SUMMARY
Prokaryotic CRISPR-Cas adaptive immune systems utilize sequence specific RNA-guided endonucleases to defend against infection by viruses, bacteriophages and mobile elements, while these foreign genetic elements evolve diverse anti-CRISPR proteins to overcome the CRISPR-Cas-mediated defense of the host. Recently, AcrIIA2 and AcrIIA4, encoded by Listeria monocytogenes prophages were shown to block the endonuclease activity of Type II-A Streptococcus pyogenes Cas9 (SpyCas9). We now report the crystal structure of AcrIIA4 in complex with single-guide RNA-bound SpyCas9, thereby establishing that AcrIIA4 preferentially targets critical residues essential for PAM duplex recognition, as well as blocks access to key catalytic residues lining the RuvC pocket. These structural insights, validated by biochemical assays on key mutants, demonstrate that AcrIIA4 competitively occupies both PAM-interacting and non-target DNA strand cleavage catalytic pockets. Our studies provide insights into anti-CRISPR mediated suppression mechanisms for inactivating SpyCas9, thereby broadening the applicability of CRISPR-Cas regulatory tools for genome editing.
TOC Image
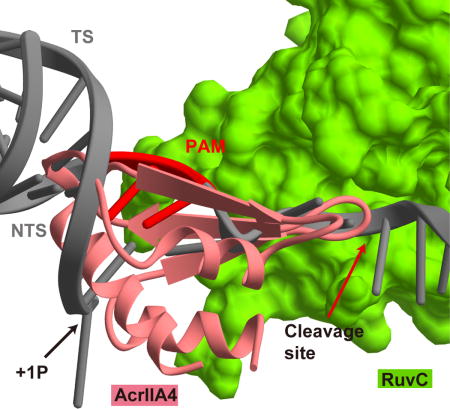
INTRODUCTION
In bacteria, defense against phage predation relies on restriction-modification (R-M) systems, as well as clustered regularly interspaced short palindromic repeats and CRISPR associated (CRISPR-Cas) systems (Dupuis et al., 2013; Hille and Charpentier, 2016; Labrie et al., 2010; Marraffini, 2015; Mohanraju et al., 2016; Nishimasu and Nureki, 2016; Wright et al., 2016). The R-M systems provide ubiquitous and innate protection against any invaders not possessing countermeasures. The CRISPR-Cas systems were found in almost all archaea and about 50% of bacteria and function as the only adaptive immune system, which utilizes sequence specific RNA-guided endonucleases to cut foreign genetic elements. The CRISPR-Cas immunity pathway relies on divergent Cas proteins and CRISPR RNA (crRNA) and can be divided into three stages: spacer acquisition, crRNA biogenesis and target interference (van der Oost et al., 2009; Wright et al., 2016). The Cas proteins and crRNA are encoded by the CRISPR locus, which is composed of variable cas genes and a CRISPR array comprising short direct repeats separated by spacers. Based on the phylogenetic classification, the CRISPR-Cas systems can be grouped into two classes and subdivided into 6 types and 19 subtypes: Class 1 systems (Type I, III and IV) employ multi-subunit surveillance ribonucleoprotein complexes termed Cascade (CRISPR-associated complex for antiviral defense), while Class 2 systems (Type II, V and VI) rely on single effector proteins (Makarova et al., 2011; Makarova et al., 2015; Shmakov et al., 2015).
In turn, phages and mobile genetic elements have developed divergent strategies to overcome the immune defense systems in host bacteria, that include adsorption inhibition, abortive infection, R-M systems, and CRISPR-Cas systems (Samson et al., 2013). Recently, a range of phage-encoded ‘anti-CRISPR’ proteins have been identified that suppress different CRISPR-Cas systems. Specially, ten Type I-F and four Type I-E anti-CRISPR proteins were discovered to inactivate Pseudomonas aeruginosa CRISPR-Cas systems (Bondy-Denomy et al., 2015; Bondy-Denomy et al., 2013; Pawluk et al., 2014; Pawluk et al., 2016b). Four Type II-A anti-CRISPR proteins encoded by L. monocytogenes prophages were identified and two of them blocked both S. pyogenes Cas9 (SpyCas9) and L. monocytogenes Cas9 (LmoCas9) (Rauch et al., 2017). Three families of Type II-C anti-CRISPR proteins targeting N. meningitidis Cas9 (NmeCas9) have also been reported (Pawluk et al., 2016a). Moreover, a Type VI-B anti- Cas13b protein Csx27 was identified as an inhibitor for both B. zoohelcum Cas13b (Type VI-B1) and P. buccae Cas13b (Type VI-B2) systems, while an activator Csx28 for P. buccae Cas13b was also identified (Smargon et al., 2017).
The programmability of CRISPR-Cas systems have been widely developed as genetic tools and provide immense promise for therapeutic applications (Barrangou and Doudna, 2016; Hsu et al., 2014). Two class 2 CRISPR-Cas effectors, namely Cas9 (type II) (Barrangou and Doudna, 2016; Hsu et al., 2014; Jiang and Marraffini, 2015; Sternberg and Doudna, 2015; Wright et al., 2016) and Cas13a (also known as Cpf1, type V) (Zetsche et al., 2015) that cleave dsDNA targets have been successfully harnessed for genome editing. Newly identified Cas13a (also known as C2c2, Type VI), which cleaves RNA targets (Abudayyeh et al., 2016; East-Seletsky et al., 2017; East-Seletsky et al., 2016; Liu et al., 2017), has been developed as a rapid detection tool for diagnosis of pathogens and genotyping (Gootenberg et al., 2017). Cas9 requires either a pair of RNA molecules, namely crRNA and tracrRNA (trans-activating crRNA), or a synthetic single-guide RNA (sgRNA) involving a tetra loop covalently-linking the 3′ end of crRNA to the 5′ end of tracrRNA (Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012), as well as a short 3′-G-rich protospacer adjacent motif (PAM) sequence on the non-target strand proximal to the cleavage site (Deveau et al., 2008; Mojica et al., 2009). Cas9 uses two nuclease domains, namely HNH and RuvC domains to generate blunt-end double-strand breaks. The HNH domain cleaves the target DNA strand that is base-paired with the crRNA guide, while the RuvC domain cleaves the non-base-paired single-stranded non-target DNA strand (Barrangou et al., 2007; Deltcheva et al., 2011; Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012). To date, SpyCas9 remains the most commonly used and powerful genome-editing tool. Besides, a catalytically dead mutant SpyCas9 (dSpyCas9, D10A/H840A) has served as an effective and specific RNA-guided genome-binding platform (Dominguez et al., 2016; Qi et al., 2013; Wang et al., 2016a). Although the principles underlying SpyCas9 cleavage specificity are well understood and the technology underlying its commercial applicability are well developed, further knowledge is required to minimize side effects resulting from alternate cleavage patterns, thereby insuring effective and safe genome editing in the clinic. The discovery of anti-CRISPR proteins has provided the prospect of introducing robust, specific and genetically encodable ‘off-switch’ tools for modulating Cas9 activity. Till very recently, the suppression mechanisms of two reported anti-SpyCas9 proteins, AcrIIA2 and AcrIIA4, which block the activity of SpyCas9 in bacteria and human cells (Rauch et al., 2017), have remained unknown.
To address this goal, we investigated the structural principles underlying the mode of action of these AcrII anti-CRISPR proteins in overcoming CRISPR-Cas mediated host defense. We observed direct interactions between sgRNA-bound SpyCas9 and AcrIIA4, as well as AcrIIA2. To better understand the inhibition mechanism of AcrIIA4, we determined the crystal structure of AcrIIA4 bound to the SpyCas9-sgRNA binary complex. The structural comparisons reveal that AcrIIA4 preferentially interacts with SpyCas9-sgRNA binary complex and competitively occupies the binding surface for the PAM duplex, as well as blocks access to the active site in the RuvC pocket, thereby further contributing to the inactivation of SpyCas9. Our structural studies and biochemical characterization of interfacial mutants for AcrIIA4 bound to sgRNA-bound SpyCas9 provides guidelines for the further development of natural anti-CRISPR ‘off-switch’ tools for genome engineering and related biotechnology applications.
RESULTS
AcrIIA2 and AcrIIA4 Physically Interact with SpyCas9
It has been reported previously that anti-CRISPR protein AcrIIA2 partially blocks dSpyCas9 function in Escherichia coli, while AcrIIA4 shows almost completely blockage. Both AcrIIA2 and AcrIIA4 also suppress the Cas9-based genome editing in HEK293T cells (Rauch et al., 2017). We tested for whether AcrIIA2 and AcrIIA4 can suppress SpyCas9 activity for a linear dsDNA target containing a 5′-TGG-3′ PAM sequence in vitro in the presence of a 85-nt sgRNA, as described previously (Jinek et al., 2012). With increasing amount of AcrIIA2 and AcrIIA4, the enzymatic activity of SpyCas9 was blocked in a dose-dependent manner, with AcrIIA4 exhibiting a relatively higher blockage effect (Figure 1A, left panel) than AcrIIA2 (Figure S1A), consistent with reported in vivo data (Rauch et al., 2017). Indeed, availability of sufficient AcrIIA2 and AcrIIA4 results in the complete blockage of the catalytic activity of SpyCas9 (Figure 1A, right panel and S1A,), similar to the effect observed for mutations in dSpyCas9 (Figure 1A, right panel).
Figure 1. AcrIIA4 directly interacts with sgRNA-bound SpyCas9 and inactivates SpyCas9.
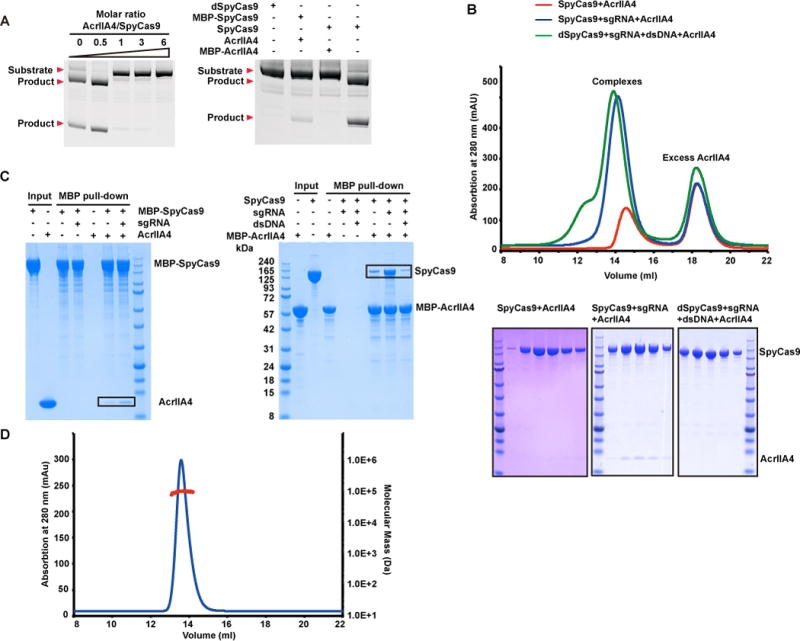
(A) In vitro enzymatic assay monitoring cleavage of linear dsDNA by SpyCas9 and sgRNA in the presence of AcrIIA4. The molar ratios of AcrIIA4:SpyCas9 are shown at the top of each lanes (left panel). The inhibition between MBP-tagged SpyCas9 and AcrIIA4 are also detected and compared with dSpyCas9 (right panel).
(B) AcrIIA4 selectively forms a stable complex with sgRNA-bound SpyCas9 rather than apo or DNA-bound SpyCas9-sgRNA in solution. SEC was performed using SpyCas9 in the presence or absence of sgRNA and sgRNA-dsDNA.
(C) AcrIIA4 physically interacts with sgRNA-bound SpyCas9. MBP pull-down assays were performed using MBP-tagged AcrIIA4 and SpyCas9 in presence or absence of sgRNA and sgRNA-dsDNA.
(D) Oligomeric state of AcrIIA4 in solution detected by SEC-MALS. The horizontal red line represents the SEC-MALS calculated mass for AcrIIA4. The calculated and theoretical molecular masses are 10.4 kDa and 10.2 kDa, respectively, indicating that AcrIIA4 exists as a monomer in solution.
See also Figure S1.
We then investigated whether AcrIIA2 and AcrIIA4 inactivate SpyCas9 by direct interaction. Previous structural data showed that large conformational changes occur during the transitions between apo-SpyCas9, sgRNA-bound, and dsDNA target-bound states (Anders et al., 2014; Jiang et al., 2016; Jiang et al., 2015; Jinek et al., 2014; Yamano et al., 2016). We performed size-exclusion chromatography (SEC) to detect whether AcrIIA2 and AcrIIA4 specifically bind to one state or universally to more than one state of SpyCas9 and also whether they form stable complexes with SpyCas9 in solution (Figure 1B and S1B, top panels). To avoid potential cleavage of the target dsDNA, we used dSpyCas9 to generate dsDNA target-bound ternary complex. Notably, in the presence of sgRNA, AcrIIA2 and AcrIIA4 form stable complexes with SpyCas9, whereas no band or very weak bands were detected when using either apo-SpyCas9 or dSpyCas9-sgRNA-dsDNA ternary complex (Figure 1B and S1B, bottom panels).
In addition, AcrIIA2 and AcrIIA4 show strong binding to sgRNA-bound MBP- SpyCas9 binary complex, while very weak interactions were detected between apo MBP-SpyCas9 with AcrIIA2, and with AcrIIA4 (Figure 1C, left panel and S1C). Similarly, MBP-AcrIIA4 only shows strong binding to sgRNA-bound SpyCas9, while dsDNA target-bound dSpyCas9-sgRNA shows significantly reduced binding affinity to AcrIIA4 (Figure 1C, right panel). The attachment of an MBP tag at either SpyCas9 or AcrIIA4 has minimal effect on the blockage of enzymatic activity (Figure 1A, right panel). The data outlined in Figure 1A–C demonstrate that AcrIIA2 and AcrIIA4 specifically recognize sgRNA-bound rather than either apo or dsDNA target-bound states of SpyCas9.
We analyzed whether AcrIIA2 and AcrIIA4 formed monomers or higher oligomers in the free state in solution by size-exclusion chromatography coupled with in-line multiangle light scattering analysis (SEC-MALLS). The measured molecular masses are close to the theoretical molecular mass for monomers (14.4 kDa vs 14.2 kDa for AcrIIA2, and 10.4 kDa vs 10.2 kDa for AcrIIA4), indicating that AcrIIA2 and AcrIIA4 in the free state exist as monomers in solution (Figure 1D and S1D).
Structure of SpyCas9-sgRNA-AcrIIA4 Ternary Complex
The color-coded domain architecture of SpyCas9 is shown in Figure 2A, while the color-coded secondary structure of the sgRNA composed of covalently-linked crRNA and tracrRNA is shown in Figure S2A. We solved the crystal structure of AcrIIA4 bound to dSpyCas9-sgRNA at 2.6 Å resolution (x-ray statistics summarized in Table 1). SpyCas9 resembles an overall ‘Crab Claw’ fold consisting of an α-helical recognition (REC) lobe composed of Helical-I and Helical-II domains and a NUC lobe composed of RuvC, HNH, Topo, and CTD domains, as described previously (Figure 2B) (Anders et al., 2014; Jiang et al., 2016; Jiang et al., 2015; Jinek et al., 2014; Nishimasu et al., 2014). The arginine-rich bridge helix (BH) motif connects the two lobes. The tertiary fold of the sgRNA in the AcrIIA4-bound ternary complex is shown in Figure S2B. AcrIIA4 is positioned on one face of SpyCas9 formed by Topo, RuvC, and CTD domains, with one AcrIIA4 bound per SpyCas9-sgRNA complex (see circled region, Figure 2B and S3A, B). AcrIIA4 adopts a fold composed of a 3-stranded β-sheet with 3 α-helices positioned along one face of this β-sheet (Figure S3C), with the electrostatics of its SpyCas9-interacting surface shown in Figure S3D. A DALI search detected no structural similarity between AcrIIA4 and reported structures, indicating that AcrIIA4 adopts an unidentified fold in the ternary complex.
Figure 2. Overall structure of AcrIIA4-SpyCas9-sgRNA complex.
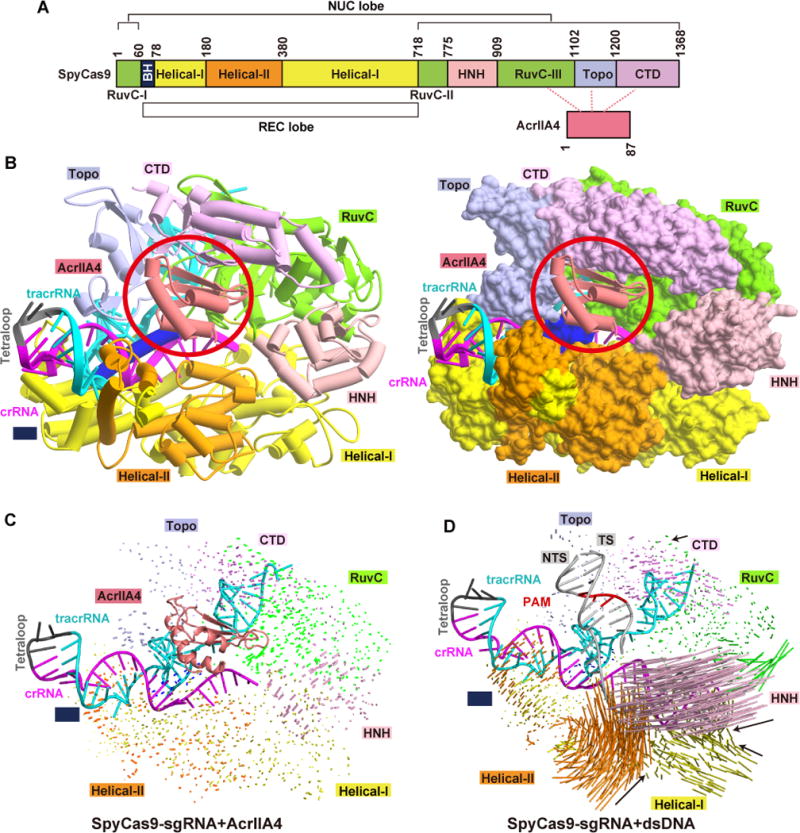
(A) Domain organization of SpyCas9 and AcrIIA4.
(B) Ribbon and surface representations of AcrIIA4-SpyCas9-sgRNA ternary complex, color-coded as defined in panel A. The AcrIIA4 molecule in each view is highlighted by a red circle.
(C, D) Structural comparison of domain movement on proceeding from the SpyCas9-sgRNA binary complex to the SpyCas9-sgRNA-AcrIIA4 ternary complex (panel C) and to the SpyCas9-sgRNA-dsDNA ternary complex (panel D). Vector lengths correlate with the domain motion scales. TS and NTS represent target and non-target DNA strands, respectively.
See also Figures S2 and S4.
Table 1.
| Beam line | APS-ID24E | |
| Wavelength (Å) | 0.9792 | |
| Space group | P22A | |
| Cell parameters a, b, c (Å) | ||
| a, b, c (Å) | 72.19, 101.24, 303.56 | |
| Resolution (Å) | 50.0–2.60 (2.69–2.60)a | |
| Rmerge(%) | 9.2 (49.7) | |
| Average I/σ(I) | 14.3 (1.6) | |
| Completeness (%) | 94.6 (72.1) | |
| CC(1/2) | 0.805 | |
| Average redundancy | 4.3 (3.4) | |
| No. of unique reflections | 65,587 | |
| Refinement and structure model | ||
| Rwork/Rfree(%) | 17.8/22.6 | |
| No. of non-H atoms | 13,175 | |
| Average B factor(Å2) | ||
| SpyCas9 | 60.1 | |
| AcrIIA4 | 53.6 | |
| RNA | 60.6 | |
| RMS deviations | ||
| Bond lengths (Å) | 1.081 | |
| Bond angles (°) | 0.016 | |
| Ramachandran plot (%) | ||
| Favored | 95.3 | |
| Allowed | 4.7 | |
| Outliers | 0 | |
Highest resolution shell (in Å) shown in parentheses.
The overall structure of SpyCas9 in the AcrIIA4-bound SpyCas9-sgRNA ternary complex is almost identical to that in the SpyCas9-sgRNA binary complex (Figure 2C). By contrast, addition of dsDNA to the SpyCas9-sgRNA binary complex results in large conformational changes in the Helical and HNH domains of SpyCas9 (Figure 2D). These results establish that binding of AcrIIA4, in contrast to binding of dsDNA, induces minimal conformational changes of SpCas9 within the SpyCas9-sgRNA context.
Recognition of sgRNA-bound SpyCas9 by AcrIIA4
AcrIIA4 interacts with a positively charged surface of SpyCas9 formed by the Topo, RuvC, and CTD domains (Figure S3B) via extensive hydrophilic contacts. The contacts with the Topo domain involve the α1–β1 loop (connecting helix α1 and β strand β1) and β2–β3 loop of bound AcrIIA4 (Figure 3A, B). The side chains of Asp14 and Asn36 of bound AcrIIA4 are involved in a hydrogen-bonding network with the main chains of Glu1108, Ser1109 and Ser1136, as well as the side chain of Ser1109 of the Topo domain (Figure 3B). It should be noted that the same Glu1108 and Ser1109 are key residues that stabilize the +1 phosphate, while key residue Ser1136 contacts the base of a PAM nucleotide in the non-target DNA strand via a water molecule in the structure of the corresponding SpyCas9-sgRNA-dsDNA ternary complex (Anders et al., 2014).
Figure 3. Detailed Interactions of AcrIIA4 with sgRNA-bound SpyCas9.
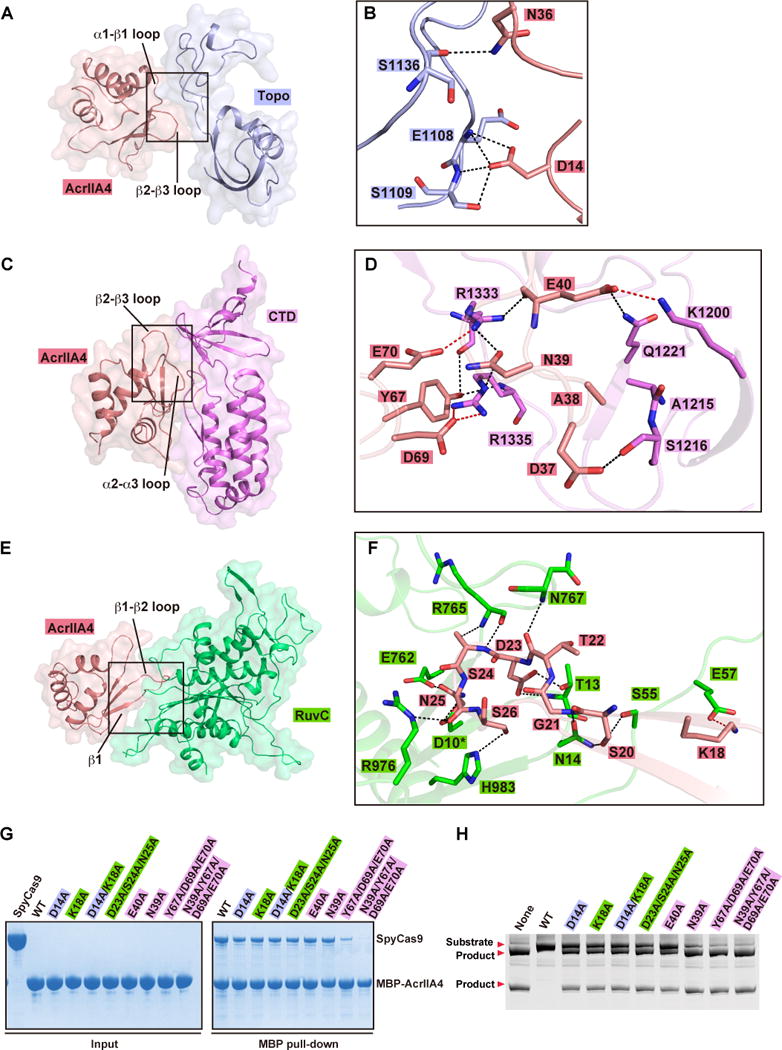
(A, C, E) Surface views of the interfaces between AcrIIA4 and Topo domain (panel A), CTD domain (panel C), and RuvC domain (panel E). The interface segments are highlighted by black boxes.
(B, D, F) Detailed interactions at the interfaces between AcrIIA4 and Topo domain (panel B), CTD domain (panel D), and RuvC domain (panel F) are shown in stick representations. The color code is the same as Figure 2A. Hydrogen bonds and salt bridges are colored as black and red dashed lines, respectively.
(G) Mutation analysis of AcrIIA4 residues involving in the binding to sgRNA-bound SpyCas9 by MBP pull-down assay of MBP-tagged AcrIIA4.
(H) In vitro enzymatic assay of Ala mutations of AcrIIA4 residues that impaired or abolished binding of AcrIIA4 to sgRNA-bound SpyCas9.
See also Figure S3.
The interface between AcrIIA4 (through its β2–β3 and α2–α3 loops) and the CTD domain consists of several hydrophilic interactions (Figure 3C, D). Residues Asp37 to Glu40 in the β2–β3 loop, and the following β-strand (β3) of bound AcrIIA4 interact with residues Lys1200, Ala1215, Ser1216, and Gln1221 in the CTD domain. It is worth noting that these residues in the CTD domain interact with the backbones of the PAM duplex in the structure of the corresponding SpyCas9-sgRNA-dsDNA ternary complex (Anders et al., 2014). Interestingly, residues Arg1333 and Arg1335 in the β-hairpin of the CTD domain, that are essential for the recognition of PAM sequence in the SpyCas9-sgRNA-dsDNA ternary complex (Anders et al., 2014), are instead recognized by residues Tyr67, Asp69 and Glu70 in the α2–α3 loop and residue Asn39 in β3 of bound AcrIIA4 (Figure 3D).
At the interface with the RuvC domain, an extensive intermolecular hydrogen-bond network is formed involving Lys18 in β1 and Ser20 to Ser26 in the β1–β2 loop (Figure 3E, F). Remarkably, the protruding β1–β2 loop of AcrIIA4 inserts into the active site of RuvC domain and hydrogen bonds with key residues (Glu762 and His 983) lining the RuvC catalytic pocket (Figure 3F). We used dSpyCas9 (D10A/H840A) for crystallization, so we modeled the side chain of Asp10 in the catalytic pocket based on reported structures (Anders et al., 2014; Jiang et al., 2016; Jiang et al., 2015; Jinek et al., 2014; Nishimasu et al., 2014) and found that the side chain of key catalytic residue Asp10 could potentially hydrogen bond with Asn25 in the β1–β2 loop of bound AcrIIA4. These interactions between AcrIIA4 and key residues (Asp10, Glu762, and His 983) lining the RuvC catalytic pocket result in the shielding of the active site and abolishing the access of non-target dsDNA strand into the active site.
We generated single or multiple Ala mutations of AcrIIA4 residues involved in intermolecular contacts and investigated the impact of these mutations on binding to SpyCas9 and inhibition of catalytic activity. These AcrIIA4 Ala mutants show reduced binding affinity to SpyCas9, with the triple mutant Y67A/D69A/E70A and its tetra mutant counterpart containing also N39A resulting in essentially complete loss in binding affinity (Figure 3G). Notably, in the related MBP pull-down assay, the same Ala mutants show reduced or complete loss in the inhibition capacity as monitored by cleavage of linear dsDNA (Figure 3H). Due to the extended hydrophilic intermolecular interactions at the interface between AcrIIA4 and SpyCas9, single alanine mutations at these sites show a relatively reduced impact (Figure 3G, H) when compared to single charge reversal mutations (Figure S3E, F). In addition, double or multiple alanine mutations at the interface (Figure 3G, H) show comparable impact to those observed for single charge reversal mutations (Figure S3E, F) (see also Dong et al. 2017).
Competitive Binding by AcrIIA4 for PAM Duplex-interacting Surface
AcrIIA4 targets the concave surface formed by the Topo, CTD and RuvC domains in the SpyCas9-sgRNA binary complex and buries an area of 1,693 Å2 on ternary complex formation (Figure 4A). By contrast, positioning of the PAM duplex of dsDNA on the surface formed by Topo and CTD domains of the SpyCas9-sgRNA binary complex buries an area of 553 Å2 on ternary complex formation (Figure 4B). These results are indicative of AcrIIA4 having higher binding affinity than dsDNA for sgRNA-bound SpyCas9 binary complex. We investigated the extent to which AcrIIA4 and target dsDNA could competitively bind to the binary complex by electrophoretic mobility shift (EMSA) assays. Indeed, when supplied with AcrIIA4 and target dsDNA simultaneously, AcrIIA4 competed with dsDNA target to preferentially bind to the dSpyCas9-sgRNA binary complex (Figure 4C, left panel).
Figure 4. Inhibition mechanism of SpyCas9 by AcrIIA4.
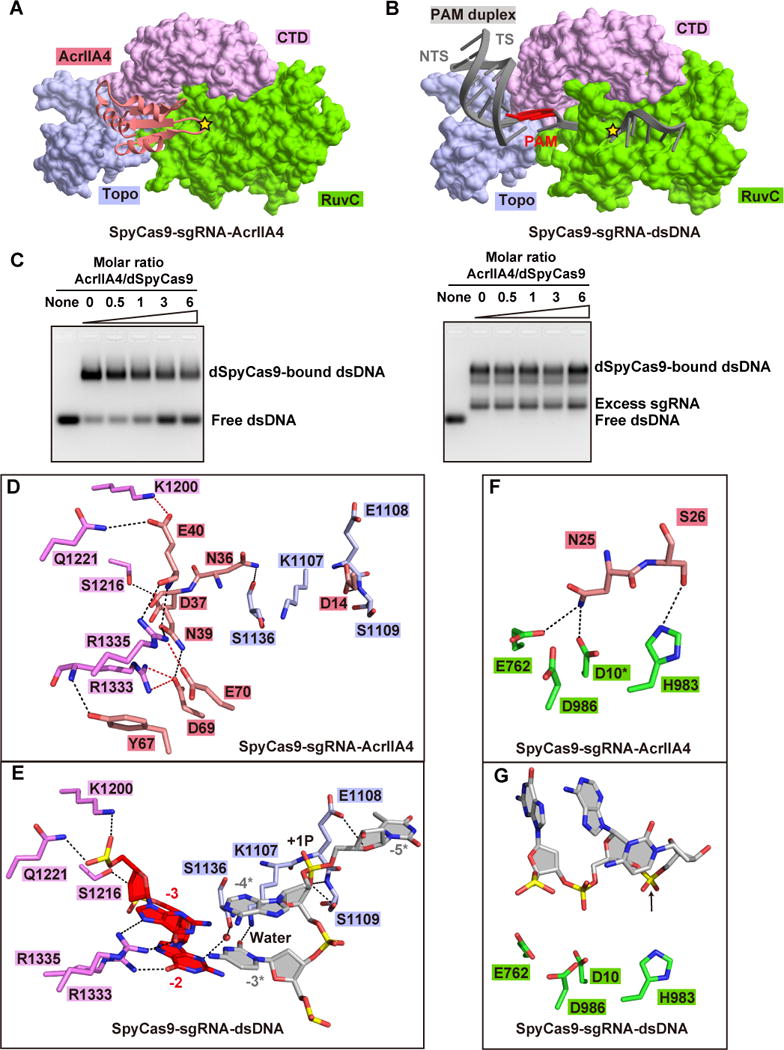
(A, B) Recognition of AcrIIA4 (panel A) and the PAM duplex (panel B) by Topo, CTD, and RuvC domains of SpyCas9. The active site of RuvC domain for non-target DNA strand in each panel is highlighted by a yellow star. TS and NTS represent target and non-target DNA strands, respectively.
(C) AcrIIA4 competitively binding to preformed SpyCas9-sgRNA binary complex (left panel) rather and SpyCas9-sgRNA-dsDNA ternary complex (right panel) in EMSA assays. The molar ratios of AcrIIA4:SpyCas9 are shown at the top of the gel.
(D, E) Detailed interactions between AcrIIA4 (panel D) and PAM duplex (panel E) with the Topo and CTD domains of SpyCas9, respectively.
(F, G) Recognition of key catalytic residues in the RuvC active site by AcrIIA4 (panel F) and non-target DNA strand (panel F). The modeled side chain of catalytic residue Asp10 is marked by asterisk. The position of cleavage site in non-target DNA strand is pointed by a black arrow.
See also Figure S4.
AcrIIA4 Cannot Replace Bound dsDNA in its Preformed Ternary Complex
On addition of AcrIIA4 to the preformed dSpyCas9-sgRNA-dsDNA ternary complex, no release of free DNA band was observed in EMSA assays (Figure 4C, right panel), suggesting that AcrIIA4 cannot access and replace the bound dsDNA substrate. We also investigated whether AcrIIA2 shows the same behavior as AcrIIA4 and found that AcrIIA2 is also able to compete with dsDNA target for binding to dSpyCas9-sgRNA binary complex, but unable to release bound dsDNA from its preformed ternary complex (Figure S4A).
DISCUSSION
Blockage of PAM Duplex Recognition Interface by Bound AcrIIA4
The interface between AcrIIA4 and the Topo and CTD domains (Figure 4A) almost overlaps with that reported previously between dsDNA and these two domains (Figure 4B). Residues Asp37 to Glu40 in the β2–β3 loop, and Asp69 to Glu70 in the α2–α3 loop of bound AcrIIA4 (Figure 4D and S4B) occupy similar positions to those of the backbones of the PAM duplex (Figure 4E and S4B) and form extensive interactions with residues associated with PAM duplex binding (Figure 4D, E).
Asp14 of the α1–β1 loop of AcrIIA4 (Figure 4D and S4B) is positioned at the corresponding position occupied by the +1 phosphate group of target dsDNA (Figure 4E and S4B) and interacts with Glu1108 and Ser1109 in the phosphate lock loop (Figure 4E). Separation of the double strands of target dsDNA and zipping up of the guide:target heteroduplex has been observed to start from the first nucleotide of the target DNA strand, with stabilization of +1 phosphate group important for kink formation in the target DNA strand at the junction between the PAM and guide-target duplexes (Anders et al., 2014). Hence, occupation of the phosphate lock site by bound AcrIIA4 would perturb formation of the guide:target heteroduplex. Moreover, Arg1333 and Arg1335 in the CTD domain that are critical for readout of the 5′-NGG PAM sequence (Figure 4E and S4B) are recognized by AcrIIA4 (Figure 4D and S4B), resulting in inhibition of readout of the PAM sequence.
Taken together, the relatively larger and overlapped interface ensures that AcrIIA4 could competitively interact with and occupy the PAM duplex-interaction surface, thereby blocking the binding of dsDNA substrate. In summary, AcrIIA4 ‘locks’ and ‘covers’ all the key residues involved in PAM duplex and +1 phosphate group recognition.
Blockage of RuvC Active Site by AcrIIA4
At the interface with the RuvC domain, the protruding β1–β2 loop and β1 of AcrIIA4 are directed towards the active site within the RuvC domain, as well as the tunnel for entrance of the non-target strand into RuvC active site (Figure 4A, B, and S4B). Ser20 to Ser26 of the β1–β2 loop and Lys18 of β1 of AcrIIA4 form contacts with residues within and adjacent to the active site (Figure 3F), especially key catalytic residues Glu762, His983, and the modeled side chain of Asp 10 (Figure 4F, G), thereby preventing the entrance of non-target DNA strand into the active site of the RuvC domain.
Selective Binding of AcrIIA4 to sgRNA-bound SpyCas9
During the transition from the apo state to the pre-target sgRNA-bound binary complex, the REC lobe undergoes large conformational changes, while small conformational changes are observed for the NUC lobe (Figure S4C) (Jinek et al., 2014; Jiang et al., 2015). In addition, several regions in the NUC lobe become ordered upon sgRNA binding. Some of these regions in the binary complex are targeted by AcrIIA4 on ternary complex formation. Specifically, the Gly1103 to Ser1136 segment of the Topo domain undergoes a disordered to ordered transition on binary complex formation, thereby defining a surface targeted by the α1–β1 and β2–β3 loops of bound AcrIIA4 (Figure 3B). The Arg765 to Gln774 segment connecting RuvC and HNH domains is also disordered in the apo state, becomes ordered on binary complex formation, such that the backbone of these two arginines forms hydrogen bonds with the β1–β2 loop of bound AcrIIA4 (Figure 3F). The movements of Helical-I on proceeding from apo to sgRNA-bound states also alters the orientation of the loop (Ser55 to Glu60) between BH and RuvC-I domains, which results in the formation of hydrogen contacts between sgRNA-bound SpyCas9 and bound AcrIIA4 (Figure 3F). In addition, small movements of the CTD domain on binary complex formation may also provide a better shape-complementary surface for AcrIIA4. These structural results provide a possible explanation why AcrIIA4 has higher binding affinity to sgRNA-bound SpyCas9 than apo-SpyCas9 and as a result selectively recognizes the SpyCas9 binary complex.
Upon dsDNA binding to form the ternary complex, the Helical-I and Helical-II domains of the REC lobe undergo further domain movements, as does the HNH domain of the NUC lobe (Figure 2D) (Anders et al., 2014; Jiang et al., 2016; Jiang et al., 2015; Nishimasu et al., 2014). The HNH domain moves towards the target strand cleavage site that is adjacent to the PAM duplex, thereby resulting in the formation of a narrow channel, which limits access by AcrIIA4 and its replacement of bound dsDNA. This appears to explain why AcrIIA4 shows either extremely weak or no binding to the SpyCas9-sgRNA-DNA ternary complex (Figure 1B, C).
It should be noted that blockage of dsDNA binding is distinct from inhibition of nuclease activity, given our current understanding of Cas9 cleavage mechanism is that its nuclease activity is only activated upon proper DNA target recognition and R-loop formation (Sternberg et al., 2015; Sternberg et al., 2014).
Comparison of anti-CRISPR proteins targeting Cas9 versus Cascade complexes
To date, there are three reported structures of anti-CRISPR proteins, including AcrF3 that targets P. aeruginosa Type I-F Cas3 (Wang et al., 2016b; Wang et al., 2016c), and AcrF1 and AcrF2 that target P. aeruginosa Type I-F Csy cascade (Chowdhury et al., 2017). All these inhibitors, as well as AcrIIA4, share low sequence identify and adopt divergent overall structures (Figure S3C and Figure S4D, E, F, upper panels). AcrF1, AcrF2, and AcrIIA4 are composed of one variant β-sheet and several a-helices, whereas AcrF3 is composed of six a-helices. In addition, these inhibitors employ distinct inhibition mechanisms, with AcrF1 and AcrF2 targeting distinct sites on Csy cascade. AcrF2 contacts the PAM-interacting region of Csy cascade through its negatively charged surface (Figure S3D and S4D, lower panel), while AcrF1 blocks the hybridization of target DNA strand with crRNA. AcrF3 functions as a dimer and prevents target DNA access to its binding site, as well as prevents recognition of Cas3 by upstream Cascade, and activation by ATP. AcrIIA4, like AcrF2, prevents PAM duplex access to its binding site and also blocks non-target DNA strand access to the RuvC catalytic pocket. However, no common interaction model could be formulated involving these four inhibitors, suggesting that divergent anti-CRISPR proteins are likely to have evolved independently, thereby targeting variant CRISPR-Cas systems.
Summary
Anti-CRISPR AcrIIA4 protein antagonizes the CRISPR-Cas9 host defense system through abolishment of all possible contributors governing the activity of SpyCas9, that includes prohibiting the recognition of and access to the PAM duplex, blocking the tunnel for non-target DNA strand access, and through occupancy of the catalytic pocket in the RuvC domain. In addition, AcrIIA4 exhibits higher binding affinity than dsDNA for sgRNA-bound SpyCas9 and is able to competitively bind to the binary complex resulting in the inactivation of SpyCas9. AcrIIA4 specifically binds to sgRNA-bound SpyCas9 rather than either apo or dsDNA-bound SpyCas9-sgRNA ternary complex, indicative of high efficiency for the inhibition. The pre-target sgRNA-bound state is ready for loading and cleavage of dsDNA target; however, apo-SpyCas9 requires either sgRNA or crRNA/tracrRNA binding in order to achieve the conformational changes needed for dsDNA target loading. For the dsDNA target-bound SpyCas9-sgRNA ternary complex, displacement of the PAM duplex or melting of the guide:target heteroduplex requires overcoming a high energy barrier. In addition, domain rearrangement during pre-target-bound to target-dsDNA-bound generates a narrow surface for optimal recognition of the PAM duplex, which is not suitable for AcrIIA4 association.
Competitive Publication
During preparation of our manuscript for submission, a related paper has appeared on line in Nature outlining structural studies of AcrIIA4 bound to sgRNA-bound SpyCas9 (Dong et al. 2017), with this study and our contribution reaching common conclusions on anti-CRISPR mediated targeting of the cleavage activity of Cas9 endonucleases.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents could be directed to, and will be fulfilled by Dinshaw Patel (pateld@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plasmid DNA for in vitro transcription was amplified in Escherichia coli DH5α strain in Lysogeny broth (LB) medium at 37 °C overnight.
Recombinant proteins were overexpressed in Escherichia coli BL21 (DE3) strain in Lysogeny broth (LB) medium. The cells were grown at 37 °C until OD600 reached 0.8 and then induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (GoldBio) at 18 °C for 20 hr.
METHOD DETAILS
Protein Expression and Purification
The gene encoding full-length Streptococcus pyogenes cas9 was synthesized and inserted into a modified pRSF-Duet-1 vector (Novagen) with N-terminal His6-SUMO tag following an ubiquitin-like protease (ULP1) cleavage site. Recombinant protein was overexpressed in Escherichia coli BL21 (DE3) strain in Lysogeny broth (LB) medium. The cells were grown at 37 °C until OD600 reached 0.8 and then induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (GoldBio) at 18 °C for 20 hr. Cell pellets was resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride), lysed by the EmulsiFlex-C3 homogenizer (Avestin), and centrifuged at 20,000 rpm for 1 hr at 4 °C in a JA-20 fixed angle rotor (Avanti J-E series centrifuge, Beckman Coulter). The supernatant containing SpyCas9 protein was loaded to 5 ml HisTrap Fastflow column (GE Healthcare) preequilibrated in buffer A and eluted with buffer A supplemented with 480 mM imidazole. The His6-SUMO tag was removed by ULP1 and during dialysis against buffer A and then separated by re-loading to HisTrap column. The flow-through fraction was further dialyzed against buffer B (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5 mM β-mercaptoethanol) and loaded on 5 ml HiTrap Heparin HP sepharose column (GE Healthcare) pre-equilibrated in buffer B and eluted by a linear gradient from 300 mM to 1 M. The recombinant protein was further purified by Superdex 200 16/60 column (GE Healthcare) in buffer C (20 mM HEPES, pH 7.2, 300 mM NaCl, 2 mM MgCl2, 5 mM DTT). The relevant fractions were concentrated to 20 mg/ml and flash-frozen in liquid nitrogen and stored in -80 °C.
The genes encoded full-length acrIIA2 and acrIIA4 were synthesized and subcloned into a modified pRSF-Duet-1 vector with N-terminal His6-SUMO tag, respectively. The proteins were overexpressed in Escherichia coli BL21 (DE3) strain and affinity purified using HisTrap Fast flow column by the same method as described above. After removing the His6-SUMO tag, the flow-through fractions containing the recombinant proteins were concentrated and loaded on Superdex 200 16/60 column pre-equilibrated in buffer B (20 mM HEPES, pH 7.2, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT). Different mutations were generated based on PCR-based method. The mutants were purified by the same method as described above. For MBP pull-down assay, the gene encoded SpyCas9 and acrIIA4 were inserted into a modified pMAL vector with a N-terminal His6-MBP tag following a Tobacco Etch Virus (TEV) cleavage site. The MBP-tagged proteins was purified using HisTrap Fast flow column by the same method as described above.
To assemble the SpyCas9-sgRNA-AcrIIA4 ternary complex, the purified doubly mutated D10A/H840A protein was mixed with sgRNA and AcrIIA4 at the molar ratio of 1:1.1:10 and incubated on ice for 30 min. The reconstituted binary and ternary complexes were purified by gel filtration chromatography on a Superdex 200 10/300 column pre-equilibrated in buffer D (20 mM HEPES, pH 7.2, 200 mM NaCl, 2 mM MgCl2, 5 mM DTT).
Crystallization, Data Collection, and Structure Determination
Crystallization was performed using the hanging drop vapor diffusion method at 20 °C. Crystals of SpyCas9-sgRNA-AcrIIA4 ternary complex was grown from drops consisting of 1 μl protein solution (about 8 mg/ml) and 1 μl reservoir solution containing 0.1 M HEPES (pH 7.0), and 15% PEG3350 (v/v). The crystals were cryopretected by the reservoir solution supplemented with 30% ethylene glycol. The data set was collected at 100 K at the Advanced Photo Source (APS) at the Argonne National Laboratory. The diffraction data was processed with HKL2000 (Otwinowski and Minor, 1997). The statistics of the diffraction data are summarized in Table 1.
The structure SpyCas9-sgRNA-AcrIIA4 ternary complex was solved by the molecular repalcement (MR) method using Phenix (Adams et al., 2010) using SpyCas9-sgRNA binary complex (PDB code: 4ZT0) (Jiang et al., 2015) as a search template. AcrIIA4 was manually built by using Coot (Emsley et al., 2010). The structural model was refined using Phenix (Adams et al., 2010). The statistics of the structure refinement and the quality of the final structure model are also summarized in Table 1. All molecular graphics were generated by PyMOL (http://www.pymol.org) and CueMol (http://www.cuemol.org).
In vitro Transcription and Purification of sgRNA
The sgRNA followed by the hammerhead ribozyme was transcribed in vitro using T7 RNA polymerase. Large scale transcription reaction (20 ml) was performed in buffer 100 mM Tris-HCl, pH 7.9, 30 mM DTT, 15 mM MgCl2, 2 mM spermidine, 4 mM each NTP, 50 μg/ml DNA template, 2.5 μg home-made T7 RNA polymerase. The mixture was incubated at 37°C for 3 hr and then supplemented by MgCl2 to final concentration of 50 mM and incubated for another 30 min. The transcribed sgRNA was purified by 10% denaturing TBE-urea PAGE, extracted from gel by electroelution using Elutrap (GE Healthcare), and then further purified by ion-exchange using HiTrap Q Fastflow sepharose column (GE Healthcare) pre-equilibrated by buffer E (20 mM Tris-HCl, pH 7.0). Elution of sgRNA was achieved by a linear gradient from 0 mM to 1 M NaCl in 20 column volumes. The RNA was denatured at 95 °C for 5 min and slowly cooling to room temperature.
Template of sgRNA for in vitro transcription (from 5′ to 3′):
GGATAACTCAATTTGTAAAAAAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGTCGACGGAGTCTAGACTCCGTCCTGATGAGTCCGTGAGGACGAAACACTTTTTCAAGTTG
In vitro Cleavage Assay
The ~600 bp linear target DNA containing the 5′-TGG-3′ PAM was synthesized and amplified by PCR. The cleavage reaction was performed by mixing target DNA, sgRNA and SpyCas9 proteins at the molar ratio of 1:1:1, in cleavage buffer (20 mM Tris-HCl, pH7.5, 100 mM KCl, 5 mM MgCl2, 5 mM DTT, 5% glycerol). AcrIIA2 or AcrIIA4 was added to the reaction system simultaneously. Reaction was performed at 37°C for 15 min and cleaned up by PCR purification columns (Roche) and then run in 10% TBE Urea gel.
Target DNA sequence (from 5′ to 3′):
ACCCATGGGAGCAGCTGGTCAGAGGGGACCCCGGCCTGGGGCCCCTAACCCTATGTAGCCTCAGTCTTCCCATCAGGCTCTCAGCTCAGCCTGAGTGTTGAGGCCCCAGTGGCTGCTCTGGGGGCCTCCTGAGTTTCTCATCTGTGCCCCTCCCTCCCTGGCCCAGGTGAAGGTGTAAACTATAACTCAATTTGTAAAAAATGGTATTGGCAGAAGCTGGAGGAGGAAGGGCCTGAGTCCGAGCAGAAGAAGAAGGGCTCCCATCACATCAACCGGTGGCGCATTGCCACGAAGCAGGCCAATGGGGAGGACATCGATGTCACCTCCAATGACTAGGGTGGGCAACCACAAACCCACGAGGGCAGAGTGCTGCTTGCTGCTGGCCAGGCCCCTGCGTGGGCCCAAGCTGGACTCTGGCCACTCCCTGGCCAGGCTTTGGGGAGGCCTGGAGTCATGGCCCCACAGGGCTTGAAGCCCGGGGCCGCCATTGACAGAGGGACAAGCAATGGGCTGGCTGAGGCCTGGGACCACTTGGCCTTCTCCTCGGAGAGCCTGCCTGCCTGGGCGGGCCCGCCCGCCACCGCAGCCTCCCAGCTGCTCTCCGTGTCTCCAATCTCCCTTTTGTTTTGATGCATTTCTGTTTTAATT
PAM is highlighted in bold, and the target sequence is underlined.
SEC-MALLS
SEC-MALLS experiments were performed by using a Superdex 75 10/300 column (GE Healthcare) and a Shimadzu HPLC System. Protein samples (100 μL) at a concentration of 2 mg/mL were loaded onto a gel filtration column and eluted with one column volume (24 mL) in buffer B (20 mM HEPES, pH 7.2, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT) at a flow rate of 0.3 mL/min. The eluting fractions were monitored using a DAWN HELEOS-II 18-angle light scattering detector (Wyatt Technologies), a SPD20A UV/Vis detector (Shimadzu), and an Optilab rEX refractive index monitor (Wyatt Technologies). Data were analyzed by using Astra (Wyatt Technologies).
Size-Exclusion Chromatography Assay
The target and non-target DNA strands were purchased from IDT (Integrated DNA Technologies) and dissolved in buffer E consisting of 20 mM Tris-HCl, pH 7.5, 50 mM NaCl. The target and non-target DNA strands were mixed together with a molar ratio of 1:1. The mixture of two DNA strands were denatured at 95 °C for 5 min and then annealed by slowly cooling to room temperature. For the assays of the apo-SpyCas9 and AcrIIA2/AcrIIA4, the purified wild-type SpyCas9 and AcrIIA2/AcrIIA4 were mixed at the molar ratio of 1:10 and incubated on ice for 30 min. For the assays of the sgRNA-bound SpyCas9, wild-type SpyCas9, sgRNA, and AcrIIA2/AcrIIA4 were mixed at the molar ratio of 1:1.1:10 and incubated on ice for 30 min. For the assays of the DNA-bound SpyCas9, dSpyCas9, sgRNA, dsDNA and AcrIIA2/AcrIIA4 were mixed at the molar ratio of 1:1.1:1.5:10 and incubated on ice for 30 min. All the mixture was loaded on Superdex 200 10/300 column (GE Healthcare) in buffer D (20 mM HEPES, pH 7.2, 200 mM NaCl, 2 mM MgCl2, 5 mM DTT). The fractions are detected by SDS-PAGE.
MBP Pull-down Assay
100 μg MBP-SpyCas9 and 100 μg AcrIIA2/AcrIIA4, in presence or absence of sgRNA (molar ratio, 1:1.1), and 30 μl amylose resin (New England Biolabs) were mixed at 4°C for 1 hr in buffer D (20 mM HEPES, pH 7.2, 200 mM NaCl, 2 mM MgCh, 5 mM DTT). Alternately, 100 μg MBP-AcrIIA4 and 100 μg SpyCas9, in presence or absence of sgRNA (molar ratio, 1:1.1) and sgRNA-dsDNA (molar ratio, 1:1.1:1.5), and 30 μl amylose resin (NEB) were mixed at 4°C for 1 hr in buffer D. The resin was washed three times using buffer D and detected by SDS-PAGE.
Electrophoretic Mobility Shift Assay (EMSA)
The purified dSpyCas9 (0.1 nM) and sgRNA at a molar ratio of 1:1.1 were mixed in the absence or presence of dsDNA at a molar ratio of 1:1.5, and then incubated on ice for 30 min in buffer D (20 mM HEPES, pH 7.2, 200 mM NaCl, 2 mM MgCl2, 5 mM DTT) to generate dSpyCas9-sgRNA binary and dSpyCas9-sgRNA-dsDNA ternary complexes. The purified AcrIIA2/AcrIIA4 and dsDNA (molar ratio, 1:1.5) were simultaneously mixed with dSpyCas9-sgRNA binary complex and incubated on ice for 30 min in buffer D. Alternately, the purified AcrIIA2/AcrIIA4 was mixed with dSpyCas9-sgRNA-dsDNA ternary complex and incubated on ice for 30 min in buffer D. The mixture was run on 2% aragose gel at 4°C and visualized by ethidium bromi de (Sigma).
QUANTIFICATION AND STATISTICAL ANALYSIS
In vitro cleavage, SEC-MALLS, SEC, and MBP pull-down experiments were repeated three times, and representative results were shown.
DATA AND SOFTWARE AVAILABILITY
The atomic coordinate has been deposited in the Protein Data Bank with accession number 5VW1 (SpyCas9-sgRNA-AcrIIA4 ternary complex). Original unprocessed gel images in this manuscript have been deposited to Mendeley Data and are available at https://dx.doi.org/10.17632/34cnpysb7k.1
Supplementary Material
HIGHLIGHTS.
Crystal structure of S. pyogenes Cas9 in complex with sgRNA and suppressor AcrIIA4
Selective recognition of pre-target bound SpyCas9 binary complex by AcrIIA4
Competitive binding of AcrIIA4 over dsDNA for SpyCas9-sgRNA binary complex
Mechanistic insights into blockage of SpyCas9 preventing dsDNA cleavage by AcrIIA4
Acknowledgments
X-ray diffraction studies were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIGMS grant P41GM103403 and U.S. Department of Energy grant DE-AC02-06CH11357. The Pilatus 6M detector on 24-ID-C beam line is funded by a NIHORIP HEI grant (S10 RR029205). The research was supported by NIH grant GM104962 to D.J.P., and by the Memorial Sloan Kettering Cancer Center Core Grant (P30CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Codes
The atomic coordinate and structure factors have been deposited for the following SpyCas9-sgRNA-AcrIIA4 ternary complex (5VW1).
Author Contributions
H.Y. undertook all the biochemical and structural studies under the supervision of D.J.P.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN, Hoffmann C, Nosaka L, Bondy-Denomy J, Maxwell KL, Davidson AR, et al. Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Cell. 2017;169:47–57 e11. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR- encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017 doi: 10.1038/nature22377. on line, http://dx.doi.org/10.1038/nature22377. [DOI] [PubMed]
- Dupuis ME, Villion M, Magadan AH, Moineau S. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun. 2013;4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell. 2017;66:373–383 e373. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jiang W, Marraffini LA. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu Rev Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA- mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, Wang Y. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell. 2017;168:121–134 e112. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Nureki O. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr Opin Struct Biol. 2016;43:68–78. doi: 10.1016/j.sbi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016a;167:1829–1838 e1829. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio. 2014;5:e00896. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Staals RH, Taylor C, Watson BN, Saha S, Fineran PC, Maxwell KL, Davidson AR. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol. 2016b;1:16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2017;168:150–158 e110. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DB, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017 doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016a;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma J, Cheng Z, Meng X, You L, Wang M, Zhang X, Wang Y. A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses. Cell Res. 2016b;26:1165–1168. doi: 10.1038/cr.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yao D, Xu JG, Li AR, Xu J, Fu P, Zhou Y, Zhu Y. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol. 2016c;23:868–870. doi: 10.1038/nsmb.3269. [DOI] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


