Abstract
The fission yeast Schizosaccharomyces pombe expresses the CCAAT-binding factor Php4 in response to iron deprivation. Php4 forms a transcription complex with Php2, Php3, and Php5 to repress the expression of iron proteins as a means to economize iron usage. Previous in vivo results demonstrate that the function and location of Php4 are regulated in an iron-dependent manner by the cytosolic CGFS type glutaredoxin Grx4. In this study, we aimed to biochemically define these protein-protein and protein-metal interactions. Grx4 was found to bind a [2Fe-2S] cluster with spectroscopic features similar to other CGFS glutaredoxins. Grx4 and Php4 also copurify as a complex with a [2Fe-2S] cluster that is spectroscopically distinct from the cluster on Grx4 alone. In vitro titration experiments suggest that these Fe-S complexes may not be interconvertible in the absence of additional factors. Furthermore, conserved cysteines in Grx4 (Cys172) and Php4 (Cys221 and Cys227) are necessary for Fe-S cluster binding and stable complex formation. Together, these results show that Grx4 controls Php4 function through binding of a bridging [2Fe-2S] cluster.
TOC image
Grx4 forms a cysteine-ligated [2Fe-2S] binding complex with the transcriptional repressor Php4 to regulate transcription of iron utilization genes.
Introduction
Maintenance of iron homeostasis is an essential function for most organisms given the critical role for this metal in cell metabolism. Low intracellular iron levels hamper the activity of iron-dependent enzymes, while iron overaccumulation can disrupt other metal trafficking pathways and catalyze formation of reactive oxygen species. Consequently, cells must closely regulate iron uptake and usage to maintain a balance between sufficient and excess levels.
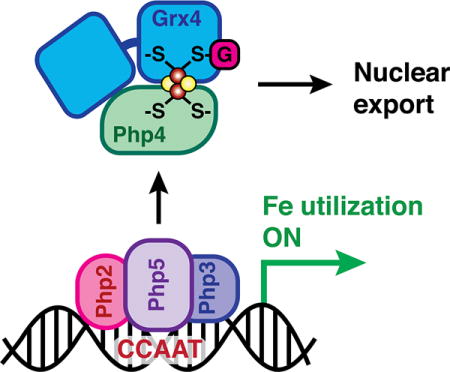
The fission yeast Schizosaccharomyces pombe employs two transcription factors to control cellular iron levels, Fep1 under iron replete conditions, and Php4 under iron deprivation.1 The actions of Php4 and Fep1 are coordinated at the transcriptional level since Fep1 and Php4 reciprocally regulate expression of each other in an iron-dependent manner.2 Fep1 is a GATA-type transcriptional repressor that mainly controls genes involved in iron acquisition.3 When cells are iron-replete, Fep1 binds to GATA sequences in these genes and represses their transcription, downregulating iron transport systems. When cells are iron deficient, Php4 acts as a transcriptional repressor with the CCAAT-binding complex.4 In S. pombe, Php2, Php3 and Php5 form a heterotrimer that binds CCAAT cis-acting elements to activate gene expression. Genes regulated by this complex encode for proteins involved in iron-dependent metabolic pathways such as the TCA cycle, mitochondrial respiration, amino acid biosynthesis, heme biosynthesis, and iron-sulfur cluster assembly.5 Under iron-deplete conditions, Php4 is recruited to the CCAAT-binding complex, causing it to switch from an activator to a repressor. The main function of Php4 is therefore iron conservation: when cellular iron is limiting, expression of genes encoding iron-dependent proteins is repressed.
While expression of Php4 at the transcriptional level is controlled by Fep1, Php4 is also regulated in an iron-dependent manner at the protein level. The cytosolic CGFS glutaredoxin Grx4 controls both Fep1 and Php4 function since Fep1- and Php4-regulated genes are constitutively repressed in a grx4Δ strain, causing sensitivity to iron depletion.6–9 In the case of Php4, Grx4 regulates interaction with the Php2/3/5 complex as well as the exportin Crm1.8 Under iron-deplete conditions, Php4 accumulates in the nucleus and binds to the CCAAT-binding complex (Php2/3/5) to repress iron utilization genes. When iron levels increase, Php4 dissociates from Php2/3/5 and is relocalized to the cytosol by Crm1. Yeast two-hybrid mapping experiments revealed that Grx4 physically interacts with the C-terminal region of Php4, through the N-terminal thioredoxin-like (TRX) and C-terminal glutaredoxin-like (GRX) domains of Grx4.10 Association with the TRX domain is relatively weak and constitutive, while association with the GRX domain is iron-dependent and requires Cys172 of the conserved CGFS active site. Transcriptional analysis of Php4-regulated genes in strains expressing C172S/A Grx4 mutants indicates that Cys172 is absolutely required for Grx4-dependent inhibition of Php4 activity.6,9,10 In Php4, Cys221 and Cys227 in the C-terminus are specifically required for interaction with the GRX domain of Grx4, and may be involved in iron-mediated inactivation of Php4.10
It has been well-established that both prokaryotic and eukaryotic CGFS Grxs form [2Fe-2S]-bridged homodimers with the CGFS active site Cys and two glutathione (GSH) molecules serving as the iron ligands.11–22 Furthermore, the S. cerevisiae orthologs of S. pombe Grx4, namely Grx3/4, form [2Fe-2S]-bridged heterodimers with the BolA protein Fra2, which function in delivering an inhibitory Fe-S cluster to the transcriptional activators Aft1 and Aft2.23 Given this precedent for S. cerevisiae Grx3/4 forming [2Fe-2S]-bridged complexes with iron regulatory proteins, and the previous studies in S. pombe documenting the in vivo Cys-dependent physical and functional interactions between Grx4 and Php4, these two proteins are proposed to form a Fe-S binding complex that inhibits Php4 activity in iron-replete conditions.10,24 However, characterization of this complex at the molecular level has not yet been reported.
In order to understand how Grx4 physically interacts with and regulates Php4, we expressed and characterized both proteins using biochemical, analytical, and spectroscopic techniques. We determined that as-purified Grx4 binds a [2Fe-2S] cluster similar to other CGFS glutaredoxins. While Php4 expressed alone does not co-purify with iron or an Fe-S cluster, it does bind a [2Fe-2S] cluster in complex with Grx4. In addition, we found that the conserved Cys residues in Grx4 (C172) and Php4 (C221/C227) are required for Fe-S cluster binding and strengthen the protein-protein interaction between Php4 and Grx4. Taken together, these results suggest that Grx4 regulates Php4 function through complex formation with a bridging [2Fe-2S] cluster.
Experimental
Plasmid construction
The open reading frame (ORF) of S. pombe Php4 was amplified from S. pombe genomic DNA by PCR and cloned into the NcoI and SalI sites of pRSFDuet-1 (Novagen) to create pRSFDuet-1-Php4, subsequently generating 6xHis-tagged Php4. The ORF of S. pombe Grx4 was amplified from S. pombe genomic DNA by PCR and cloned into the NdeI and XhoI sites of pRSFDuet-1 to create pRSFDuet-1-Grx4. Dual expression plasmids for Php4-Grx4 were made by inserting the Php4 ORF into the pRSFDuet-1-Grx4 plasmid. Php4 (C221A, C227A) and Grx4 (C172A) mutants were created by site-directed mutagenesis of the above plasmids.
Protein Expression and Purification
Overexpression of Grx4 was performed in the E. coli BL21(DE3) strain in LB media at 37 °C. Cells were grown to A600 = 0.6–0.8, then were induced with 20 μM isopropyl β-D-thiogalactosidase (IPTG) and incubated at 25 °C. Cells were collected 18 h after induction and resuspended in 50 mM Tris-HCl, pH 8.0, followed by sonication and centrifugation to remove cell debris. The cell-free extract was subjected to ammonium sulfate precipitation with the protein salting out in the 40% cut. The protein pellet was resuspended in 50 mM Tris-HCl, pH 8.0, 750 mM (NH4)2SO4 and loaded onto a Phenyl Sepharose column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 8.0, 750 mM (NH4)2SO4, 100 mM NaCl. The protein was eluted with a 750-0 mM (NH4)2SO4 gradient, and fractions containing Grx4 as judged by SDS-PAGE and UV-visible spectroscopy were collected and concentrated. Concentrated protein was loaded onto a HiLoad Superdex 75 gel filtration column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 8.0, 150 mM NaCl. The purest fractions of [2Fe-2S] Grx4 as judged by SDS-PAGE and UV-visible spectroscopy were collected, concentrated, and stored at −80 °C. Coexpression of Php4 with Grx4 was performed with the pRSFDuet-1-Php4-Grx4 expression plasmid transformed into E. coli BL21(DE3) using the procedure described above for Grx4.
Overexpression of Php4 was performed in the E. coli BL21(DE3) strain in LB media at 37 °C. Mid-logarithmic-phase cells (A600 = 0.6–0.8) were induced with 1 mM IPTG and grown at 16 °C. Cells were collected 18 h after induction and resuspended in 50 mM MES, pH 6.0, followed by sonication and centrifugation to remove cell debris. The cell-free extract was loaded onto a SP FF cation-exchange column (GE Healthcare) equilibrated with 50 mM MES, pH 6.0. Most of the protein eluted in the wash step and was collected and exchanged into 50 mM Tris-HCl, pH 8.0 buffer. This sample was loaded to a DEAE anion-exchange column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 8.0. The protein was eluted with a 0–1 M NaCl gradient using 50 mM Tris-HCl, pH 8.0, 1 M NaCl. Fractions containing Php4 as judged by SDS-PAGE were collected, concentrated, and loaded onto a HiLoad Superdex 75 gel filtration column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 8.0, 150 mM NaCl. The purest fractions of Php4 as judged by SDS-PAGE were collected, concentrated, and stored at −80 °C. All Php4 and Grx4 mutants (either alone or coexpressed) were purified using the same procedure as the wild-type forms. All purifications were carried out under anaerobic conditions (O2 < 5 ppm) in a glovebox (Coy Laboratory Products), with the exception of Php4 WT and mutants (expressed without Grx4). S. cerevisiae Fra2Δ1–35 was overexpressed in E. coli and purified as previously described.25
Biochemical Analyses
Protein concentrations were determined by the Bradford Assay (Bio-Rad) using bovine serum albumin as the standard. Iron and acid-labile sulfur concentrations were measured as previously described.13 For GSH measurements, the purified Fe-S protein complexes were denatured and precipitated with 1% 5-sulfosalicylic acid, and GSH in the supernatant was measured by the 5, 5′-dithiobis(2-nitrobenzoic acid) -GSSG reductase cycling assay as described previously.26 We note that buffers used for Grx4 and Php4 purification did not contain any GSH, so any GSH bound to the protein complexes is derived from the E. coli cells.
Analytical and Spectroscopic Methods
Analytical gel filtration analyses were performed on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM GSH and calibrated with a high molecular weight calibration kit (GE Healthcare). The same set of fractions was collected for all proteins eluted from gel filtration, and were analyzed by SDS-PAGE in order to compare relative elution times.
UV-visible absorption spectra were recorded using a Beckman DU-800 spectrophotometer. CD spectra were recorded under anaerobic conditions on identical samples using a Jasco J-815 spectropolarimeter (Jasco, Easton, MD). EPR spectra were recorded using a Bruker EMX plus spectrometer (Bruker, Billerica, MA), equipped with an Oxford ESR900 continuous flow cryostat (Oxford Instruments, Oxfordshire, UK), and quantified under nonsaturating conditions by double integration against a 1.0 mM CuEDTA standard. The samples were reduced under anaerobic conditions by addition of stoichiometric sodium dithionite and frozen within five minutes in liquid nitrogen. EPR conditions were as follows: microwave frequency, ~9.6 GHz, modulation frequency, 100 kHz, modulation amplitude, 10 G, microwave power, 10 mW, and temperature 10–20 K.
Iron-sulfur cluster reconstitutions
Purified Grx4 or Php4-Grx4 was incubated with 20-fold excess ferrous ammonium sulfate, 20-fold excess L-cysteine, a catalytic amount of IscS, and 5 mM GSH in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl under anaerobic conditions on ice for 2 h. Excess reagents were purified out using a Phenyl Sepharose column (GE Healthcare) with a decreasing linear gradient from 750 to 0 mM (NH4)2SO4, and the red-colored fractions were pooled and concentrated.
Surface plasmon resonance
Binding kinetics of apo Php4-Grx4 complexes were determined by surface plasmon resonance (SPR), using a Biacore 3000 (Biacore, GE Healthcare). His-Php4 (WT) was immobilized on a Sensor Chip NTA activated with Ni2+ according to the manufacturer’s instructions. Grx4 (WT or C172A) was injected over the flow cell for 9.3 min at 9–10 different concentrations between 25 μg/ml (0.9 μM) and 20 mg/ml (740 μM) using a flow rate of 35 μl/min. The assays were run at 25° C in 50 mM Tris-HCl, pH 8.0, 0.005% Tween-20 with a blank injection (buffer only) between each run. Bound protein was removed with a 60-sec wash of 25 mM NaOH using a flow rate of 15 μl/min. The NTA sensor chip was regenerated with 300 mM EDTA. Global data analysis was performed using BIAEvaluation 4.1 (Biacore Software) applying a Langmuir 1:1 binding model with baseline drift to calculate kon (association rate constant) and koff (dissociation rate constant). A linear baseline drift was included to account for leaching of His-Php4 from the sensor surface during the association and dissociation phases.
CD-monitored titrations of Php4 and Grx4
Several titration methods were used in an attempt to convert between Grx4 homodimer and Php4-Grx4 heterocomplexes. All samples were prepared and scanned under anaerobic conditions with 1 mM GSH, and the [2Fe-2S] content was kept constant at 50 μM. Php4 was pre-incubated with 5 mM DTT to reduce any disulfides formed during purification, and then desalted anaerobically before mixing with [2Fe-2S] Grx4. Method 1: Increasing amounts of Php4 were added to [2Fe-2S] Grx4 up to a 2.5 : 1 ratio of Php4 to [2Fe-2S]. Method 2: [2Fe-2S] Grx4 was incubated first with 2-fold molar excess S. cerevisiae Fra2 to generate a [2Fe-2S] Grx4-Fra2 heterodimer. Php4 was added to this mixture in 2-fold and 5-fold molar excesses relative to [2Fe-2S]. A 1:2 mixture of [2Fe-2S] Grx4:Fra2 was used as a control.
Results and discussion
Grx4 binds a [2Fe-2S]2+ cluster
In a recent report, recombinant S. pombe Grx4 was reconstituted with an Fe-S cluster, which requires GSH and is sensitive to the presence of oxygen.6 However, only the UV-visible spectrum of this Fe-S cluster complex was provided, which showed a single peak ~400 nm, as well as two shoulders between 500–700 nm, indicative of a mixture of [2Fe-2S] and linear [3Fe-4S] clusters.27 Previously published spectra of [2Fe-2S] Grxs purified under strictly anaerobic conditions typically have two distinct peaks at ~320 and ~410 nm, with a characteristic shoulder at ~445 nm.11,13,28,29 To confirm that S. pombe Grx4 binds a bona fide [2Fe-2S]2+ cluster similar to previously characterized Grxs, we overexpressed and purified recombinant S. pombe Grx4 from E. coli. The purified protein had a reddish-brown color, and was investigated by spectroscopic and biochemical methods. The UV-visible absorption spectrum of as-purified Grx4 is nearly identical to other monothiol glutaredoxins from S. cerevisiae (Sc Grx3) and E. coli (Ec Grx4), with peaks at 323, 413, and 445 nm, indicative of an [2Fe-2S]2+ cluster (Fig. 1). As expected, the CD spectrum for S. pombe Grx4 was also comparable to Sc Grx3 and Ec Grx4. Positive peaks at 312, 460, and 550–590 nm and negative peaks at 348, 408, and 518 nm are characteristic of [2Fe-2S]2+-bridged Grx homodimers.
Fig. 1.
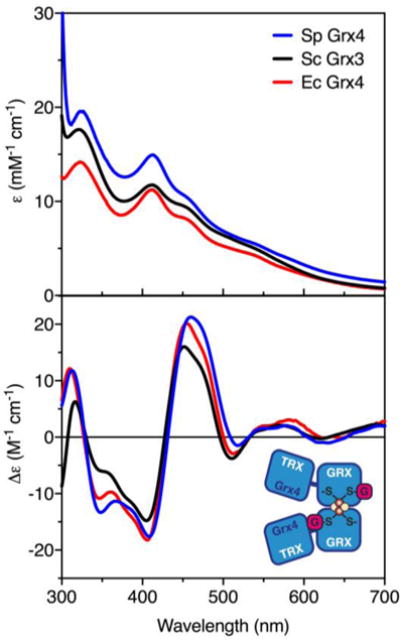
UV-visible absorption (top) and CD spectra (bottom) of [2Fe-2S] Grx4 from S. pombe (Sp, blue line), [2Fe-2S] Grx3 from S. cerevisiae (Sc, black line),13 and [2Fe-2S] Grx4 from E. coli (Ec, red line).27 ε and Δε values are based on [2Fe-2S] concentration. Inset, model for [2Fe-2S] Grx4 demonstrating ligation of the cluster by the GRX domain.
Grx4 and Php4 copurify as a complex
To characterize the interactions between Php4 and Grx4 in vitro, Php4 was initially expressed and purified separately. Soluble recombinant Php4 was extracted from E. coli, yielding apo, monomeric protein, with no apparent color in the protein extract. To determine if Grx4 forms a stable complex with Php4, the proteins were coexpressed in E. coli. The induced cells had a distinct reddish-brown color, similar to cells overexpressing Grx4 alone, which was not observed when Php4 was expressed alone. Moreover, Php4 and Grx4 were found to copurify as a stable complex with a reddish-brown color. The individual proteins and the Php4-Grx4 complex were subjected to size-exclusion chromatography to confirm complex formation. The same fractions were collected from the elution of each protein or complex, and analyzed by SDS-PAGE (Fig. 2). Php4 expressed alone runs as a monomer in fractions 5–7 (theoretical mass: 34.4 kD, apparent mass: 30–40 kD). [2Fe-2S] Grx4 elutes in fractions 4–5 (theoretical [2Fe-2S] homodimer mass: 55.0 kD, apparent mass: 60–90 kD) and higher order oligomers in fractions 1–3 (>90 kD). Coexpression of Php4 with Grx4 shows no sign of the Php4 monomer but instead results in co-elution of the two proteins in fractions 4–6 (theoretical heterodimer mass 63.1 kD; apparent mass, 60–90 kD) and higher order oligomers in fractions 2–3 (>90 kD). We note that the apparent molecular masses for Grx4 expressed alone and the Grx4-Php4 heterocomplex were difficult to determine since samples with Grx4 tended to aggregate and/or adsorb to the column during the experiment, resulting in Grx4 elution across a broad range of fractions. Nevertheless, comparison of Php4 expressed alone versus with Grx4 shows a clear shift in the elution time, indicating formation of a complex between Php4 and Grx4 with a higher apparent molecular mass than Php4 alone.
Fig. 2.
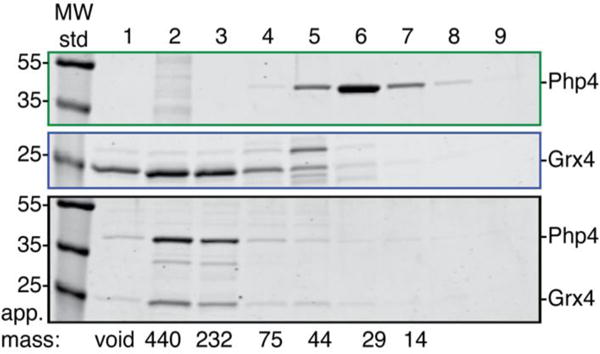
Php4 and Grx4 coelute as a complex. SDS-PAGE analysis of gel filtration fractions collected for purified Php4 expressed alone (top), Grx4 expressed alone (middle), and Php4 coexpressed with Grx4 (bottom). Identical fractions from each analytical gel filtration experiment were loaded for comparison of elution volumes. In this experiment, larger proteins/complexes elute earlier and smaller ones later. The theoretical masses of Php4 and Grx4 are 34.3 and 27.1 kD, respectively. Masses (in kD) of MW standards used in the SDS-PAGE gel are shown on the left. The elution position and apparent mass (in kD) of the molecular weight standards used to calibrate the gel filtration column are shown across the bottom.
Php4-Grx4 binds a [2Fe-2S] cluster with a coordination environment distinct from [2Fe-2S] Grx4
The identity of the chromophore on the purified Php4-Grx4 complex was investigated and compared to Grx4. Both the absorption and CD spectra of the Php4-Grx4 complex suggest the presence of a [2Fe-2S] cluster; however, the spectra of the Fe-S cluster on Php4-Grx4 are significantly different from those of the Fe-S center on Grx4 alone (Fig. 3A). In particular, the dominant visible absorption band for Php4-Grx4 is blue shifted to 408 nm compared to the equivalent band for Grx4 at 413 nm. In addition, Php4-Grx4 has a shoulder ~ 320 nm instead of a distinct peak as seen for Grx4 (Fig. 3A, top). These differences are reflected in the CD spectrum of Php4-Grx4, which exhibits changes in the intensity and wavelengths of the visible CD bands (Fig. 3A, bottom). Overall, comparison of the excited state electronic structures of the [2Fe-2S]2+ centers in Php4-Grx4 and Grx4 alone suggest differences in the coordination environment and/or chirality of the [2Fe-2S] centers.
Fig. 3.
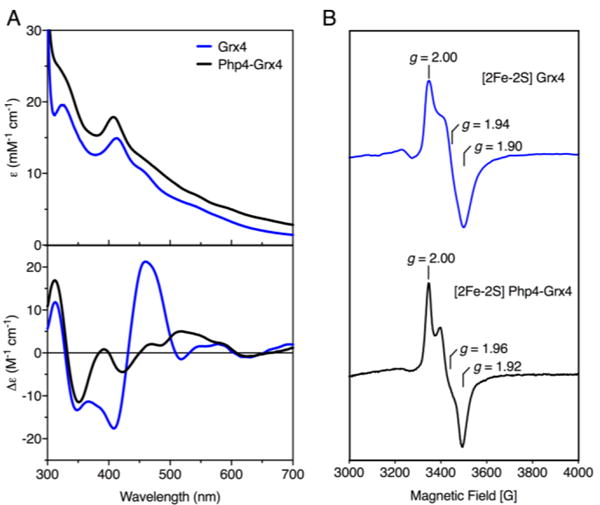
[2Fe-2S] Grx4 and [2Fe-2S] Php4-Grx4 complexes are distinct. (A) Comparison of UV-visible absorption (top) and CD spectra (bottom) of [2Fe-2S] Php4-Grx4 heterodimer (black line) with [2Fe-2S] Grx4 homodimer (blue line). ε and Δε values are based on [2Fe-2S] concentration. (B) Comparison of the X-band EPR spectra of dithionite-reduced [2Fe-2S] Grx4 (top) with [2Fe-2S] Php4-Grx4 (bottom). Spectra recorded at 10–20 K and 10 mW microwave power with a microwave frequency of 9.60 GHz and modulation amplitude of 10 G.
To confirm binding of an Fe-S cluster in the Php4-Grx4 complex and investigate the role of GSH in cluster ligation, we measured iron and acid-labile sulfide in the Grx4 and Php4-Grx4 complexes. These analyses show an Fe:S ratio close to 1:1 for Grx4 and Php4-Grx4 with ~0.42 [2Fe-2S] clusters per Grx4 dimer and ~0.41 [2Fe-2S] clusters per Php4-Grx4 (Table 1). Interestingly, the GSH measurement of the Php4-Grx4 complex suggests only 1 GSH bound per [2Fe-2S] cluster (Fe:S:GSH ratio of 1:1:0.4) in comparison to the Grx4 alone, which is closer to 2 GSH per cluster (Fe:S:GSH ratio of 1:1:0.8). These clear differences in GSH coordination to the clusters in Grx4 and Php4-Grx4 may reflect the distinct coordination environments observed in the UV-visible absorption and CD spectra.
Table 1.
Fe, S2−, and GSH measurements for purified Fe-S cluster proteins. Values are reported per dimer. Data are the average of three independent samples.
| Protein | Fe | S2- | GSH | Fe:S:GSH |
|---|---|---|---|---|
| Grx4 | 0.84 ± 0.18 | 0.83 ± 0.10 | 0.70 ± 0.17 | 1 : 1 : 0.8 |
| Php4-Grx4 | 0.81 ± 0.05 | 0.78 ± 0.08 | 0.32 ± 0.06 | 1 : 1 : 0.4 |
The EPR spectra of Grx4 and Php4-Grx4 provide information about ligand identity, redox properties and stability of the reduced [2Fe-2S]+ cluster in each complex (Fig. 3B). Both samples were unstable to reduction as evidenced by the absence of an EPR signal upon addition of excess sodium dithionite. Thus, samples were reduced with stoichiometric amounts of dithionite under anaerobic conditions, and frozen under liquid nitrogen within five minutes. The [2Fe-2S]+ Grx4 homodimer gave a rhombic S = ½ EPR signal, g1,2,3 = 2.00, 1.94, and 1.90 (gav ~ 1.95), accounting for less than 10% of the cluster content (Fig. 3B, top). This signal is similar to other [2Fe-2S]-bridged CGFS Grxs, which are also known to be reductively labile, resulting in a low number of spins per cluster.13,28,29 [2Fe-2S]+ Php4-Grx4 also gives a rhombic S = ½ EPR signal, g1,2,3 = 2.00, 1.96, and 1.92 (gav = 1.96), accounting for ~20% of the cluster content (Fig. 3B, bottom). Therefore, as with [2Fe-2S] Grx4, the [2Fe-2S] cluster on Php4-Grx4 is reductively labile. The gav values and g-value anisotropy provide information about the ligation of [2Fe-2S]+ clusters, which are sensitive to ligation changes at the Fe(II) site.13 [2Fe-2S]+ clusters with all-cysteine ligation exhibit gav ~1.95–1.97, whereas gav decreases to ~1.93–1.94 when one cysteine is replaced by a histidine.13 Given these trends, S. pombe Grx4 likely binds its [2Fe-2S] cluster through all-cysteine ligation from the active site CGFS cysteines and the cysteine moiety of two GSH molecules, as demonstrated for other CGFS Grxs. Similarly, the signal obtained for Php4-Grx4 is also characteristic of a [2Fe-2S]+ cluster with all-cysteine ligation. Taken together, these results confirm that Php4 binds a bridging [2Fe-2S]2+ cluster with Grx4.
[2Fe-2S] Php4-Grx4 and [2Fe-2S] Grx4 are not interconvertible in vitro
We previously demonstrated that S. cerevisiae [2Fe-2S] Grx3/Grx4 homodimers interact with the cytosolic protein Fra2 to form heterodimeric complexes.13,25,30 Fra2 is a member of the BolA family of proteins that serve as functional bindingpartners for CGFS Grxs in a variety of organisms.14 Grx3/4-Fra2 heterodimers, in turn, regulate activity of the S. cerevisiae Fe- responsive transcription factors Aft1/Aft2 via specific transfer of the [2Fe-2S] cluster from Grx3-Fra2 to Aft2.23 The stoichiometric conversion of [2Fe-2S] Grx3 homodimer to [2Fe-2S] Grx3-Fra2 heterodimer upon titration with Fra2 as well as Fe-S cluster transfer from Grx3-Fra2 to Aft2 was monitored by CD spectroscopy, providing a convenient handle to observe changes in Fe-S cluster coordination.23,25 Since S. pombe [2Fe-2S] Grx4 and [2Fe-2S] Grx4-Php4 complexes have distinct CD spectra, we used a similar approach to determine if [2Fe-2S] Grx4 is converted to the [2Fe-2S] Php4-Grx4 complex upon titration with purified Php4. Addition of up to 2.5-fold molar excess of Php4 to [2Fe-2S] Grx4 did not yield any significant changes in the CD spectrum (Fig. 4A), suggesting that Php4 does not bind the cluster or alter the Fe-S cluster coordination environment under these conditions. The addition of 5 mM DTT to this mixture, which has been shown to accelerate Fe-S cluster transfer reactions,31 also did not facilitate formation of the [2Fe-2S] Php4-Grx4 heterocomplex (data not shown). These results demonstrate that [2Fe-2S] Grx4 cannot be converted to the [2Fe-2S] Php4-Grx4 complex by simple titration with Php4.
Fig. 4.
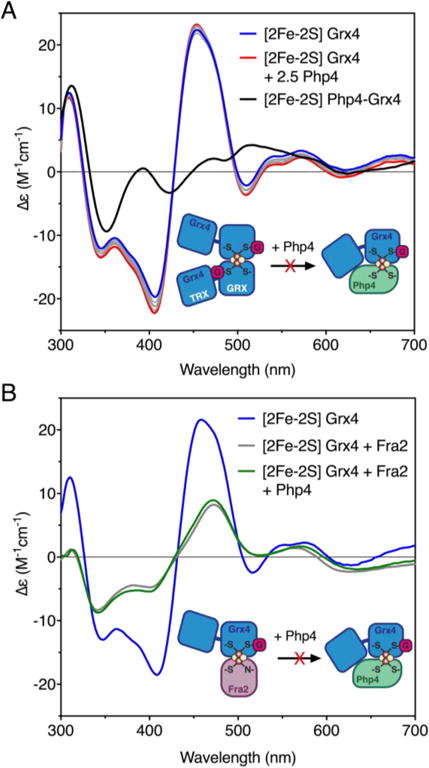
CD-monitored titrations of [2Fe-2S] Grx4 with Php4 and S. cerevisiae Fra2. (A) CD spectra for [2Fe-2S] Grx4 (blue line) titrated with 0.1 – 2.0 molar excess Php4 (gray lines). The red line corresponds to 2.5 molar excess Php4 added to [2Fe-2S] Grx4. As-purified [2Fe-2S] Php4-Grx4 is shown as a black line. (B) CD spectra for as-purified [2Fe-2S] Grx4 (blue line) incubated with 2-fold molar excess of Sc Fra2 (gray line), or 2-fold excess of Sc Fra2 then 5-fold excess of Php4 (green line). Δε values are based on the [2Fe-2S]2+ cluster concentration [75 μM for (A) and 50 μM for (B)].
In S. cerevisiae, the iron-responsive transcription factor Aft2 preferentially interacts with and accepts an Fe-S cluster from [2Fe-2S] Grx3-Fra2 heterodimers rather than [2Fe-2S] Grx3 homodimers.23 To determine whether Fra2 has a similar role in facilitating interaction between S. pombe Php4 and Grx4, we tested whether a Grx4-Fra2 heterodimer is capable of interacting with Php4. Although in vivo data argue against a role for S. pombe Fra2 in the Php4-Grx4 interaction,32 we tested the Grx4-Php4-Fra2 interaction by mixing [2Fe-2S] Grx4 with Fra2 prior to adding Php4 (Fig. 4B). The S. cerevisiae (Sc) and S. pombe (Sp) Fra2 proteins are orthologs (39% identical, 63% similar), and Sc Fra2 can complement Sp Fra2 function under iron depletion conditions in vivo (Fig. S1, ESI†). Therefore, Sc Fra2 was used in the titration with Sp Grx4 since this purified protein was available. Mixing [2Fe-2S] Sp Grx4 with a two-fold excess of Sc Fra2 resulted in distinct changes in the CD spectrum (Fig. 4B, gray line), demonstrating that interaction of Fra2 with [2Fe-2S] Grx4 alters the coordination environment of the Fe-S cluster, similar to the effect observed upon titration of [2Fe-2S] Sc Grx3 with Sc Fra2.25 However, addition of Php4 to this mixture in both 2- and 5-fold excesses did not yield any further changes compared to the [2Fe-2S] Grx4 + Fra2 spectrum (Fig. 4B, compare gray and green lines). Thus, these results suggest that Fra2 does not facilitate interaction of Php4 with Grx4 under these conditions, consistent with the in vivo findings previously reported.32
We next asked if Fe-S complex formation occurred in the reverse direction by testing whether the [2Fe-2S] Php4-Grx4 heterocomplex could be converted to the [2Fe-2S] Grx4 homodimer by titration with apo-Grx4 (Fig. S2A, ESI†). Addition of excess Grx4 did not convert [2Fe-2S] Php4-Grx4 to the [2Fe-2S] Grx4 homodimer, indicating that these forms are not interconvertible under these conditions. Furthermore, we tested whether pre-mixed apo Php4-Grx4 accepts a cluster from [2Fe-2S] Grx4 homodimer. In vivo, Php4 and Grx4 interact under iron deficiency conditions, thus the apo complex may need to be formed before cluster transfer.10 However, by titrating apo Php4-Grx4 into [2Fe-2S] Grx4, we saw no evidence for Fe-S cluster transfer (Fig. S2B, ESI†). These results suggest that the conversion of [2Fe-2S] Grx4 to [2Fe-2S] Php4-Grx4 may require some other protein or factor found in cells.
Finally, we tested whether formation of the Fe-S-bridged Grx4 homodimer or Php4-Grx4 complex is preferred upon anaerobic cysteine desulfurase-mediated Fe-S reconstitution of apo Php4-Grx4. As a control, a sample of apo-Grx4 was subjected to reconstitution in the absence of Php4, generating a CD signal that closely overlapped the spectrum for as-purified [2Fe-2S] Grx4 (Fig. 5, blue lines). However, Fe-S reconstitution of a 1:1 mixture of Php4 and apo-Grx4 produced a sample with CD peaks that more closely matched as-purified [2Fe-2S] Php4-Grx4 (with some intensity differences), with little indication of [2Fe-2S] Grx4 homodimer formation (Fig. 5, black lines). These results suggest that [2Fe-2S] Php4-Grx4 is the thermodynamically preferred cluster-bound complex under these conditions.
Fig. 5.
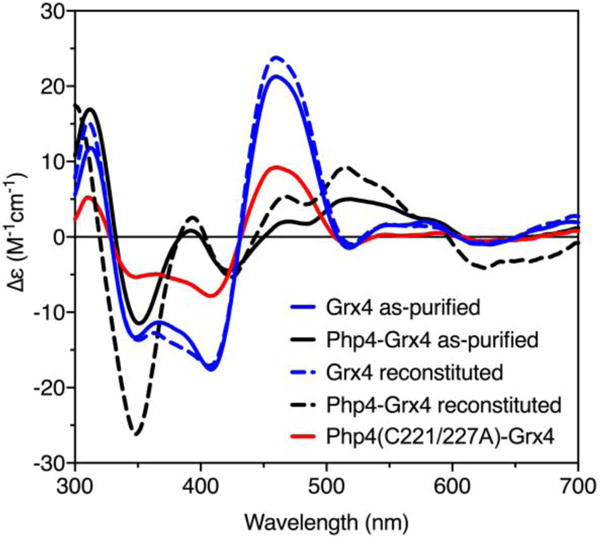
Comparison of CD spectra of as-purified [2Fe-2S] Grx4 (blue solid line) and [2Fe-2S] Php4-Grx4 (black solid line) with reconstituted [2Fe-2S] Grx4 (blue dotted line), [2Fe-2S] Php4-Grx4 (black dotted line), and [2Fe-2S] Php4(C221/227A)-Grx4 (red line).
Grx4 C172 and Php4 C221/C227 are required for [2Fe-2S] cluster binding and stable complex formation
The conserved CGFS active site in S. pombe Grx4 includes the cysteine residue (Cys172) that serves as an Fe-S cluster ligand in previously characterized CGFS Grxs from other organisms.11–13 A recent study demonstrated that S. pombe Grx4 cannot be reconstituted with an Fe-S cluster when this cysteine is changed to a serine (C172S).6 To confirm this result, we tested whether a C172A substitution would affect Fe-S cluster-binding in the as-purified Grx4 homodimer alone or in complex with Php4. Grx4 (C172A) overexpressed alone or with Php4 was found to be predominantly insoluble. Insoluble protein pellets did not have any color to indicate Fe or Fe-S cluster binding. Protein was extracted from inclusion bodies with urea, followed by a stepwise dialysis to remove urea and allow for protein refolding. This sample purified without an Fe-S cluster and was found to be relatively unstable. Although Grx4 (C172A) still copurified with Php4 to some extent (Fig. S3), the ratio of Php4 to Grx4 is visibly reduced compared to the WT complex (compare relative Php4 and Grx4 band intensities in Php4-Grx4 complexes in Fig. 2 and Fig. S3, ESI†). Presumably, the Php4-Grx4 interaction is not completely abolished by mutating this Cys due to the presence of the TRX domain-dependent interaction detected in vivo.10
As mentioned previously, Php4 contains two conserved cysteines, Cys221 and Cys227, which are required for iron-dependent interaction with Grx4 in vivo.10 These cysteines are hypothesized to act as Fe-S cluster ligands in the Php4-Grx4 complex.1 We tested this by substituting both cysteines with alanine (C221/227A) and determining if Php4 still interacted with Grx4. Php4(C221/227A) overexpressed and purified alone is a relatively soluble protein. When coexpressed with Grx4, Php4(C221/227A) copurifies with Grx4 to a certain extent as shown by SDS-PAGE analysis (Fig. S3, ESI†), although the sample elutes from the analytical gel filtration column across several fractions, suggesting multiple aggregation states. Due to this heterogeneity, we were unable to reliably determine the stoichiometry of the complex by analytical gel filtration. The Fe-S cluster yield from this as-purified complex was low, so the sample was subjected to Fe-S reconstitution using the as-purified complex or a 1:1 ratio of Php4 (C221/227A) and Grx4 purified separately and mixed together. Both approaches yielded a CD spectrum with features almost identical to that of [2Fe-2S] Grx4 (Fig. 5, compare red and blue lines), although with lower peak intensities. These results suggest that while Php4 (C221/227A) can still interact with Grx4, it no longer participates in binding the [2Fe-2S] cluster. Taken together, these data provide evidence that Grx4 Cys172 and Php4 Cys221 and Cys227 are required for Fe-S cluster binding and complex stability.
In order to quantify the stability of the Php4-Grx4 complex and measure the effects of substituting key Cys residues in Grx4 or Php4, we measured the kinetic parameters for the binding interaction between Grx4 and Php4 using surface plasmon resonance (SPR) (Fig. 6, Table 2). The apo forms of the proteins were used for these experiments due to the sensitivity of the Fe-S clusters to oxygen-induced degradation. With His-Php4(WT) as the immobilized ligand and Grx4(WT) or Grx4(C172A) as the analyte, we measured the association (kon) and dissociation (koff) rate constants for the interaction in order to calculate the equilibrium dissociation constant (KD = koff/kon). These experiments demonstrated that the Grx4(C172A) variant binds to Php4 with a 26-fold lower affinity than WT Grx4 (897 μM vs. 34.1 μM) (Table 2). This result is consistent with yeast two-hybrid data indicating that Cys172 in Grx4 plays a key role in stabilizing the Grx4-Php4 interaction in vivo.10 As stated earlier, this previous yeast two-hybrid study also demonstrated that Cys221 and Cys227 in Php4 are required for interaction with the GRX domain of Grx4. We attempted to measure the in vitro binding interaction between His-Php4(C221,227A) and Grx4 using SPR, but were unable to obtain consistent results with this Php4 variant as the immobilized ligand, possibly due to heterogeneity in the purified Php4(C221,227A) sample. In any case, the SPR results clearly show a significant role for Grx4 Cys172 in stabilizing the Php4-Grx4 interaction, even in the absence of Fe-S cluster binding.
Fig. 6.
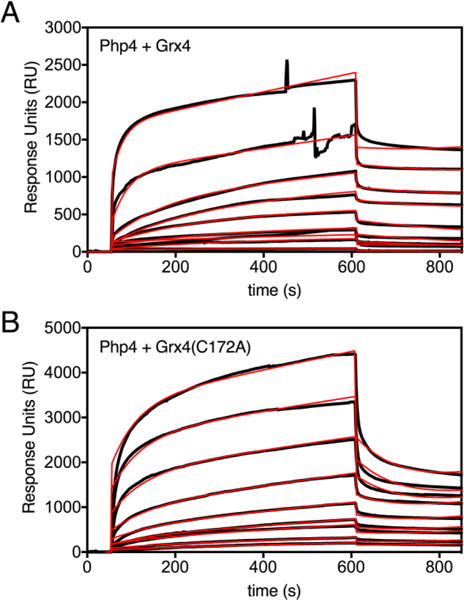
Biacore response data for WT (A) and C172A (B) Grx4 binding to His-Php4 captured on a biosensor surface. The association phase, in which Grx4 samples were flowed across immobilized His-Php4, occurs from 50 to 610 s (9.3 min injection time), while the dissociation phase was recorded from 610 to 860 s. Experimental data (shown in black) represent the responses of varying concentrations of Grx4; 0.92 μM to 553 μM in (A), and 9.23 μM to 738 μM in (B). Global fit analyses of the sensorgrams to a 1:1 binding model with pseudo first order kinetics are shown in red (Table 2).
Table 2.
SPR kinetic measurements for the interaction between His-Php4 and WT or C172A Grx4. The rate constants kon and koff (and standard error of the fit) were determined by global fitting of sensorgrams in Fig. 6 using BIAevaluation software. KD = koff/kon.
| Ligand | Analyte | kon (M−1s−1) | koff (s−1) | KD (μM) |
|---|---|---|---|---|
| His-Php4 | Grx4 | 64.3 (0.6) | 2.19 (0.04) × 10−3 | 34.1 (0.08) |
| His-Php4 | Grx4(C172A) | 10.2 (0.1) | 9.16 (0.10) × 10−3 | 897 (16) |
Conclusions
Previous in vivo studies in S. pombe established that the CGFS glutaredoxin Grx4 binds to and regulates Php4 function in response to iron bioavailability.6,8,10 The strength of this interaction is iron-responsive and dependent on several conserved Cys residues, Cys172 in Grx4 and Cys221 and Cys227 in Php4. Based on these data, we sought to characterize the iron-dependent interaction between Grx4 and Php4 in vitro. In this study we report that recombinant Grx4 expressed alone binds a [2Fe-2S]2+ cluster with spectroscopic features almost identical to previously characterized CGFS Grxs. When coexpressed, Php4 and Grx4 copurify as a complex with a bound [2Fe-2S]2+ cluster. Analysis via UV-visible absorption, CD, and EPR spectroscopies indicate the presence of a [2Fe-2S]2+ cluster in the Php4-Grx4 complex that is distinct from that of [2Fe-2S]2+ Grx4. However, gav values from the EPR signals suggest complete cysteine ligation for both cluster types. Furthermore, both of these clusters are relatively labile based on overall cluster-loading on the proteins after purification (only ~40% for each), as well as spin quantitation of their EPR signals upon reduction.
It was shown previously that E. coli, S. cerevisiae, and human [2Fe-2S] Grxs can be converted to [2Fe-2S] Grx-BolA heterocomplexes upon addition of the BolA protein binding partner.15,25,28,29,33,34 Furthermore, [2Fe-2S]-bridged Grx homodimers and [2Fe-2S]-bridged Grx-BolA heterocomplexes can transfer Fe-S clusters to specific apo acceptor proteins.11,15,18,23,27,33,35,36 Of note, the [2Fe-2S] Grx3-Fra2 complex in S. cerevisiae specifically transfers its cluster to the transcription factor Aft2 when these proteins are mixed.23 Given these previous findings, we sought to determine if [2Fe-2S] Grx4 would convert to [2Fe-2S] Php4-Grx4, or if there was a cluster transfer in a similar fashion. However, contrary to previous studies demonstrating facile conversion of [2Fe-2S] Grx homodimers to [2Fe-2S]-bridged heterocomplexes, several attempts to produce a transition between complexes were not successful. Addition of Php4 to [2Fe-2S] Grx4 did not allow formation of the [2Fe-2S] Php4-Grx4 complex. Similarly, addition of apo-Grx4 to [2Fe-2S] Php4-Grx4 did not drive formation of the [2Fe-2S] Grx4 homodimer. Mixing [2Fe-2S] Grx4 with apo-Php4-Grx4 complex did not facilitate cluster transfer, nor did mixing [2Fe-2S] Grx4 with Php4 in the presence of Fra2 protein. Intriguingly, Fe-S cluster reconstitution of a mixture of Php4 and Grx4 primarily produces the [2Fe-2S] Php4-Grx4 complex rather than [2Fe-2S] Grx4. Although the [2Fe-2S] Php4-Grx4 complex appears to be thermodynamically favored under the Fe-S reconstitution conditions we tested, it is possible that there is a kinetic barrier to interconversion from [2Fe-2S] Grx4 to [2Fe-2S] Grx4-Php4 that requires additional factors and/or specific conditions to overcome. We note that this phenomenon was observed for the delivery of an Fe-S cluster to Aft2 in S. cerevisiae. Although the Fe-S cluster in the S. cerevisiae [2Fe-2S] Grx3 homodimer is relatively labile, this complex was inefficient at delivering an Fe-S cluster to Aft2, while the [2Fe-2S] Grx3-Fra2 heterodimer performed this task effectively. In S. pombe, as Php4 and Grx4 constitutively interact regardless of iron binding, it seems likely that a transfer protein delivers the [2Fe-2S] cluster to apo Php4-Grx4 in the nucleus.10 This possibility is supported by in vivo fluorescence imaging experiments demonstrating predominant localization of functional GFP-tagged Grx4 to the nucleus in both iron-starved and iron-replete cells.7,32 Presumably, Grx4 is in the apo form and associates with Php4 at the Php2/3/5 complex in iron-starved conditions.
The results presented herein provide further evidence for CGFS Grxs as dynamic sensors of cellular iron status and build on the current model for Fe-dependent inhibition of Php4 via Grx4 binding (Fig. 7).1,2,8,10,24 The in vitro studies reported here taken together with the following previously published in vivo observations support this proposed regulation mechanism: (1) deletion of grx4 or depletion of GSH results in constitutive repression of Php4-regulated genes suggesting regulation of Php4 by both Grx4 and GSH,5,8 (2) export of GFP-Php4 from the nucleus to the cytosol visualized by fluorescence microscopy is iron-dependent and requires Grx4,8 and (3) the iron-dependent interaction between Grx4 and Php4 detected by yeast two-hybrid and bimolecular fluorescence complementation assays is dependent on Cys172 in Grx4 and Cys221/227 in Php4.10 Our in vitro protein-protein and Fe-S binding analyses support this model and provide additional molecular details about the coordination chemistry of the binding interaction. In the proposed model shown in Fig. 7, Grx4 and Php4 constitutively interact through the TRX domain of Grx4 under iron-deplete conditions.8,10 Php4 is able to bind to and inactivate the Php2/3/5 transcriptional complex (Fig. 7, top). When iron levels are sufficient, the Php4-Grx4 complex binds a [2Fe-2S] cluster. Since Grx4 and Php4 interact constitutively in vivo, it seems likely that a [2Fe-2S] cluster is delivered to this complex to convert it from apo to holo, rather than Php4 binding to and converting [2Fe-2S] Grx4. Fe-S cluster binding and complex stability are dependent on conserved cysteines in these proteins: Cys172 in Grx4 and Cys221 and Cys227 in Php4. Binding of this Fe-S cluster facilitates inactivation and relocalization of Php4 to the cytosol, allowing the Php2/3/5 complex to activate transcription of iron utilization genes (Fig. 7, bottom). This mode of interaction is somewhat different from S. cerevisiae Aft2 that receives an Fe-S cluster from the Fra2-Grx3 complex to form a [2Fe-2S]-bridged Aft2 homodimer.23 To our knowledge, this study is the first report of a CGFS Grx regulating a transcription factor through direct binding without an accessory protein such as BolA/Fra2. Thus, the CGFS Grxs in these two evolutionarily divergent organisms apparently have a common role in iron regulation, albeit via different mechanisms. Further studies are needed to determine the identity of the [2Fe-2S] cluster donor protein for the Php4-Grx4 complex and the mechanism of transfer.
Fig. 7.
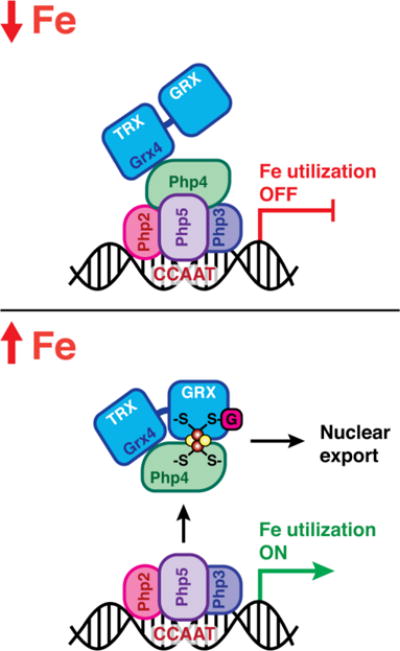
Proposed model for regulation of Php4 via Grx4. During iron starvation conditions, Grx4 interacts with Php4 through the TRX domain, independent of iron. Php4 is free to bind and modify the function of the Php2/3/5 transcriptional complex in the nucleus, repressing iron utilization genes. Under iron replete conditions, the Php4-Grx4 complex binds a [2Fe-2S] cluster. [2Fe-2S] Php4-Grx4 is released from the Php2/3/5 complex and exported from the nucleus, allowing transcriptional activation of iron utilization genes.
Supplementary Material
Significance to metallomics.
In the fission yeast Schizosaccharomyces pombe, the transcription factor Php4 acts to limit iron utilization under conditions of iron deficiency by repressing expression of genes involved in iron-rich metabolic pathways. The glutaredoxin Grx4 binds to Php4 in vivo and regulates its repressor activity and subcellular localization in an iron-dependent manner. We report here the molecular details of this Php4-Grx4 interaction, providing insight into the mechanisms of iron regulation in this model eukaryote.
Acknowledgments
We thank Ariane Brault from the S. Labbé research group for generation of the homologous recombination cassettes used for expression of S. cerevisiae FRA2 in S. pombe. This work was supported by grants GM100069 (to C.E.O.) and GM118164 (to C.E.O.) from the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) and by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant RGPIN-2015/2020-04878 (to S.L.).
Footnotes
Electronic Supplementary Information (ESI) available. See DOI:
References
- 1.Brault A, Mourer T, Labbé S. Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts. IUBMB Life. 2015;67:801–815. doi: 10.1002/iub.1441. [DOI] [PubMed] [Google Scholar]
- 2.Labbé S, Khan MG, Jacques JF. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2013;16:669–676. doi: 10.1016/j.mib.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Pelletier B, Beaudoin J, Mukai Y, Labbé S. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J Biol Chem. 2002;277:22950–22958. doi: 10.1074/jbc.M202682200. [DOI] [PubMed] [Google Scholar]
- 4.Mercier A, Pelletier B, Labbé S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercier A, Watt S, Bahler J, Labbé S. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot Cell. 2008;7:493–508. doi: 10.1128/EC.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedo J Encinar del, Gabrielli N, Carmona M, Ayte J, Hidalgo E. A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast. PLoS Genet. 2015;11:e1005106. doi: 10.1371/journal.pgen.1005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jbel M, Mercier A, Labbé S. Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1. Eukaryot Cell. 2011;10:629–645. doi: 10.1128/EC.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercier A, Labbé S. Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem. 2009;284:20249–20262. doi: 10.1074/jbc.M109.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KD, Kim HJ, Lee KC, Roe JH. Multi-domain CGFS-type glutaredoxin Grx4 regulates iron homeostasis via direct interaction with a repressor Fep1 in fission yeast. Biochem Biophys Res Commun. 2011;408:609–614. doi: 10.1016/j.bbrc.2011.04.069. [DOI] [PubMed] [Google Scholar]
- 10.Vachon P, Mercier A, Jbel M, Labbé S. The monothiol glutaredoxin Grx4 exerts an iron-dependent inhibitory effect on Php4 function. Eukaryot Cell. 2012;11:806–819. doi: 10.1128/EC.00060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G. The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry. 2011;50:8957–8969. doi: 10.1021/bi2008883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banci L, Brancaccio D, Ciofi-Baffoni S, Del Conte R, Gadepalli R, Mikolajczyk M, Neri S, Piccioli M, Winkelmann J. [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis. Proc Natl Acad Sci USA. 2014;111:6203–6208. doi: 10.1073/pnas.1400102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banci L, Ciofi-Baffoni S, Gajda K, Muzzioli R, Peruzzini R, Winkelmann J. N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat Chem Biol. 2015;11:772–778. doi: 10.1038/nchembio.1892. [DOI] [PubMed] [Google Scholar]
- 19.Haunhorst P, Berndt C, Eitner S, Godoy JR, Lillig CH. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem Biophys Res Commun. 2010;394:372–376. doi: 10.1016/j.bbrc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 21.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh KL. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 23.Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci USA. 2014;111:4043–4048. doi: 10.1073/pnas.1318869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Outten CE. In: Iron-Sulfur Clusters in Chemistry and Biology. 1. Rouault T, editor. Walter de Gruyter GmbH & Co.; Berlin: 2014. p. xxiv, 648. ch 19. [Google Scholar]
- 25.Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem. 2011;286:867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Outten CE, Culotta VC. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J Biol Chem. 2004;279:7785–7791. doi: 10.1074/jbc.M312421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Bandyopadhyay S, Shakamuri P, Naik SG, Huynh BH, Couturier J, Rouhier N, Johnson MK. Monothiol glutaredoxins can bind linear [Fe3S4]+ and [Fe4S4]2+ clusters in addition to [Fe2S2]2+ clusters: spectroscopic characterization and functional implications. J Am Chem Soc. 2013;135:15153–15164. doi: 10.1021/ja407059n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dlouhy AC, Li H, Albetel AN, Zhang B, Mapolelo DT, Randeniya S, Holland AA, Johnson MK, Outten CE. The Escherichia coli BolA protein IbaG forms a histidine-ligated [2Fe-2S]-bridged complex with Grx4. Biochemistry. 2016;55:6869–6879. doi: 10.1021/acs.biochem.6b00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Mapolelo DT, Randeniya S, Johnson MK, Outten CE. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry. 2012;51:1687–1696. doi: 10.1021/bi2019089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vranish JN, Russell WK, Yu LE, Cox RM, Russell DH, Barondeau DP. Fluorescent probes for tracking the transfer of iron-sulfur cluster and other metal cofactors in biosynthetic reaction pathways. J Am Chem Soc. 2015;137:390–398. doi: 10.1021/ja510998s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacques JF, Mercier A, Brault A, Mourer T, Labbé S. Fra2 is a co-regulator of Fep1 inhibition in response to iron starvation. PLoS One. 2014;9:e98959. doi: 10.1371/journal.pone.0098959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banci L, Camponeschi F, Ciofi-Baffoni S, Muzzioli R. Elucidating the molecular function of human BOLA2 in GRX3-dependent anamorsin maturation pathway. J Am Chem Soc. 2015;137:16133–16143. doi: 10.1021/jacs.5b10592. [DOI] [PubMed] [Google Scholar]
- 34.Nasta V, Giachetti A, Ciofi-Baffoni S, Banci L. Structural insights into the molecular function of human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 complexes. Biochim Biophys Acta. 2017;1861:2119–2131. doi: 10.1016/j.bbagen.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Frey AG, Palenchar DJ, Wildemann JD, Philpott CC. A glutaredoxin-BolA complex serves as an iron-sulfur cluster chaperone for the cytosolic cluster assembly machinery. J Biol Chem. 2016;291:22344–22356. doi: 10.1074/jbc.M116.744946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vranish JN, Das D, Barondeau DP. Real-time kinetic probes support monothiol glutaredoxins as intermediate carriers in Fe-S cluster biosynthetic pathways. ACS Chem Biol. 2016;11:3114–3121. doi: 10.1021/acschembio.6b00632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


