Abstract
Background
Glycated hemoglobin (HbA1c) is used to diagnose type 2 diabetes (T2D) and assess glycemic control in patients with diabetes. Previous genome-wide association studies (GWAS) have identified 18 HbA1c-associated genetic variants. These variants proved to be classifiable by their likely biological action as erythrocytic (also associated with erythrocyte traits) or glycemic (associated with other glucose-related traits). In this study, we tested the hypotheses that, in a very large scale GWAS, we would identify more genetic variants associated with HbA1c and that HbA1c variants implicated in erythrocytic biology would affect the diagnostic accuracy of HbA1c. We therefore expanded the number of HbA1c-associated loci and tested the effect of genetic risk-scores comprised of erythrocytic or glycemic variants on incident diabetes prediction and on prevalent diabetes screening performance. Throughout this multiancestry study, we kept a focus on interancestry differences in HbA1c genetics performance that might influence race-ancestry differences in health outcomes.
Methods & findings
Using genome-wide association meta-analyses in up to 159,940 individuals from 82 cohorts of European, African, East Asian, and South Asian ancestry, we identified 60 common genetic variants associated with HbA1c. We classified variants as implicated in glycemic, erythrocytic, or unclassified biology and tested whether additive genetic scores of erythrocytic variants (GS-E) or glycemic variants (GS-G) were associated with higher T2D incidence in multiethnic longitudinal cohorts (N = 33,241). Nineteen glycemic and 22 erythrocytic variants were associated with HbA1c at genome-wide significance. GS-G was associated with higher T2D risk (incidence OR = 1.05, 95% CI 1.04–1.06, per HbA1c-raising allele, p = 3 × 10−29); whereas GS-E was not (OR = 1.00, 95% CI 0.99–1.01, p = 0.60). In Europeans and Asians, erythrocytic variants in aggregate had only modest effects on the diagnostic accuracy of HbA1c. Yet, in African Americans, the X-linked G6PD G202A variant (T-allele frequency 11%) was associated with an absolute decrease in HbA1c of 0.81%-units (95% CI 0.66–0.96) per allele in hemizygous men, and 0.68%-units (95% CI 0.38–0.97) in homozygous women. The G6PD variant may cause approximately 2% (N = 0.65 million, 95% CI 0.55–0.74) of African American adults with T2D to remain undiagnosed when screened with HbA1c. Limitations include the smaller sample sizes for non-European ancestries and the inability to classify approximately one-third of the variants. Further studies in large multiethnic cohorts with HbA1c, glycemic, and erythrocytic traits are required to better determine the biological action of the unclassified variants.
Conclusions
As G6PD deficiency can be clinically silent until illness strikes, we recommend investigation of the possible benefits of screening for the G6PD genotype along with using HbA1c to diagnose T2D in populations of African ancestry or groups where G6PD deficiency is common. Screening with direct glucose measurements, or genetically-informed HbA1c diagnostic thresholds in people with G6PD deficiency, may be required to avoid missed or delayed diagnoses.
Ines Barroso and colleagues identify a genetic variant that leads to reduced levels of HbA1c in African American adults; 2% of this population are at risk of missed diagnosis for diabetes.
Author summary
Why was this study done?
Blood glucose binds in an irreversible manner to circulating hemoglobin in red blood cells (RBCs), generating “glycated hemoglobin,” called HbA1c. HbA1c is used to diagnose and monitor diabetes.
Previous large-scale human genetic studies have demonstrated that HbA1c is influenced by genetic variants. Some variants are thought to influence the function, structure, and lifespan of the red blood itself (“erythrocytic variants”), while others are thought to influence blood glucose control (“glycemic variants”). This study aimed to identify additional variants influencing HbA1c levels, and investigate the extent to which variants affecting this measurement independently of blood glucose concentration may lead to misdiagnosis, mistreatment, and human health disparities.
What did the researchers do and find?
We studied genetic variants and their association with HbA1c levels in almost 160,000 people from European, African, East Asian, and South Asian ancestry from 82 separate studies worldwide. We found 60 genetic variants influencing HbA1c, of which 42 variants were new. Of the 60 variants, we found 19 glycemic variants and 22 erythrocytic variants.
In approximately 33,000 people from 5 ancestry groups followed carefully over time, we found that the more glycemic variants a person had, the higher their risk to get diabetes (OR = 1.05 per HbA1c-raising allele, p = 3 × 10−29). However, more erythrocytic variants did not lead to a higher risk of diabetes, meaning erythrocytic variants that lower HbA1c levels independently from glucose concentration could lead to missed diagnosis of diabetes.
Next, we found that in everyone but those of African ancestry, those with more versus those with less of the 60 HbA1c genetic variants had a fairly small difference in HbA1c (about 0.2 units), while those of African ancestry had a larger difference (about 0.8 units, a fairly large number for this medical test).
This difference in African ancestry was explained by one erythrocytic variant on the X chromosome. This variant mutates the protein made by the gene “glucose-6-phosphate dehydrogenase” (G6PD), which can shorten RBC lifespan, and thus lower HbA1c levels, no matter the blood glucose level.
About 11% of people of African American ancestry carry at least one copy of this G6PD variant, while almost no one of any other ancestry does. We estimated that if we tested all Americans for diabetes using HbA1c, about 650,000 African Americans would be missed because of these genetically lowered HbA1c levels.
What do these findings mean?
We may want to investigate the benefits of screening for the G6PD genotype in specific communities or perform additional diagnostic tests to avoid health disparities between communities.
It will also be important to follow up with additional studies to check whether new standardized thresholds for diagnoses should be recommended for those that have this G6PD variant.
Introduction
Type 2 diabetes (T2D) is a health scourge rising unabated worldwide, escaping all past and current control measures, in part because only half of prevalent T2D worldwide has been clinically diagnosed [1]. Glycated hemoglobin (HbA1c) is an accepted diagnostic test for T2D and a principal clinical measure of glycemic control in individuals with diabetes. T2D arises from the environment interacting with genetics. Studies investigating genetic contributions to HbA1c in individuals of European [2–4] and Asian ancestry [5–7] have identified 18 loci influencing HbA1c through glycemic and nonglycemic pathways, the latter primarily reflecting erythrocytic biology. Alterations in HbA1c that are due to genetic variation acting through nonglycemic pathways may not accurately reflect ambient glycemia or T2D risk and could affect the validity of HbA1c as a diagnostic test and measure of glycemic control in some individuals or populations. Some genetic variants (e.g., the sickle cell variant HbS) that vary in frequency across ancestries can interfere with the accuracy of certain assays [8]. Further, certain hematologic conditions associated with shortened erythrocyte lifespan (e.g., hemolytic anemias) lower HbA1c values irrespective of the assay performed. HbA1c values in such patients may no longer accurately reflect ambient glycemia [9].
Epidemiologic studies have reported ethnic differences in HbA1c, with African Americans having, on average, higher HbA1c than European ancestry Americans [10]. While these differences are largely due to demographic and metabolic factors [11,12], genetic factors associated with hematologic conditions that impact erythrocyte turnover may confound the relationship between HbA1c and glycemia, causing misclassification of T2D diagnosis [8,13].
This study had 3 aims, the first was to expand genetic discovery efforts to larger sample sizes, including populations of ancestries not previously studied, to uncover novel loci influencing HbA1c and that might capture a greater fraction of the variability in HbA1c. Second, as done in previous studies, we aimed to classify the variants as acting through glycemic or erythrocytic biology. Thirdly, as erythrocytic variants may influence HbA1c due to effects on the red blood cell (RBC), we wished to explore whether this might lead to HbA1c values that no longer reflected ambient glycemia. To do this, we specifically tested the hypothesis that HbA1c-associated genetic variants, particularly those that act through erythrocytic pathways, influence the performance of HbA1c for diabetes risk prediction and diabetes diagnoses (S1 Fig).
Methods
Analysis plans were followed and can be found in S1 Analysis Plans.
Genetic discovery study participants
In the genetic discovery analysis, we combined data from up to 159,940 participants (maximum number available for any variant) of European, African American, East Asian, and South Asian ancestry, including subsets from previous publications [4,5] (S1 Table, S2 Fig). All participants were free of diabetes defined by physician diagnosis, medication use, or fasting glucose (FG) ≥ 7 mmol/L. A small number of cohorts also removed individuals with 2hr glucose (2hrGlu) ≥ 11.1 mmol/L, or HbA1c ≥ 6.5%, where FG was not available (details of exclusions by individual cohorts, S1 Table). Analysis followed the details in S1 Analysis Plans (Hemoglobin A1c Genetic Discovery Analysis Plan).
HbA1c measurement
Where possible, studies reported HbA1c as a National Glycohemoglobin Standardization Program (NGSP) percent [14] (S1 Table).
Genotyping and quality control
Each cohort was genotyped on commercially available genome-wide arrays (for instance, the Affymetrix Genome-Wide Human SNP Assay 6.0 or the Illumina Human610-Quad BeadChip) or the Illumina CardioMetabochip (Metabochip) [15]. Variant and sample quality control (QC) was conducted within each cohort following a shared analysis plan (S1 Analysis Plans). Cohorts were advised to keep SNPs with hardy-weinberg-disequilibrium p-value ≥ 1 × 10−6, SNP genotyping call rate ≥ 95% and minor allele frequency (MAF) ≥ 1% (full details of SNP and sample QC can be found in S1 Table). Following QC, studies with genome-wide array data were imputed (primarily using the Phase 2 of the International HapMap Project reference panel [16], see S1 Table, row 40), and poorly imputed variants (variants which could not reliably be inferred from surrounding variants) were excluded based on standard imputation quality thresholds (R-sq < 0.3, INFO < 0.4). Approximately 2.5 million SNPs were available for analysis after imputation and QC (S1 Table). QC of the Metabochip data is described elsewhere, but included filtering out poorly genotyped individuals or low-quality SNPs [17]. Variant association testing in men and women combined was conducted under an additive model adjusting for study-specific covariates and was limited to variants with MAF of at least 1% in each cohort. Details of the study cohorts, genotyping platforms and QC criteria, imputation reference panel, covariates in the analysis, and software used are provided for each study in S1 Table. Our study followed STREGA guidelines (S1 Checklist).
Genetic discovery using ancestry-specific and trans-ancestry meta-analyses
Association data were combined within each ancestry group using a fixed-effects meta-analysis in METAL, which assumes the SNP effect is the same for each study within an ancestry [18]. Results for each cohort were corrected for any systematic biases, such as residual population structure using the genomic control inflation factor, λGC [17,19]. We excluded variants from further follow-up if they had an ancestry-specific sample size N < 20,000 in Europeans, N < 3,000 in African Americans, N < 7,000 in East Asians, and N < 3,000 in South Asians (minimum number of samples, where the threshold was chosen to minimize signals driven by a single cohort), or evidence of significant within-ancestry heterogeneity, suggesting effect size significantly differs between cohorts of the same ancestry (Cochran’s Q-test heterogeneity p-value < 0.0001). We retained the lead variant in the X-chromosome analysis of the African American ancestry data (rs1050828, G202A in G6PD) despite significant heterogeneity, as it was a strong biological candidate.
Ancestry-specific meta-analysis results were conservatively corrected for a second round of genomic control by ancestry: European (λGC = 1.072); African American (λGC = 1.020); East Asian (λGC = 1.027); South Asian (λGC = 1.004); and combined using the Meta-Analysis of Transethnic Association (MANTRA) software that accounts for allelic heterogeneity across ancestry groups [20].
Identification of primary and secondary distinct HbA1c-associated signals
Variants were considered to be significantly associated with HbA1c when they met standard genome-wide significant thresholds (based on p = 0.05 divided by the estimated number of independent tests across the genome), of p < 5 × 10−8 in the European and Asian, or p < 2.5 × 10−8 in African American [21] ancestry-specific meta-analyses, or a log10 Bayes Factor ≥6 in the transancestry meta-analysis. All significant variants within 500 kb of a lead (most significantly associated) variant were grouped into a single locus. Novel loci were by definition >500 kb from previously reported HbA1c-associated variants. We ran approximate conditional analyses using the Genome-wide Complex Trait Analysis (GCTA) software [22,23] (following analysis plans detailed in S1 Analysis Plans, Conditional analyses in GCTA) using the Women’s Genome Health Study (WGHS, Europeans), Jackson Heart Study (JHS, African Americans), Singapore Malay Eye Study (SiMES, East Asians), and the London Life Sciences Prospective Population Study (LOLIPOP, South Asians) as reference populations for linkage disequilibrium (LD) estimates, to confirm the lead variants on the autosomes (within 1 Mb) were distinct, and similarly used exact conditional regression for the African-American signals on the X-chromosome in JHS.
To identify distinct signals at associated loci (that is, secondary signals), we performed approximate conditional analyses using GCTA, conditioning on lead variants identified in the transancestry MANTRA analysis. Where the lead variant was absent in a cohort, an exact proxy (r2 = 1) was used, unless the variant was very low frequency or monomorphic.
Classification of variants as glycemic or erythrocytic
We extracted summary association statistics from publicly available meta-analysis results for glycemic [17,24–26] and blood-cell [27] traits and asked a subset of the genome-wide discovery cohorts to repeat association analyses for each lead variant, conditioning on any one of FG, 2hrGlu, hemoglobin level (Hb), mean corpuscular volume (MCV), or mean corpuscular hemoglobin (MCH), where available (S3 Fig, S2 Table and S3 Table).
Variants were classified as “glycemic” if they were associated (p < 0.0001) with any of the glycemic traits from publicly available results or had ≥25% attenuation of variant HbA1c effect size in association models conditioned on fasting or 2hrGlu. That is, evidence of being associated with any of the glycemic traits or a reduction in the effect of the variant on HbA1c after repeated association analysis in a model additionally adjusting for fasting/2hrGlu, suggested the observed association with HbA1c was being driven through an association with fasting/2hrGlu. Variants not classified as glycemic were classified as “erythrocytic” if they were associated (p < 0.0001) with Hb, MCH, MCV, PCV, RBC, or MCHC in the publicly available results or, as above, had ≥25% attenuation of effect size in Hb-, MCV-, or MCH-conditioned models (suggesting the observed association with HbA1c was being driven through an association with these blood cell traits). The 25% attenuation threshold was chosen as the optimal balance between specificity and sensitivity based on comparisons with the classification based only on association with any of the glycemic/erythrocytic traits. Two SNPs were classified based on evidence from the literature, rs12132919 (TMEM79) was classified as erythrocytic based on association with MCHC in Japanese individuals [28] and rs7616006 (SYN2) was classified as erythrocytic based on association with platelet count in Europeans [29].
Variants associated with HbA1c but not glycemic or erythrocytic traits remained “unclassified” (S3 Fig). A single variant (rs579459 near ABO) was classified as both glycemic and erythrocytic, but as we were primarily concerned about variants that might affect HbA1c without reflecting ambient glycemia and this variant also affected glycemia, we treated it as glycemic in all analyses.
Effect of HbA1c genetic scores on reclassification of prevalent undiagnosed T2D for population screening using HbA1c
Analyses on the reclassification of prevalent T2D around the HbA1c 6.5% threshold before and after accounting for the contribution of erythrocytic variants were conducted in up to 19,380 individuals and incident T2D prediction analyses in up to 33,241 individuals from European, African, and East Asian ancestry cohorts (derived in part from discovery cohorts; in S4 Table, and following the details in the S1 Analysis Plans, Net-reclassification analysis). We acknowledge that nonindependent GWAS discovery and application cohorts can lead to inflated effect estimates [30]; however, this was not evident in our study, and effect estimates across all cohorts were similar with low heterogeneity.
We estimated reclassification of prevalent T2D status by HbA1c after accounting for the contribution of erythrocytic loci in 5 population-based cohorts with 3 ancestries partially overlapping with the discovery GWAS: the Framingham Heart Study (FHS), the Atherosclerosis Risk in Communities Study (ARIC), and the Multiethnic Study of Atherosclerosis (MESA) in individuals of European ancestry; ARIC and MESA in African Americans; and MESA, the Taiwan-Metabochip Study for Cardiovascular Disease (TAICHI), and the Singapore Prospective Study (SP2) in East Asians (N = 19,380). Variant-adjusted HbA1c was calculated as:
where Yi was the measured HbA1c for individual, i, is the ancestry-specific, meta-analytic β coefficient for the kth erythrocytic SNP, gki is the dosage (estimated number of HbA1c-raising alleles), and E(gki) was two times the HbA1c-raising allele frequency. When the less frequent (minor) allele was associated with higher HbA1c, it was coded as the HbA1c-raising allele, when it was associated with lower HbA1c, the more frequent (major) allele was coded as the HbA1c-raising allele. As some HbA1c-raising alleles in one ancestry could be HbA1c-lowering in a different ancestry, we coded HbA1c-raising alleles by ancestry.
Participants on antidiabetic therapy were excluded, and screen-detected T2D was defined as FG ≥ 7 mmol/L. For the reclassification analysis, we constructed 2-by-2 tables showing the proportion of participants reclassified around the HbA1c 6.5% diagnostic threshold, with and without adjusting measured HbA1c for the contribution of erythrocytic loci.
Calculation of genetic risk scores
Genetic risk scores of erythrocytic variants and glycemic variants (GS-E and GS-G, respectively) were calculated as detailed in S1 Analysis Plans (Investigate the Effect of Glycemic and Erythrocytic Hemoglobin A1c (HbA1c) Genetic Variants on Diabetes Prediction), as standard in the field, by summing the number of ancestry-specific HbA1c-raising alleles at each variant (0, 1, 2, or expected number of alleles based on the probability of each genotype), multiplied by their ancestry-specific β coefficients for HbA1c from the genome-wide association study (GWAS) meta-analysis multiplied by the number of variants and divided by the sum of β coefficients [31]. This means the contribution of each associated variant to the trait, in a given individual, is influenced by the number of “risk alleles” (or in this case HbA1c-raising alleles) and the effect of the variant on the trait (increase in HbA1c estimated from the meta-analysis).
Effect of HbA1c genetic scores on prediction of incident T2D
We tested the hypothesis that glycemic and erythrocytic HbA1c loci predicted incident T2D differently in Europeans, East Asians, and African Americans from 5 cohorts (partially overlapping with the discovery GWAS) with prospective follow-up: FHS, the European Prospective Investigation into Cancer and Nutrition InterAct project (EPIC-InterAct), ARIC, MESA, and the Singapore Chinese Health Study (SCHS) (N = 33,241). Using age- and sex-adjusted regression models, we tested the association between the genetic scores GS-E or GS-G and incident T2D, defined by FG ≥ 7 mmol/L, 2hrGlu ≥ 11.1 mmol/L, antidiabetic medication use, or a physician diagnosis for T2D, accrued over a 10-to-15-year follow-up period. Clinical practice guidelines did not include HbA1c as a diagnostic test until 2010. As the majority of incident T2D cases were accrued before 2010, participants are very unlikely to have received a T2D diagnosis based only on HbA1c measurements. To test whether individuals with higher GS-E, compared to those with lower GS-E, had lower T2D risk for the same HbA1c, we adjusted models for baseline HbA1c. We meta-analyzed results using a fixed-effects meta-analysis and assessed heterogeneity using Higgin's I-squared. See S1 Analysis Plans (Investigate the Effect of Glycemic and Erythrocytic Hemoglobin A1c (HbA1c) Genetic Variants on Diabetes Prediction) for analysis plan.
Ancestral differences in the genetic architecture of HbA1c
In FHS, ARIC, MESA, and SCHS, we calculated the difference in HbA1c of individuals at the bottom and top 5% of the distribution of an ancestry-specific GS composed of all 60 variants (GS-Total) and an equivalent analysis using GS-E.
We also pursued additional analyses at chromosome X rs1050828 because this single variant showed the largest effect on HbA1c in African Americans and was monomorphic in the other ancestries. The T allele is known to be associated with glucose-6-phosphate dehydrogenase (G6PD) deficiency, an enzymatic defect causing hemolytic anemia [32,33]. Imperfect correlation between HbA1c and glycemia may indicate the impact of reduced erythrocyte lifespan on HbA1c in individuals with the T allele. Fructosamine, a measure of serum protein glycation not influenced by erythrocyte-related factors, reflects average glycemia over the previous 2–3 weeks. Following the analysis plan detailed in S1 Analysis Plans (The Difference Between Fructosamine-inferred HbA1c and Measured HbA1c) we thus calculated the estimated residuals from a linear regression of HbA1c on fructosamine in ARIC African Americans (N = 1,676) to determine whether the T allele was associated with lower HbA1c than predicted by fructosamine, suggesting that the T allele artificially lowered HbA1c through a reduction in the average erythrocytic lifespan. We then reported the mean estimated residuals by genotype (women: CC, CT, TT; men: C, T).
Estimated number of African Americans with T2D in the United States whose diagnosis would be missed due to the G6PD variant if screened with HbA1c
Using publicly-available data from the National Health and Nutrition Examination Survey (NHANES) 2013–2014 [34], a nationally representative sample of US residents, we calculated the proportion of African American adults (aged ≥ 18 years) with T2D who would be missed by not accounting for rs1050828 when using a single HbA1c diagnostic threshold of 6.5%, assuming the observed effect size of rs1050828, allele frequency of 11% and accounting for NHANES sampling design. The study sample was restricted to 1,133 adults, aged ≥ 18 years, who self-identified as non-Hispanic black with measured HbA1c in 2013–2014. We defined known T2D by self-reported physician diagnosis or medication use. Assuming Hardy-Weinberg Equilibrium and a T allele frequency of 11% for the G6PD variant in our sample, we lowered the diagnostic threshold from the widely accepted 6.5%-units cut-point to 5.7%-units in men with the T genotype, 5.8%-units in women with the TT genotype, and 6.2%-units in women with the CT genotype. We then calculated the proportion of African American individuals with missed T2D diagnosis if screened with HbA1c using the 6.5% diagnostic threshold. We applied procedures to account for sampling probabilities and complex sampling design to enable population-level inferences. Data analysis was performed using SAS (version 9.2 or 9.3; SAS Institute, Cary, NC).
Results
HbA1c-associated genetic variants and classification into glycemic and nonglycemic pathways
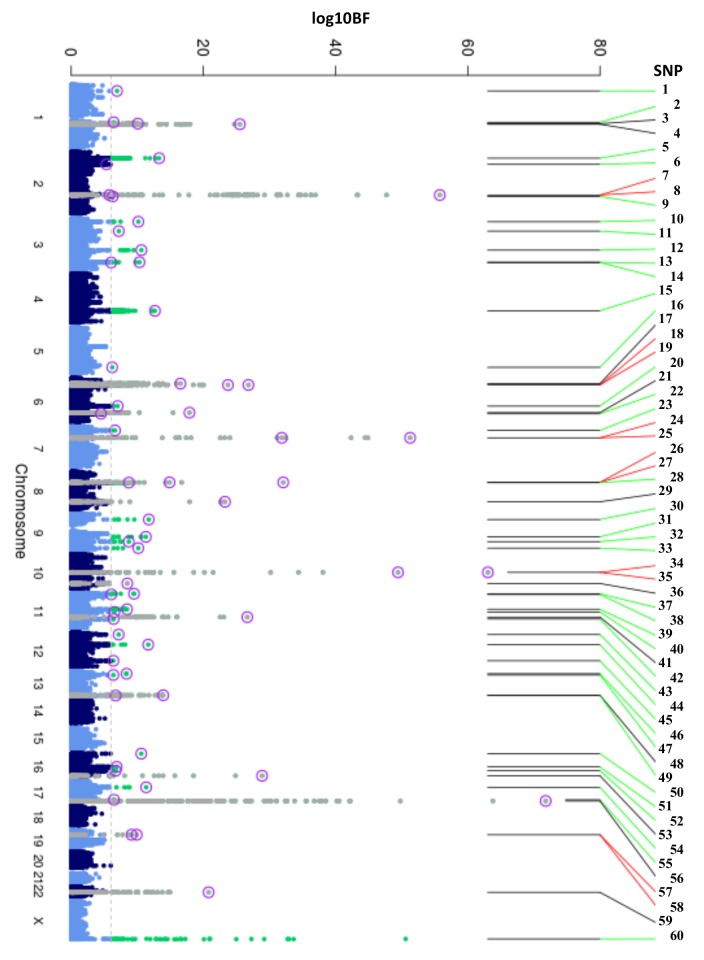
To discover new genetic loci influencing HbA1c in populations from 4 different ancestries (European, African American, East Asian, and South Asian), we performed within-ancestry fixed-effects genome-wide association meta-analyses and transancestry meta-analyses using a model that allowed for different effects between ancestry groups (Methods, S2 Fig). Using this approach in up to 159,940 participants without diabetes, we identified 60 variants associated with HbA1c at genome-wide significance (Fig 1, Table 1 and S5 Table). Of 60, 18 have been previously reported, and 42 were novel, including distinct secondary signals at 5 known loci. To classify the associated loci into groups reflecting their likely mode of action on HbA1c, we repeated association analyses conditioning on erythrocytic or glycemic traits and performed lookups in publicly-available association results summary statistics for additional glycemic and erythrocytic traits (Methods, S3 Fig, S2 Table and S3 Table). Based on the combined results from conditional and lookup results, we were able to classify 22 variants as erythrocytic and 19 as glycemic, with 19 remaining unclassified (Fig 1, Table 1 and S5 Table).
Fig 1. Manhattan plot of HbA1c associated variants.
Manhattan plot of the transethnic meta-analysis results in MANTRA. The dashed grey line indicates log10BF = 6. Grey and green points denote known/novel loci, respectively. The lead HbA1c-associated variants identified through the ancestry-specific/transethnic analyses are circled in purple (the G6PD variant was not included in the MANTRA analysis, but the locus on the X-chromosome is indicated in the figure). Lines joining the plot & SNP number denote known loci (black), novel loci (green), and loci with a secondary distinct signal (red). MANTRA, Meta-Analysis of Transethnic Association.
Table 1. Table of HbA1c associated variants.
Table with results and classification of the 60 HbA1c-associated variants. SNP number corresponds to number in Fig 1.
| SNP | Markername | Chr. | Position (bp) | Effect Allele | Other Allele | Gene | Status | Signals | Classification | European ancestry METAL p-value |
Trans-ethnic MANTRA log10BF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs2375278 | 1 | 25401625 | A | G | SYF2 | Novel | Single | Unclassified | 2.03 × 10−7 | 6.93 |
| 2 | rs267738 | 1 | 149207249 | T | G | CERS2 | Novel | Single | Unclassified | 2.59 × 10−9 | 6.41 |
| 3 | rs12132919 | 1 | 154584765 | A | C | TMEM79 | Known | Single | Erythrocytic | 0.0169 | 10.08 |
| 4 | rs857691 | 1 | 156893002 | T | C | SPTA1 | Known | Single | Erythrocytic | 3.97 × 10−25 | 25.52 |
| 5 | rs17509001 | 2 | 23874735 | C | T | ATAD2B | Novel | Single | Unclassified | 1.94 × 10−15 | 13.30 |
| 6 | rs12621844 | 2 | 48268239 | T | C | FOXN2 | Novel | Single | Unclassified | 1.87 × 10−8 | 5.32 |
| 7 | rs13387347 | 2 | 169463092 | T | C | G6PC2 | Novel | Multiple | Glycemic | 0.308 | 5.77 |
| 8 | rs560887 | 2 | 169471394 | C | T | G6PC2 | Known | Multiple | Glycemic | 1.48 × 10−58 | 55.77 |
| 9 | rs17256082 | 2 | 175000610 | C | T | SCRN3 | Novel | Single | Unclassified | 0.00112 | 6.27 |
| 10 | rs7616006 | 3 | 12242648 | A | G | SYN2 | Novel | Single | Erythrocytic | 5.07 × 10−10 | 10.16 |
| 11 | rs9818758 | 3 | 49357929 | A | G | USP4 | Novel | Single | Unclassified | 7.74 × 10−10 | 7.20 |
| 12 | rs11708067 | 3 | 124548468 | A | G | ADCY5 | Novel | Single | Glycemic | 1.42 × 10−12 | 10.62 |
| 13 | rs8192675 | 3 | 172207577 | T | C | SLC2A2 | Novel | Single | Glycemic | 1.38 × 10−11 | 10.33 |
| 14 | rs4894799 | 3 | 173278234 | A | G | FNDC3B | Novel | Single | Unclassified | 1.80 × 10−6 | 6.05 |
| 15 | rs13134327 | 4 | 144879245 | A | G | FREM3 | Novel | Single | Glycemic | 2.64 × 10−15 | 12.66 |
| 16 | rs11954649 | 5 | 156988069 | G | C | SOX30 | Novel | Single | Unclassified | NA | 6.20 |
| 17 | rs7756992 | 6 | 20787688 | G | A | CDKAL1 | Known | Single | Glycemic | 2.80 × 10−12 | 16.53 |
| 18 | rs1800562 | 6 | 26201120 | G | A | HFE | Known | Multiple | Erythrocytic | 4.67 × 10−28 | 26.81 |
| 19 | rs198846 | 6 | 26215442 | G | A | HFE | Novel | Multiple | Erythrocytic | 1.18 × 10−23 | 23.72 |
| 20 | rs11964178 | 6 | 109668728 | A | G | C6orf183 | Novel | Single | Erythrocytic | 6.38 × 10−10 | 7.03 |
| 21 | rs11154792 | 6 | 135473333 | T | C | MYB | Known | Single | Erythrocytic | 7.45 × 10−18 | 17.89 |
| 22 | rs592423 | 6 | 139882386 | A | C | CITED2 | Novel | Single | Erythrocytic | 3.96 × 10−8 | 4.50 |
| 23 | rs2191349 | 7 | 15030834 | T | G | DGKB | Novel | Single | Glycemic | 2.09 × 10−7 | 6.63 |
| 24 | rs4607517 | 7 | 44202193 | A | G | GCK | Known | Multiple | Glycemic | 8.76 × 10−38 | 51.28 |
| 25 | rs3824065 | 7 | 44213783 | C | T | GCK | Novel | Multiple | Glycemic | 4.22 × 10−35 | 31.87 |
| 26 | rs6474359 | 8 | 41668351 | T | C | ANK1 | Known | Multiple | Unclassified | 1.50 × 10−16 | 14.88 |
| 27 | rs4737009 | 8 | 41749562 | A | G | ANK1 | Known | Multiple | Erythrocytic | 4.48 × 10−27 | 32.08 |
| 28 | rs6980507 | 8 | 42502241 | A | G | SLC20A2 | Novel | Single | Erythrocytic | 3.58 × 10−8 | 8.73 |
| 29 | rs11558471 | 8 | 118254914 | A | G | SLC30A8 | Known | Single | Glycemic | 1.38 × 10−19 | 23.26 |
| 30 | rs2383208 | 9 | 22122076 | A | G | MTAP | Novel | Single | Glycemic | 7.04 × 10−12 | 11.74 |
| 31 | rs7040409 | 9 | 90693056 | C | G | C9orf47 | Novel | Single | Erythrocytic | 2.56 × 10−14 | 11.29 |
| 32 | rs1467311 | 9 | 109576753 | G | A | KLF4 | Novel | Single | Unclassified | 2.09 × 10−7 | 8.72 |
| 33 | rs579459 | 9 | 135143989 | C | T | ABO | Novel | Single | Glycemic | 9.42 × 10−9 | 10.14 |
| 34 | rs4745982 | 10 | 70759849 | T | G | HK1 | Known | Multiple | Erythrocytic | 2.87 × 10−65 | 63.05 |
| 35 | rs10823343 | 10 | 70761019 | A | G | HK1 | Novel | Multiple | Unclassified | 1.68 × 10−55 | 49.45 |
| 36 | rs17747324 | 10 | 114742493 | C | T | TCF7L2 | Known | Single | Glycemic | 6.12 × 10−11 | 8.49 |
| 37 | rs3782123 | 11 | 195198 | C | A | BET1L | Novel | Single | Unclassified | 1.51 × 10−10 | 9.51 |
| 38 | rs2237896 | 11 | 2815016 | G | A | KCNQ1 | Novel | Single | Glycemic | 0.00246 | 6.07 |
| 39 | rs174577 | 11 | 61361390 | C | A | FADS2 | Novel | Single | Glycemic | 5.45 × 10−7 | 8.45 |
| 40 | rs11603334 | 11 | 72110633 | G | A | ARAP1 | Novel | Single | Glycemic | 6.85 × 10−9 | 6.53 |
| 41 | rs10830963 | 11 | 92348358 | G | C | MTNR1B | Known | Single | Glycemic | 2.23 × 10−23 | 26.64 |
| 42 | rs11224302 | 11 | 99961814 | C | T | CNTN5 | Novel | Single | Erythrocytic | 4.76 × 10−7 | 6.40 |
| 43 | rs2110073 | 12 | 6946143 | T | C | PHB2 | Novel | Single | Unclassified | 4.44 × 10−8 | 7.18 |
| 44 | rs2408955 | 12 | 46785398 | T | G | SENP1 | Novel | Single | Erythrocytic | 1.42 × 10−15 | 11.65 |
| 45 | rs10774625 | 12 | 110394602 | G | A | ATXN2 | Novel | Single | Erythrocytic | 1.46 × 10−8 | 6.38 |
| 46 | rs11619319 | 13 | 27385599 | G | A | PDX1 | Novel | Single | Glycemic | 4.58 × 10−7 | 8.38 |
| 47 | rs576674 | 13 | 32452302 | G | A | KL | Novel | Single | Glycemic | 1.39 × 10−5 | 6.38 |
| 48 | rs282587 | 13 | 112399663 | G | A | ATP11A | Known | Single | Unclassified | 1.70 × 10−12 | 13.92 |
| 49 | rs9604573 | 13 | 113571085 | T | C | GAS6 | Novel | Single | Unclassified | 9.60 × 10−9 | 6.72 |
| 50 | rs11248914 | 16 | 233563 | T | C | ITFG3 | Novel | Single | Erythrocytic | 2.56 × 10−14 | 10.60 |
| 51 | rs1558902 | 16 | 52361075 | A | T | FTO | Novel | Single | Unclassified | 3.27 × 10−8 | 6.88 |
| 52 | rs4783565 | 16 | 67307691 | A | G | CDH3 | Novel | Single | Erythrocytic | 1.73 × 10−7 | 6.73 |
| 53 | rs837763 | 16 | 87381230 | T | C | CDT1 | Known | Single | Erythrocytic | 1.68 × 10−28 | 28.89 |
| 54 | rs9914988 | 17 | 24207230 | A | G | ERAL1 | Novel | Single | Erythrocytic | 2.77 × 10−11 | 11.34 |
| 55 | rs2073285 | 17 | 73628956 | C | T | TMC6 | Novel | Single | Unclassified | 1.27 × 10−4 | 6.47 |
| 56 | rs1046896 | 17 | 78278822 | T | C | FN3KRP | Known | Single | Unclassified | 4.46 × 10−64 | 71.79 |
| 57 | rs11086054 | 19 | 17107737 | A | T | MYO9B | Novel | Multiple | Unclassified | 8.16 × 10−6 | 9.12 |
| 58 | rs17533903 | 19 | 17117523 | A | G | MYO9B | Known | Multiple | Erythrocytic | 5.27 × 10−12 | 9.912 |
| 59 | rs4820268 | 22 | 35799537 | G | A | TMPRSS6 | Known | Single | Erythrocytic | 1.40 × 10−22 | 20.79 |
| 60 | rs1050828 | X | 153417411 | T | C | G6PD | Novel | Single | Erythrocytic | NA* | NA |
*African American meta-analysis p-value for the G6PD variant (rs1050828) = 8.23 × 10−135. Chr, chromosome; MANTRA, Meta-Analysis of Transethnic Association
Effect of HbA1c genetic scores on reclassification of prevalent undiagnosed T2D in population screening using HbA1c
Next, we tested whether erythrocytic variants influenced the ability of HbA1c to accurately classify individuals with diabetes when screening populations using a single HbA1c measurement. In 5 cohorts, among the 767 individuals with undiagnosed T2D by FG ≥ 7 mmol/L, 390 (50.8%) had measured HbA1c < 6.5% and would remain undiagnosed based on HbA1c. After accounting for the effect of erythrocytic variants, 5 (1.3%) of these individuals were correctly reclassified to having a HbA1c ≥ 6.5%. Among the 18,613 individuals without T2D by FG < 7 mmol/L, 266 (0.3%) had measured HbA1c ≥ 6.5% and would be incorrectly diagnosed with T2D by HbA1c. After accounting for the effect of erythrocytic variants, 50 (18.8%) of these individuals [13 of 80 (16.3%) European ancestry, 28 of 109 (25.7%) African ancestry, 9 of 77 (11.7%) Asian ancestry] were correctly reclassified to having a HbA1c<6.5% (Table 2, S6 Table). While adjusting for the effect of erythrocytic variants improved reclassification for individuals diagnosed with T2D by only HbA1c and not FG, it caused wrong reclassification for individuals diagnosed with T2D by both FG and HbA1c (S6 Table), suggesting that accounting for the contribution of erythrocytic variants may not be relevant for individuals who already meet diagnostic thresholds using both FG and HbA1c.
Table 2. Reclassification of individuals with discordant T2D status based on prevailing diagnostic thresholds for FG and HbA1c before and after accounting for the effect of erythrocytic variants.
| FG ≥ 7 mmol/L but HbA1c < 6.5% (N = 390) | FG < 7mmol/L but HbA1c ≥ 6.5% (N = 266) | |||
|---|---|---|---|---|
| Not reclassified after accounting for the effect of erythrocytic variants | Reclassified to HbA1c ≥ 6.5% after accounting for the effect of erythrocytic variants | Not reclassified after accounting for the effect of erythrocytic variants | Reclassified to HbA1c < 6.5% after accounting for the effect of erythrocytic variants | |
| European | 314 | 1 (0.3%) | 67 | 13 (16.3%) |
| African | 64 | 4 (6.3%) | 81 | 28 (25.7%) |
| Asian | 7 | 0 (0.0%) | 68 | 9 (11.7%) |
| Total | 385 | 5 (1.3%) | 216 | 50 (18.8%) |
FG, fasting glucose; HbA1c, glycated hemoglobin
Effect of HbA1c genetic scores on prediction of incident T2D
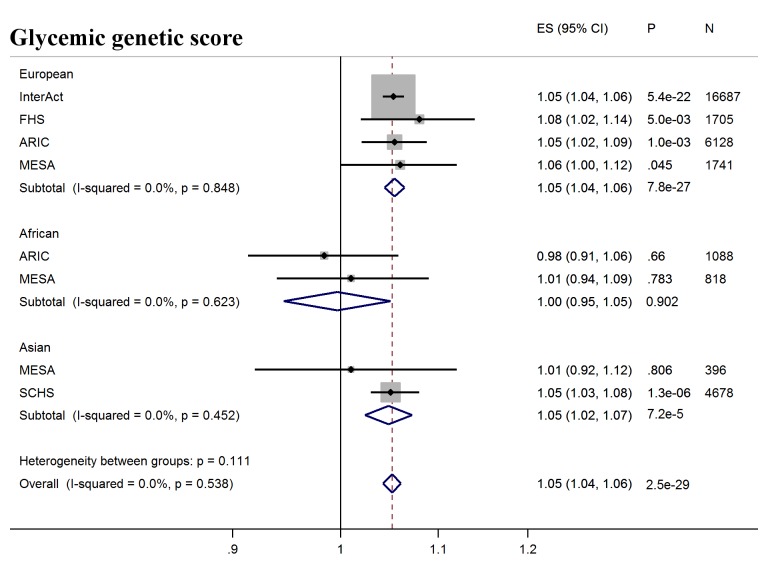
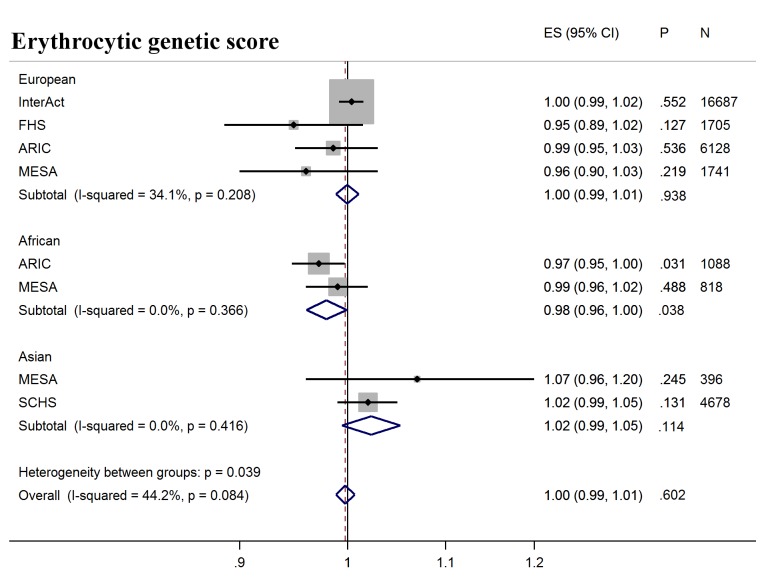
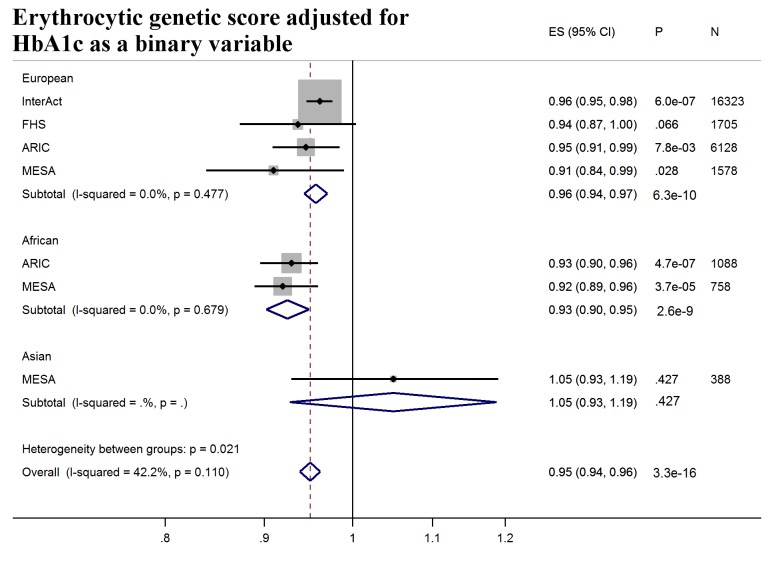
Next, we tested whether erythrocytic variants influenced the ability of HbA1c to predict incident diabetes in initially nondiabetic populations. GS-G was associated with increased incidence of T2D (odds ratio [OR] per weighted allele 1.05, 95% CI 1.04–1.06 p = 2.5 × 10−29) overall, although not in African Americans (Fig 2, S7 Table). GS-E was not associated overall with incident T2D (OR 1.00 95% CI 0.99–1.01, p = 0.60) (Fig 3, S7 Table), but was negatively associated with incident T2D in Europeans and African Americans after adjusting for HbA1c (OR 0.95, 95% CI, 0.94–0.96, p = 3.3 × 10−16) (Fig 4, S4 Fig and S7 Table), meaning individuals with a higher GS-E will have a lower risk of developing T2D given the same HbA1c value, suggesting that despite having the same HbA1c value, this does not reflect the same level of glycemia.
Fig 2. T2D prediction, glycemic genetic score.
Forest plot of association between glycemic genetic score with incident T2D over a decade-long follow-up period, by ancestry. MESA (European and Asian ancestry) and the G6PD variant (rs1050828) in ARIC (European and African American) were not included in the discovery GWAS analysis. Effect estimates were combined in a fixed effects meta-analysis. Overall effect estimate: 1.05, 95% CI 1.04–1.06, p = 2.5 × 10−29. ARIC, Atherosclerosis Risk in Communities Study; ES, Effect Size; FHS, Framingham Heart Study; GWAS, genome-wide association study; G6PD, glucose-6-phosphate dehydrogenase; I-Squared, Higgin's I-squared statistic, a measure of heterogeneity; MESA, Multiethnic Study of Atherosclerosis; SCHS, Singapore Chinese Health Study; T2D, type 2 diabetes.
Fig 3. T2D prediction, erythrocytic genetic score.
Forest plot of association between erythrocytic genetic score with incident T2D over a decade-long follow-up period, by ancestry. MESA (European and Asian ancestry) and the G6PD variant (rs1050828) in ARIC (European and African American) were not included in the discovery GWAS analysis. Effect estimates were combined in a fixed effects meta-analysis. Overall effect estimate: 1.00, 95% CI 0.99–1.01, p = 0.60. ARIC, Atherosclerosis Risk in Communities Study; ES, Effect Size, FHS, Framingham Heart Study; GWAS, genome-wide association study; G6PD, glucose-6-phosphate dehydrogenase; I-Squared, Higgin's I-squared statistic, a measure of heterogeneity; MESA, Multiethnic Study of Atherosclerosis; SCHS, Singapore Chinese Health Study; T2D, type 2 diabetes.
Fig 4. T2D prediction, erythrocytic genetic score adjusted for HbA1c as a binary variable.
Forest plot of association between erythrocytic genetic score with incident T2D over a decade-long follow-up period adjusted for HbA1c as a binary variable (≥5.7% versus <5.7%), by ancestry. HbA1c at baseline was not available in SCHS and was excluded from the meta-analysis. MESA (European and Asian ancestry) and the G6PD variant (rs1050828) in ARIC (European and African American) were not included in the discovery GWAS analysis. Effect estimates were combined in a fixed effects meta-analysis. Overall effect estimate: 0.95, 95% CI 0.94–0.96, p = 3.3 × 10−16. ARIC, Atherosclerosis Risk in Communities Study; ES, Effect Size; GWAS, genome-wide association study; FHS, Framingham Heart Study; G6PD, glucose-6-phosphate dehydrogenase; HbA1c, glycated hemoglobin; I-Squared, Higgin's I-squared statistic, a measure of heterogeneity; MESA, multiethnic study of atherosclerosis; SCHS, Singapore Chinese Health Study; T2D, type 2 diabetes.
Ancestral differences in the genetic architecture of HbA1c
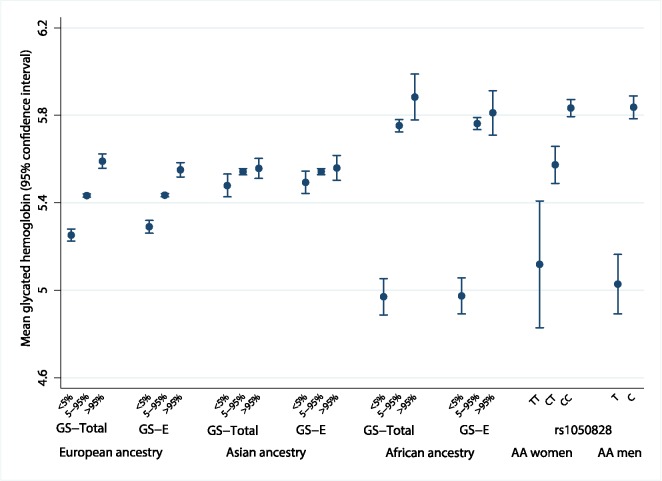
The population genetic history of African ancestry groups has undergone selective pressure due to the effects of malaria and other infectious diseases on erythrocytes, unlike in most European ancestry populations [35]. This led us to seek ancestral differences in the genetic determinants of HbA1c. The variance in HbA1c levels explained by all 60 genetic variants over a basic regression model including age and sex was 4.2%–5.8% in Europeans, 6.0%–14.3% in East Asians, and 8.9%–9.7% in African Americans (S8 Table). In addition, compared to Europeans and East Asians, African Americans had the largest difference in mean HbA1c between the bottom and top 5% of the GS-Total distribution (0.91%-units, 95% CI 0.78–1.05; Fig 5 and S9 Table).
Fig 5. Mean HbA1c of individuals at the bottom 5% and top 5% of the distribution of ancestry-specific genetic scores and rs1050828 by genotype.
The difference in measured HbA1c of individuals at the bottom 5% and top 5% of the distribution of an ancestry-specific additive GS composed of all 60 variants (GS-Total), and the equivalent calculation for an ancestry-specific GS composed of up to 20 erythrocytic variants (GS-E). Far right of the figure shows the mean HbA1c by genotype for chromosome X rs1050828. AA men, African American men; AA women, African American women; HbA1c, glycated hemoglobin; GS, genetic scores.
Erythrocytic variants alone explained around one-fifth to three-quarters of the total explained genetic variance in HbA1c (S8 Table). The absolute differences in mean HbA1c from the bottom and top 5% of the GS-E distribution were similar to GS-Total, implying that genetically-induced differences in HbA1c may be largely driven by erythrocytic variants (S9 Table and S10 Table). In African Americans, this difference was largely driven by the C-to-T missense variant (G202A) in G6PD, rs1050828 on chromosome X. This variant alone explained 14.4% of variance in HbA1c (MESA; 9.6% in women; 19.9% in men). Men with the T allele had, on average, an absolute 0.81%-units (95% CI 0.66–0.96) lower HbA1c than those with the C allele. Homozygous TT women had, on average, an absolute 0.68%-units (95% CI 0.38–0.97) lower HbA1c compared to CC homozygous women. The effect size was similar after excluding those with anemia (Hb < 12 g/dL in women and < 13 g/dL in men, S11 Table).
Fructosamine is another measure of serum protein glycation, which reflects glycemia over a 2–3 week window, but unlike HbA1c it is not influenced by RBC traits; therefore, we sought to explore the difference between fructosamine-inferred HbA1c and measured HbA1c (Methods, S1 Analysis Plans) to test the hypothesis that the G6PD variant might be influencing HbA1c levels independently of ambient glycemia. Among African Americans, the T allele at rs1050828 was associated with measured HbA1c that was lower than fructosamine-predicted HbA1c (0.31%-units, 95% CI 0.25–0.37, p = 6.4 × 10−19). Among men with the C allele, measured HbA1c was similar to fructosamine-predicted HbA1c (residuals, 0.04%-units, 95% CI −0.04 to 0.12, N = 351). This suggested that only the T allele was associated with markedly lower HbA1c than expected from glycemic measurements (S11 Table).
Public health implications of the G6PD variant on T2D screening
Given the large effects of the G6PD G202A variant on HbA1c levels, we sought to investigate the impact this variant would have on diabetes detection if using HbA1c as a screening tool. To do this, we used publicly-available data from NHANES 2013–2014 [34], a nationally representative sample of the US, to calculate the proportion of African Americans adults with T2D who would be missed by not accounting for rs1050828 when using a single HbA1c diagnostic threshold of 6.5%, assuming the observed effect size of rs1050828, allele frequency of 11%, and accounting for NHANES sampling design. In the NHANES sample of African Americans (N = 1,133), the mean age was 44.2 years (standard error 0.9), 55.2% were women, and mean HbA1c, excluding those with physician-diagnosed T2D, was 5.5%-units (standard error 0.02). 13.45% of African American adults aged ≥ 18 years had physician-diagnosed T2D with an additional 2.50% with undiagnosed T2D by HbA1c ≥ 6.5%. An additional estimated 2.17% (95% CI 1.88–2.46) with HbA1c < 6.5% may be considered to have T2D if the effect of rs1050828 was accounted for by using genotype-specific diagnostic thresholds of 5.7% for T in men, 5.8% for TT, and 6.2% for TC in women. According to the 2014 United States Census Bureau, approximately 29.9 million adults identified themselves as African American [36], suggesting that 0.65 (95% CI 0.55–0.74) million adults with T2D would remain undiagnosed when screened by a single HbA1c measurement if this genetic information were not taken into account (S12 Table).
Discussion
In a very large transancestry GWAS of HbA1c, we identified 42 novel and 18 known genetic variants associated with HbA1c, explaining 4%–14% of the trait variance. Genetic variants influencing HbA1c through erythrocytic pathways did not predict future T2D, and adjusting for their contribution to HbA1c led to a moderate misclassification of T2D by adjusted HbA1c. Notably, we detected strong ancestral differences in the contribution of genetic variants to HbA1c that substantially altered the performance of HbA1c as a diagnostic test for T2D in African Americans compared with Europeans and East Asians.
Our findings elucidate the contribution of common genetic variants to the genetic architecture of HbA1c and identify an important interface of modern human genetics with clinical and public health. In people of European and Asian ancestry, we found multiple genetic loci with small-to-modest effects, whereas, in African American ancestry, the genetic architecture was dominated by a single variant at G6PD (G202A). This variant was responsible for 0.81%-units HbA1c difference in men and 0.68%-units in homozygous TT women, corresponding to adjusted T2D diagnosis thresholds of 5.7 (95% CI 5.5–5.8) and 5.8 (95% CI 5.5–6.1), respectively. To meet the NGSP certification criteria, laboratory-reported HbA1c ought to be within 6% of the standard reference laboratory mean values (e.g., 6.5%-units ± 0.4%-units) for the majority of patient samples [14]. The limits of acceptable analytic variability were exceeded by this G6PD variant. This may also have important implications for the management of diabetes, with carriers of the HbA1c-lowering G6PD allele requiring adjusted (lower) HbA1c treatment targets. Previous epidemiologic studies have shown that a 1%-unit increase in HbA1c in individuals without T2D was associated with a more than 2-fold increase in risk of future T2D and a 20%–50% increased risk of cardiovascular disease (CVD) [37]. HbA1c ≥ 6.5% compared to those with HbA1c < 5.7% had a higher risk of kidney disease and retinopathy [38].
Only one other African-specific variant, rs11954649, located in the intron of SOX30, reached genome-wide significance in African Americans. However, this variant had a relatively small effect size (β = 0.12 per G allele) on HbA1c and was not classified as glycemic or erythrocytic. The variant was thus not included in the genetic scores and, unlike G6PD, the causal transcript and biological mechanism through which it influences HbA1c remains unclear. Future studies on larger sample sizes of ethnic minorities can focus on dissecting the genomic and biological implications of novel HbA1c-related variants.
When considering all ethnicities, both glycemic and erythrocytic variants influence measured HbA1c; yet, only glycemic variants were associated with increased T2D risk (5% per allele) over a decade-long follow-up period. For an equivalent HbA1c, individuals carrying more erythrocytic HbA1c-raising alleles, or fewer HbA1c-lowering alleles, had lower incident T2D risk (−5% per allele), implying that for the same HbA1c level those individuals with the greater number of erythrocytic HbA1c- raising alleles have artificially higher HbA1c values that do not reflect ambient glycemia. Thus, the influence of erythrocytic HbA1c variants may partly explain why some individuals with the same HbA1c may have different risks of future T2D. We note that the estimates of variance explained by genetic variants underlying HbA1c were comparable with those for FG in Europeans (4.8%) [17].
Our results on the reclassification of prevalent T2D were consistent with previous reports indicating that a diagnostic cut-point at 6.5% for HbA1c classified fewer cases than FG ≥ 7 mmol/L [39,40]. Adjusting for the contribution of erythrocytic variants correctly reclassified approximately 1 in 5 individuals with FG < 7 mmol/L who were incorrectly diagnosed as having T2D (HbA1c ≥ 6.5%) to having HbA1c < 6.5%, suggesting that a subset of these individuals may have artificially elevated HbA1c due to the contribution of the erythrocytic variants.
Though the specific G6PD variant we identified is monomorphic in Asian and European ancestry, other diverse G6PD variant alleles have reached polymorphic frequencies in malarial endemic regions around the world [35]. G6PD deficiency is unlikely to be identified through routine screening for anemia in healthy individuals, and universal screening for G6PD deficiency is not currently recommended worldwide [32,41]. Testing for G6PD deficiency is only performed on individuals before being prescribed specific drugs, such an antimalarial medications, or in patients with clinical presentation consistent with the disease; for instance, prolonged neonatal jaundice or hemolytic crisis following exposure to specific drugs, infections, or foods [32]. Thus, asymptomatic individuals often remain unaware of their G6PD genotype status and screening for the G6PD genotype before using HbA1c to diagnose T2D may be warranted in populations or ethnic groups where G6PD deficiency is common. Similarly, a recent study identified a significant hemolytic risk in women heterozygous for the G6PD Mahidol variant when treated with primaquine who were not detected by current screening methods [42]. Rarer hematologic conditions that reduce erythrocyte lifespan, e.g., hereditary hemolytic anemias, hereditary spherocytosis, and hemoglobinopathies have also been shown to lower HbA1c [9,43], and should also be considered before using HbA1c in these patients. We recommend additional testing using direct glucose measurements (e.g., FG or oral glucose tolerance testing) or other erythrocyte-independent methods to diagnose T2D. This supports the use of a combination of HbA1c and FG to confirm T2D diagnosis in routine screening [44]. Future studies could also explore G6PD effect modification by HbA1c assay type.
Further studies in large cohorts with HbA1c, glycemic, and erythrocytic traits are required to better determine the biological action of genetic variants that have yet to be classified. Similarly, future analyses conditional on RBC distribution width or reticulocyte count will help to better understand the effects of erythrocytic HbA1c-associated variants, should such data become available. The relatively small sample size for Asian and African ancestry cohorts limited the discovery of ancestry-specific genetic variants, beyond the African-specific G6PD variant, and could explain why GS-G was associated with higher incident T2D in European, but not other, ancestries. This underscores the need to extend such studies to non-European populations, particularly those with a high prevalence of some hemoglobinopathies or iron deficiency anemias. Epidemiologic studies have reported higher mean HbA1c in African Americans compared to European ancestry individuals in the US [45,46]. While our genetic findings could not determine whether this difference was completely attributable to relative hyperglycemia, accounting for the effect of the G6PD variant that lowers HbA1c only in African Americans would further widen this disparity.
In conclusion, HbA1c remains an appropriate diagnostic test for the majority of people of diverse genetic backgrounds, having lower intraindividual variability compared to FG with the ability to capture chronic hyperglycemia, and robust associations with T2D-related complications [37]. Nevertheless, nonglycemic lowering of measured HbA1c for 1 in 10 African American men who carry this G6PD variant, and 1 in a 100 African American women homozygous for this variant, could amount to 0.65 (95% CI 0.56–0.74) million African American adults in the US with a missed T2D diagnosis using HbA1c as a screening test. We therefore recommend investigation of the possible benefits of screening for the G6PD genotype along with using HbA1c to diagnose T2D in populations of African ancestry or groups where G6PD deficiency is common, and screening with direct glucose measurements, or genetically-informed HbA1c diagnostic thresholds in people with G6PD deficiency. This work supports a role for a precision medicine application to reduce race-ethnic health disparities using HbA1c genetics to improve T2D diagnosis and prediction and to inform screening strategies for T2D across the African continent where the prevalence of the G6PD variant can reach 20%.
Supporting information
(DOC)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(DOCX)
Acknowledgments
Published data on glycemic traits were contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. The ARIC authors thank the staff and participants of the ARIC study for their important contributions. The CAGE authors thank the participants who made this work possible and who gave it value. The CAGE authors would like to thank Drs. Toshio Ogihara, Yukio Yamori, Akihiro Fujioka, Chikanori Makibayashi, Sekiharu Katsuya, Ken Sugimoto, Kei Kamide, and Ryuichi Morishita and the many physicians of the participating hospitals and medical institutions in Amagasaki Medical Association for their assistance in collecting the DNA samples and accompanying clinical information. The CROATIA authors would like to acknowledge the invaluable contributions of the Institute for Antropological Research, Zagreb, Croatia, the administrative team in Split, and the people of Vis and Korcula. The D.E.S.I.R. Study Group would like to acknowledge its members: B. Balkau, P. Ducimetière, E. Eschwège (INSERM U1018); F. Alhenc-Gelas (INSERM U367); A. Girault (CHU D’Angers); F. Fumeron, M. Marre, R Roussel (Bichat Hospital); F. Bonnet (CHU de Rennes); S. Cauchi, P. Froguel (CNRS UMR8090, Lille); Alençon, Angers, Blois, Caen, Chateauroux, Chartres, Cholet, Le Mans, Orléans, Tours (Centres d’Examens de Santé); J. Cogneau (Institute de Recherche Médecine Générale); General practitioners of the region; C. Born, E. Caces, M. Cailleau, O. Lantieri, J. G. Moreau, F. Rakotozafy, J. Tichet, and S. Vol. (Institute inter-Regional pour la Santé). Analyses contributed by FHS/MGH/BU reflect intellectual input and resource development from the FHS investigators participating in the SNP Health Association Resource (SHARe) project. The DIAGEN authors are grateful to all of the patients who cooperated in this study and to their referring physicians and diabetologists in Saxony. The InterAct authors thank all EPIC participants and staff for their contribution to the study. The InterAct authors also thank staff from the Technical, Field Epidemiology and Data Functional Group Teams of the MRC Epidemiology Unit in Cambridge, UK, for carrying out sample preparation, DNA provision and QC, genotyping and data-handling work. The JHS authors thank the JHS participants and staff for their contributions to this work. The KORA authors are grateful to all members of the Helmholtz Zentrum München, the field staff in Augsburg, and the Augsburg registry team who were involved in the planning, organization, and conduct of the KORA studies. In addition, the authors express their appreciation to all study participants. The Leipzig-adult authors thank all those who participated in the study. The Leipzig-kid authors are grateful to all the patients and families for contributing to the study. They also highly appreciate the support of the Obesity Team and Auxo Team of the Leipzig University Children’s Hospital for management of the patients and to the Pediatric Research Center Lab Team for support with DNA banking. The Lifelines authors thank Behrooz Alizadeh, Annemieke Boesjes, Marcel Bruinenberg, Noortje Festen, Pim van der Harst, Ilja Nolte, Lude Franke, Mitra Valimohammadi for their help in creating the GWAS database, and Rob Bieringa, Joost Keers, René Oostergo, Rosalie Visser, Judith Vonk for their work related to data collection and validation. The Lifelines authors are also grateful to the study participants, the staff from the Lifelines Cohort Study and the contributing research centers delivering data to Lifelines and the participating general practitioners and pharmacists. The LOLIPOP authors thank the participants and research staff who made the study possible. The LURIC authors extend their appreciation to the participants of the LURIC study and thank the LURIC study team who were either temporarily or permanently involved in patient recruitment as well as sample and data handling, in addition to the laboratory staff at the Ludwigshafen General Hospital and the Universities of Freiburg and Ulm, Germany. The MESA authors thank the investigators and participants of the MESA study for their significant and ongoing contributions. The NHAPC authors are grateful to all participants of the NHAPC and also thank their colleagues at the laboratory and local CDC staffs of Beijing and Shanghai for their assistance with data collection. The NSHD authors are very grateful to the members of NSHD birth cohort for their continuing interest and participation in the study. The NSHD authors would also like to acknowledge the Swallow group, UCL, who performed the DNA extractions (Rousseau, et al 2006). DOI: 10.1111/j.1469-1809.2006.00250. The ORCADES authors would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh, and the people of Orkney. The SardiNIA authors are grateful to all the volunteers who generously participated in the study, as well as the Lanusei team for their continuous work. The SCHS-CHD (cases and controls) authors thank Siew-Hong Low of the National University of Singapore for supervising the field work of the SCHS and the Ministry of Health in Singapore for assistance with the identification of AMI cases via database linkages. They also acknowledge the founding, longstanding principal investigator of the SCHS, Mimi C. Yu. The SHIP authors are grateful to the contribution of Ravi Kumar Chilukoti, Florian Ernst, Anja Hoffmann, and Astrid Petersmann in generating the SNP data. The contributions of the SHIP staff and participants are gratefully acknowledged. The Sorbs authors thank all those who participated in the study. The Sorbs authors would also like to thank Knut Krohn (Microarray Core Facility, University of Leipzig, Institute of Pharmacology) for the genotyping support and Joachim Thiery (Institute of Laboratory Medicine, Clinical Chemistry and Molecular Diagnostics, University of Leipzig) for clinical chemistry services. The Sorbs authors thank Inga Prokopenko (Imperial College London, UK) and Anubha Mahajan (WTCHG, University of Oxford, UK) for statistical analyses. The TRAILS authors are grateful to all adolescents who participated in this research and to everyone who worked on this project and made it possible. The Twingene authors thank Tomas Axelsson, Ann-Christine Wiman, and Caisa Pöntinen at the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se) for their excellent assistance with genotyping. The TWT2D authors thank the Taiwan Diabetes Consortium for phenotypes assessment and the National Center for Genome Medicine of the National Core Facility Program for Biotechnology, Ministry of Science and Technology for the technical/bioinformatics support.
Disclaimer: The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or NHSBT.
Members of the EPIC-CVD Consortium
Adam Butterworth, John Danesh.
Members of the EPIC-InterAct Consortium
Claudia Langenberg, Robert A. Scott, Stephen J. Sharp, Nita G. Forouhi, Nicola D. Kerrison, Matt Sims, Debora ME. Lucarelli, Inês Barroso, Panos Deloukas, Mark I. McCarthy, Antonio Agudo, Beverley Balkau, Aurelio Barricarte, Heiner Boeing, Miren Dorronsoro, Paul W. Franks, Sara Grioni, Rudolf Kaaks, Timothy J. Key, Carmen Navarro, Peter M. Nilsson, Kim Overvad, Domenico Palli, Salvatore Panico, J. Ramón Quirós, Olov Rolandsson, Carlotta Sacerdote, María, José Sánchez, Nadia Slimani, Annemieke MW. Spijkerman, Anne Tjonneland, Rosario Tumino, Yvonne T. van der Schouw, Elio Riboli, Nicholas J. Wareham.
Members of the Lifelines Cohort Study
Behrooz Z Alizadeh, H Marike Boezen, Lude Franke, Pim van der Harst, Gerjan Navis, Marianne Rots, Harold Snieder, Morris Swertz, Bruce HR Wolffenbuttel, Cisca Wijmenga.
Abbreviations
- ARIC
Atherosclerosis Risk in Communities Study
- CVD
cardiovascular disease
- EPIC-InterAct
European Prospective Investigation into Cancer and Nutrition InterAct project
- FG
fasting glucose
- FHS
Framingham Heart Study
- GCTA
Genome-wide Complex Trait Analysis
- GS-E
genetic scores of erythrocytic variants
- GS-G
genetics scores of glycemic variants
- G6PD
glucose-6-phosphate dehydrogenase
- GWAS
genome-wide association studies
- Hb
hemoglobin level
- HbA1c
glycated hemoglobin
- JHS
Jackson Heart Study
- LD
linkage disequilibrium
- LOLIPOP
London Life Sciences Prospective Population Study
- MAF
minor allele frequency
- MANTRA
Meta-Analysis of Transethnic Association
- MCH
mean corpuscular hemoglobin
- MCV
mean corpuscular volume
- MESA
Multiethnic Study of Atherosclerosis
- NGSP
National Glycohemoglobin Standardization Program
- NHANES
National Health and Nutrition Examination Survey
- OR
odds ratio
- QC
quality control
- RBC
red blood cell
- SCHS
Singapore Chinese Health Study
- SiMES
Singapore Malay Eye Study
- SP2
Singapore Prospective Study
- TAICHI
Taiwan-Metabochip Study for Cardiovascular Disease
- T2D
type 2 diabetes
- WGHS
Women’s Genome Health Study
- 2hrGlu
2hr glucose
Data Availability
The ancestry-specific and transethnic genome-wide meta-analysis summary statistics for association with HbA1c, and published data included in this study, are available to download from the MAGIC website, www.magicinvestigators.org/downloads. Uniform analysis plan(s) showing the QC and data analysis steps in detail are provided in the supporting information file S1 Analysis Plans.
Funding Statement
Please refer to the supporting information file S1 Financial Disclosure for full information with regard to funding and financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.(2015) IDAtE. https://www.idf.org/e-library/epidemiology-research/diabetes-atlas/13-diabetes-atlas-seventh-edition.html.
- 2.Franklin CS, Aulchenko YS, Huffman JE, Vitart V, Hayward C, Polasek O, et al. The TCF7L2 diabetes risk variant is associated with HbA(1)(C) levels: a genome-wide association meta-analysis. Ann Hum Genet. 2010;74(6):471–8. doi: 10.1111/j.1469-1809.2010.00607.x . [DOI] [PubMed] [Google Scholar]
- 3.Pare G, Chasman DI, Parker AN, Nathan DM, Miletich JP, Zee RY, et al. Novel association of HK1 with glycated hemoglobin in a non-diabetic population: a genome-wide evaluation of 14,618 participants in the Women's Genome Health Study. PLoS Genet. 2008;4(12):e1000312 doi: 10.1371/journal.pgen.1000312 ; PubMed Central PMCID: PMC2596965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–39. doi: 10.2337/db10-0502 ; PubMed Central PMCID: PMC2992787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Takeuchi F, Lee JY, Li H, Wu JY, Liang J, et al. Multiple Non-glycemic Genomic Loci Are Newly Associated with Blood Level of Glycated Hemoglobin in East Asians. Diabetes. 2014. doi: 10.2337/db13-1815 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryu J, Lee C. Association of glycosylated hemoglobin with the gene encoding CDKAL1 in the Korean Association Resource (KARE) study. Hum Mutat. 2012;33(4):655–9. doi: 10.1002/humu.22040 . [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Ong RT, Tay WT, Sim X, Ali M, Xu H, et al. A study assessing the association of glycated hemoglobin A1C (HbA1C) associated variants with HbA1C, chronic kidney disease and diabetic retinopathy in populations of Asian ancestry. PLoS One. 2013;8(11):e79767 doi: 10.1371/journal.pone.0079767 ; PubMed Central PMCID: PMCPMC3820602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bry L, Chen PC, Sacks DB. Effects of hemoglobin variants and chemically modified derivatives on assays for glycohemoglobin. Clin Chem. 2001;47(2):153–63. . [PubMed] [Google Scholar]
- 9.Krauss JS, Hahn DA, Harper D, Shell S, Baisden CR. The affinity glycated hemoglobin in a family with hereditary spherocytosis and in other non-hemoglobinopathic hemolytic anemias. Ann Clin Lab Sci. 1987;17(5):331–8. . [PubMed] [Google Scholar]
- 10.Menke A, Rust KF, Savage PJ, Cowie CC. Hemoglobin A1c, fasting plasma glucose, and 2-hour plasma glucose distributions in U.S. population subgroups: NHANES 2005–2010. Ann Epidemiol. 2014;24(2):83–9. doi: 10.1016/j.annepidem.2013.10.008 ; PubMed Central PMCID: PMC3946694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruthur NM, Kao WH, Clark JM, Brancati FL, Cheng CY, Pankow JS, et al. Does genetic ancestry explain higher values of glycated hemoglobin in African Americans? Diabetes. 2011;60(9):2434–8. doi: 10.2337/db11-0319 ; PubMed Central PMCID: PMC3161314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkataraman K, Kao SL, Thai AC, Salim A, Lee JJ, Heng D, et al. Ethnicity modifies the relation between fasting plasma glucose and HbA1c in Indians, Malays and Chinese. Diabet Med. 2012;29(7):911–7. doi: 10.1111/j.1464-5491.2012.03599.x ; PubMed Central PMCID: PMC3504343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacy ME, Wellenius GA, Sumner AE, Correa A, Carnethon MR, Liem RI, et al. Association of Sickle Cell Trait With Hemoglobin A1c in African Americans. JAMA. 2017;317(5):507–15. doi: 10.1001/jama.2016.21035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little RR, Rohlfing CL, Sacks DB, National Glycohemoglobin Standardization Program Steering C. Status of hemoglobin A1c measurement and goals for improvement: from chaos to order for improving diabetes care. Clin Chem. 2011;57(2):205–14. doi: 10.1373/clinchem.2010.148841 . [DOI] [PubMed] [Google Scholar]
- 15.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793 doi: 10.1371/journal.pgen.1002793 ; PubMed Central PMCID: PMC3410907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International HapMap C, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258 ; PubMed Central PMCID: PMC2689609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. doi: 10.1038/ng.2385 ; PubMed Central PMCID: PMC3433394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340 ; PubMed Central PMCID: PMC2922887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. . [DOI] [PubMed] [Google Scholar]
- 20.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–22. doi: 10.1002/gepi.20630 ; PubMed Central PMCID: PMC3460225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–5. doi: 10.1002/gepi.20303 . [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011 ; PubMed Central PMCID: PMC3014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ATC, Replication DIG, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–75, S1-3. doi: 10.1038/ng.2213 ; PubMed Central PMCID: PMC3593158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–69. doi: 10.1038/ng.2274 ; PubMed Central PMCID: PMC3613127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142–8. doi: 10.1038/ng.521 ; PubMed Central PMCID: PMC2922003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60(10):2624–34. doi: 10.2337/db11-0415 ; PubMed Central PMCID: PMC3178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–75. doi: 10.1038/nature11677 ; PubMed Central PMCID: PMC3623669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42(3):210–5. doi: 10.1038/ng.531 . [DOI] [PubMed] [Google Scholar]
- 29.Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480(7376):201–8. doi: 10.1038/nature10659 ; PubMed Central PMCID: PMCPMC3335296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–15. doi: 10.1038/nrg3457 ; PubMed Central PMCID: PMCPMC4096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornelis MC, Qi L, Zhang C, Kraft P, Manson J, Cai T, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med. 2009;150(8):541–50. ; PubMed Central PMCID: PMC3825275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leong A. Is there a need for neonatal screening of glucose-6-phosphate dehydrogenase deficiency in Canada? Mcgill J Med. 2007;10(1):31–4. ; PubMed Central PMCID: PMC2323536. [PMC free article] [PubMed] [Google Scholar]
- 33.Motulsky AG, Stamatoyannopoulos G. Clinical implications of glucose-6-phosphate dehydrogenase deficiency. Ann Intern Med. 1966;65(6):1329–34. . [DOI] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention (CDC). National Center for Health Statistics (NCHS). National Health and Nutrition Examination Survey Data. Hyattsville, MD: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2013–2014. [Google Scholar]
- 35.Luzzatto L, Usanga FA, Reddy S. Glucose-6-phosphate dehydrogenase deficient red cells: resistance to infection by malarial parasites. Science. 1969;164(3881):839–42. . [DOI] [PubMed] [Google Scholar]
- 36.United States Census Bureau. American FactFinder. Available from: http://factfinder.census.gov/faces/tableservices/jsf/pages/productview.xhtml?pid=ACS_14_1YR_S0201&prodType=table.
- 37.Selvin E, Steffes MW, Zhu H, Matsushita K, Wagenknecht L, Pankow J, et al. Glycated hemoglobin, diabetes, and cardiovascular risk in nondiabetic adults. N Engl J Med. 2010;362(9):800–11. doi: 10.1056/NEJMoa0908359 ; PubMed Central PMCID: PMC2872990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selvin E, Ning Y, Steffes MW, Bash LD, Klein R, Wong TY, et al. Glycated hemoglobin and the risk of kidney disease and retinopathy in adults with and without diabetes. Diabetes. 2011;60(1):298–305. doi: 10.2337/db10-1198 ; PubMed Central PMCID: PMCPMC3012185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowie CC, Rust KF, Byrd-Holt DD, Gregg EW, Ford ES, Geiss LS, et al. Prevalence of diabetes and high risk for diabetes using A1C criteria in the U.S. population in 1988–2006. Diabetes Care. 2010;33(3):562–8. doi: 10.2337/dc09-1524 ; PubMed Central PMCID: PMC2827508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selvin E, Parrinello CM, Sacks DB, Coresh J. Trends in prevalence and control of diabetes in the United States, 1988–1994 and 1999–2010. Ann Intern Med. 2014;160(8):517–25. doi: 10.7326/M13-2411 ; PubMed Central PMCID: PMC4442608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watchko JF, Kaplan M, Stark AR, Stevenson DK, Bhutani VK. Should we screen newborns for glucose-6-phosphate dehydrogenase deficiency in the United States? J Perinatol. 2013;33(7):499–504. doi: 10.1038/jp.2013.14 . [DOI] [PubMed] [Google Scholar]
- 42.Chu CS, Bancone G, Moore KA, Win HH, Thitipanawan N, Po C, et al. Haemolysis in G6PD Heterozygous Females Treated with Primaquine for Plasmodium vivax Malaria: A Nested Cohort in a Trial of Radical Curative Regimens. PLoS Med. 2017;14(2):e1002224 doi: 10.1371/journal.pmed.1002224 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danescu LG, Levy S, Levy J. Markedly low hemoglobin A1c in a patient with an unusual presentation of beta-thalassemia minor. Endocr Pract. 2010;16(1):89–92. doi: 10.4158/EP09160.CR . [DOI] [PubMed] [Google Scholar]
- 44.American Diabetes A. 2. Classification and Diagnosis of Diabetes. Diabetes Care. 2017;40(Suppl 1):S11–S24. doi: 10.2337/dc17-S005 . [DOI] [PubMed] [Google Scholar]
- 45.Ziemer DC, Kolm P, Weintraub WS, Vaccarino V, Rhee MK, Twombly JG, et al. Glucose-independent, black-white differences in hemoglobin A1c levels: a cross-sectional analysis of 2 studies. Ann Intern Med. 2010;152(12):770–7. doi: 10.7326/0003-4819-152-12-201006150-00004 . [DOI] [PubMed] [Google Scholar]
- 46.Herman WH, Ma Y, Uwaifo G, Haffner S, Kahn SE, Horton ES, et al. Differences in A1C by race and ethnicity among patients with impaired glucose tolerance in the Diabetes Prevention Program. Diabetes Care. 2007;30(10):2453–7. doi: 10.2337/dc06-2003 ; PubMed Central PMCID: PMC2373980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(DOCX)
Data Availability Statement
The ancestry-specific and transethnic genome-wide meta-analysis summary statistics for association with HbA1c, and published data included in this study, are available to download from the MAGIC website, www.magicinvestigators.org/downloads. Uniform analysis plan(s) showing the QC and data analysis steps in detail are provided in the supporting information file S1 Analysis Plans.