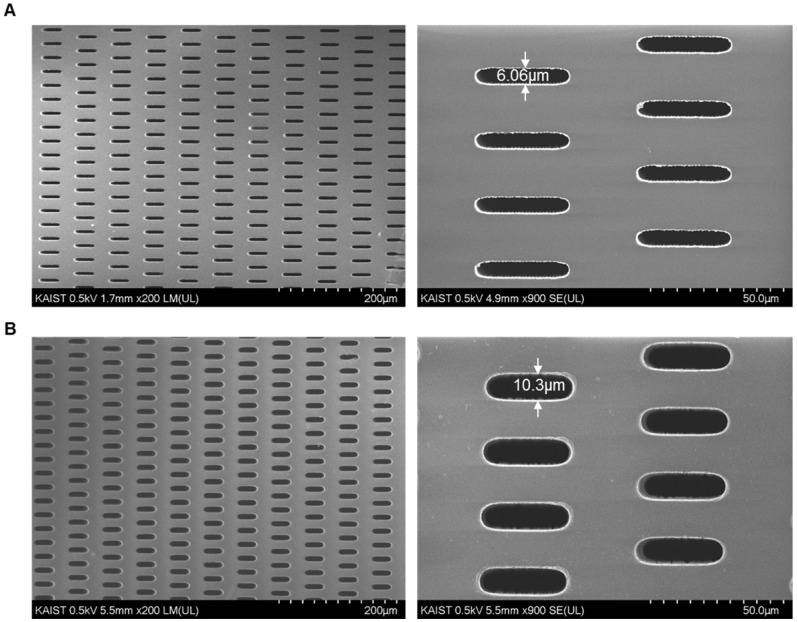
Our previous membrane CTC filter device showed high capture efficiency and improved throughput (5ml/h) compared to that of previous micropillar typed device 24 while maintaining the high viability of captured cells. However, this device had limited to process large amount of sample volume up to 5 milliliter of blood without clogging, so it was not suitable to use it in real clinical samples exceeding 5ml. In addition, the fabricated filter need to be aligned and interconnected with two PDMS layers for flow guiding and silicon tubes, which make it difficult for user to simple and rapid clinical use. In order to use this device in much simpler way and clinical use, we have optimized and developed the overall filter designs and their slit density; circular filter shape, much denser slit formation with the acrylate-zig for elimination alignment process. For the fabrication of microfilters, one of the widely utilized photosensitive polymer, SU8, but rarely used for solid structures, has chosen with the merits of single mask fabrication and its solidity. The overall fabrication procedure (Fig. 2A) using SU8 is similar to that applied for previous device fabrication, however the lithography conditions have been modulated and optimized for high-aspect ratio and the denser design of the present work. Owing to the optimized lithographic conditions including exposure times and air gap, we successfully fabricated circular-shaped microfilter device with at least 2-fold higher tapered-slit density while maintaining their mechanical strength, thus achieving high-throughput CTC isolation without clogging of cells and fracture of the device. The tapered-slit formation was confirmed using Field Emission Scanning Electron Microscopy (FE-SEM). The images, as shown in Fig. 3, indicate that the present microfilters succefully fabricated with regular formation of tapered-slits, which is one of the most characretistic design distinguished from the previous straight-hole or slit designs. The robustness of design was examined by manual mode of CTC filtration with model sample and showed that ther present device is able to endure even in approximately 100-300ml/h of sample flow rate, which means that present device is easy to operatae without special precaution (Fig. S1). Two conventional syringe-compatible acrylate zig made sample processing easy to operate for anyone who is unfamiliar to those operations. Figure 2B, C are the photographs of the fabricated and prepared photosensitive polymer-based filter and their compositions. By help of those compartments and simple experimental setup (Fig. 4A), present microfilter device facilitate the simple and rapid CTC capture and release in an easy-to-follow way.
Figure 2.
Fabrication of present photosensitive polymer-based microfilter device for circulating tumor cell isolation: (A) fabrication procedure of the microfilter using photosensitive polymer; (B) the photo of the fabricated microfilter with enlarged view of the tapered slits; (C) the photo of components for the present device.
Figure 3.
Confirmation of the tapered-slit formation using photosensitive polymer: (A) the front side (outlet) of the present photosensitive polymer-based microfilter with an image magnified by a factor of 200 (left) and 900 (right), respectively; (B) the back side (inlet) of the present microfilter with an image magnified by a factor of 200 (left) and 900 (right), respectively. (All images were taken an acceleration voltage of 0.5)
Figure 4.
Label-free rapid viable enrichment of circulating tumor cell: (A) the experimental setup of the present microfilter; (B) the isolated GFP labeled-lung cancer cells (A549G) captured by the present microfilter (top) and isolated colorectal cancer cells (SW620) and leukocytes after immunofluorescence staining (bottom)
2.2. Device verification using model samples including lung/colorectal cancer cells at the high/low cell concentration
We confirmed the performance of present photosensitive polymer-based microfilter using 2 different spiked cancer cell conditions; 1) high-concentration of lung (A549G) /colorectal GFP cancer cells (DLD-1G), 2) low-concentration of lung (A549) /colorectal non-GFP cancer cells (DLD-1). All blood samples were diluted to 1:2(Blood:PBS), and introduced into the present device using negative pressure of syringe pump. The prepared blood samples were passed through at the flow rate of 10ml/h after the experimental verification of optimal flow rate (Supplementary Material S2), and all experiments at each condition repeated twice.
For the experiment at 1) conditions, first we demonstrated the performance of present device in terms of capture efficiency, release rate and viability using 3ml of blood sample containing the 2 different GFP lung/colorectal cancer cell at the higher cell concentration (50,100cells/ml). With the merit of autonomously fluorescent GFP cells, we can test the device performance independent to antibody staining. We enumerated the entire fluorescent cells captured in microfilter under fluorescence microscope. Figure 4B (top) shows the captured A549G in the filter. At the highest cell centration of 100(cells/ml), present device captures 72.9% and 91.7% of A549G and DLD-1G, respectively. At the 2 times less diluted cell concentration of 50(cells/ml), our device captures 83.5% and 93.1% of A549G and DLD-1G, respectively (Fig. 5A). Averagely, 78.2% and 92.4% of A549G and DLD-1G cells are captured by our device. Compared to A549G, average diameter of DLD-1G is 2.5μm larger. Higher capture efficiency for DLD-1G might be resulting from this larger diameter, higher chance to be captured from the microfilter. The capturing ability tests were followed by release ability test. After releasing the captured cells using 5ml of PBS, we carefully counted the released cancer cell number, and calculated the release rate at the different cell concentrations. For the experiment at the cell concentration of 100, present device released the 56.4% and 33.1% of A549G and DLD-1G, respectively. At the 50, 73.3% and 38.2% of A549G and DLD-1G are released from the device, respectively (Fig. 5B). The average release rate for A549G and DLD-1G are 64.8% and 35.7%, respectively. The larger cancer cell, DLD-1G, showed lower released rate. Since larger cell might be tightly captured on the microfilter, it could be released less efficiently compared to smaller cell. The present device shows average 50.2% release rate using two types of cancer cell (Fig. S5), however it might be more improved by surface functionalization or reducing non-specific binding. Followed by verifying the capture/release performance, we verified the viability changes before and after capturing experiments. Before the capturing experiments, the 96.4% and 94.7% of A549G and DLD-1G showed the viability, respectively. After capturing experiments, their viability was reduced to 80.9% and 80.3%, respectively (Fig. 5C). We verified that average 80.6% of captured cancer cells remained their viability during filtration using the present microfilter. We further examined the purifying performance of the present microfilter and verified that the purity of the present device is 17.44%. The detailed procedure is described in Supplementary Material S4.
Figure 5.
The performance of the present microfilter: (A) the capture efficiency of the A549G and DLD-1G cells depending on the cell concentrations (50-100cells/ml); (B) the release rate of the A549G and DLD-1G cells depending on the cell concentrations; (C) the viability of the A549G and DLD-1G cells before and after cell isolation; (D) the average capture efficiency of the lung and colorectal cancer cells at the low cell concentration (5-15cells/ml); (E) the capture efficiency of 4 different cancer cells depending on the cell concentrations(5-15cells/ml); (F) the average capture efficiency of 7 different cancer cells from 7 different organs at the low cell concentration (5-15cells/ml).
For the experiment at 2) conditions, we verified the capture efficiency using 9ml of blood sample containing non-GFP cancer cells. Before the examination using clinical specimens, we verified the device performance at more realistic conditions; non-fluorescent cancer cells at the lower cell concentration of 5-15 cells/ml reflecting the CTC frequency from the previous studies. 17, 26 For these experiments, 4 different non-GFP cancer cells were used. A549 and NCI-H23 are originated from lung and DLD-1 and SW620 are from colorectal. The average capture efficiency for 4 different cancer cells at the cell concentration of 5-15cells/ml was shown in Figure 5D. The present device captures average 76.8% and 82.8% of lung and colorectal cancer cells, respectively. Fluorescent images in Fig. 4B (bottom) shows the captured non-GFP cancer cells and leukocyte after immunostaining. The total average capture efficiency is 5% lower than that of experiments using GFP cancer cell at higher concentration. Depending on expression level of cytokeratin, some cancer cells cannot be stained sufficiently enough to count. Considering these effects, the capture efficiency at lower concentrations is similar to that at higher concentrations. Next, we evaluated the CTC detection sensitivity at various cancer cell concentrations of 5-15cell/ml (Fig. 5E). At every cell concentration, over 70% of spiked cancer cells were captured. In addition, even in extremely rare cell concentration, 5 cells/ml, our device successfully captured over 79% of spiked cancer cells for 4 different cancer cells, thus verifying CTC detection potentials at the extremely rare cell concentration as low to only a few cells per 1ml of whole blood.
2.3. Device verification using model samples including various types of cancer cells
In order to verify the applicability of present device independent to types of cancers, we extended our studies to 7 more types of cancers, such as breast, stomach, pancreas, prostate, ovary, cervix and kidney. We selected 7 different cell lines, MDA-MB-231 for breast, SNU1 for stomach, Panc03.27 for pancreas, LNCaP for prostate, SKOV3 for ovary, HeLa for Cervix, and SN12C for kidney to make model CTC samples, then used for verifying capture efficiency of present microfilter. All conditions for experiments were identical to that of experiments at lower cell concentrations, however, we fixed the spiked cell concentration as 10(cells/ml) for direct comparison at same condition. All experiments using each cell lines repeated three times. Figure 5F shows the average capture efficiencies of present microfilter for 7 different cancer cells. Depending on the type of cancers, the average capture efficiency varied from 62.52% (for HeLa) to 91.44% (for LNCaP). We enumerated the cancer cells as CK+/CD45-/DAPI+, thus low expression of cytokeratin cannot be counted, resulting in low capture efficiency. In addition, we filtered the cancer cells based on their size and deformability, so cancer cells having smaller and more deformable phenotype might be passed through our device. In spite of those limitations, average capture efficiency for 7 different cancer cells is 76.55±12.1%. Considering 4 more cancer cell lines of lung and colorectal studied in 2.2, our device isolates average 77.7±10.0% of cancer cells in 9 cancers, 11 different cancer cell lines without any other pretreatment or chemical lysis temps, which is comparable or even higher performance than previously presented CTC filtration methods. 20, 25
2.4. Circulating tumor cell isolation from cancer patient bloods
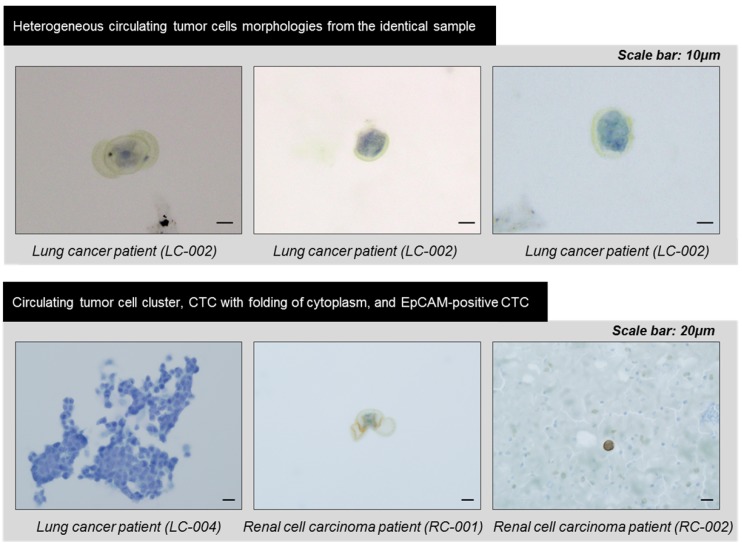
In order to verify the device's potential for clinical use, we applied the devices to blood samples from 18 cancer patients with lung (n=6), colorectal (n=6), pancreatic (n=4), and renal cancers (n=2) and 2 healthy donors. Similar to experiments using model samples including cancer cell lines, 5ml of unprocessed patient's blood was diluted to 10ml of PBS, and this 15ml of samples were directly processed through present device. After the sample processing, the captured cells were released and followed by immunofluorescence staining. Figure 6 shows the representative image of circulating tumor cells including EpCAM positive and negative CTCs. Because the present method does isolate the CTC regardless of their surface protein expression, considerable amount (34.7%) of the EpCAM negative CTCs were succefully isolated, showing that the present device is capable to isolate heterogeneous subtypes of CTCs. In addition, mega-sized(>30μm) undefined CTC-like cells also detected in some cases (Fig.6 bottom). Those cells were usually expressing positive to both cytokeratin and CD45. We did not counted those cells as CTCs but we found that cells with similar morphology were also discovered by immunohistochemical staining. The overall results obtained from the clinical blood samples are summarized in Table 1. Of the 18 cancer cases, 11 (61.1%) cases showed at least one CTC by immunofluorescence staining and their average EpCAM positivity was 65.3%. Depending on cancer types, their EpCAM positivity varied from 62.5% (pancreatic cancer) to 100% (colorectal cancer). By immunofluorescence staining, the average number of CTCs was 2.5 and ranged from 1 to 8 in the all types of cancers. Meanwhile, no CTC was detected from two healthy controls. Of the 18 samples of cancer patients, 6 samples of lung cancer patients and 2 samples of renal cancer patients were independentaly processed by the device additionally and processed samples were identified by immunohistochemical staining for pathological evaluation of CTCs. Seven (87.5%) out of 8 cases evaluated showed at least one CTC and their count ranged from 3 to 172. Those CTCs includes morphologically characteristic cells and clustered CTCs composed of over 100 individual cancer cells. Especially, the CTCs and clustered CTCs remained intact, demonstrating that the present device maintains not only their viability but also their original morphology. The CTCs from lung cancer patients were immunohistochemically negative for typical lung cancer phenotypic marekers of TTF-1 and p63, and the previous study of CTCs from liver cancer patints also showed no expression of typical hepatocelluar carcinoma phynotypic markers.27 Our results suggest that CTCs are diffrent from the primary tumor tissue in phenotype, and that CTC can show loss of tissue specific differentiation, probably due to epithelial-mesenchymal transition. The CTCs also revealed various cell morphology even in the same samples, and the heterogeneous morphology may be due to cellular plemorphism of malignant tumor cells, in addition to deformability of CTCs caused by processing. Three cases, two lung cancers (LC-001, 002) and the other renal cancer (RCC-01) showed unusual morphology of CTCs with folding of cytoplasm (Fig. 7), probably due to partial capture of cells in the pores of the device. An interesting finding was the detection of CTC clusters in lung cancer cell patients (LC-004 and LC-006), and these CTC clusters were not secondary (clogging during the processing) but may be derived from primary lung lesion by vascular invasion into pulmonary vessels, since the cells in the cluster were arranged in lumen-like structure with round outline, suggesting glandular differentiation.
Figure 6.
The immunofluorescent images of the captured circulating tumor cells from the cancer patients with pancreatic cancer, colorectal cancer and lung cancer.
Table 1.
The verification of the circulating tumor cells from the enriched by the present microfilter devices. IF (Immunofluorescence), IHC (immunohistochemistry) and RT-PCR analysis of CTCs from 18 patients and 2 control samples.
| Cancer Type |
Sample ID | Sample description | IF (Immunofluorescence) |
IHC (Immunohistochemistry) |
RT-PCR (Reverse-transcription PCR) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of CTCs (per mL) | No. of CTCs | Remarks | CK-19 | EpCAM | MUC1 | ||||||||
| Sex | Age | Stage | Others | EpCAM (+) | EpCAM (-) | Total | |||||||
| Lung Cancer |
LC-001 | F | 67 | 1a | Adenocarcinoma | 0 | 0 | 0 | 3 | ||||
| LC-002 | M | 67 | - | HD | 1 | 2 | 3 | 3 | |||||
| LC-003 | F | 75 | 1a | Adenocarcinoma | 1 | 1 | 2 | 0 | |||||
| LC-004 | M | 81 | 1a | Adenocarcinoma | 0 | 0 | 0 | 172 | Clustered CTC | ||||
| LC-005 | F | 78 | 1a | Adenocarcinoma | 1 | 0 | 1 | 3 | |||||
| LC-006 | F | 48 | 1b | Adenocarcinoma | 0 | 0 | 0 | 27 | Clustered CTC | ||||
| Colorectal Cancer | CC-001 | F | 51 | 4 | Adenocarcinoma | 0 | 1 | 1 | + | + | |||
| CC-002 | F | 56 | 1 | Adenocarcinoma, recurrence case | 1 | 0 | 1 | + | + | ||||
| CC-003 | F | 65 | 4 | Adenocarcinoma | 1 | 0 | 1 | + | + | ||||
| CC-004 | M | 79 | 4 | Adenocarcinoma | 0 | 1 | 1 | + | + | ||||
| CC-005 | F | 70 | 4 | Adenocarcinoma | 0 | 0 | 0 | - | + | ||||
| CC-006 | F | 76 | 4 | Adenocarcinoma | 0 | 0 | 0 | + | + | ||||
| Pancreatic Cancer | PC-001 | M | 68 | 2 | CA19-9: 6 | 2 | 5> | 8> | + | + | + | ||
| PC-002 | M | 68 | 2 | CA19-9: 8 | 3 | 1 | 4 | + | + | + | |||
| PC-003 | M | 50 | 1 | CA19-9: 15 | 1 | 0 | 1 | + | + | + | |||
| PC-004 | F | 77 | 2 | CA19-9: 18 | 3 | 3 | 6 | + | + | + | |||
| Renal Cell Carcinoma | RC-001 | M | 46 | 3 | Rt. RCC | 0 | 0 | 0 | 1 | ||||
| RC-002 | F | 54 | Lt. RCC | 2 | 0 | 2 | 1 | EpCAM (+) | |||||
| Control | Ctr-001 | M | 40 | - | - | 0 | 0 | 0 | - | - | N/A | - | - |
| Ctr-002 | M | 21 | - | - | 0 | 0 | 0 | - | - | N/A | - | - | |
Figure 7.
The heterogeneous morphologies of the circulating tumor cells captured by the present filter and identified by immunohistochemical staining.
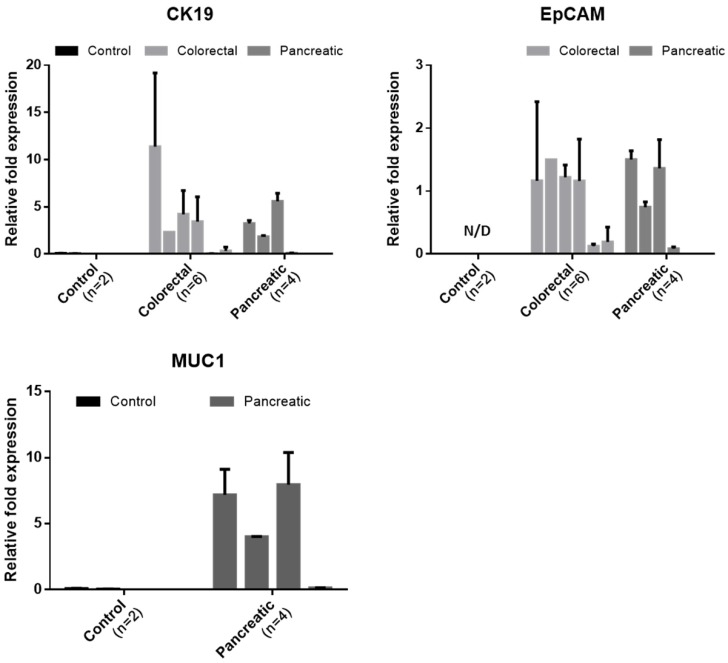
Of the 20 blood samples, 6 samples from colorectal cancer patient, 4 from pancreatic cancer patient, and 2 from healthy donor were independently processed by another device and studied to quantify the tumor associated mRNA expressions in captured CTCs. We measured the expression level of two or three cancer-associated markers. For colorectal sample, the expression level of cytokeratin and EpCAM were analyzed. For sample from pancreatic cancer, the expression level of cytokeratin 19, EpCAM, MUC1 were analyzed. For sample from healthy control, the expression level of all primer used were analyzed. The relative fold expression of each genes are described in Figure 8. Those all cancer cases showing at least 1 CTC expressed the cytokeratin 19 and EpCAM in molecular level. For colorectal cancer cases, all cases showing at least 1 CTC showed both EpCAM and CK19 expression. The CC005 and 6, whose CTC was not detected by IF, showed negligible CK19 expression. However, these cases showed few amount of EpCAM expression. For pancreatic cancer cases, all cases expressed the EpCAM. However, PC-004 showed negligible level of CK19 and MUC1. Especially, MUC1 expression of pancreatic cancer patients was significant. On the other hand, the sample from two healthy donors did not show any tumor-associated genes in detectable level. Especially, healthy donors' Ct value for EpCAM was over 35 cycles, which imply that EpCAM is highly specific to circulating tumor cell. Because the set of experiment involved was too small to conclude our results, however this result showed that our device succefully isolate and release the cells and those harvested cells are applicable to further downstream analysis to see their heterogeniety and ambiguity.
Figure 8.
The relative expression levels of the cancer-associated genes (CK19, EpCAM, and MUC1) in the captured cells by the present microfilters, normalized to GAPDH
3. Conclusion
Here, we report a new means of capturing viable CTCs rapidly and easily. Many researches revealed the potentials of CTCs to see the stage of disease and and prognostic value after treatment, however, little studies have been proposed for enriching the CTCs from cancer patient's bloods in label-free and simple manners. Our photosensitive polymer-based microfilter device, using tapered-slit filter and connected to commercial syringe, propose a useful alternatives for non-experts in microfluidics to get the CTCs for their clinical or molecular study without biased-view of CTCs. In addition, those comprehensive studies and reliable CTC capturing performance (77.7±10.0%) using 9 types of cancer cells at 5 different spiked cell concentrations (5-100cells/ml) give strong evidence for wide use of the present device to any cancer types, independent to their surface marker expression such as EpCAM. At the clinical studies using patient samples from 4 different cancers, the present device captured 1 to 172 CTCs including CTC cluster and the previously undiscovered CTC-like cells, indicating that the present microfilter have huge potential to be used in in-depth CTC researches and CTC-based diagnosis. In addition, while we used the syringe pump for generating steady flow to verify our device at the same condition, the equipment-free mode with a hand could be applied for capturing CTCs. We believed that these simple CTC isolation device will be helpful for rapid cancer diagnosis and prognosis at the developing country under limited equipment and power resources.
4. Experimental Section
Device Design and Fabrication
A schematic diagram of present photosensitive polymer-based microfilter device is shown in Fig. 1. The present device consists of one photosensitive polymer (SU8)based circular-shaped microfilter and two acryl filter cassettes for interconnecting to commercial syringe. We designed the circular tapered-slit filter having the diameter of 20mm contains 625x625 (=165,625) tapered-slits at the slit density of 52,727/cm2, which is sufficient for high throughput isolation. Each oval-shaped tapered-slits designed with wider inlet and the narrower outlet in depth facilitate not only viable isolation with less stress reduced to 18% than that of previous straight-hole filter, but also selective CTC isolation from other blood cells, having the identical size but different ability to deform. Furthermore, those oval-shaped slit is much more efficient to isolate CTC clusters with maintaining their original formations. Those effects were verified theoretically and experimentally in our previous works. 22 The single slit design and an outlet width of tapered-slit were confirmed from previous our works, however, the slit density and device composition has been improved and optimized for the high throughput handheld device application.
Forming tapered-slits on SU8 membrane is accomplished by our simple and novel single mask UV lithography method. The fabrication procedure is shown in Fig. 2A. The tapered slit angle of 2 degree is successfully formed from the adjustment of the UV exposure dose on the spin coated SU8 layer and the air gap between photomask and SU8 filter layer with high reproducibility. Intended manipulations of those factors make the tapered-slit uniformly as a result of diffraction, reflection, and refraction effects during UV lithography 23. The fabricated circular microfilter is inserted between the filter cassettes with O-rings and connected to commercial syringe for ready to use.
Cancer Cell Preparation
In order to evaluate the present device's performance, we used both 6 different cancer cell lines for lung/colorectal cancer and 6 typical cell lines for 6 different cancers (breast, stomach, pancreas, prostate, ovary, cervical cancer). Each cell line was purchased or donated from ATCC and Samsung Medical Center (SMC), respectively. Cells were incubated under general cell culture conditions, however, their media compositions varies depending on type of cells. Typically, cells were cultured in RPMI or DMEM media containing 1% penicillin, 10% FBS for 3 days at 37ºC with 5% CO2 supplied incubator (SANYO, MCO18AIC). Prior to experiments using cancer cells, each cell line was trypsinized and detached cells were repeatedly spiked in 1x PBS buffer before it ready to use.
During cell culture, we measured the size of 6 different cancer cell lines for lung/colorectal cancer, then calculated the average excluding the biggest and smallest cell size. The average diameter of cell lines were 20.43μm for A549, 24.75μm for H23, 27.87μm for DLD-1, 22.56μm for SW620. The GFP-transfected cells had a diameter of 27.21μm for A549G and 29.71μm for DLD-1G. The size variation between GFP and non-GFP cells was 1.3μm to 4.8μm.
Blood Sample Preparation
Cancer cells spiked blood samples and cancer patients blood sample were used to evaluate the device performance and clinical use, respectively. The blood samples of healthy volunteers were collected under protocols approved by institutional review board (IRB). All blood samples were collected in BD Vacutainer tube containing ehylenediaminetetraacetic acid (EDTA) to prevent blood coagulation, and followed by dilution with ratio of 1:2 (Blood:PBS). The incubated and trypsinized cancer cells were carefully replaced in 96-well plate and enumerated under microscope. After then, known number of cancer cells were spiked in prepared diluted blood samples. In order to reduce the counting error, we enumerate the spiked cancer cell number 3 times, and used average number. The remained number of cancer cells on well plate was subtracted from number of spiked cancer cells. Depending on the aim of our experiments, high concentration sample (50 and 100 cancer cells/1ml of whole blood) containing GFP cancer cells or low-concentration sample (5, 10, and 15 cancer cells/1ml of whole blood) containing non-GFP cancer cells were prepared.
The blood samples from 18 patients with lung, colorectal cancer, pancreatic, and renal cell carcinoma were recruited from Samsung Medical Center (SMC) and Seoul National University Hospital (SNUH), Daegu Catholoic University Medical Center (DCUMC), and Samsung Medical Center (SMC), respectively, according to a protocol approved by IRB. Each blood sample (5ml for AMC or 10ml for SMC) was collected in BD Vacutainer® tube, and then used within 12 hours after sampling. As like cancer cell spiked blood sample, patient blood samples diluted to 1:2 without any pretreatment, such as erythrocyte lysis or density gradient isolation for minimizing the cell loss during any other process.
Experimental setup and sample processing
One syringe pump (KdS Legato 180, USA) was additionally prepared to generate stable and fixed negative flow in our microfilter device. The experimental setup is shown in Fig. 4A. We successfully captured the considerable number of cancer cells at the mode of manual sample injection by hands (Supplementary Material S1), however, we fixed the experimental setup using syringe pump for all experiments in order to verify our performance in controlled flow rate conditions. After connecting our device into syringe pump, the prepared samples were processed into our device under withdraw mode of syringe pump at the flow rate of 10ml/h. The captured cancer cells were enumerated to calculate the capture efficiency. After captured cancer cell enumeration, 5ml of PBS were injected reversely into device at the flow rate of 50ml/h in order to release the captured cancer cells. We calculated the release efficiency using the ratio of released cancer cell number to captured cancer cell number. For the experiments using non-GFP cancer cells, further immunofluorescence staining was conducted just before the enumeration of captured and released cancer cells. The viability of the captured cells was also verified using live/dead kit (Calcein AM/Eth-D). We enumerated the live/dead cells before and after capturing experiments and those viability test was performed independently of capture/release experiments. For the clinical samples, each sample was passed through the present microfilter, then washed out the remained cells with PBS. All captured cells were released and cytospinned on slide glass using cytocentrifuge (Thermo Scientific, USA). The cell-cytospinned slide glasses were followed by immunostaining.
Field Emission Scanning Electron Microscopy (FE-SEM)
The confirmation of the tapered-slit formation on the filter was examined with a SU5000 (Hitachi, Japan). FE-SEM was operated at an acceleration voltage of 0.5kV and with working distance of 1.7-5.5mm. The present filter was attached to double-sided adhesive carbon tape mounted on SEM stub and coated with osmium of 2.5nm thickness. The front side (outlet) and back side (inlet) of the microfilter were examined with an image magnified by a factor of 200 and 900.
Immunofluorescence staining protocol
To identify the non-GFP cancer cells and circulating tumor cells from patient bloods, we additionally stained the captured cells using cytokeratin, CD45, and DAPI. First, we fixed the captured cells on the present microfilter using 500μl of 4% paraformaldehyde solution at the flow rate of 5ml/h, then washed out excess fixation solution using 1ml of PBS buffer solution. To penetrate the staining dyes into target cells over the cell membrane, we loaded 500μl of 0.5% Triton X-100 solutions, and then added 500μl of 1% bovine serum albumin (BSA) solution to eliminate the non-specific binding of dye, which generates the false positive results. After blocking, we stained the captured cells using the 600μl of staining dye composed of 10μl of PE(or FITC)-conjugated cytokeratin dyes for epithelial cell staining, FITC(or PE)-conjugated CD45 dyes for WBC staining, and 5μl of DAPI staining dye for nucleus staining. We washed out the excess dye using 1ml of PBS at the flow rate of 5ml/h, disassembled the microfilter device, and then enumerated the epithelial cells under fluorescent microscope. The stained cells on the microfilter were scanned and manually enumerated under the automated microscope (Eclipse Ti, Nikon, Japan) equipped with 3 different fluorescence filter and MetaMorph system. All stained cells were carefully examined considering both staining criteria (CK+/CD45-/DAPI+) and morphological features, such as size, nucleus-to-cytoplasm ratio and irregularity. For the clinical samples, we additionally used the Cy-5-conjugated EpCAM dyes for epithelial CTC examination.
Immunohistochemical staining
For pathological confirmation of CTCs, immunohistochemical staining was conducted for lung and renal cancer cases independently. The additional 3ml of bloood sample was used and all captured cells from the present photosensitive polymer-based filter device were gently cytospinned on slide glasses, and fixed with 4% paraformaldehyde in 15 minutes. After fixation, each slide were immunostained by using the BenchMark XT Slide Preparation System (Ventana Medical Systems, Inc., Tucson, AZ, USA), with two to three antibodies. For lung cancer, TTF-1 (clone 8G7G3/1, 1:200, DAKO, Glostrup, Denmark) and p63 (clone 4A4, 1:100, Biocare Medical, Concord, CA, USA) were stained to two slides. For renal cancer , EpCAM (clone VU-1D9, 1:2,000, Calbiochem, San Diego, CA, USA), CD10 (clone 56C6, 1:400, Leica Biosystems, Newcastle, UK), and cytokeratin (clone AE1/AE3, 1:100, DAKO, Glostrup, Denmark) were stained to three slides. Positive expression was defined as the unequivocal brownish staining in the cytoplasm and/or cell membrane. Cytologic criteria for tumor cells were as follow: large cell size (1.5 times larger than white blood cells), large nuclear size with high nuclear/cytoplasmic ratio, irregular nuclear membrane, and presence of cytoplasm.27 The presence of tumor cells regardless of tumor-associated antibody expression was considered as CTC, and the total number of CTCs in entire slide was counted. All slides were examined by one pathologist (Chang, H.J.).
Real-time PCR
All captured cells were gently released from the device and anlalyzed using RT-PCR (BIO RAD, USA). We used the SuperPrep® Cell lysis & RT kit for qPCR (TOYOBO, Japan), which is optimized for rare cell RT-PCR, following the manufacturer's instructions, including lysis, reverse transcription, and real-time PCR. Briefly, we first lysed the captured cells using lysis solution and stopped enzymatic reaction by adding stop solution. Complimentary DNAs (cDNA) were synthesized from cell lysates, and additional qPCR was conducted with prepared specific target primers for quantitative assay. The qPCR reactions were performed using an C1000 thermal cycler (Bio-Rad, Hercules, CA) and underwent the following cycles: heat activation at 95°C for 15 min, 45 cycles of denaturation at 94°C for 15 s, annealing at 58°C and extension at 72°C. Three cancer-related genes (EpCAM, CK19, and MUC1) were used and each gene expression levels were normalized to expression level of GAPDH (housekeeping gene). We contained at least one no template control (NTC) and data were analyzed with Bio-Rad CFX Manager.
The primers used were: hEpCAM, sense: 5'-AATGTGTGTGCGTGGGA-3', antisense: 5'-TTCAAGATTGGTAAAGCCAGT-3'; hCK19, sense: 5'-AACGGCGAGCTAGAGGTGA-3', antisense: 5'-GGATGGTCGTGTAGTAGTGGC-3'; hMUC1, sense: 5'-TGCCGCCGAAAGAACTACG-3', antisense: 5'-TGGGGTACTCGCTCATAGGAT-3'; and hGAPDH, sense: 5'-CTTCACCACCATGGAGGAGGC-3', antisense: 5'-GGCATGGACTGTGGTCATGAG-3'.
Acknowledgments
This research was supported by the Fundamental R&D Programs for Core Technology of Materials funded by the Ministry of Trade, Industry and Energy (Project No. 10078295)
Supplementary Material
Supplementary figures and tables.
References
- 1.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayer DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S, Stathopoulos EN, Chlouverakis G, Lianidou E, Kakolyris S, Georgoulias V, Mavroudis D. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27(13):2177–84. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 3.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–39. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. P Natl Acad Sci. USA. 2010;107:18392–97. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, OuYang WH, Xu X, Zhang Z, Song M, Lee T, Garcia MA, Xiong B, Hou S, Tseng HR, Fang X. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013;25:2368–73. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Bréchot C, Paterlini-Bréchot P. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156(1):57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–61. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 8.Tan SJ, Yobas L, Lee G, Ong C, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microcevices. 2009;11:883–92. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 9.Huang T, Jia CP, Yang J, Sun WJ, Wang WT, Zhang HL, Cong H, Jing FX, Mao HJ, Jin QH, Zhang Z, Chen YJ, Li G, Mao GX, Zhao JL. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron. 2014;51:213–18. doi: 10.1016/j.bios.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Sim TS, Kwon K, Park JC, Lee JG, Jung HI. Multistage-multiorifice flow fractionation (MS-MOFF): continuous size-based separation of microspheres using multiple series of contraction/expansion microchannels. Lab Chip. 2011;11:93–99. doi: 10.1039/c0lc00109k. [DOI] [PubMed] [Google Scholar]
- 11.Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AAS, Chaudhuri PK, Tan DSW, Lim WT, Lee SC, Chen PCY, Lim CT, Han J. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14:128–37. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Lu W, Tang C, Liu Y, Sun J, Mu X, Zhang L, Dai B, Li X, Zhuo H, Jiang X. Label-Free Isolation and mRNA Detection of Circulating Tumor Cells from Patients with Metastatic Lung Cancer for Disease Diagnosis and Monitoring Therapeutic Efficacy. Anal Chem. 2015;87:11893–900. doi: 10.1021/acs.analchem.5b03484. [DOI] [PubMed] [Google Scholar]
- 13.Lee MG, Shin JH, Choi S, Park JK. Enhanced blood plasma separation by modulation of inertial lift force. Sens Actuators B Chem. 2014;190:311–17. [Google Scholar]
- 14.Warkiani ME, Khoo BL, Wu L, Tay AKP, Bhagat AAS, Han J, Lim CT. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc. 2016;11(1):134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 15.Fan X, Jia C, Yang J, Li G, Mao H, Jin Q, Zhao J. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens Bioelectron. 2015;71:380–386. doi: 10.1016/j.bios.2015.04.080. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, Shi J, Li S, Wang L, Cayre YE, Chen Y. Microfluidic device with integrated microfilter of conical-shaped holes for high efficiency and high purity capture of circulating tumor cells. Sci Rep. 2014;4:6052. doi: 10.1038/srep06052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin HK, Zheng SY, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Portable Filter-Based Microdevice for Detection and Characterization of Circulating Tumor Cells. Clin Cancer Res. 2010;16:5011–18. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, Moulec SL, André F, Fizazi K, Soria JC, Vielh P. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–53. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams DL, Stefansson S, Haudenschild C, Martin SS, Charentier M, Chumsri S, Cristofanilli M, Tang CM, Alpaugh RK. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch(®) CTC test. Cytometry A. 2015;87:137–44. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 20.Zheng SY, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13:203–13. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou MD, Hao S, Williams AJ, Harouaka RA, Schrand B, Rawal S, Ao Z, Brenneman R, Gilboa E, Lu B, Wang S, Zhu J, Datar R, Cote R, Tai YC, Zheng SY. Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells. Sci Rep. 2014;4:7392. doi: 10.1038/srep07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang YT, Doh I, Cho YH. Tapered-slit membrane filters for high-throughput viable circulating tumor cell isolation. Biomed Microdevices. 2015;17(2):45. doi: 10.1007/s10544-015-9949-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Chan-Park MB, Conner SR. Effect of exposure dose on the replication fidelity and profile of very high aspect ratio microchannels in SU-8. Lab Chip. 2004;4(6):646–653. doi: 10.1039/b403304c. [DOI] [PubMed] [Google Scholar]
- 24.Doh I, Yoo HI, Cho YH, Lee J, Kim HK. Viable capture and release of cancer cells in human whole blood. Appl Phys Lett. 2012;101:043701. [Google Scholar]
- 25.Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, Takahashi T, Murakami H, Nakamura Y, Tsuya A, Shukuya T, Ono A, Akamatsu H, Watanabe R, Ono S, Mori K, Kanbara H, Yamaguchi K, Tanaka T, Matsunaga T, Yamamoto N. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. Plos One. 2013;8(6):e67466. doi: 10.1371/journal.pone.0067466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T, Matsunaga T. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal Chem. 2010;82:6629–35. doi: 10.1021/ac101222x. [DOI] [PubMed] [Google Scholar]
- 27.Nam SJ, Yeo HY, Chang HJ, Kim BH, Hong EK, Park JW. A New Cell Block Method for Multiple Immunohistochemical Analysis of Circulating Tumor Cells in Patients with Liver Cancer. Cancer Res Treat. 2016;48(4):1229–1242. doi: 10.4143/crt.2015.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.