Abstract
The protein disulfide isomerase (PDI) family is a group of multifunctional endoplasmic reticulum (ER) enzymes that mediate the formation of disulfide bonds, catalyze the cysteine-based redox reactions and assist the quality control of client proteins. Recent structural and functional studies have demonstrated that PDI members not only play an essential role in the proteostasis in the ER but also exert diverse effects in numerous human disorders including cancer and neurodege-nerative diseases. Increasing evidence suggests that PDI is actively involved in the proliferation, survival, and metastasis of several types of cancer cells. Although the molecular mechanism by which PDI contributes to tumorigenesis and metastasis remains to be understood, PDI is now emerging as a new therapeutic target for cancer treatment. In fact, several attempts have been made to develop PDI inhibitors as anti-cancer drugs. In this review, we discuss the properties and diverse functions of human PDI proteins and focus on recent findings regarding their roles in the state of diseases including cancer and neurodegeneration.
Keywords: Cancer, Chaperone, Protein disulfide isomerase, Proteostasis
INTRODUCTION
Formation of disulfide bond (S-S bond) between cysteine residues is a crucial and rate-limiting step for the correct folding of nascent polypeptide chains in the ER (1). Nearly one-third of all eukaryotic proteins, including the majority of secretory proteins (~80%), contain at least one S-S bond (2). During oxidative folding in the ER, S-S bonds are formed by oxidation of thiol groups of cysteines and then rearranged (isomerized) until the correct conformation is achieved (3). Although the formation of S-S bonds also occurs in the intermembrane space (IMS) of mitochondria, this post-translational modification takes place primarily in the ER and is critical for the maturation and stabilization of a majority of secretory and membrane proteins (4). The PDI family is a group of ER enzymes responsible for the formation, breakage, and rearrangement of protein disulfide bond. PDI, the first ER protein discovered as a folding catalyst, is a highly abundant ER protein constituting approximately 0.8% of a total protein (5). Inside cells, S-S bond is formed by thiol-disulfide exchange reactions in which electrons are initially transferred from the reduced substrates to oxidized PDI. Reduced PDI is then regenerated (oxidized) by the FAD-linked sulfhydryl oxidase Ero1, which generates H2O2 by transferring electrons directly onto oxygen (4, 6). The chemical aspects of thiol-disulfide exchange reactions and the enzymatic properties of PDI and its partners in oxidative protein folding are well-explained in several review articles (6–8). In this review, we will focus on the domain organizations and diverse functions of human PDI proteins as well as their relevance to disease states, especially cancer and neurodegenerative diseases.
THE PDI GENE FAMILY
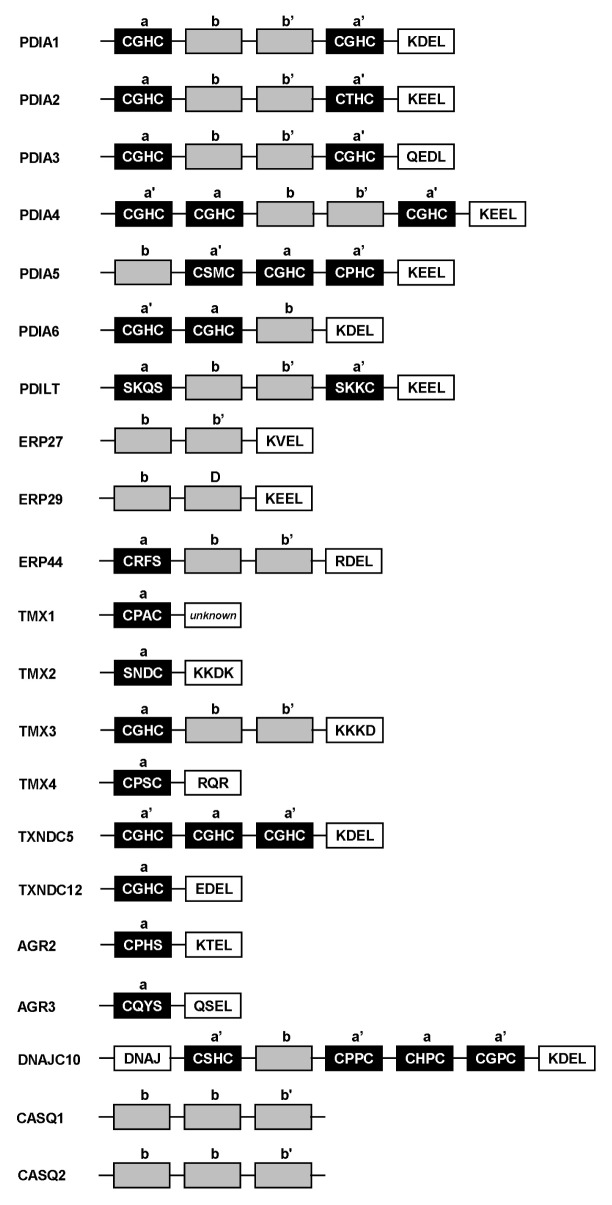
The PDI family is a part of the thioredoxin (TRX) superfamily which also includes TRXs, peroxiredoxins, and glutaredoxins. Most of the enzymes catalyzing the formation of S-S bonds belong to the TRX superfamily (9). According to the HUGO gene nomenclature committee (HGNC) database, the human PDI gene family is comprised of 21 members (http://www.genenames.org/cgi-bin/genefamilies/set/692), which vary in length, domain arrangement and substrate specificity but share a common structural feature - the TRX-like domain (Table 1, Fig. 1). PDIA1 (usually referred as PDI) is the archetype PDI protein and originally identified as the β-subunit of prolyl-4-hydroxylase (P4H) that catalyzes the formation of 4-hydroxyproline in collagen (10). PDIA1 is a central mediator of the oxidative folding in the ER and catalyzes the oxidation, reduction, and isomerization of S-S bonds in the substrates. In addition, PDIA1 and other PDI proteins function as molecular chaperones, which are usually thiol-independent (11). PDIA1 has a multi-domain structure consisting of catalytic domains (i.e., TRX-like domains) and the C-terminal ER-retention sequence. Other PDI proteins also show a similar modular composition of TRX-like domains followed by the C-terminal ER-retention sequence (Fig. 1). All human PDI proteins contain at least one TRX-like domain, which can be divided into two types (a and b) depending on the presence of catalytic motif. The consensus sequence for those catalytic motives is CXXC and the most conserved motif is CGHC (12, 13). Catalytically active a-type domains (a and a’ domains) contain cysteines in the active sites that are thiol-reactive (7). Interestingly, a-type domains found in several PDI proteins (e.g., AGR2, AGR3, and ERp44) are lacking one cysteine in the active site motif (which is usually replaced with serine) and PDILT has no cysteine at the N-terminal a-type domain (13, 14). Despite their structural similarity to TRX, catalytically inactive b-type domains (b and b’ domains) do not contain cysteines and thus cannot mediate S-S bond formation. Instead, the b-type domains are involved in the protein recruitment, which is important for chaperone activity (13).
Table 1.
Members of the human PDI gene family
| Gene name | Protein size | Other aliases | TRX-like domain(s) | Catalytic-site sequence | ER retention sequence |
|---|---|---|---|---|---|
| PDIA1 | 508aa | PDI, P4HB, PHDB | a–b–b′–a′ | CGHC, CGHC | KDEL |
| PDIA2 | 525aa | PDIp, PDA2 | a–b–b′–a′ | CGHC, CTHC | KEEL |
| PDIA3 | 505aa | ERp57, ERp60, GRP57, GRP58, P58 | a–b–b′–a′ | CGHC, CGHC | QEDL |
| PDIA4 | 645aa | ERp70, ERp72 | a′–a–b–b′–a′ | CGHC, CGHC, CGHC | KEEL |
| PDIA5 | 519aa | PDIR | b–a′–a–a′ | CSMC, CGHC, CPHC | KEEL |
| PDIA6 | 440aa | ERp5, P5, TXNDC7 | a′–a–b | CGHC, CGHC | KDEL |
| PDILT | 584aa | PDIA7 | a–b–b′–a′ | SKQS, SKKC | KEEL |
| ERP27 | 273aa | PDIA8 | b–b′ | - | KVEL |
| ERP29 | 262aa | PDIA9, ERp28, ERp31 | b | - | KEEL |
| ERP44 | 406aa | PDIA10, TXNDC4 | a–b–b′ | CRFS | RDEL |
| TMX1 | 280aa | PDIA11, TXNDC1 | a | CPAC | - |
| TMX2 | 296aa | PDIA12, TXNDC14, | a | SNDC | KKDK |
| TMX3 | 454aa | PDIA13, TXNDC10 | a–b–b′ | CGHC | KKKD |
| TMX4 | 349aa | PDIA14, TXNDC13 | a | CPSC | RQR |
| TXNDC5 | 432aa | PDIA15, ERp46, Endo-PDI | a′–a–a′ | CGHC, CGHC, CGHC | KDEL |
| TXNDC12 | 172aa | PDIA16, AGR1, ERp16, ERp18, ERp19 | a | CGHC | EDEL |
| AGR2 | 175aa | PDIA17, XAG-2, HAG-2 | a | CPHS | KTEL |
| AGR3 | 166aa | PDIA18, HAG-3, BCMP11 | a | CQYS | QSEL |
| DNAJC10 | 793aa | PDIA19, ERdj5, JDPI, | a′–b–a′–a–a′ | CSHC, CPPC, CHPC, CGPC | KDEL |
| CASQ1 | 396aa | PDIB1 | b–b–b′ | - | - |
| CASQ2 | 399aa | PDIB2 | b–b–b′ | - | - |
Fig. 1.
Domain organization of human PDI proteins. Arrangement of TRX-like domains and ER-retention sequence in 21 members of the human PDI gene family are shown here. The catalytically active a-type domains (a and a’) are depicted as black boxes with the active site motif sequences shown inside and the catalytically inactive b-type domains (b and b’) are depicted as gray boxes. ER-retention sequences are depicted as white boxes with the motif sequences shown inside. The N-terminal signal sequences and other functional domains present in certain PDI proteins are not shown.
Although PDI members invariably contain the TRX-like domain, which is likely resulted from the duplications of a single TRX-like domain in the ancestral protein, they differ considerably in size, domain composition outside of the TRX-like domain and enzymatic properties. Phylogenetic analysis actually revealed the existence of subfamilies within the PDI family. Among the PDI members, AGR and TMX subfamilies are unique because they have only a-type domain (Fig. 1). Contribution of PDI proteins carrying non-canonical catalytic motifs in their a-type domains (e.g., AGR2, AGR3, PDILT, and ERp44) to thiol-reactive oxidative folding in vivo is unclear although they are capable of forming the mixed S-S bonds with client proteins (13, 15). More interestingly, ERp27 and ERp29 carry b-type domains only (16, 17). Presumably, these atypical PDI members function primarily as molecular chaperones rather than as oxidoreductases for a specific set of substrates. It reiterates that the grouping of human PDI proteins is primarily based on sequence and structural similarity rather than the enzymatic properties. Detailed information on the human PDI gene family can be found in a recent review article and references therein (1).
The CASQ subfamily is somewhat enigmatic because of its unusual domain organization and specialized functions. CASQ1 and CASQ2, the main calcium-binding proteins of the sarcoplasmic reticulum and the regulator of calcium release in muscle (18), have only b-type domains. Surprisingly, CASQ1 and CASQ2 are the only PDI proteins without ER-retention sequence (Fig. 1); instead, there are C-terminal Asp/Glu-rich (acidic) Ca2+ binding regions involved in Ca2+ flux via regulation of the Ca2+ channel and direct interaction with Ca2+ (19). Functions of CASQ proteins in non-muscle tissues are unknown and they could be functionally irrelevant to other PDI proteins despite the presence of b-type domains.
The AGR subfamily (AGR2, AGR3 and TXNDC12) was identified as novel members of PDI in 2005 (20). In contrast to CASQ proteins, AGR proteins contain single a-type domain plus an ER-retention sequence but no b-type domain. AGR proteins are thus the smallest PDI (> 200 amino acids vs. 400–650 amino acids for typical PDI proteins) (1). Although AGR2 protein contains a non-canonical catalytic motif lacking the C-terminal cysteine (CPHS), it is still capable of forming a mixed disulfide bond with mucin and the mutation of the conserved cysteine (C81S) abolishes their association (15). Functional analyses of each cysteine in the consensus CXXC motif demonstrated that the N-terminal cysteine is important for the formation of a transient S-S bond with the substrate whereas the C-terminal cysteine is involved in the substrate release (21). An explanation is that the PDI members carrying the non-canonical catalytic motif (e.g., AGR2, AGR3, and ERp44) may have different (and possibly very limited) substrate specificities.
AGR2 and AGR3 were first identified as human orthologs of X. laevis secreted protein XAG-2 which expressed during the development of the mucus-secreting cement gland. Studies have revealed that AGR2 is highly expressed in estrogen receptor α (ERα)-positive malignant breast cancer cells as well as many other adenocarcinomas including colorectal, ovarian, pancreatic and prostate cancers (22). AGR3, originally discovered from a screening of membrane-associated proteins as breast cancer membrane protein 11 (BCMP11), is similarly over-expressed in ERα-positive breast cancer cells (23). The expression of AGR2 and AGR3 genes, both located at 7p21.1, is not always correlated. In fact, the uncoupled expression in prostate and ovarian cancer is also reported (24). Accumulating evidence indicates that AGR2 could function as an oncoprotein that can stimulate the proliferation and promote the metastasis of cancer cells. Clinical studies have focused on the exploitation of this PDI protein as a potential tumor biomarker and prognostic factor. The involvement of PDI proteins, especially AGR proteins, in oncogenesis will be discussed again (see below).
BIOLOGICAL FUNCTIONS OF PDI PROTEINS
The catalytic properties and structural features of PDI proteins and the regulation of redox balance by PDI members have been extensively studied and details can be found in a number of review articles (5, 8, 13, 14). In addition to their principal roles as catalysts for the formation and rearrangement of S-S bonds, PDI proteins are also functioning as chaperones (25). Molecular chaperones assist protein folding/refolding by inhibiting the non-productive folding and aggregation of partially folded intermediates or damaged polypeptides through their ability to recognize and interact with proteins in non-native conformation. As explained above, PDI (PDIA1) is the β-subunit of P4H and the chaperone activity rather than the enzyme activity of PDI is necessary for the assembly of P4H molecules (8, 10). Intriguingly, PDI helps the refolding of misfolded proteins without S-S bonds and PDI with no isomerase activity also increases the refolding of substrates (e.g., proinsulin) to a certain extent, suggesting that the catalytic activity and chaperone function of PDI can be dissociated (26). PDI also acts as a molecular chaperone for ERα, altering its conformation and affecting the ERα-ER-responsive element (ERE) interaction. PDI mutant lacking isomerase activity was as effective as normal PDI in enhancing ERα-ERE interaction, indicating that the chaperone activity is independent of catalytic activity of the PDI protein (27). In vascular smooth muscle cells, PDI interacts with NAD(P)H oxidase, the source of reactive oxygen species, and acts as a redox-sensitive regulatory factor by modulating the RhoGTPase activation (28, 29). These results demonstrate that the chaperone activities of PDI are crucial for many client proteins.
While a wealth of information on the functions of archetypal PDI (PDIA1) - as a general catalyst of native S-S bond formation and as a molecular chaperone assisting protein folding - is available, less is known about the functions of other PDI members despite their similarities in sequence and domain organization. Analysis of mixed disulfide bonds formed by major PDI proteins (e.g., PDIA1, ERp57, and ERp72) revealed distinct substrate specificities and showed that each PDI member is specialized for different sets of substrates (30). There are several members (e.g., PDIp, PDIR, P5, PDILT, ERp44, and ERdj5) also possessing chaperone activities (31). PDIA2 (PDIp), originally identified as a pancreas-specific PDI protein, plays a similar role as PDIA1 but is less effective in oxidation/reduction (1). Moreover, the substrate specificity (i.e., the binding motif) of PDIA2 is different from that of PDIA1 (32). PDIA2 also functions as a chaperone of denatured substrates, which is independent of its enzymatic activity (33). ERp57 (PDIA3), a glycoprotein-specific PDI protein, plays a central role in the peptide loading onto major histocompatibility complex (MHC) class I molecules. In fact, ERp57 is a component of the peptide-loading complex (PLC) which includes TAP1, TAP2, tapasin and calreticulin (34). The redox activity of ERp57 is not required for the peptide loading and the stability of MHC class I molecules is also unaffected by the mutation of the catalytic motif of ERp57 (35).
ERp57 is also a major player in the quality control of newly synthesized glycoproteins in conjunction with another ER chaperones calnexin (CNX) and calreticulin (CRT). ERp57 is protecting the substrates from ER-associated degradation (ERAD) at the early stage of chaperone-mediated sorting and, at the later stage, promoting the maturation of substrates that eventually exit to the cell surface via Golgi apparatus (36). ERp57 is up-regulated in the neurons of prion-related disorders (PrDs) and physically interacts with the cellular prion protein (PrPc) (37). Subsequent studies demonstrated that ERp57 controls the maturation and total levels of PrP suggesting that this PDI protein is a cellular factor regulating the biosynthesis and folding of PrP (38). In contrast, PDIA1 promotes the ERAD of misfolded proteins by recognizing and targeting them for degradation (39). The degradation of dislocated MHC class I heavy chain molecules induced by the human cytomegalovirus US2 also requires the substrate binding activity of PDIA1 (40). These results establish that PDI proteins, especially PDIA1 and ERp57, are important for the quality control and proper degradation of proteins in the ER (termed as ‘proteostasis’).
Physiological roles of other PDI family members carrying catalytic a-type domains are poorly understood and data on their functions on ER proteostasis is limited. ERp72 (PDIA4) is one of the largest PDI members (645 aa) and contains five TRX-like domains (Table 1). ERp72 is a component of the multiprotein chaperone complex that includes ER Hsp70 (BiP), Grp94, PDI and ERp29 (41). ERp72 is shown to associate with thyroglobulin and the cell surface NAD(P)H oxidase Nox-1 (13). PDIR (PDIA5) is structurally unique because a N-terminal non-catalytic b-type domain precedes three catalytic a-type domains (Fig. 1). Functions of PDIR are largely unknown, although its expression pattern indicates a role in the folding of glycoproteins. Interestingly, the b-type domain of PDIR has a binding surface for ERp72 and CRT suggesting a functional relationship with these ER proteins (42). In contrast, P5 (PDIA6) cooperates with BiP and acts as a key reductase for the ERAD of misfolded proteins (e.g., proinsulin carrying a mutation). Although P5 shows narrow substrate specificity, it associates preferentially with the proteins requiring BiP function (30, 43, 44). PDILT (PDIA7) is an interesting member of the PDI family because it is expressed specifically in testis and contains unusual catalytic site motives (SKQS and SKKC) (45). A recent study demonstrated that PDILT, together with the testis-specific CRT-like protein calsperin, is involved in the S-S bond formation and maturation of a membrane-bound metalloprotease ADAM3, which is required for sperm migration (46). ERp44 (PDIA10), a PDI protein in secretory cells, plays a regulatory role in the assembly of IgM and adiponectin in the early secretory pathway (47, 48). ERp44 is a special member of PDI family since its activity is regulated by pH change which occurs when it shuttles between the ER and cis-Golgi compartment. This pH change enables this PDI to act as a post-ER quality control factor for the assembly of secretory proteins (49, 50). ERp46 (PDIA16, Endo-PDI) was originally reported as a PDI protein preferentially expressed in endothelial cells and functions as a survival factor against hypoxic stress (51). Interestingly, ERp46 contains multiple a-type domains linked by unusually long loops but no b-type domain (Fig. 1). This arrangement is not observed in any other PDI proteins and may account for the functional difference between ERp46 and archetype PDI (52). ERdj5 (PDIA19, DNAJC10) is the largest member of the PDI family (793 aa) and also belongs to the DnaJ (Hsp40) heat shock protein family. ERdj5 possesses a N-terminal J-domain that mediates the interaction with BiP. It also associates with EDEM (ER degradation–enhancing α-mannosidase–like protein) and reduces S-S bonds in the client proteins. ERdj5 is a reductase component of the ERAD supramolecular complex that facilitates the accelerated degradation of misfolded proteins (53). A recent study revealed that ERdj5 acts as an ER reductase not only for the proteins to be dislocated to the cytosol for degradation (ERAD substrates) but also for the proteins (e.g., LDL receptor) undergoing productive folding (54). TMX proteins (PDIA11–14) carrying a single a-type domain are the only PDI proteins with a transmembrane domain. Although their enzymatic activities and the expression levels in a variety of tissues have been reported, little is known about their physiological functions (1).
ROLES OF PDI PROTEINS IN THE DISEASE STATES
The ER is a central compartment for folding, post-translational modifications, and quality control of proteins, which are collectively termed as ‘proteostasis’. Dysregulation of proteostasis could exert adverse effects and is potentially detrimental to the cells. As the main catalyst for intra- and intermolecular S-S bonds and as the molecular chaperones for client proteins, PDI proteins play a key role in the maintenance and regulation of proteostasis. Disturbance of proteostasis can lead to the activation of ER stress response known as ‘unfolded protein response (UPR)’. At earlier stages, UPR alleviates the ER stress by up-regulating the chaperones for protein folding and by inducing ERAD and autophagy to eliminate misfolded proteins (55). UPR and ER stress are increasingly implicated in many disease states and accumulating evidence indicates that PDI proteins, as a vital component of UPR, play important roles in the pathophysiology of many disorders. However, recent studies suggest that the impacts of PDI proteins on diseases are much more complex and the unconventional roles of PDI in pathophysiology have also been reported.
PDI and Neurodegenerative Diseases
Neurodegenerative disorders, such as Alzheimer’s diseases (AD), Parkinson’s disease (PD), Huntington’s diseases (HD), amyotrophic lateral sclerosis (ALS) and prion-related disorders, are classified as protein misfolding disorders (PMD) because a hallmark of these diseases is the accumulation of misfolded proteins within affected tissues (56). Special attention has been given to the importance of UPR in PMD and roles of the regulatory factors, including PDI, have been investigated (57). PDI proteins are up-regulated in many PMD and associate with disease proteins in that they are often recruited to the insoluble aggregates (58). PDI proteins are usually cytoprotective and even prevent the aggregation of misfolded proteins (59). However, the opposite effects are also reported and the PDI inhibitor could provide protection against toxic effects (e.g., apoptotic cell death) (60). Interestingly, a number of studies reported S-nitrosylation of PDI in the brain tissues derived from AD and PD. This post-translational modification of PDI, triggered by reactive nitrogen species (RNS), inactivates the catalytic activity of PDI and perturbs proteostasis, which eventually leads to neuronal cell death (61). More information on the roles of each PDI protein in neurodegenerative diseases can be found in a recent review article and references therein (62).
PDI and Cancer
The need to both correct the folding defects and also eliminate the aggregated proteins may explain easily why PDI proteins play important roles in PMD. In contrast, the functions of PDI proteins in oncogenesis and cancer progression are much more complex. There is ample evidence supporting that PDI proteins are strongly associated with a variety of cancer. Although they are among the most abundant cellular proteins, PDI proteins are frequently up-regulated in cancers. Analysis of microarray data sets revealed that PDI (PDIA1) is significantly and universally over-expressed in a wide variety of cancer types including brain, lymphoma, kidney, ovarian, prostate and lung cancers (21). Similar results were also obtained from the analysis of cytosolic and cell surface proteomes derived from various cancers (63, 64). Moreover, over-expressed PDI is frequently correlated with metastasis and invasiveness (65, 66). Other PDI members are also up-regulated in various types of cancer, including metastatic cancer. Analyses of transcriptomes and proteomes revealed that ERp57, ERp72, and P5 are all highly expressed in breast, thyroid, rectal, gastric and liver cancers (67–69). Over-expressed ERp57 and P5 even confer the resistance to cisplatin-induced death of lung cancer cells (70). Accordingly, efforts have been made to use PDI proteins as a prognostic factor for clinical use (71).
PDI proteins are often concomitantly up-regulated with other UPR proteins (e.g., BiP and Grp94), which stresses the importance of UPR in controlling the cell survival (72). Given these findings, the oncogenic effects of PDI proteins are apparently mediated by their role in the UPR signaling pathway. A recent finding showed that PDIR (PDIA5) can activate ATF6α, a membrane-anchored transcription factor that modulates UPR signaling. PDIA5 mediates the rearrangement of S-S bonds in ATF6α and as a consequence promotes the packaging of ATF6α into COPII vesicles (73). Observations that PDI inhibitor(s) can enhance apoptosis in cancer cells or sensitize them to anti-cancer agents suggest a pro-survival role of PDI via regulation of apoptosis (74). Previously we reported that cytosolic PDI is cleaved by caspase-3 and -7 during apoptosis and over-expressed PDI can suppress apoptotic cell death (75). Knockdown of PDI contrarily reduces the growth of ovarian cancer cells (76). On the other hand, the pro-apoptotic effects of PDI were also reported. An organometallic anti-cancer agent causes cytotoxicity by induction of ER stress and up-regulation of several key UPR components including PDI. Similarly, sodium butyrate induces apoptosis in colorectal cancer cells via the up-regulation of UPR proteins (PDI included). Such effects are presumably caused by prolonged activation of UPR via the action of pro-apoptotic transcription factors, especially CHOP (77, 78).
While PDI plays a central role in UPR signaling and the control of cancer cell survival, PDI also facilitates the activation of metalloproteases at the cell surface (e.g., ADAM17) that modulate cell signaling by catalyzing the shedding of membrane-associated proteins (79). The activation of ADAM17 is regulated by redox change and such an effect must be important for the growth factor signaling in cancer cells. It is intriguing that PDI proteins, normally confined to the ER, are also localized at the cell surface and catalyze the thiol-disulfide exchange in extracellular proteins. The presence of PDI proteins at the cell surface and its functional implications have been debated for a while. As mentioned above, surface PDI proteins interacting with client proteins (e.g., metalloproteases, selectins and integrins) catalyze the formation/isomerization of S-S bonds in the clients (80). These effects are particularly interesting because PDI proteins also promote cancer invasion and metastasis. Thiol-disulfide exchange catalyzed by extracellular PDI may activate proteolytic enzymes (e.g., MMPs) or membrane proteins mediating cell adhesion/migration (e.g., integrins), both of which are well-known metastatic factors (21).
In addition to their roles in UPR signaling, control of apoptosis and the catalysis of external thiol-disulfide exchange, PDI proteins also contribute to cancer via different pathways. PDI interacts with a wide variety of proteins (a partial list of PDI interactome can be found in reference #11). The chaperone activity of PDI, which is usually thiol-independent, is crucial for many client proteins, some of which are located at the cell surface, for proper folding and/or quality control. Moreover, different PDI proteins have non-redundant functions and the involvement of each member is mechanistically distinct. TXNDC5 (endo-PDI), for example, is required for angiogenesis that involves over-expressed cathepsin B and MMP-9, which in turn specifically activate the Ras/Raf/MEK/ERK pathway (81). ERp29 is even more surprising because it has no catalytic a-type domain; this PDI member is nevertheless involved in diverse aspects of cancer - progression, metastasis and apoptosis. ERp29 also functions in UPR signaling pathway and, more importantly, regulates the epithelial-mesenchymal transition (EMT) which is critical for cancer invasion and metastasis. In mesenchymal-like breast cancer cells, over-expressed ERp29 increases E-cadherin by inactivation of ERK signaling, which promotes transition into epithelial properties (82). ERp29 also inhibits apoptosis in breast cancer cells through the up-regulation of Hsp27 which is caused by down-regulation of eIF2α (83).
Among human PDI proteins, AGR proteins are probably the most closely related to oncogenesis and several lines of investigations have established their importance in the tumor growth and metastasis of cancer. AGR proteins (mostly AGR2 and AGR3) are now considered as oncogenic and pro-metastatic factors for most of the adenocarcinomas. Clinical attempts have been made to use AGR2 as a cancer marker for circulating tumor cells and metastatic cells in lymph nodes (23). AGR2 is the most-studied member of AGR subfamily and the elevated expression of AGR2 is frequently used as a prognostic factor for patient outcome. In contrast, the roles of AGR3 in tumor biology are relatively unknown despite high sequence identity (~70%) with AGR2 (84). For simplicity, we hereafter discuss mostly AGR2. Expression of AGR2 can be induced by various stimuli including sex hormones, hypoxia, and ER stress. Estrogen treatment increases AGR2 expression in breast cancer cells (ER can directly bind to AGR2) and androgen is also capable of inducing AGR2 in prostate cancer cells (AGR2 has AR-binding sites in the promoter region) (23, 84, 85).
Mechanistically AGR2 demonstrates several distinct characteristics. AGR2, plays a role in the regulation of UPR signaling and control of apoptosis (similar to archetype PDI), which also requires its dimerization (86, 87). However, the exact roles of AGR2 in the thiol exchange reactions in the ER are not clearly understood. Until now, mucin is the only known substrate requiring AGR2 as a thiol-disulfide exchanger for its processing (15). A recent report showed that AGR2 also promotes the presentation of EGF receptor at the plasma membrane by forming mixed S-S bonds indicating that other physiological substrates requiring AGR2 function may exist (88). Functional redundancy of AGR2 with other PDIs in protein folding and UPR signaling also needs to be addressed. Similar to other PDI proteins, AGR2 can be localized extracellularly and secreted (89). A recent study claimed that extracellular AGR2, functionally independent of ER luminal AGR2, is critical for the tumorigenesis (90). Yeast two-hybrid assays show that several membrane proteins can interact with AGR2 (therefore proposed as AGR2 receptors); however, their actual association inside cells remains to be determined and the biological consequence of such associations should also be examined (89). In addition, AGR2 interacts with a number of proteins with diverse functions, raising a possibility that it can also act as a molecular chaperone (89). In the future studies, it should be investigated whether AGR2 also functions as a molecular chaperone and influences the folding or assembly of the interacting proteins. Finally, the requirement of AGR2 in proteostasis or ERAD pathways also needs to be further studied.
CONCLUSIONS
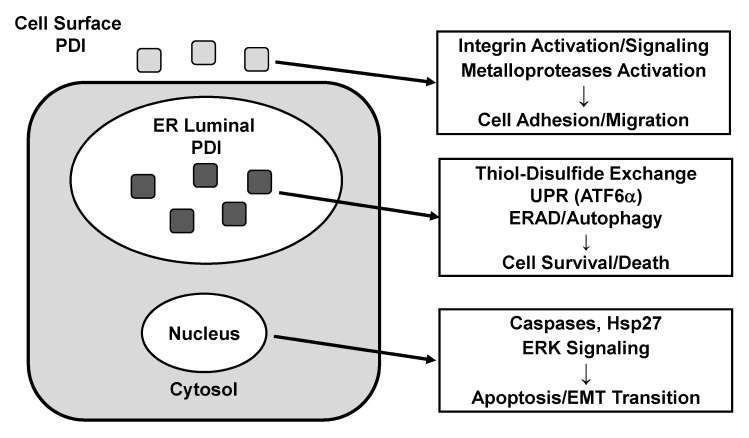
In this review, we briefly explain the domain architecture of members of the human PDI gene family and discuss their biological functions with special attention to the involvement in human diseases, e.g., neurodegenerative diseases and cancer. Although PDI proteins serve primarily as ER enzymes catalyzing the formation/isomerization of disulfide bonds and thus assisting the proper folding of proteins, they also have additional functions as molecular chaperones as well as regulatory factors for protein homeostasis in the ER. Multiple functions mediated by PDI proteins probably allow them to actively participate in diverse cellular processes including UPR signaling and apoptosis, which are critical for survival or death of cells. Increased expression of PDI observed in many types of cancer also indicates that cancer cells, which synthesize more proteins than normal cells to sustain rapid growth, may need higher ER capacity for proper protein folding. This phenomenon may render cancer cells more vulnerable to PDI inhibition than the normal cells, which makes PDI proteins a potential drug target for the treatment of cancers. Subcellular localization of PDI proteins and the proposed function(s) related to oncogenesis and metastasis are summarized in Fig. 2. However, the exact functions of PDIs at different localizations, their roles in cellular signaling pathways other than UPR and, functional difference (or redundancy) between the many PDI proteins need to be extensively studied before we can fully utilize these interesting proteins as an important target for intervention.
Fig. 2.
Sub-cellular localizations and functions of PDI proteins related to cancer. PDI proteins located at the cell surface are involved in the cell adhesion/migration via activation of metallo-proteases and integrins. In the ER, PDI proteins are required for UPR which determines the survival/death of cells via ATF6α activation or ERAD/autophagy induction. Although the localization of PDI in the cytosol and/or nucleus are not clearly understood, PDI proteins control apoptosis via caspase activation and Hsp27 induction. PDI proteins also play a role in the regulation of the epithelial-mesenchymal transition (EMT). See text for more details.
ACKNOWLEDGEMENTS
This work is supported by the grant from National Research Foundation of Korea (grant number 2013R1A1A2011628) and the sabbatical year research grant from Seoul Women’s University (2014).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosnjak I, Bojovic V, Segvic-Bubic T, et al. Occurrence of protein disulfide bonds in different domains of life: a comparison of proteins from the Protein Data Bank. Prot Eng Des Selec. 2014;27:65–72. doi: 10.1093/protein/gzt063. [DOI] [PubMed] [Google Scholar]
- 3.Depuydt M, Messens J, Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- 4.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: Two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari DM, Söling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339:1–10. doi: 10.1042/bj3390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson DA, Gannon SA, Thorpe C. Oxidative protein folding: From thiol–disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic Biol Med. 2015;80:171–182. doi: 10.1016/j.freeradbiomed.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatahet F, Ruddock LW. Protein disulfide isomerase: A critical evaluation of Its function in disulfide bond formation. Antoxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Wang X, Wang CC. Protein disulfide–isomerase, a folding catalyst and a redox-regulated chaperone. Free Radic Biol Med. 2015;83:305–313. doi: 10.1016/j.freeradbiomed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Yoshioka J. Thioredoxin superfamily and its effects on cardiac physiology and pathology. Compr Physiol. 2015;5:513–530. doi: 10.1002/cphy.c140042. [DOI] [PubMed] [Google Scholar]
- 10.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/S0945-053X(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 11.Soares Moretti AI, Martins Laurindo FR. Protein disulfide isomerases: Redox connections in and out of the endoplasmic reticulum. Arch Biochem Biophys. 2017;617:106–119. doi: 10.1016/j.abb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Alanen HI, Williamson RA, Howard MJ, et al. Functional characterization of ERp18, a new endoplasmic reticulum-located thioredoxin superfamily member. J Biol Chem. 2003;278:28912–28920. doi: 10.1074/jbc.M304598200. [DOI] [PubMed] [Google Scholar]
- 13.Kozlov G, Maattanen P, Thomas DY, et al. A structural overview of the PDI family of proteins. FEBS J. 2010;277:3924–3936. doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- 14.Appenzeller-Herzog C, Ellgaard L. The human PDI family: Versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Park SW, Zhen G, Verhaeghe C, et al. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci U S A. 2009;106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alanen HI, Williamson RA, Howard MJ, et al. ERp27, a new non-catalytic endoplasmic reticulum-located human protein disulfide isomerase family member Interacts with ERp57. J Biol Chem. 2006;281:33727–33738. doi: 10.1074/jbc.M604314200. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari DM, Nguyen Van P, Kratzin HD, et al. ERp28, a human endoplasmic-reticulum-lumenal protein, is a member of the protein disulfide isomerase family but lacks a CXXC thioredoxin-box motif. Eur J Biochem. 1998;255:570–579. doi: 10.1046/j.1432-1327.1998.2550570.x. [DOI] [PubMed] [Google Scholar]
- 18.Novák P, Soukup T. Calsequestrin distribution, structure and function, its role in normal and pathological situations and the effect of thyroid hormones. Physiol Res. 2011;60:439–452. doi: 10.33549/physiolres.931989. [DOI] [PubMed] [Google Scholar]
- 19.Shin DW, Ma J, Kim DH. The asp-rich region at the carboxyl-terminus of calsequestrin binds to Ca2+ and interacts with triadin. FEBS Lett. 2003;486:178–182. doi: 10.1016/S0014-5793(00)02246-8. [DOI] [PubMed] [Google Scholar]
- 20.Persson S, Rosenquist M, Knoblach B, et al. Diversity of the protein disulfide isomerase family: identification of breast tumor induced Hag2 and Hag3 as novel members of the protein family. Mol Phylogenet Evol. 2005;36:734–740. doi: 10.1016/j.ympev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Xu S, Sankar S, Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov Today. 2014;19:1359–6446. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 22.Brychtova V, Vojtesek B, Hrstka R. Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett. 2011;304:1–7. doi: 10.1016/j.canlet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res. 2013;15:204. doi: 10.1186/bcr3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray TA, MacLaine NJ, Michie CO, et al. Anterior Gradient-3: a novel biomarker for ovarian cancer that mediates cisplatin resistance in xenograft models. J Immunol Methods. 2012;378:20–32. doi: 10.1016/j.jim.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274:5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- 26.Winter J, Klappa P, Freedman RB, et al. Catalytic activity and chaperone function of human protein-disulfide isomerase are required for the efficient refolding of proinsulin. J Biol Chem. 2002;277:310–317. doi: 10.1074/jbc.M107832200. [DOI] [PubMed] [Google Scholar]
- 27.Schultz-Norton JR, McDonald WH, Yates JR, et al. Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor α structure and function. Mol Endocrinol. 2006;20:1982–1995. doi: 10.1210/me.2006-0006. [DOI] [PubMed] [Google Scholar]
- 28.Janiszewski M, Lopes LR, Carmo AO, et al. Regulation of NAD(P)H oxidase by associated protein disulfide isomerase in vascular smooth muscle cells. J Biol Chem. 2005;280:40813–40819. doi: 10.1074/jbc.M509255200. [DOI] [PubMed] [Google Scholar]
- 29.Pescatore LA, Bonatto D, Forti FL, et al. Protein Disulfide Isomerase Is Required for Platelet-derived Growth Factor-induced Vascular Smooth Muscle Cell Migration, Nox1 NADPH Oxidase Expression, and RhoGTPase Activation. J Biol Chem. 2012;287:29290–29300. doi: 10.1074/jbc.M112.394551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jessop CE, Watkins RH, Simmons JJ, et al. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maattanen P, Gehring K, Bergeron JJ, et al. Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Ruddock LW, Freedman RB, Klappa P. Specificity in substrate binding by protein folding catalysts: Tyrosine and tryptophan residues are the recognition motifs for the binding of peptides to the pancreas-specific protein disulfide isomerase PDIp. Protein Sci. 2000;9:758–764. doi: 10.1110/ps.9.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu XM, Zhu BT. Human pancreas-specific protein disulfide-isomerase (PDIp) can function as a chaperone independently of its enzymatic activity by forming stable complexes with denatured substrate proteins. Biochem J. 2010;429:157–169. doi: 10.1042/BJ20091954. [DOI] [PubMed] [Google Scholar]
- 34.Dong GI, Wearsch PA, Peaper DR, et al. Insights into MHC Class I peptide loading from the structure of the Tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peaper DR, Cressewell P. The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc Natl Acad Sci U S A. 2008;105:10477–10482. doi: 10.1073/pnas.0805044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frenkel Z, Shenkman M, Kondratyev M, et al. Separate roles and different routing of Calnexin and ERp57 in endoplasmic reticulum quality control revealed by interactions with asialoglycoprotein receptor chains. Mol Biol Cell. 2004;15:2133–2142. doi: 10.1091/mbc.E03-12-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watts JC, Huo H, Bai Y, et al. Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones. PLoS Pathog. 2009;5:e1000608. doi: 10.1371/journal.ppat.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres M, Medinas DB, Matamala JM, et al. The Protein-disulfide Isomerase ERp57 Regulates the Steady-state Levels of the Prion Protein. J Biol Chem. 2015;290:23631–23645. doi: 10.1074/jbc.M114.635565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molinari M, Galli C, Piccaluga, et al. V Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SO, Cho KM, Cho SL, et al. Protein disulphide isomerase is required for signal peptide peptidase-mediated protein degradation. EMBO J. 2010;29:363–375. doi: 10.1038/emboj.2009.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meunier L, Usherwood YK, Chung KT, et al. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinaik R, Kozlov G, Gehring K. Structure of the non-catalytic domain of the protein disulfide isomerase-related protein (PDIR) reveals function in protein binding. PLoS One. 2013;8:e62021. doi: 10.1371/journal.pone.0062021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutkevich LA, Cohen-Doyle MF, Brockmeier U, et al. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell. 2010;21:3093–3105. doi: 10.1091/mbc.E10-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorasia DG, Dudek NL, Safavi-Hemami H, et al. A prominent role of PDIA6 in processing of misfolded proinsulin. Biochim Biophys Acta. 2016;1864:715–723. doi: 10.1016/j.bbapap.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 45.van Lith M, Hartigan N, Hatch J, et al. PDILT, a divergent testis-specific protein disulfide isomerase with a non-classical SXXC motif that engages in disulfide-dependent interactions in the endoplasmic reticulum. J Biol Chem. 2005;280:1376–1383. doi: 10.1074/jbc.M408651200. [DOI] [PubMed] [Google Scholar]
- 46.Tokuhiro K, Ikawa M, Benham AM, et al. Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility. Proc Natl Acad Sci U S A. 2012;109:3850–3855. doi: 10.1073/pnas.1117963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anelli T, Stefania Ceppi S, Bergamelli L, et al. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26:4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hampe L, Radjainia M, Xu C, et al. Regulation and quality control of adiponectin assembly by endoplasmic reticulum chaperone ERp44. J Biol Chem. 2015;290:18111–18123. doi: 10.1074/jbc.M115.663088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vavassori S, Cortini M, Masui S, et al. A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol Cell. 2013;50:783–792. doi: 10.1016/j.molcel.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anelli T, Sannino S, Sitia R. Proteostasis and “redoxtasis” in the secretory pathway: Tales of tails from ERp44 and immunoglobulins. Free Radic Biol Med. 2015;83:323–330. doi: 10.1016/j.freeradbiomed.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan DC, Huminiecki L, Moore JW, et al. EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem. 2003;278:47079–47088. doi: 10.1074/jbc.M308124200. [DOI] [PubMed] [Google Scholar]
- 52.Okumura M, Kadokura H, Inaba K. Structures and functions of protein disulfide isomerase family members involved in proteostasis in the endoplasmic reticulum. Free Radic Biol Med. 2015;83:314–322. doi: 10.1016/j.freeradbiomed.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 53.Ushioda R, Hoseki J, Araki K, et al. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 54.Oka OB, Pringle MA, Schopp IM, et al. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol Cell. 2013;50:793–804. doi: 10.1016/j.molcel.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perri ER, Thomas CJ, Parakh S, et al. The Unfolded Protein Response and the Role of Protein Disulfide Isomerase in Neurodegeneration. Front Cell Dev Biol. 2016;3:80. doi: 10.3389/fcell.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skovronsky DM, Lee VM, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 57.Parakh S, Atkin JD. Novel roles for protein disulphide isomerase in disease states: a double edged sword? Front Cell Dev Biol. 2015;3:1–11. doi: 10.3389/fcell.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang SB, Shi Q, Xu Y, et al. Protein Disulfide Isomerase Regulates Endoplasmic Reticulum Stress and the Apoptotic Process during Prion Infection and PrP Mutant-Induced Cytotoxicity. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0038221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu LR, Liu XL, Chen J, et al. Protein disulfide isomerase interacts with tau protein and inhibits its fibrillization. PLoS ONE. 2013;8:e76657–e76657. doi: 10.1371/journal.pone.0076657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffstrom BG, Kaplan A, Letso R, et al. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2012;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forrester MT, Benhar M, Stamler JS. Nitrosative stress in the ER: a new role for S-nitrosylation in neuro-degenerative diseases. ACS Chem Biol. 2006;1:355–358. doi: 10.1021/cb600244c. [DOI] [PubMed] [Google Scholar]
- 62.Andreu CI, Woehlbier U, Torres M, et al. Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett. 2012;586:2826–2834. doi: 10.1016/j.febslet.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 63.Shin BK, Wang H, Yim AM, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
- 64.Rho JH, Roehrl MH, Wang JY. Glycoproteomic analysis of human lung adenocarcinomas using glycoarrays and tandem mass spectrometry: differential expression and glycosylation patterns of vimentin and fetuin A isoforms. Protein J. 2009;28:148–160. doi: 10.1007/s10930-009-9177-0. [DOI] [PubMed] [Google Scholar]
- 65.Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4:1686–1696. doi: 10.1074/mcp.M400221-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Zong J, Guo C, Liu S, et al. Proteomic research progress in lymphatic metastases of cancers. Clin Transl Oncol. 2012;14:21–30. doi: 10.1007/s12094-012-0757-7. [DOI] [PubMed] [Google Scholar]
- 67.Ramos FS, Serino LT, Carvalho CM, et al. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet Mol Res. 2015;14:6960–6967. doi: 10.4238/2015.June.26.4. [DOI] [PubMed] [Google Scholar]
- 68.Xu X, Wei X, Ling Q, et al. Identification of two portal vein tumor thrombosis associated proteins in hepatocellular carcinoma: protein disulfide-isomerase A6 and apolipoprotein A-I. J Gastroenterol Hepatol. 2011;26:1787–1794. doi: 10.1111/j.1440-1746.2011.06796.x. [DOI] [PubMed] [Google Scholar]
- 69.Uyy E, Suica VI, Boteanu RM, et al. Endoplasmic Reticulum Chaperones Are Potential Active Factors in Thyroid Tumorigenesis. J Proteome Res. 2016;15:3377–3387. doi: 10.1021/acs.jproteome.6b00567. [DOI] [PubMed] [Google Scholar]
- 70.Tufo G, Jones AW, Wang Z, et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014;21:685–695. doi: 10.1038/cdd.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu SJ, Won JK, Ryu HS, et al. A novel prognostic factor for hepatocellular carcinoma: protein disulfide isomerase. Korean J Intern Med. 2014;29:580–587. doi: 10.3904/kjim.2014.29.5.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartkowiak K, Effenberger KE, Harder S, et al. Discovery of a novel unfolded protein response phenotype of cancer stem/progenitor cells from the bone marrow of breast cancer patients. J Proteome Res. 2010;9:3158–3168. doi: 10.1021/pr100039d. [DOI] [PubMed] [Google Scholar]
- 73.Higa A, Taouji S, Lhomond S, et al. Endoplasmic reticulum stress-activated transcription factor ATF6α requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2016;34:1839–1849. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lovat PE, Corazzari M, Armstrong JL, et al. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 2008;68:5363–5369. doi: 10.1158/0008-5472.CAN-08-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Na KS, Park BC, Jang M, et al. Protein disulfide isomerase is cleaved by caspase-3 and -7 during apoptosis. Mol Cells. 2007;24:261–267. [PubMed] [Google Scholar]
- 76.Xu S, Butkevich AN, Yamada R, et al. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc Natl Acad Sci U S A. 2012;109:16348–16353. doi: 10.1073/pnas.1205226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng X, Leyva ML, Jenny M, et al. A ruthenium-containing organometallic compound reduces tumor growth through induction of the endoplasmic reticulum stress gene CHOP. Cancer Res. 2009;69:5458–5466. doi: 10.1158/0008-5472.CAN-08-4408. [DOI] [PubMed] [Google Scholar]
- 78.Zhang J, Yi M, Zha L, et al. Sodium Butyrate Induces Endoplasmic Reticulum Stress and Autophagy in Colorectal Cells: Implications for Apoptosis. PLoS One. 2016;11:e0147218. doi: 10.1371/journal.pone.0147218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willems SH, Tape CJ, Stanley PL, et al. Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem J. 2010;428:439–450. doi: 10.1042/BJ20100179. [DOI] [PubMed] [Google Scholar]
- 80.Benham AM. The protein disulfide isomerase family: key players in health and disease. Antioxid Redox Signal. 2012;16:781–789. doi: 10.1089/ars.2011.4439. [DOI] [PubMed] [Google Scholar]
- 81.Camargo LDL, Babelova A, Mieth A, et al. Endo-PDI is required for TNFα-induced angiogenesis. Free Radic Biol Med. 2013;65:1398–1407. doi: 10.1016/j.freeradbiomed.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 82.Zhang D, Richardson DR. Endoplasmic reticulum protein 29 (ERp29): An emerging role in cancer. Int J Biochem Cell Biol. 2011;43:33–36. doi: 10.1016/j.biocel.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 83.Zhang D, Putti TC. Over-expression of ERp29 attenuates doxorubicin-induced cell apoptosis through up-regulation of Hsp27 in breast cancer cells. Exp Cell Res. 2010;316:3522–3531. doi: 10.1016/j.yexcr.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 84.Obacz J, Takacova M, Brychtova V, et al. The role of AGR2 and AGR3 in cancer: similar but not identical. Eur J Cell Biol. 2015;94:139–147. doi: 10.1016/j.ejcb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 85.Brychtova V, Vojtesek B, Hrstka R. Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett. 2011;304:1–7. doi: 10.1016/j.canlet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 86.Higa A, Mulot A, Delom F, et al. Role of pro-oncogenic protein disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) in the control of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286:44855–44868. doi: 10.1074/jbc.M111.275529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryu J, Park SG, Lee PY, et al. Dimerization of pro-oncogenic protein Anterior Gradient 2 is required for the interaction with BiP/GRP78. Biochem Biophys Res Commun. 2013;430:610–615. doi: 10.1016/j.bbrc.2012.11.105. [DOI] [PubMed] [Google Scholar]
- 88.Dong A, Wodziak D, Lowe AW. Epidermal growth factor receptor (EGFR) signaling requires a specific endoplasmic reticulum thioredoxin for the post-translational control of receptor presentation to the cell surface. J Biol Chem. 2015;290:8016–8027. doi: 10.1074/jbc.M114.623207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chevet E, Fessart D, Delom F, et al. Emerging roles for the pro-oncogenic anterior gradient-2 in cancer development. Oncogene. 2013;32:2499–2509. doi: 10.1038/onc.2012.346. [DOI] [PubMed] [Google Scholar]
- 90.Fessart D, Domblides C, Avril T. Secretion of protein disulphide isomerase AGR2 confers tumorigenic properties. eLife. 2016;30:5. doi: 10.7554/eLife.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]