Abstract
SRSF2, a Serine-Arginine rich (SR) protein, is a splicing activator that mediates exon inclusion and exclusion events equally well. Here we show SRSF2 directly suppresses intron splicing to suppress cassette exon inclusion in SMN pre-mRNA. Through a serial mutagenesis, we demonstrate that a 10 nt RNA sequence surrounding the branch-point (BP), is important for SRSF2-mediated inhibition of cassette exon inclusion through directly interacting with SRSF2. We conclude that SRSF2 inhibits intron splicing to promote exon exclusion.
Keywords: Exon exclusion, Intron retention, Pre-mRNA splicing, SRSF2
INTRODUCTION
Pre-mRNA splicing is a necessary process for gene expression in higher eukaryotes whereby non-coding introns are removed from pre-mRNA and coding exons are ligated to produce mRNA (1). The sequences that trigger pre-mRNA splicing, called splicing signals, include a GU dinucleotide near the 5′ splice-site (5′SS) at the beginning of the intron. Towards the 3′ end of the intron are three sequence elements including an AG dinucleotide at the 3′ splice-site (3′SS), with the branch-point (BP) and polypyrimidine tract (PPT) located upstream of the 3′SS.
Alternative splicing (AS) is a fundamental biological process in higher eukaryotic cells, producing different mRNA isoforms with different levels of coding potential and stability (2, 3). In addition to splicing signals, there are other cis-acting RNA elements having regulatory roles by providing target sites for trans-acting factors. Aberrant splicing is known to be the cause of various genetic diseases and cancer (4–6). For example, in spinal muscular atrophy (SMA), the survival of motor neuron (SMN1) gene is deleted in SMA patients but SMN2, a duplicate of SMN1, is present. The problem is that a few point mutations within SMN2 cause cassette exon skipping, thereby expressing non-functional proteins (7, 8).
SRSF2 is a 35-kDa serine/arginine-rich (SR) protein belonging to a family of highly conserved splicing factors in higher eukaryotes that facilitate constitutive and alternative splicing (9–11). All SR proteins have a similar bipartite structure including two functional domains: an RNA binding domain, comprising multiple RNA recognition motifs (RRMs), and an arginine-serine-rich (RS) domain (12–14). The consensus binding site for SRSF2 is more diverse than that of SRSF1. Although a recent structure-based study revealed SSNG (S = C / G) to be a highly consensus binding sequence (15), in vitro binding with SELEX and CLIP-seq analysis indicate that SRSF2 binds to GA-rich or purine-rich sequences (16). Although SR proteins generally function as splicing activators, there are reports demonstrating they are capable of being splicing suppressors (17–19). In particular, SRSF9 interacts with the silencer element and directly down-regulates exon recognition (18, 20). A recent study revealed that a similar frequency of exon skipping events and exon inclusion events are stimulated by SRSF1 or SRSF2 (21). However, the mechanism by which SR proteins mediate exon exclusion remains unclear.
Here, we demonstrate that SRSF2 functions as a splicing inhibitor that contributes directly to exon exclusion. Analysis of serial deletion mutants and chimeric minigenes suggest that a 10 nt sequence surrounding BP is the regulatory element responsible for inhibiting intron splicing. Furthermore, the 10 nt regulatory sequence allows SRSF2 to inhibit constitutive splicing. Our results reveal that inhibition of intron splicing contributes to regulation of cassette exon skipping.
RESULTS
SRSF2 promotes exon exclusion
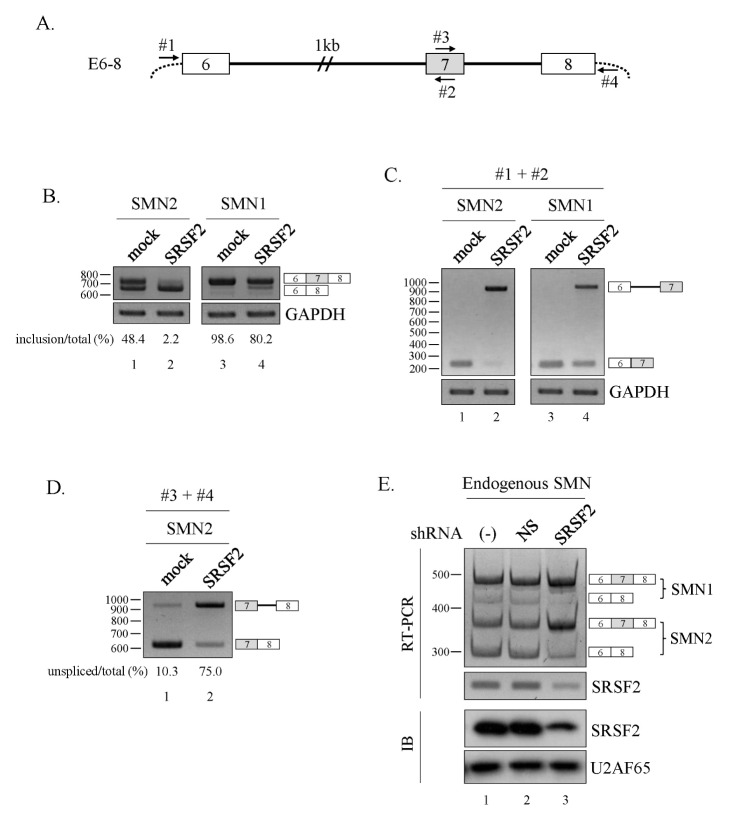
An interesting study by Pandit et al. (21), using genome-wide RNA-seq and analysis of alternative splicing events, demonstrated that SRSF2 stimulates both exon skipping and exon inclusion events equally well. To understand the role of SRSF2 in cassette exon exclusion, we wanted to test a gene whose exon skipping is mediated by SRSF2. Wee et al. (22) reported that reduced SRSF2 expression increased cassette exon7 inclusion of SMN pre-mRNA. Therefore, we co-expressed SRSF2 and the SMN1 or SMN2 minigene (E6–8) described earlier, which includes exon6 through exon8 with a shortened intron6 (Fig. 1A) (7, 23). Interestingly, over-expression of SRSF2 favored cassette exon7 exclusion in the SMN2, SMN1 minigenes, as evidenced by greater production of the exon7 skipped isoform (Fig. 1B, lane 2 and 4).
Fig. 1.
SRSF2 promotes exon exclusion. (A) Schematic diagram of the E6–8 minigene. Exons are depicted as numbered boxes, introns as solid lines. (B) RT-PCR analysis of the E6–8 minigene from the SMN1/2 locus in SRSF2-expressing cells. (C) RT-PCR analysis of intron6 splicing within the E6–8 minigene using primers #1 and #2. (D) RT-PCR analysis of intron7 splicing within the E6–8 minigene using primers #3 and #4. (E) RT-PCR analysis to detect alternative splicing of endogenous SMN1 and SMN2 using RNA extracted from cells infected with lentiviruses with SRSF2-targeting shRNA (SRSF2) or non-silencing shRNAs (NS).
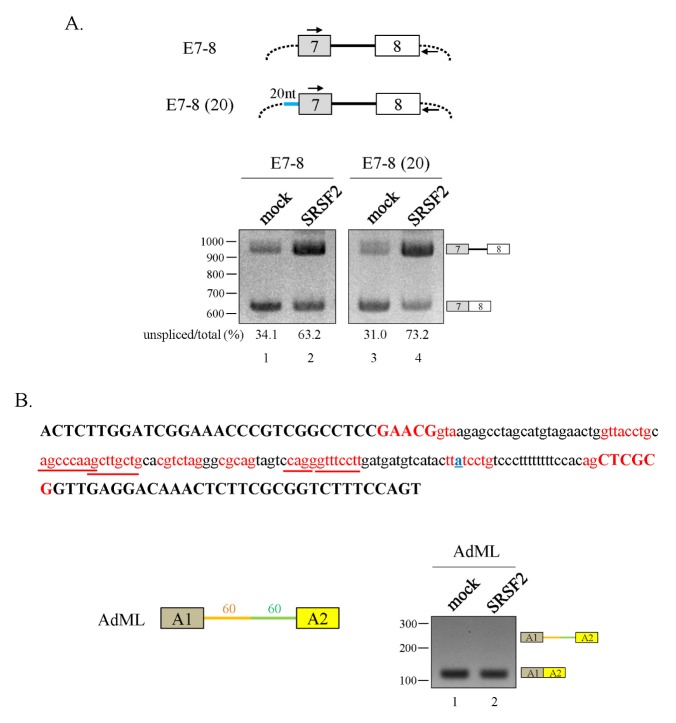
Given these observations, our goal was to determine the mechanism of SRSF2-mediated exon skipping by first demonstrating how SRSF2 regulates intron6 splicing. Thus, we designed oligonucleotide primers capable of hybridizing with our minigene expression vector (#1) and exon7 (#2) (Fig. 1A). Interestingly, intron6 splicing was much lower for both SMN1 and SMN2 minigene when SRSF2 was overexpressed (Fig. 1C, lanes 2 and 4). Surprisingly, a cryptic 3′SS located 682 nt upstream of the canonical 3′AG of exon7 was activated when SRSF2 was overexpressed. Thus intron6 splicing was suppressed by SRSF2, and in the meantime activating a cryptic 3′SS. Another unknown function of SRSF2 is whether it affects splicing at a second, nearby intron. We designed another set of oligonucleotide primers to anneal with exon7 (#3) and the plasmid vector (#4) such that we could detect splicing of intron7 (Fig. 1A). In mock-transfected cells, we observed mostly fully spliced exons (E7+8) whereas overexpression of SRSF2 favored transcripts that did not fully splice out intron7 (Fig. 1D). This contrast with our observation that lentivirus-mediated downregulation of SRSF2 expression stimulates endogenous exon7 inclusion (Fig. 1E). We also considered the possibility that SRSF2 mediates intron retention only in the presence of three exons. To test this idea, we constructed a two-exon minigene, E7–8, where all other upstream exons and introns were deleted (Fig. 2A, upper panel). Here, intron7 retention was still high for the E7–8 minigene when SRSF2 was overexpressed (Fig. 2A, lanes 1 and 2), although to a lesser extent than found in the E6–8 minigene (Fig. 1D). Next, it was also possible that adding the 3′SS sequences to the upstream exon (exon7) could modulate SRSF2 activity. Therefore, we constructed another minigene [E7–8 (20)] where a 20 nt sequence encompassing the 3′SS was added to the end of exon7 (Fig. 2A, upper panel). As seen with E7–8, intron7 splicing was greatly reduced when SRSF2 was overexpressed (Fig. 2A, lanes 3 and 4). Thus, SRSF2 promotes intron retention in the absence or presence of the upstream 20 nt RNA. Others have reported that SR proteins inhibit splicing, and a series of evidence supports the notion that SRSF2 stimulates splicing; however, it has not been reported yet that SRSF2 directly suppresses intron splicing (24, 25).
Fig. 2.
SRSF2 promotes SMN intron7 retention. (A) (Upper panel) Schematic diagram of the E7–8 and E7–8 (20) minigenes. The primer binding sites are indicated with arrows. (Lower panel) RT-PCR analysis of the E7–8 and E7–8 (20) minigenes in SRSF2-expressing cells. (B) (Upper panel) Nucleotide sequence of a portion of the AdML pre-mRNA. The two exons are shown as dark yellow and yellow boxes, the upstream intron is indicated in orange, and the downstream intron is indicated in green. (Right panel) RT-PCR analysis of the AdML minigene using RNAs in SRSF2-expressing cells.
Given these observations, it was important to determine whether SRSF2 suppresses splicing in other pre-mRNAs. Here, we used the AdML pre-mRNA which is known to be a sensitive test for the inhibitory function of the U2AF65 protein (7, 26). Although AdML pre-mRNA includes several potential SRSF2-binding sequences, our results indicate that SRSF2 is unable to suppress intron (Fig. 2B). Therefore, it appears that SRSF2 mediates intron retention in a pre-mRNA-sequence-specific manner.
A 10 nt RNA sequence surrounding the BP of intron7 is required for splicing inhibition by SRSF2
Because SRSF2 inhibits splicing in an RNA-sequence-specific manner, we attempted to identify the RNA sequences important for this function. As SRSF2 has degenerate binding specificity, SMN pre-mRNA includes many potential SRSF2 binding sites. Initially, we attempted to produce SRSF2-binding mutants where an intron tethered to an SR protein inhibits splicing. However, we could not identify any mutants that disrupted SRSF2 activity (data not shown). Therefore, we used an alternate approach and generated random deletion mutations. It was possible that exon8 expresses a crucial RNA element necessary for SRSF2 function. We constructed serial exon8 deletion mutants of exon8 (Supplementary Fig. 1A, top panel). Although minigenes with longer exon8 were slightly more capable of inhibiting intron7 splicing, all mutants displayed similar ability to inhibit intron7 splicing (Supplementary Fig. 1A, bottom panel). Next, we analyzed intron7 to determine if its sequences are responsible for inhibiting splicing. We produced three upstream-intron deletion mutants of intron7 nearest to exon8 (Supplementary Fig. 1B, top panel). Intron7 splicing was greatly affected in all mutant constructs as we observed mostly unspliced transcripts (Supplementary Fig. 1B, bottom panel). Thus, it seems that neither exon8 nor intron7 is required for SRSF2-mediated inhibition.
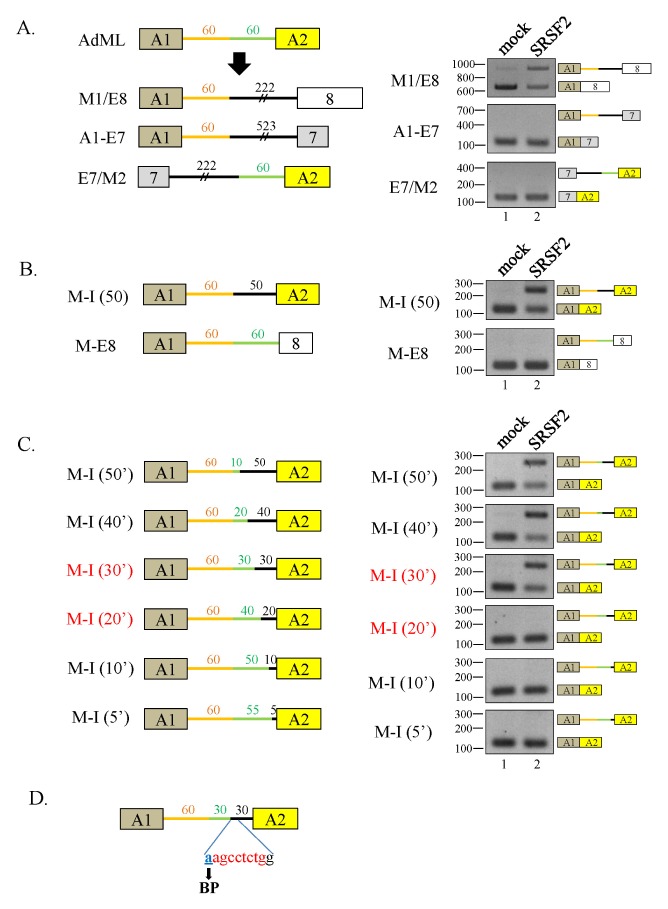
To find the regulatory elements for splicing inhibition, we produced chimeric minigenes of SMN and AdML to include the upstream half of AdML and the downstream half of the E7–8 (M1/E8) (Fig. 3A, left panel). Experiments with the M1/M8 minigene demonstrated that SRSF2 significantly repressed splicing, suggesting that the downstream half of intron7 contains important regulatory sequences for SRSF2 (Fig. 3A, right panel). Similarly, the A1–E7 minigene, where the downstream half of AdML was replaced with the downstream half of E6–7, showed no inhibition of splicing as the fully spliced transcript was produced (Fig. 3A, right panel). Another chimera, a minigene including the upstream half of E7–8 and the downstream half of AdML, E7/M2, did not inhibit splicing, confirming that the downstream half of E7–8 contains regulatory sequences important for SRSF2 function.
Fig. 3.
A 10 nt RNA sequence surrounding the BP of intron7 is required for splicing inhibition by SRSF2. (A) (Left panel) Schematic diagram of the M1/E8, A1–E7, and E7/M2 minigenes. (Right panel) RT-PCR analysis of intron splicing in the minigenes in SRSF2-expressing cells. (B) (Left panel) Schematic diagram of M-I (50) and M-E8 minigenes. (Right panel) RT-PCR analysis of the minigenes in SRSF2-expressing cells. (C) (Left panel) Schematic diagram of M-I (50′, 40′, 30′, 20′, 10′ and 5′) minigenes. Green lines represent intron sequences from AdML and black lines represent introns from SMN. The exon and intron lengths are also shown. (Right panel) RT-PCR analysis of intron splicing of the minigenes in the SRSF2-expressing cells. (D) The 10 nt regulatory sequence and its location within the AdML minigene are indicated.
It became clearer that the intronic sequences located toward the 3′ half of intron7 contained regulatory sequences important for SRSF2 function. We generated two more minigenes M-I (50) (where the 3′ half of the AdML intron was replaced with a 50 nt sequence from the 3′ SMN intron7) and M-E8 (where the second exon from AdML was replaced with exon8 from SMN) (Fig. 3B, left panel). Interestingly, SRSF2 could inhibit intron splicing from the M-I (50), but not the M-E8, demonstrating the importance of the 50 nt putative regulatory sequence for SRSF2 function (Fig. 3B, right panel). To further identify the nucleotide sequences important for regulating splicing, we created larger substitutions by inserting increasingly large AdML downstream intron sequences within the 50 nt regulatory sequence using the M-I (50) as a template (Fig. 3C, left panel). While two mutants that included 40 [M-I (40′)] or 30 nt [M-I (30′)] substitutions showed significant splicing inhibition in the presence of SRSF2, the other three mutants containing 20-[M-I (20′)], 10–[M-I (10′)], or 5 nt [M-I (5′)] substitutions did not significantly inhibit splicing (Fig. 3C, right panel). It is important to note that the 10 nt putative regulatory sequence includes a BP and other surrounding sequences (Fig. 3D). Therefore, this is strong evidence that the 10 nt RNA sequences between [M-I (30′)] and [M-I (20′)] is crucial for SRSF2-mediated inhibition of intron splicing.
A 10 nt RNA sequence physically interacts with SRSF2 and inhibits constitutive splicing
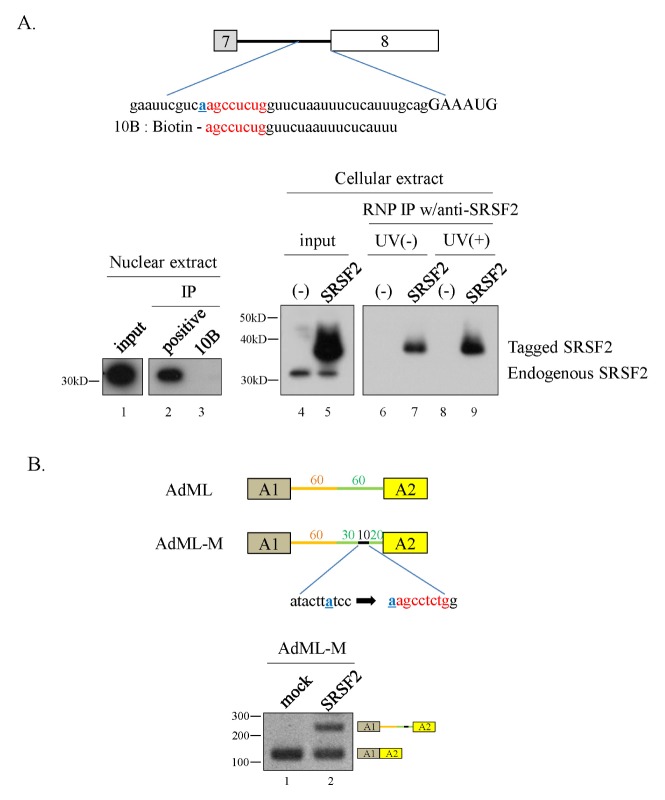
Using a bioinformatics-based approach (ESE finder) (27), we predicted that the above-mentioned 10 nt RNA sequence could interact with SRSF2. To confirm this prediction, we synthesized a biotin-labeled RNA having the 10 nt sequence and including the adjacent nucleotides from the 5′-end of exon8 (10B) (Fig. 4A). RNA pulldown and immunoprecipitation assays with SRSF2-specific antibodies demonstrated that the 10B RNA does not interact with SRSF2 from HeLa cell nuclear extracts (Fig. 4A, lower panel, lanes 2 and 3). Moreover, 10B RNA does not interact with SRSF2 from cellular extracts (Fig. 4A, lower panel, lane 6). Although endogenous SRSF2 was unable to bind to 10B, the tagged-SRSF2 showed strong interaction with 10B RNA both in the absence and presence of UV light (Fig. 4A, lower panel, lanes 7 and 9). Therefore, the 10 nt RNA interacts with SRSF2 only at high protein concentrations.
Fig. 4.
A 10 nt RNA sequence physically interacts with SRSF2 and inhibits constitutive splicing. (A) (Upper panel) The 10 nt regulatory sequence with surrounding sequences and biotin-labeled RNA sequence (10B) are shown. (Lower panel) RNA-pulldown and immunoprecipitation analyses with SRSF2-specific antibody are shown. (B) (Upper panel) Schematic diagram of the AdML-M minigene. (Lower panel) RT-PCR analysis of intron splicing in the AdML-M minigene.
One remaining question is whether the 10 nt RNA regulatory element inhibits constitutive splicing. To address this question, we substituted the 10 nt sequence from AdML pre-mRNA, located 20 nt upstream of the 3′SS, with the 10 nt RNA from SMN minigene (AdML-M) (Fig. 4B). As expected, SRSF2 inhibited splicing of AdML-M pre-mRNA in cells overexpressing SRSF2 (Fig 4B, lower panel). Therefore, the 10 nt sequence, perhaps upon binding SRSF2, inhibits constitutive splicing.
DISCUSSION
We used SMN pre-mRNA as a model system to study inhibition of exon inclusion by SRSF2. We observed SRSF2 was able to suppress splicing of flanking introns. In addition, we provided evidence that SRSF2 directly inhibits intron splicing. Unlike U2AF65, SRSF2 did not inhibit the splicing of AdML pre-mRNA, suggesting that SRSF2-mediated inhibition of splicing is RNA-sequence specific. Experiments with serial deletion mutants and chimeric SMN-AdML minigenes, revealed that exon8, exon7, and the upstream half of intron7 (the portion nearest to exon7) are dispensable for SRSF2 function. Furthermore, a 10 nt sequence surrounding the BP of SMN pre-mRNA is crucial regulatory element for SRSF2-mediated splicing inhibition. As the 10 nt RNA sequence provides relatively weak binding potential, it can interact only with overexpressed SRSF2 but not endogenous SRSF2. Importantly, the 10 nt also inhibited splicing in AdML pre-mRNA when SRSF2 was overexpressed.
Although many reports have demonstrated that SR proteins are splicing activators, inhibition of splicing by the SR proteins has been reported in a few cases. For example, SRSF10 inhibits splicing in the dephosphorylated state (17), while SRSF9 suppresses 3′ splicing by interacting with the upstream intron (18). Using in vitro splicing experiments with the Hβ-globin pre-mRNA, it has been shown that SRSF2 is required for the first step of splicing (24, 28). Here, we have presented the first evidence that SRSF2 suppresses intron splicing. There are two possible explanations for these contradictory observations. First, the inhibitory activity of SRSF2 can overcome its promoting activity in spliceosome assembly. Second, the pre-mRNAs used in these experiments are different, and assay systems are also different depending on in vitro or cell-based assays. For example, U2AF65 was shown to be an important splicing activator (29), but our cell-based experiments demonstrated that U2AF65 inhibits pre-mRNA splicing (7).
SRSF2 has degenerate RNA sequence specificity, resulting in many potential SRSF2-binding sites on SMN pre-mRNA. Our several chimeric minigenes revealed that a 10 nt sequence surrounding a BP are crucial for SRSF2-mediated splicing inhibition. Why the physical interaction between some SR proteins and RNAs are important remains to be determined. One possibility is that some steric hindrance caused by SRSF2 could be overcome by the general splicing activity of SRSF2, so that functional inhibition does not occur.
SR proteins are thought to promote exon skipping through activities mediated by the constitutive exon (30). For instance, SRSF1 contacts downstream constitutive exons to promote exon skipping (31). However, the detailed mechanisms for SR protein-induced exon skipping remain largely unknown. We propose that SRSF2 does not suppress constitutive splice-site pairing. The available evidence suggest that SR proteins enhance splicing when located upstream of 5′SS, but inhibit splicing when located downstream of 5′SS (32). We observed a potential SRSF2-binding sequence immediately upstream of the exon in AdML pre-mRNA, but splicing was not inhibited. Thus, location dependency was not observed in this case. One explanation is that while Erkelenz et al. performed the experiments with artificial MS2-tethered SR protein (32), we used naturally occurring SR proteins. Another possibility is that location dependency does not exist in 3′SS because, in addition to 3′AG, both PPT and BP are also involved in 3′SS selection.
MATERIALS AND METHODS
Plasmid construction
The E6–8 minigene was generated as described previously (7). E7–8, E7–8 (20) were generated using the E6–8 construct as a template. The M1/E8, A1–E7, E7/M2, M-I (50), M-E8, and M-I (50′, 40′, 30′, 20′, 10′, 5′) constructs were generated using the E6–8 and AdML constructs as templates and inserted into the pcDNA3.1 vector using the NheI, KpnI, and EcoRI sites.
Cell culture and transfection
293T cells were cultured in DMEM media supplemented with antibiotics and 10% FBS. Cells were maintained at 37°C and in 5% CO2. Plasmid transfections were performed using polyethyleneimide (PEI) according to the manufacturer’s protocol.
RT-PCR
Total cellular RNA was extracted using the RiboEx reagent (GeneAll) according to the manufacturer’s instructions. 1 μg of total RNA was reverse transcribed using M-MLV reverse transcriptase (ELPiS) and amplified by PCR using G-Taq polymerase (Cosmo Genetech). The following primers were used for splicing studies: Exon6.F (5′-ATAATTCCCCCACC ACCTCC-3′), Exon8.R (5′-ACTACAACACCCTTCTCACAG-3′), pcDNA.F (5′-CACTGCTTACTGGCTTATCGAA-3′), pcDNA.R (5′-CTAGAAGGCACAGTCGAGGCT-3′), AdML.F (5′-ACTCTT GGATCGGAAACCCGT-3′).
RNA immunoprecipitation assay
RNA immunoprecipitation assays were performed as described previously (33). Briefly, 100 pmole of 5′-biotinylated RNA was mixed with nuclear or cellular extracts prepared from HeLa cells in Buffer D at 4°C. The mixtures were precipitated with streptavidin-conjugated agarose beads. Proteins resolved by SDS-PAGE were transferred to a PDVF membrane and blocked with 5% skim milk. The membrane was probed with an anti-SRSF2 antibody (Millipore) and the antibody-SRSF2 conjugates were detected by enhanced chemiluminescence (ECL).
Knockdown of SRSF2 with lentivirus-mediated shRNA
Knockdown of SRSF2 was performed as described previously (34). Briefly, shRNA lentivirus was generated by co-transfection of SRSF2 mRNA silencing sequence plasmid (Open Biosystems) with helper plasmids into 293T cells using polyethyleneimide (PEI) reagent. The lentiviruses were added to the cells for 72 h supplemented with 10 μg/ml polybrene.
Immunoblotting
Immunoblotting with anti-SRSF2 antibody was performed as previously described (34). Briefly, cells were harvested and lysed with lysis buffer for 30 min at 4°C. The supernatants were collected and separated on 12% SDS-PAGE, and then transferred to PDVF membrane. After blocking with 5% skim milk, membrane was incubated anti-SRSF2 (Millipore) or anti-U2AF65 antibodies. The signal was detected by enhanced chemiluminescence (ECL).
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by an NRF-2015R1A2A1A15054247 grant awarded to Haihong Shen, an NRF-2016R1A2B1007135 grant awarded to Xuexiu Zheng, the Cell Logistics Research Center (2016R1A5A1007318) fund of the National Research Foundation (NRF) of Korea, and an GIST Research Institute (GRI) grant funded by the Gwangju Institute of Science and Technology (GIST).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SA, Ahn SI, Gallo JM. Tau mis-splicing in the pathogenesis of neurodegenerative disorders. BMB Rep. 2016;49:405–413. doi: 10.5483/BMBRep.2016.49.8.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Moon H, Loh TJ, et al. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc Natl Acad Sci U S A. 2015;112:9926–9931. doi: 10.1073/pnas.1500639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 9.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol. 1999;77:277–291. doi: 10.1139/o99-048. [DOI] [PubMed] [Google Scholar]
- 10.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 11.Loh TJ, Moon H, Jang HN, et al. SR proteins regulate V6 exon splicing of CD44 pre-mRNA. BMB Rep. 2016;49:612–616. doi: 10.5483/BMBRep.2016.49.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branch-point to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/S1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 14.Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/S0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 18.Simard MJ, Chabot B. SRp30c is a repressor of 3′ splice site utilization. Mol Cell Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E, Stuani C, De Prato G, Baralle FE. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandit S, Zhou Y, Shiue L, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS One. 2014;9:e115205. doi: 10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho S, Moon H, Loh TJ, et al. hnRNP M facilitates exon 7 inclusion of SMN2 pre-mRNA in spinal muscular atrophy by targeting an enhancer on exon 7. Biochim Biophys Acta. 20141839:306–315. doi: 10.1016/j.bbagrm.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 26.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24:2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 29.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Ding JH, Byeon CW, et al. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghigna C, Giordano S, Shen H, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 32.Erkelenz S, Mueller WF, Evans MS, et al. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X, Cho S, Moon H, Loh TJ, Jang HN, Shen H. Detecting RNA-Protein Interaction Using End-Labeled Biotinylated RNA Oligonucleotides and Immunoblotting. Methods Mol Biol. 2016;1421:35–44. doi: 10.1007/978-1-4939-3591-8_4. [DOI] [PubMed] [Google Scholar]
- 34.Moon H, Cho S, Loh TJ, et al. SRSF2 promotes splicing and transcription of exon 11 included isoform in Ron proto-oncogene. Biochim Biophys Acta. 20141839:1132–1140. doi: 10.1016/j.bbagrm.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.