Abstract
A method has been developed for three-dimensional image reconstruction of symmetry-mismatched components in tailed phages. Although the method described here addresses the specific case where differing symmetry axes are coincident, the method is more generally applicable, for instance, to the reconstruction of images of viral particles that deviate from icosahedral symmetry. Particles are initially oriented according to their dominant symmetry, thus reducing the search space for determining the orientation of the less dominant, symmetry-mismatched component. This procedure produced an improved reconstruction of the sixfold-symmetric tail assembly that is attached to the fivefold-symmetric prolate head of ϕ29, demonstrating that this method is capable of detecting and reconstructing an object that included a symmetry mismatch. A reconstruction of ϕ29 prohead particles using the methods described here establishes that the pRNA molecule has fivefold symmetry when attached to the prohead, consistent with its proposed role as a component of the stator in the ϕ29 DNA packaging motor.
Keywords: cryo-EM image reconstruction, ϕ29, pRNA, symmetry mismatch, tail appendages
INTRODUCTION
Double-stranded DNA (dsDNA) bacteriophages consist of a capsid, or head, that contains the viral genome. Attached to the head is a tail structure that binds to the host cell to initiate infection and through which DNA passes upon injection into the host cell. Bacteriophage tails typically possess 6-fold rotational symmetry and attach to a specific, 5-fold-symmetric vertex of the head via a portal, or connector protein assembly (Caspar and Klug, 1962; Moody, 1965; Anderson et al., 1966; Crowther and Klug, 1975; Serwer, 1976; Bazinet and King, 1985; Tao et al., 1998). The connector assemblies have been shown to have 12- or 13-fold rotational symmetry (Dube et al., 1993; Prevelige and King, 1993; Müller et al., 1997; Guasch et al., 1998; Badasso et al., 2000). The potential significance of symmetry mismatches in genome packaging by tailed phages was first recognized by Hendrix (1978). The Hendrix model proposes that the connector rotates with respect to the head as DNA passes through the central pore of the connector and enters the capsid and that this rotation is made energetically possible by the symmetry mismatch between the 5-fold head and the 12- or 13-fold connector. Although variations of this basic theme have been proposed (Dube et al., 1993; Grimes and Anderson, 1997; Simpson et al., 2000), these mechanisms typically invoke connector rotation facilitated by a symmetry mismatch.
The large size and complexity of DNA packaging machines put them out of reach of X-ray crystallography and nuclear magnetic resonance spectroscopy. The icosahedral symmetry averaging procedures utilized in the image processing of cryoelectron-microscopy (cryo-EM) images of viruses result in the loss of structural information that does not conform to the imposed symmetry, such as a symmetry mismatch. Though robust procedures exist for reconstructing asymmetric objects, such as the ribosome, they fail to utilize the large amount of symmetry present in tailed phages, thus requiring a larger number of particles. Real and reciprocal space methods based on the concept of noncrystallographic symmetry averaging (Rossmann and Tao, 1999), which specifically address issues related to the re-construction of partially symmetric objects, have been proposed, but they have yet to be fully developed and tested. Here, we describe a method for the detection and reconstruction of symmetry mismatches in tailed phages. This procedure has been applied to the dsDNA bacteriophage ϕ29, a phage with a well-characterized morphogenetic pathway (Fig. 1) (Anderson and Reilly, 1993). A reconstruction of mature, DNA-emptied phage particles, in which both the fivefold symmetry of the prolate head and the sixfold symmetry of the tail assembly are preserved, demonstrates that the method is capable of detecting and reconstructing symmetry mismatches. In addition, this procedure was applied to ϕ29 prohead particles to evaluate the possibility of a symmetry mismatch between the virus head and the oligomeric prohead RNA (pRNA).
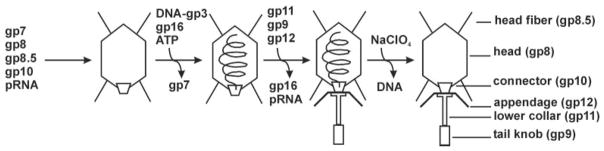
FIG. 1.
Morphogenesis of bacteriophage ϕ29. Proheads, the first particles assembled in the ϕ29 morphogenetic pathway, consist of a head–tail connector protein (gp10), a scaffolding protein (gp7), a major capsid protein (gp8), head fibers (gp8.5), and a 174-base RNA (pRNA). The viral genome (covalently attached to the viral protein gp3) is packaged into the prohead. DNA-gp3 packaging requires the addition of a virus-encoded ATPase (gp16) and ATP. The scaffolding protein gp7 leaves during this stage. Finally, gp16 and pRNA are released, and the lower collar (gp11), tail knob (gp9), and appendage (gp12) proteins are added to form the tail of the mature virion. The DNA can be ejected by incubation with NaClO4. There are symmetry mismatches between the five-fold-symmetric capsid and the 12-fold-symmetric connector or the six-fold-symmetric tail.
MATERIALS AND METHODS
Production of ϕ29 particles and electron microscopy
Proheads and phage were produced and examined by electron microscopy as reported by Tao et al. (1998). Some of the prolate particle samples were tilted up to 9° in the microscope (Dryden et al., 1993) to alleviate the problem of uneven particle distribution. Table I lists the defocus levels for images used in three-dimensional reconstruction.
TABLE I.
Cryo-EM Data and Image Reconstruction
| Data set | Prohead + pRNA | Mature phage | Tail component of mature phage |
|---|---|---|---|
| Approximate underfocus (μm)a | 2.3 | 2.6 | 2.6 |
| Number of particlesb | 609 (1153) | 1453 (2781) | 1743 (3197) |
| Correlation coefficientc | 0.324 (0.047) | 0.263 (0.056) | 0.364 (0.088) |
| Resolution (Å) | 26 | 33 | 33 |
| Symmetry imposed | 1 | 1 | 6 |
Determined from the contrast transfer function of the microscope.
The number of particles included in each three-dimensional map. The total number of boxed particles is given in parentheses.
Averaged real-space correlation coefficient and standard deviation (in parentheses) over all radii for all particles.
Three-dimensional image reconstruction
Suitable micrographs of fields of ϕ29 particles were selected and digitized at 14-μm intervals (3.9 or 3.7 Å/pixel at the specimen) on a Zeiss PHODIS scanner. Individual particle images were boxed, floated, and preprocessed to normalize mean intensities and variances and remove linear background gradients as described previously (Belnap et al., 1996). The model-based, polar-Fourier-transform (PFT) method (Baker and Cheng, 1996) was used to determine particle orientations. Previously described reconstructions of ϕ29 prohead and emptied phage particles (Tao et al., 1998) served as initial models in the PFT orientation search for these two particle populations. Orientation refinement was monitored by means of correlation coefficients computed with real and reciprocal space data (Dryden et al., 1993). All maps were computed with Fourier–Bessel procedures, and eigenvalue spectra were used to monitor the conditioning of the least-squares equations (Fuller et al., 1996). The resolution of each map was estimated by splitting the image data into two sets and comparing structure factors in the separate reconstructions. The threshold for surface-shaded rendering was generally set about 1 standard deviation above the mean electron density level. The effect of the phase-contrast transfer function (CTF) was not compensated for in the reconstruction process, and data were rejected beyond the first node. Hence, the exact defocus level on each micrograph was not critical.
Asymmetric reconstructions of proheads and emptied phage
The procedure described here was used to determine the symmetry of ϕ29 pRNA (MW 345.6 kDa) relative to the fivefold symmetry of the head (MW 11 703 kDa). It subsequently was applied to establish the symmetry of the tail (MW 5814 kDa) with respect to the phage head. In order to average between particles, the following procedure was used in the asymmetric reconstructions of prolate and emptied ϕ29 particles to resolve fivefold ambiguity in initial particle orientations.
Particle orientations (θ,ϕ,ω, as defined by Baker and Cheng (1996)) (Fig. 2) and centers were initially found assuming fivefold symmetry.
A map was calculated without imposing any symmetry averaging. Afterward, local fivefold averaging was applied to the capsid shell only (no averaging was imposed on map regions corresponding to the pRNA in prolate particles or the tail assembly in the case of emptied phage). These maps possessed asymmetric features either in the pRNA when reconstructing the prohead particles or in the tail assembly when reconstructing emptied phage. The origin of these features arises either from an unequal distribution of particle orientations or from random noise.
The new map was used as a model for a PFT-based orientation search. However, only five projections were generated from the model, corresponding to the orientations (θ, ϕ, ω), (θ, 2π/5 + ϕ, ω), (θ, 4π/5 + ϕ, ω), (θ, 6π/5 + ϕ, ω), and (θ, 8π/5 + ϕ, ω). (θ,ϕ,ω) is the orientation from (1). The best correlation between the model and the raw image was taken to be the correct orientation. The small, possibly random features obtained in step 2 of cycle 1 were sufficient to provide increasing selection power in subsequent cycles.
A new map was calculated as in (2).
Steps (3) and (4) were repeated iteratively until the process converged (i.e., orientations became stable).
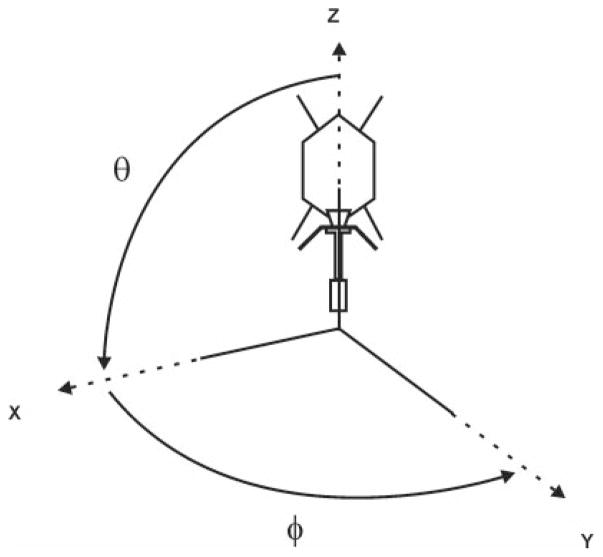
FIG. 2.
The polar angles (θ, ϕ, ω) that define particle orientation using the convention of Baker and Cheng (1996). The model used for particle orientation by PFT is placed in the polar coordinate system with its highest axis of symmetry coincident with Z. Any rotation is described by (θ, ϕ, ω), with θ and ϕ defining rotation in the planes of zx and xy, respectively. The angle ω describes the rotation about the view direction.
The logic behind this approach is that by searching ϕ in multiples of 2π/5 (multiples of 72°), each of the five equivalent positions for the head can be tested to see which one best matches the experimentally obtained raw image data. By distinguishing the previously equivalent positions around the fivefold axis, differences between these positions due to genuine deviations from fivefold symmetry are no longer averaged out. Thus, fivefold symmetry is no longer imposed on the tail (or pRNA), and its true symmetry should become apparent (Fig. 3a). It was assumed that the tail (or pRNA) portion of the structure was always positioned the same way relative to the head. Thus, there is one orientation of the head that would correctly superimpose the tail (or pRNA). Were this not the case, it would be impossible to correctly orient both the head and the tail (or pRNA) for averaging over different particles (Fig. 3b).
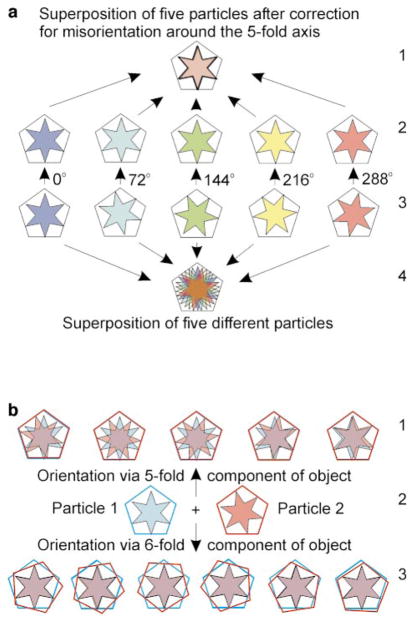
FIG. 3.
(a) Schematic of the five possible ways to align particles based on the head structure alone. The head is represented by a pentagon and the tail (or pRNA) by a six-sided star. The particles in row a3 are identical to one another, differing only in their relative rotations about the central fivefold axis (the particle on the far left is arbitrarily designated a 0° rotation). The particle shown in row a4 is the result of superimposing the five particles seen in row a3. Although the pentagons (phage heads) superimpose perfectly, the six-sided stars (tail regions) do not. In row a2, the particles from row a3 have been brought into mutual alignment by rotating each particle around the fivefold axis by a multiple of 72° (clockwise direction), as indicated by the numbers next to the arrows pointing from particles in row a3 to the corresponding particle in row a2. In row a2, both the pentagon (head) and the six-sided star (tail) are aligned for averaging between particles. Row a1 shows the result of superimposing the five particles after correction for misorientation around the fivefold axis (i.e., superposition of the five particles in row a2). (b) Schematic of two particles that differ in the relative orientations of the head (pentagon) to the tail (six-sided star). The six-sided star in particle 1 (row b2, left) is rotated 7° clockwise with respect to the six-sided star in particle 2 (b2, right). The upper row (b1) shows the five possible ways to superimpose the two particles based on alignment of the pentagon (head). Moving from left to right, particle 2 is rotated 0°, 72°, 144°, 216°, and 288° (counterclockwise) with respect to its initial orientation (b2, right). Although the pentagons are aligned in each superposition in b1, the six-sided stars remain unaligned. Row b3 illustrates the six possible ways to superimpose the two particles based on orientation of the six-sided star (tail). Initially, particle 2 (b2, right) is rotated 7° (clockwise) to align the six-sided star with the corresponding star in particle 1. Then, moving from left to right, particle 2 is rotated 0°, 60°, 120°, 180°, 240°, and 300° (counterclockwise). If particles are aligned in this manner, the six-sided stars superimpose perfectly, but the pentagons do not. Thus, when the relative orientation of the head (pentagon) with respect to the tail (six-sided star) differs between particles, it is not possible to align both the head and the tail for averaging between different particles.
The procedure was applied to both the proheads and the mature virus (Table I). The process converged after about 10 cycles using data to 26 Å for the proheads and 32 Å for the mature virus. Because the native virus contains a known symmetry mismatch (fivefold head and sixfold lower collar protein; see Fig. 1), it provided a test as to whether or not the procedure is capable of detecting a symmetry mismatch.
In the case of emptied phage particles, where the tail assembly proved to have different symmetry than the head (see Results and Discussion), additional steps were taken to utilize this sixfold symmetry to obtain an accurate reconstruction of the tail. Essentially, a new reconstruction of only the phage tail was computed in which the observed sixfold symmetry was enforced. First, particles were reboxed so that only the tail portion of the phage was included in the box. Next, an initial model of only the tail assembly was extracted from the asymmetric reconstruction of the emptied phage using the program ROBEM (R. Ashmore and T. S. Baker, unpublished). This model of the phage tail was then used in conjunction with the PFT method to determine the initial orientations of the reboxed raw images of the particle tail. As described above, the PFT method was used in subsequent rounds of orientation refinement, and all maps were calculated using Fourier–Bessel procedures. In this procedure, it was necessary to specify the limits of the tail within the model of the whole virus. This was accomplished by visual inspection of the central cross section, perpendicular to the long axis of the virus. Similarly, the center of the tails, which were seen mostly in projections perpendicular to the long axis of the virus, was chosen by eye. Sixfold symmetry was enforced for orientation determination and refinement, as well as for all map calculations. The resulting sixfold-symmetric reconstruction of the phage tail was then used to replace the corresponding tail sections in the original fivefold reconstruction of the phage, thus creating a model of the phage where both the head and the tail have been accurately reconstructed using the appropriate symmetry.
RESULTS AND DISCUSSION
Asymmetric Reconstruction of Prohead Particles
One of the more surprising results to emerge from biochemical and genetic studies of DNA packaging by bacteriophage ϕ29 is an absolute requirement for a novel, phage-encoded, 174-base RNA molecule, termed “prohead RNA” or “pRNA,” for DNA packaging (Guo et al., 1987; Hendrix, 1998). Investigations into the oligomeric state of pRNA suggested that it exists in solution as a hexamer (Guo et al., 1998; Zhang et al., 1998) stabilized by intermolecular base-pairing between adjacent monomers. Additionally, a recent reconstruction of ϕ29 proheads by Ibarra et al. (2000) located a hexameric pRNA ring at the base of the prohead. These results, however, contradict those obtained by Simpson et al. (2000) in their reconstruction of a ϕ29 prohead particle. Although Simpson et al. also located the pRNA molecule at the base of the prohead, it clearly displayed fivefold symmetry rather than sixfold symmetry (Tao et al., 1998; Simpson et al., 2000). If the pRNA had in fact possessed sixfold symmetry, application of fivefold symmetry averaging would be expected to result in 30 (or 6 ×5) smeared, low-density features rather than the observed five distinct, high-density features (Simpson et al., 2000).
If the pRNA and the capsid (Fig. 1) have different symmetries, then there is only one way to align both the head and the pRNA properly for averaging between different particles, assuming that there is a unique relationship between the head and the pRNA. Due to the fivefold symmetry used in initial orientation searches, particles would be aligned by virtue of the dominating capsid structure. Assuming that particle orientations are evenly distributed about the fivefold axis of the head, there would be 1/5 of the pRNAs in each of the five possible orientations. Thus, the pRNAs would not be properly aligned for averaging between particles. Averaging different particles with such a distribution without imposing fivefold symmetry would, nevertheless, produce a result similar to enforcing fivefold averaging (Fig. 3a).
Alternatively, it would be possible to repeat the entire reconstruction from the beginning, without ever imposing any symmetry constraints. This approach is unattractive since it does not make use of the fivefold symmetry that holds for the phage head, by far the largest portion of the object. It also seems likely that the large mass of the fivefold-symmetric head would still dominate orientation searches, again resulting in fivefold ambiguity in the particle orientations.
Ibarra et al. (2000) addressed the potential symmetry mismatch between the pRNA and the pro-head by first calculating a map with no symmetry and then computing the power spectra for different sections of the map. Using this approach, they found a 6-fold-symmetric region of the map corresponding to the pRNA. From this point on, two reconstructions were computed in parallel, one where 5-fold averaging was enforced and one where 6-fold averaging was imposed. The resulting reconstructions were then combined, such that the viral capsid was derived from the 5-fold-symmetric reconstruction, and the pRNA was derived from the 6-fold-symmetric reconstruction. This approach, however, is complicated by the fact that the connector protein, now known to possess 12-fold symmetry (Müller et al., 1997), is in close proximity to, and to a certain extent overlaps with, the pRNA density. Thus, it is possible that the small 6-fold signal that they observed had resulted from the connector rather than from the pRNA.
In an attempt to resolve the conflicting data regarding the symmetry of the pRNA molecule, we have computed a reconstruction of ϕ29 prohead particles employing a procedure designed to detect and reconstruct symmetry mismatches, while still making use of the predominant symmetry of the object (see Materials and Methods).
The results of applying the procedure to ϕ29 prohead particles are shown in Fig. 4b. As might be expected, the overall quality of the reconstruction is inferior to that seen when fivefold symmetry is enforced (Fig. 4a) (see Tao et al., 1998; Simpson et al., 2000). The pRNA appears pentameric rather than hexameric and the fivefold component of a power spectrum (Crowther and Amos, 1971) for the pRNA region of this map is clearly dominant (Fig. 5a).
FIG. 4.
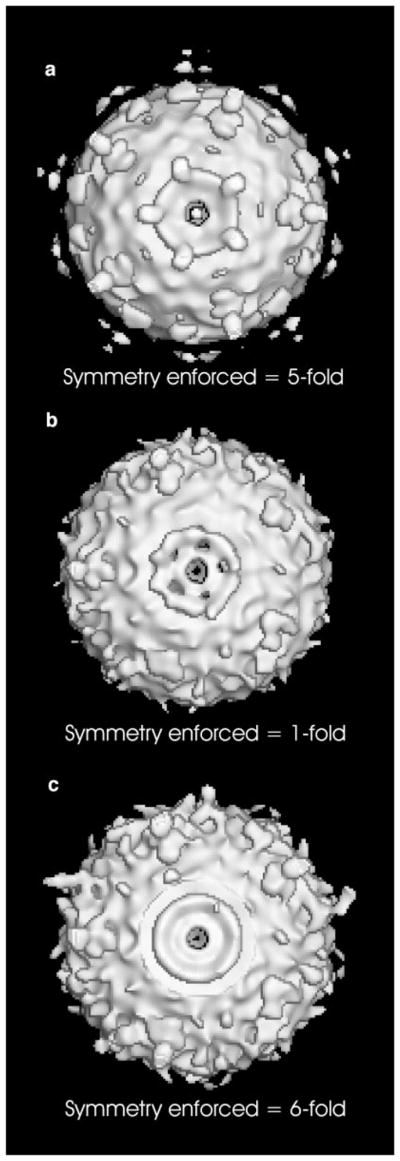
Cryo-EM reconstructions of prohead + pRNA, looking down the central fivefold axis at the pRNA density. (a) Fivefold symmetry was enforced throughout the entire procedure (Simpson et al., 2000). (b) No symmetry was enforced, with averaging occurring only between different particles. (c) Sixfold averaging was enforced for the region of density corresponding to the pRNA, resulting in smeared density.
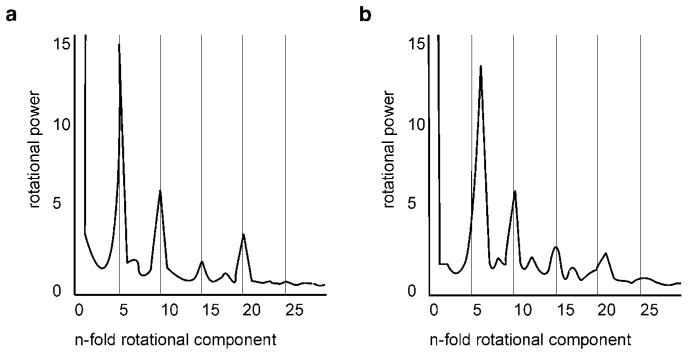
FIG. 5.
Rotational power spectra. (a) Spectrum of the cross section perpendicular to the central fivefold axis, through the pRNA, of the prohead reconstruction obtained without fivefold averaging (Fig. 4b) showing the dominant fivefold symmetry. (b) Spectrum of the cross section perpendicular to the central fivefold axis through the lower collar of the emptied phage showing the dominant sixfold symmetry.
A further refinement of the above procedure was made in order to minimize the contribution of the head to the determination of orientation and to maximize the contribution of the pRNA. The regions of the search model corresponding to the head were set to zero, and the regions of the raw particle images corresponding to the head were set to the background levels. This was readily accomplished because the long axis of the particles was roughly in the plane of the grid. Orientations obtained from the subsequent PFT search were then applied to the original, unmasked particle data. The resulting map retained fivefold symmetry for the pRNA and did not differ significantly from the map shown in Fig. 4b.
To further ensure that the pentameric pRNA is not an artifact due to imposing fivefold symmetry in the initial orientation searches, the procedure described under Materials and Methods was repeated; however, this time sixfold averaging was applied to the density region corresponding to the pRNA. The resulting smeared-out, low-density, disk-like object seen in the reconstruction (Fig. 4c) is what would be expected if incorrect symmetry averaging had been imposed on the object. The deterioration of electron density due to inappropriate symmetry averaging is a phenomenon well known to X-ray crystallographers in the application of noncrystallographic symmetry (Rossmann, 1990).
Because the end of the prohead opposite the connector possesses fivefold-symmetric fibers, positioned at approximately the same radial distance from the central fivefold as the pRNA, we had to consider the possibility that the fivefold-symmetric radial spokes that we observed for the pRNA resulted from the misorientation of individual particles (i.e., some proheads were oriented with their closed ends superimposed on the open end of other particles). This scenario seemed unlikely as the artifact would also be present in proheads lacking pRNA. However, as previously described (Simpson et al., 2000), prohead particles treated with RNase completely lack the density proposed to represent the pRNA. This scenario should have produced an image of the connector at both ends of the head, a feature that we have not observed in any of our reconstructions.
We cannot currently reconcile our results with data suggesting the pRNA is a hexamer in its active form (Guo et al., 1998; Zhang et al., 1998; Ibarra et al., 2000). It is possible that pRNA does indeed assemble as a hexamer in solution, but that one monomer is lost upon binding to the prohead, or that one molecule is displaced out of the ring. It is also possible that the oligomeric state of the pRNA on the prohead varies from preparation to preparation, in which case both our results and those obtained by Ibarra et al. could be correct.
It was previously observed by us that the prohead plus dodecameric connector, pRNA, viral ATPase (gp16), and DNA comprise a rotary motor with the prohead–pRNA–ATPase complex acting as a stator, the DNA acting as a spindle, and the connector acting as a ball-race (Simpson et al., 2000). In this model, in order for the pRNA and prohead to both be part of the stator, they must function as a single unit, thus requiring a fixed interaction that would be facilitated if the pRNA had the same symmetry as the head (i.e., fivefold symmetry). Additionally, we suggested that the viral ATPase is attached to the spokes of the pRNA.
Asymmetric Reconstruction of Mature Phage Particles
The procedure described above was also used for the reconstruction of mature phage particles that had lost their DNA. The purpose of this reconstruction was twofold. First, because the mature virus contains a known symmetry mismatch (fivefold head versus the sixfold lower collar protein), it provided a test as to whether or not the procedure described here is capable of detecting a symmetry mismatch. Second, it provided a means of obtaining an accurate reconstruction of the phage tail (previous reconstructions of the mature virus imposed fivefold averaging on the tail assembly (Tao et al., 1998)) and thus an improved reconstruction of the entire mature phage. The “tail” herein includes the connector (gp10), lower collar (gp11), and appendages (gp12), components previously called the “neck” (Anderson et al., 1966), and the tail knob (gp9).
By testing the ability of the procedure to detect a known symmetry mismatch, the legitimacy of the prohead reconstruction showing fivefold symmetry for the pRNA is also evaluated. Ideally, not only should the particle chosen to test the procedure have a known symmetry mismatch, the mass ratio of the mismatched components should be similar to the prohead/pRNA mass ratio as well. However, although the mature virus possesses a symmetry mismatch between the head and the tail, the tail is quite large when compared to the pRNA. Thus, the head/tail ratio is much smaller than the head/pRNA mass ratio. The concern is that the procedure may be capable of detecting the symmetry mismatch between the head and the tail in the mature phage, but is not sensitive enough to determine the potential symmetry mismatch between the head and the pRNA in prohead particles. To minimize this concern, the mature phage particles were initially boxed with the same box size used in the prohead reconstruction (i.e., most, but not all, of the tail was cut off). In addition to significantly reducing computation time, this ensured that the mass ratio of the symmetry-mismatched components was comparable to the mass ratio of the head/pRNA. Thus, this reconstruction serves as a control for the observation that the pRNA has fivefold symmetry when bound to the phage head.
After five cycles of the procedure described above (see Materials and Methods, Asymmetric Reconstruction of Proheads and Emptied Phage), a sixfold-symmetric density began to emerge in the region of the map corresponding to the lower collar of the tail assembly. A section through this reconstruction, perpendicular to the phage axis (Fig. 6), shows both the fivefold symmetry of the head fibers and the sixfold symmetry of the lower collar in the same section, demonstrating that the procedure did not unintentionally impose fivefold symmetry. In order to minimize the contribution of the head to orientation determination, and to maximize the contribution of the lower collar, the head was masked as described above. The subsequent PFT search determined which of the five possible orientations about the central fivefold axis was correct for each particle. These orientations were then applied to the original, unmasked particle data. Visually, the apparent sixfold symmetry of the lower collar was enhanced in the resulting reconstruction. In order to quantitatively assess the symmetry of the lower collar in this reconstruction, a rotational power spectrum (Crowther and Amos, 1971) was calculated of a section through the lower collar (Fig. 5b). It was compared to the rotational power spectrum of the same section taken from a reconstruction in which the head had not been masked out. The sixfold component of the power spectrum increased from 28 to 34%.
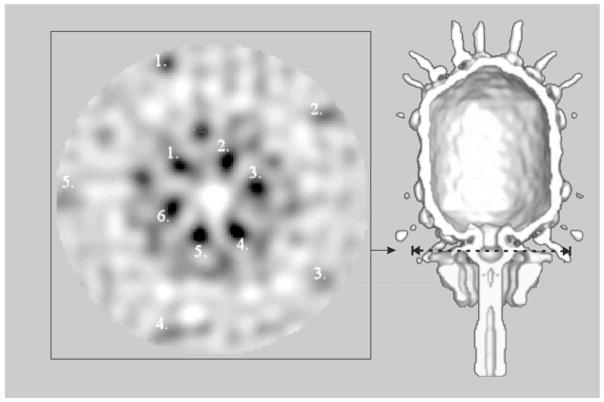
FIG. 6.
Section from a reconstruction of the mature virus. No symmetry was imposed in this reconstruction, only averaging between different particles (see Materials and Methods). In this section, perpendicular to the central fivefold axis, it is clear that the head and associated fibers have fivefold symmetry (outer circle of numbers 1–5), whereas the lower collar has approximately sixfold symmetry (inner circle of numbers, 1–6). These five- or sixfold symmetries are consistent with the known symmetries of the head and lower collar.
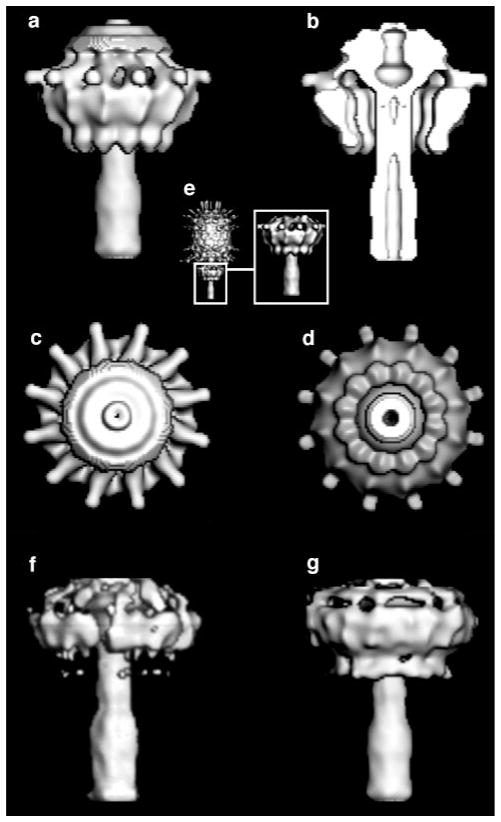
The appearance of the phage appendages (gp12) was used to assess the quality of the tail assembly in the reconstruction of the whole phage. The 12 appendages had been shown to attach to the lower collar and to function as adsorption organelles in the mature phage (Anderson et al., 1966; Tosi and Anderson, 1973; Carrascosa et al., 1982). In the micrographs of the mature phage, the appendages appear as long, spindly objects emanating from the head–tail junction. In the original reconstruction of the mature phage by Tao et al. (1998), the enforced fivefold symmetry resulted in 10, rather than 12, density features corresponding to the appendages. These density features were shortened compared to the long features present in the micrographs. In the new reconstruction of the phage tail (see Materials and Methods), the appendages appear as 12 jalapeno pepper-like objects loosely attached to the head–tail junction (Fig. 7).
FIG. 7.
Cryo-EM reconstruction of the bacteriophage ϕ29 tail structure. (a) The tail structure viewed perpendicular to the long axis of the phage showing the jalapeno pepper-like appendages. (b) Cross section of the tail; same view as in (a). (c) End-on view, looking from the head, down the tail. (d) End-on view, looking along the long axis of the phage from the bottom of the tail toward the head. (e) The tail structure in the context of the entire phage. Reconstructions of the tail imposing (f) fivefold symmetry and (g) onefold symmetry.
The length of the appendages is now consistent with their appearance in the original micrographs. Upon completion of DNA packaging in the head, addition of the tail components may provide a gating structure that retains the DNA into the head until interaction with the host triggers its release (Tao et al., 1998). Adsorption of the appendages to the Bacillus subtilis cell wall may trigger DNA release. In the cross section of the reconstruction (Fig. 7b) the narrow arms of the appendages (“vines” of the jalapeno peppers) appear to be nestled in a crevice formed by the connector, the head, and the top of the lower collar. In light of the intimate association of the appendages with the connector and the lower collar, adsorption of the appendages to the host cell might trigger a conformational change in the appendages, or apply a torque to the appendages, that is transmitted via their narrow arms to either the connector, the lower collar, or both, resulting in release of the gating mechanism and injection of the DNA through the tail into the host cell.
CONCLUSIONS
The principles demonstrated in this paper for analyzing structures that contain symmetry mismatches could be applied to numerous other problems. Examples are the special vertices of icosahedral viruses where the nucleic acid can be seen to emerge, e.g., rotaviruses (Lawton et al., 1997), and the structure of substrates in chaperones, such as GroEL (Farr et al., 2000), or in proteasomes (Zwickl et al., 2000).
Acknowledgments
We thank Cheryl Towell and Sharon Wilder for help in the preparation of the manuscript. The work was supported by grants from the National Science Foundation (M.G.R.) and the National Institutes of Health (D.L.A. and T.S.B.) and by a National Science Foundation Shared Instrumentation Grant (T.S.B. and M.G.R.).
References
- Anderson DL, Hickman DD, Reilly BE. Structure of Bacillus subtilis bacteriophage ϕ29 and the length of ϕ29 deoxyribonucleic acid. J Bacteriol. 1966;91:2081–2089. doi: 10.1128/jb.91.5.2081-2089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DL, Reilly BE. Morphogenesis of bacteriophage ϕ29. In: Sonenshein AL, Hock JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria, Biochemistry and Molecular Genetics. ASM Publications; Washington, DC: 1993. pp. 859–867. [Google Scholar]
- Badasso MO, Leiman PG, Tao Y, He Y, Ohlendorf DH, Rossmann MG, Anderson D. Purification, crystallization, and initial X-ray analysis of the head–tail connector of bacteriophage ϕ29. Acta Crystallogr Sect D. 2000;56:1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, Christensen ND, Baker TS. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascosa JL, Vinuela E, Garcia N, Santisteban A. Structure of the head–tail connector of bacteriophage ϕ29. J Mol Biol. 1982;154:311–324. doi: 10.1016/0022-2836(82)90066-3. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA. Harmonic analysis of electron microscope images with rotational symmetry. J Mol Biol. 1971;60:123–130. doi: 10.1016/0022-2836(71)90452-9. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Klug A. Structural analysis of macromolecular assemblies by image reconstruction from electron micrographs. Annu Rev Biochem. 1975;44:161–182. doi: 10.1146/annurev.bi.44.070175.001113. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: Analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: A DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GW, Furtak K, Rowland MB, Ranson NA, Saibil HR, Kirchhausen T, Horwich AL. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell. 2000;100:561–573. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles—The uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. The bacteriophage ϕ29 packaging proteins supercoil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Párraga A, Valpuesta JM, Carrascosa JL, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage ϕ29 connector particles. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage ϕ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage ϕ29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- Ibarra B, Castón JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage ϕ29 prohead. J Mol Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BVV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- Moody MF. The shape of the T-even bacteriophage head. Virology. 1965;26:567–576. doi: 10.1016/0042-6822(65)90319-3. [DOI] [PubMed] [Google Scholar]
- Müller DJ, Engel A, Carrascosa JL, Vélez M. The bacteriophage ϕ29 head–tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 1997;16:2547–2553. doi: 10.1093/emboj/16.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson NH, Baker TS. Magnification calibration and the determination of spherical virus diameters using cryo-microscopy. Ultramicroscopy. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PE, King J. Assembly of bacteriophage P22: A model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- Rossmann MG. The molecular replacement method. Acta Crystallogr Sect A. 1990;46:73–82. doi: 10.1107/s0108767389009815. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Tao Y. Cryo-electron microscopy reconstruction of partially symmetric objects. J Struct Biol. 1999;125:196–208. doi: 10.1006/jsbi.1999.4083. [DOI] [PubMed] [Google Scholar]
- Serwer P. Internal proteins of bacteriophage T7. J Mol Biol. 1976;107:271–291. doi: 10.1016/s0022-2836(76)80005-8. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi M, Anderson DL. Antigenic properties of bacteriophage ϕ29 structural proteins. J Virol. 1973;12:1548–1559. doi: 10.1128/jvi.12.6.1548-1559.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage ϕ29 DNA in vitro. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- Zwickl P, Baumeister W, Steven A. Dis-assembly lines: The proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]