SUMMARY
Cervical cancer is the second most prevalent malignant tumor among women worldwide. High-risk human papillomaviruses (HPVs) are believed to be the major causative pathogens of mucosal epithelial cancers including cervical cancer. The HPV capsid is made up of 360 copies of major (L1) and 72 copies of minor (L2) capsid proteins. To date, limited high-resolution structural information about the HPV capsid has hindered attempts to understand details concerning the mechanisms by which HPV assembles and infects cells. In this study, we have constructed a pseudo-atomic model of the HPV59 L1-only capsid and demonstrate that the C-terminal arm of L1 participates in virus-host interactions. Moreover, when conjugated to a scaffold protein, keyhole limpet hemocyanin (KLH), this arm is immunogenic in vivo. These results provide new insights that will help elucidate HPV biology, and hence pave a way for the design of next-generation HPV vaccines.
Graphical Abstract
INTRODUCTION
Human papillomaviruses (HPVs) are a family of double-stranded DNA viruses that infect human epithelial cells in a tissue-specific fashion and are implicated in several mucosal epithelial cancers (zur Hausen, 2009). There are about 170 different HPV genotypes, 15 of which (e.g., HPV16, HPV18, HPV33, HPV45, and HPV59) are considered high risk since they are the main pathogens of cervical cancers and other anogenital cancers such as anal, vulvar, and penile cancer (Munoz et al., 2003). The HPV virion is composed of multiple copies of major (L1) and minor (L2) capsid proteins that surround the 8-kbp, histone-associated, closed, circular DNA genome. In vitro, five copies of recombinant L1 assemble spontaneously into a pentamer and 72 pentamers, then self-assemble into an empty (no genome) T = 7 icosahedral shell with a maximum diameter of ~60 nm (Baker et al., 1991). The minor capsid protein, L2, is not required to form a complete capsid shell (Sasagawa et al., 1995). Disulfide bonds between multiple cysteines in L1 play an important role in binding and stabilizing L1-L1 interactions during the assembly of a complete shell (Ishii et al., 2003). These empty capsids, or virus-like particles (VLPs), present an array of antigen epitopes that mimic those found in native virions (Roldao et al., 2010).
The structure of bovine papillomavirus (BPV) was determined at ~3.6 Å resolution by cryoelectron microscopy (cryo-EM), and the reconstructed BPV density map revealed the tertiary structure of L1 and a “suspended bridge” linkage between neigh-boring pentamers (Wolf et al., 2010). In addition, the N-terminal regions were shown to form pentamer contacts in BPV capsid that might be molecular switch in determining the icosahedral triangulation lattice symmetry (T = 1 and 7) in the case of HPV16 (Chen et al., 2000; Wolf et al., 2010). Although no high-resolution structure of any HPV virion has yet been reported, the crystal structures of T = 1 VLPs of HPV16 and L1 pentamers of four different genotypes (HPV11/16/18/35) show that all L1s share a core domain structure similar to that found in BPV (Bishop et al., 2007; Chen et al., 2000; Wolf et al., 2010). Furthermore, medium-resolution cryo-EM reconstructions of several different T = 7 HPV VLPs indicate that HPVs might contain inter-pentamer contacts and inter-subunit disulfide bonds similar to those found in BPV (Zhao et al., 2014). This was verified by a 9-Å resolution, cryo-EM density map of mature HPV16 virions (Cardone et al., 2014). In addition, the cryo-EM reconstructions of an HPV16 in complex with different antibodies at resolutions between about 10 and 12 Å revealed the sites of some of the antigenic epitopes in HPV16 (Guan et al., 2015a, 2015b; Lee et al., 2014). A more accurate atomic model is needed to fully capture the structural details of inter-pentamer contacts in HPV and to better understand how surface features control antigenicity and type specificity (Chen et al., 2000; Sadeyen et al., 2003). In vitro and in a mouse model system, there is strong evidence that the HPV capsid attaches to the host cell primarily through heparan sulfate proteoglycans (HSPGs) and the non-HSPG extracellular matrix receptor, laminin-332 (formerly laminin-5), secreted by epithelial lines (Broutian et al., 2010; Culp et al., 2006a, 2006b; Giroglou et al., 2001). L1, but not L2, is the major determinant in such attachment (Roden et al., 1994). It was also found that lysine residues on surface-exposed loops of L1 were engaged in HSPG binding for HPV infection (Richards et al., 2013). Therefore, highly variable surface regions may play key roles in determining HPV tissue tropism.
In this study we employed a combination of X-ray crystallography, cryo-EM reconstruction, and molecular dynamics flexible fitting (MDFF) to construct a pseudo-atomic model of the HPV59 capsid. This model shows that the HPV59 capsomer differs mostly in the surface-loop regions from four other reported types (HPV11/16/18/35). It also reveals that the C-terminal arms of L1, responsible for interactions between neighboring pentamers, share similarity with those of BPV. In addition, to further examine virus-host interactions we designed and synthesized eight peptides that include exposed region(s) of the HPV59 VLP. Of these synthetic peptides, only those that included the “suspended bridge” segment interacted with host cells and stimulated the production of neutralizing antibody. Therefore, this linking segment might initiate, or at least participate in, the process by which HPV59 infects host cells.
RESULTS
Preparation of L1-Only HPV59 VLPs and Pentamers
A truncated construct of L1 (missing nine N-terminal residues; Figure 1A), the major capsid protein of HPV59, was expressed in Escherichia coli and maintained in soluble form under reducing conditions during purification. The purified L1 proteins were found to self-assemble into VLPs after removal of reducing agent. However, the HPV59 VLPs exhibit different sizes when examined by negative-stain transmission electron microscopy (TEM): most have a maximum diameter of ~60 nm and the rest are ≤50 nm (Figure 1B, left panel). Such size variation was further confirmed by analytical ultra-centrifugation (AUC) analysis, which revealed two particle types with sedimentation coefficient values of 118S and 139S (Figure 1C, left panel).
Figure 1. Characterization of HPV59 L1 VLPs and Pentamers.
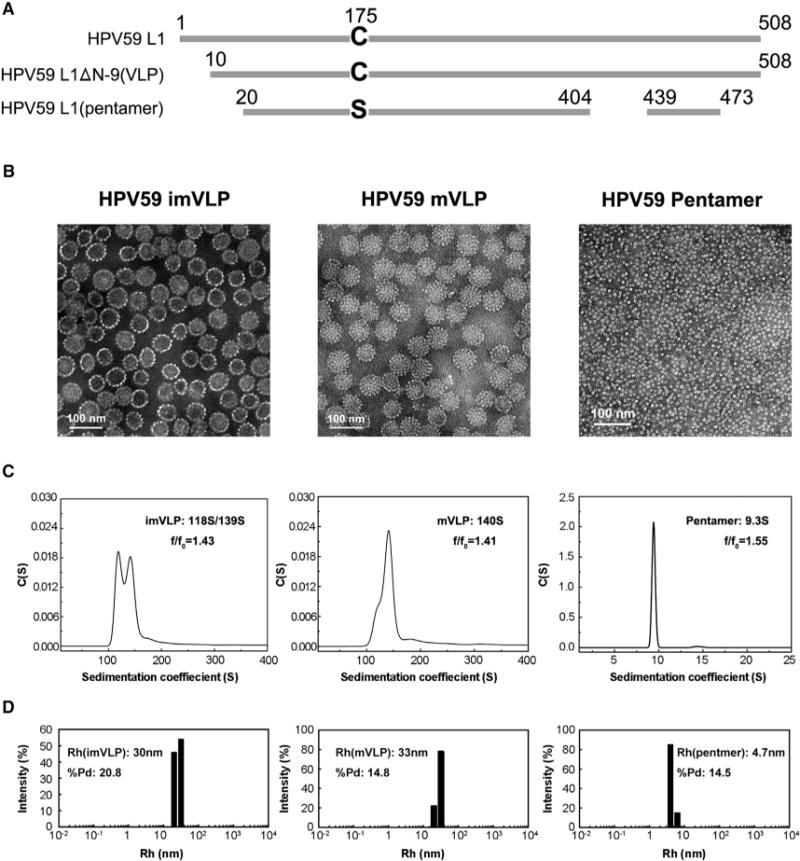
(A) Comparison of the L1 full-length sequence with that in the HPV59 VLP and the crystallized pentamer. The detailed aa sequences are shown in Figure S2A.
(B) Micrographs of purified, negatively stained samples of HPV59 L1 imVLPs, mVLPs, and pentamers. The mVLPs are more homogeneous in size and exhibit more distinct capsomeric features than the imVLPs. Scale bars, 100 nm.
(C) Sedimentation coefficient plots of HPV59 L1 imVLPs, mVLPs, and pentamers determined by analytical ultra-centrifugation provide further evidence that the mVLPs are more homogeneous in size compared with the imVLPs.
(D) DLS analyses of HPV59 L1 imVLPs, mVLPs, and pentamers.
Similar polymorphism has been observed in the VLPs of other HPV serotypes (Deschuyteneer et al., 2010; Kirnbauer et al., 1992), and such size heterogeneity has been attributed to differences in how neighboring pentamers interact (Zhao et al., 2005). It is noteworthy that such polymorphism has also been reported in the closely related simian virus 40 and polyomavirus (Salunke et al., 1989; Yan et al., 1996). The capsids of mature papillomavirus virions are known to be very stable and homogeneous in vivo owing to the specificity and strength of interactions between neighboring pentamers (Buck et al., 2005; Cardone et al., 2014). Thus, we questioned whether the homogeneity of the L1-only HPV59 VLPs could be enhanced in vitro by attempting to recapitulate the maturation process. After expressed L1s were incubated for 24 hr at room temperature in PBS buffer, this led to the self-assembly of what, in this article, we call immature VLPs or imVLPs. Mature VLPs (mVLPs) were obtained by incubating imVLPs at 37°C for 24 hr. The homogeneity and integrity of the VLPs were substantially improved, which was verified by negative-stain TEM (Figure 1B, middle panel; the diameter of most particles is ~60 nm), and AUC yielded a prominent peak at 140S (Figure 1C, middle panel). The hydrodynamic radii (particles encircled by layers of water molecules) of the imVLPs and mVLPs measured by dynamic light scattering (DLS) were 30 and 33 nm, respectively (Figure 1D, left and middle panels). In addition, the polydispersity of mVLPs (%Pd = 14.8) was notably smaller than that of imVLPs (%Pd = 20.8), which indicates that the mVLPs exhibit much better size homogeneity than the imVLPs (Figure 1D). Therefore, we conclude that incubating imVLPs at 37°C for 24 hr improved significantly the homogeneity of the VLPs. This was consistent with the fact that mVLPs contain more disulfide bonds than imVLPs (Figure S1B). However, It should be noted that this result is not consistent with previous studies on the maturation of HPV pseudo-virus in which mature capsids were smaller than immature ones (Buck et al., 2005; Cardone et al., 2014). A possible explanation for this discrepancy may be that the minichromosome in the virion exerts attractive forces on the L1 subunits, which then condenses the capsid. However, in “empty” VLPs where the L1 subunits have no DNA or minichromosome with which to interact, the capsid may achieve a stable energy-minimization state at a particle size larger than virions.
It is known that inter-pentamer disulfide bonds play a key role in the self-assembly of complete HPV particles (Ishii et al., 2003). To obtain a highly purified, homogeneous sample of HPV59 L1 pentamers for crystallization experiments, we generated the C175S HPV59 L1 mutant. This protein was digested with trypsin for the purpose of generating pentamers that could not assemble into particles (Li et al., 1997). The digested L1 proteins were analyzed with MALDI mass spectra, and the existence of two fragments (44 and 5 kDa) was confirmed (Figures S1C and S2A). Individual pentamers were directly visualized by negative-stain TEM and their integrity was confirmed by AUC and DLS (Figures 1B–1D, right panels). The c(s) profile of the pentamer sample showed a single component of 9.3S, which is consistent with a molecular mass of 242 kDa (each L1 monomer is a molecule of ~50 kDa) (Figures 1C [right panel] and S1D). Taken together, with regulatory digestion of trypsin, the HPV59 L1 mutant C175S forms highly homogeneous pentamers but could not assemble into VLPs.
HPV59 L1 Pentamer Crystal Structure
The crystal structure of the HPV59 L1 pentamer was determined at 4.0-Å resolution with final Rfactor and Rfree values of 24.3% and 26.7%, respectively (Table 1). The atomic model of the HPV18 L1 pentamer (PDB: 2R5I; Bishop et al., 2007) was used as the search model for molecular replacement, and a solution was obtained that consisted of two pentamers stacked head-to-head in the asymmetric unit. This allowed us to apply ten-fold non-crystallographic symmetry (NCS) restraints during structure refinement. The side chains of most amino acids were easily recognized in the map, and all ten monomers in the asymmetric unit consisted of two separated fragments (amino acids [aa] 20–405 and 439–473) owing to trypsin digestion and/or crystallographic disorder (Figures 1A, 2A, 2E, and S2A), which is consistent with the mass spectrometry analysis (Figure S1C).
Table 1.
Diffraction Data Collection and Refinement Statistics
| Data Collection | |
|---|---|
| Cell parameters (Å, °) |
a = 114.2, b = 161.7, c = 154.7 β = 110.3 |
| Space group | P21 |
| Resolution rangea (Å) | 50.0–4.00 (4.07–4.00) |
| Wavelength (Å) | 0.97923 |
| Observed hkl (I > σ) | 246,801 |
| Unique hkl | 44,094 |
| Redundancy | 5.6 (5.6) |
| Completeness (%) | 100.0 (100.0) |
| Overall (I/σI) | 7.8 (2.6) |
| Rsymb (%) | 28.3 (85.4) |
| Refinement | |
| Resolution range (Å) | 49.7–4.0 |
| No. of reflections | 44,074 |
| R factorc | 24.3 |
| Rfreed | 26.7 |
| RMSD bond lengths (Å) | 0.004 |
| RMSD bond angles (°) | 0.69 |
| Wilson B factor (Å2) | 85.6 |
| Average B factors (Å2) | 121.6 |
| Ramachandran Plot (%) | |
| Favored and allowed regions | 90.4 |
| Generously allowed regions | 8.7 |
| Disallowed regions | 0.9 |
Numbers in parentheses refer to the highest-resolution shell.
Rsym = Σh Σi|I1(h) − < I(h)|/Σh Σi I1(h).
R factor = Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs.
Rfree is calculated using the same equation as that for R factor, but 5.0% of reflections were chosen randomly and omitted from the refinement.
Figure 2. Crystal Structure of the HPV59 L1 Pentamer.
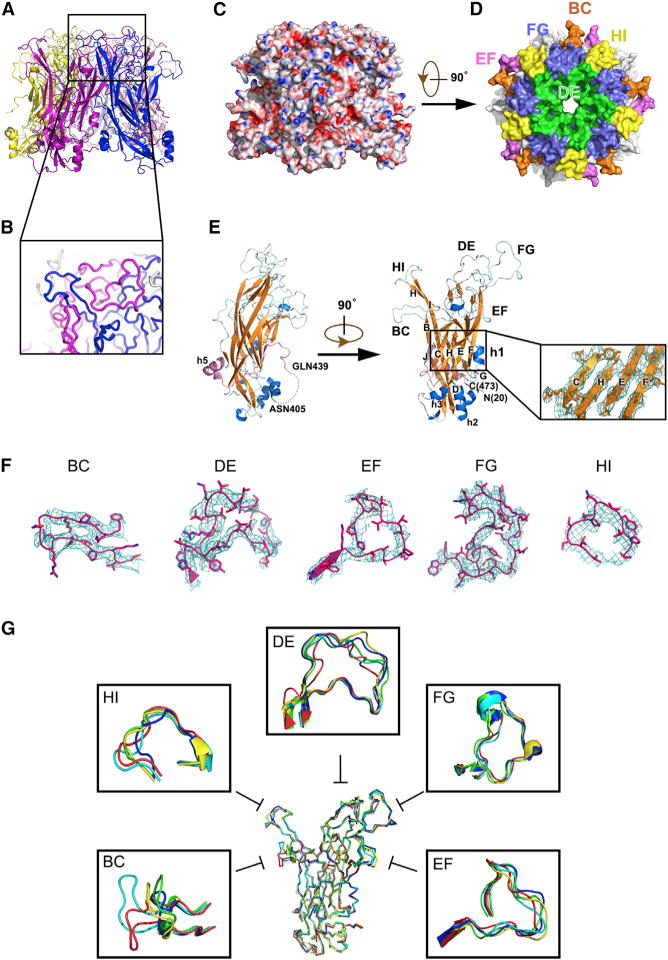
(A) Ribbon diagram of the HPV59 L1 pentamer crystal structure viewed from the side and top, respectively.
(B) Close-up view of the boxed region in (A).
(C) Side view of HPV59 L1 pentamer rendered as an electrostatic potential surface: red, blue, and white colors represent negative, positive, and neutral charge, respectively.
(D) Surface representation of the HPV59 L1 pentamer with surface loops in different colors: BC (orange), DE (green), EF (pink), FG (slate), HI (yellow).
(E) L1 monomer. The first and last residues are labeled N(20) and C(473). The β strands are colored orange and the α helices are in blue (except for h5). The connecting loops are in cyan. The C-terminal region of L1 monomer (aa 439–473) is colored pink. Strands and helices are labeled the same as the crystal structure of small HPV16 L1 VLP (PDB: 1DZL). The dotted line in the left panel signifies the missing fragment in the crystal structure between ASN405 and GLN439 due to trypsin treatment and crystallography disorder, which should be rebuilt in the complete VLP model. The inset shows the core region of L1 monomer (CHEF sheet), the 2Fo–Fc map of which is displayed at the contour level of 1.5σ.
(F) Close-up view of the five flexible surface loops of HPV59 L1. The 2Fo–Fc maps are displayed as cyan mesh at the contour level of 1.0σ.
(G) Comparison of loops in five different HPV serotypes: HPV59 (red), HPV11 (blue), HPV16 (green), HPV18 (cyan), and HPV35 (yellow).
In the crystal, the HPV59 pentamer consists of five L1 monomers and the morphology of the pentamer is characterized by a prominent, hollow lumen along the central, five-fold rotational axis (Figures 2A and 2D). Each monomer contains ~7,141 Å2 buried surface area (~31% of total surface area, calculated by PISA [Krissinel and Henrick, 2007]) and a twisted coiled coil at the interface region between two neighboring monomers (Figure 2B). The main core structure of the monomer is a canonical, eight-strand β barrel (BIDG-CHEF) with the strands joined by six loops (BC, CD, DE, EF, FG, and HI), five of which (all but CD) are located on the surface of the pentamer (Figures 2D, 2E, and S2A). The electron density of the β-barrel core domain was well defined (Figures 2E [inset] and S2B) and all of the external loops were also clearly resolvable in the electron density map (Figures 2F and S4B). Despite the discontinuity caused by trypsin treatment, the C-terminal fragment (aa 439–473) harbors a short strand (βJ) and α helix (h5, as numbered in the crystal structure of T = 1 HPV16 L1 VLP, PDB: 1DZL [Chen et al., 2000]). This fragment intimately anchors to the body of aa 20–383 via interaction between strand βJ in the C-terminal tail and strand βC of the CHEF sheet in the core domain (Figure 2E).
The crystal structure of HPV59 L1 essentially resembles the crystal structures of the L1 proteins in HPV11, 16, 18, and 35 (Bishop et al., 2007). The root-mean-square deviations (RMSDs) of the HPV59 L1 coordinates with the coordinates of the other four HPV L1 structures are 1.6, 1.5, 1.6, and 1.7 Å, respectively. The core β-sheet structures of the five pentamers share essentially identical conformations: their RMSDs of the coordinates with respect to HPV59 are 0.6, 0.7, 0.5, and 0.7 Å, respectively. However, the surface-loop regions, which are believed to be associated with type specificity and viral antigenicity (Bishop et al., 2007), differ significantly (Figure 2G and Table S1). The electrostatic map of the HPV59 L1 pentamer shows that basic amino acids are mainly located on the outer surface of the capsomers (Figure 2C), and some of them might be involved in binding to the negatively charged HSPG receptor during the initial infection process (Dasgupta et al., 2011; Richards et al., 2013).
Cryo-EM Reconstruction of HPV59 L1-Only mVLPs
Highly purified HPV59 mVLPs were obtained by means of size-exclusion chromatography. Low-dose images of unstained, vitrified mVLP samples exhibited homogeneous particles with an average diameter of ~60 nm (Figure 3A). The final 3D density map (computed from ~3,100 individual particle images), shows that the VLP capsid, like that of HPV16 and BPV virions (Baker et al., 1991; Trus et al., 1997; Wolf et al., 2010), comprises 72 capsomers arranged in a T = 7d (Baker et al., 1991) icosahedral lattice (Figure 3B).
Figure 3. Cryo-EM and 3D Image Reconstruction of HPV59 VLPs.
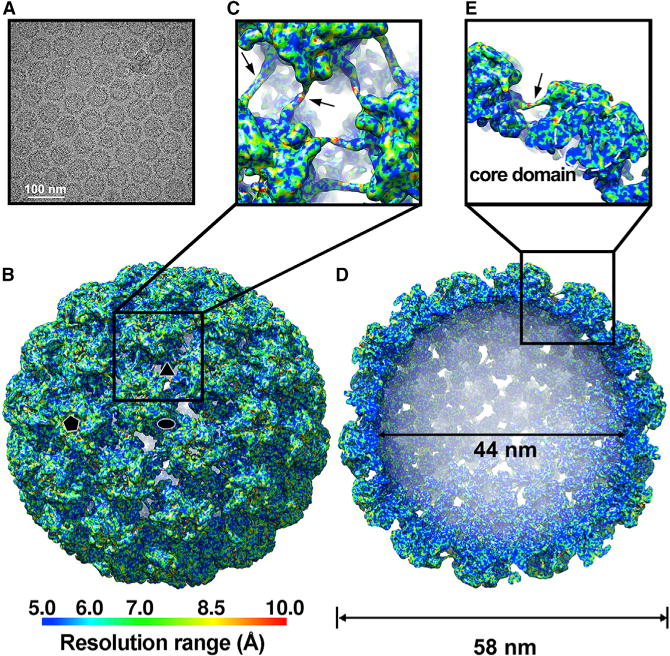
(A) Micrograph of an unstained, vitrified HPV59 VLP sample (mVLPs). Scale bar, 100 nm.
(B) Reconstructed cryo-EM map of the HPV59 VLP, viewed along the icosahedral two-fold axis. The resolution is color-coded for different regions of the HPV59 VLP. Icosahedral two-, three-, and five-fold axes are indicated by black symbols.
(C) Magnified view of the boxed region in (B).
(D) Same as (B) with the closest half of the density map removed to reveal internal features of the HPV59 VLP. The internal and outer diameters are 44 and 58 nm, respectively.
(E) Close-up view of the boxed region in (D). The dashed circle highlights the core domain of the HPV59 L1 capsomer.
Black arrows in (C) and (E) indicate the density of the “suspended bridges.”
The cryo-EM density map of HPV59 has an estimated overall resolution limit of 7.4 Å and 5.4 Å determined by gold-standard Fourier shell correlation (FSC) cutoff threshold at 0.5 and 0.143 (Figures 3B, 3D, and S3A). The local-resolution distribution of the map was estimated with the program ResMap (Kucukelbir et al., 2014) (Figures S3B and S3C). The resolution of major components of our map is around 6 Å, while a significant portion of the core region can even reach close to 5 Å (Figures 3E and S3C). The outer surface region of the capsid reaches the resolution range of 8.5–10 Å, suggesting their significant flexibility (Figures 3B, S3B, and S3C). However, the effective resolution was estimated based on the 0.5 criteria of FSC between the cryo-EM structure and the pseudo-atomic model derived from the density map. The calculated FSC goes down to 0.5 at 6 Å (Figure S3A).
The cryo-EM reconstructions of HPV VLPs and HPV16 pseudo-virion share some similar features but also some discernible differences aside from genotype distinctions. Individual L1 monomers can be identified, and the “suspended bridges” (Cardone et al., 2014) between neighboring pentamers are clearly resolved in the HPV59 VLP map (Figures 3C, 3E, and 4E). The C-terminal arm in the atomic model of T = 1 HPV16 particles contains a segment of α helix (Chen et al., 2000), but we observed no α-helix density in the C-terminal arm in the HPV59 VLP density map. This difference indicates that variability in the secondary structures of the C-terminal arm might, at least in part, direct inter-pentamer associations, and thereby the lattice arrangement (T = 1 or 7) of the VLPs.
Figure 4. Pseudo-atomic Model of the HPV59 Capsid.
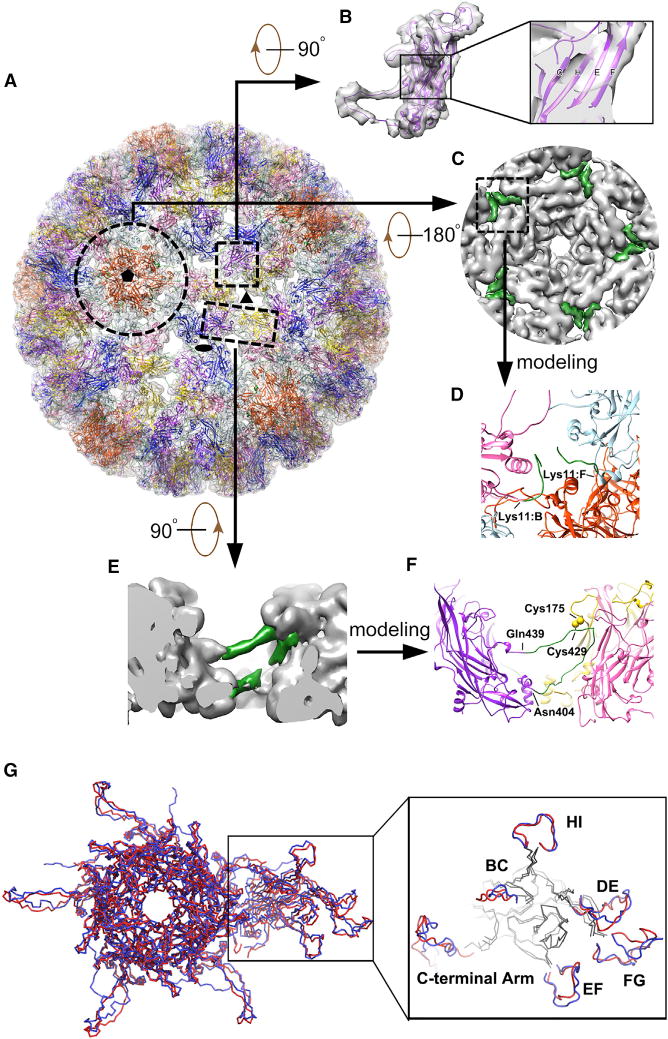
(A) Complete pseudo-atomic model of HPV59 (shown in ribbon representation) fitted into the cryo-EM density map and viewed along the axis of a 6-coordinated pentamer. Black ellipse, triangle, and pentagon symbols mark the positions of two-, three-, and five-fold axes of icosahedral symmetry, respectively. Color scheme to discriminate different L1 monomers in an asymmetric unit: chain A (light blue), chain B (pink), chain C (gold), chain D (purple), chain E (blue), and chain F (orange red).
(B) Density corresponding to one HPV59 L1 monomer and the fitted pseudo-atomic model of L1. The boxed region, shown in magnified form at the right, gives a close-up view of four β strands (C, H, E, F, respectively) in the core domain of the L1 monomer.
(C) Inside view of the 5-coordinated pentamer outlined by the broken circle in (A). Green regions, corresponding to the difference densities between the fitted crystal structures and cryo-EM map, were used to trace and build the N-terminal region of chains B and F.
(D) Close-up view of the region boxed in (C), representing the N-terminal models of chains B and F derived from the green-colored density in (C).
(E) Close-up side view of two neighboring capsomers related by icosahedral three-fold symmetry. The region colored green was the difference density between the fitted crystal structure and cryo-EM map, and was used to build the model of the C-terminal arm.
(F) The same region as (E) but shown in ribbon diagram. The part in green refers to the manually built part of the model.
(G) Comparison of the asymmetric units in models of HPV59 (red) and BPV (blue). The outlined region on the right highlights structural differences in the various surface loops between these two models. The BPV model is from a cryo-EM reconstruction (Wolf et al., 2010) (PDB: 3IYJ).
Pseudo-atomic Model of the Complete HPV59 L1 Capsid
The asymmetric unit of the HPV59 VLP contains six L1 monomers (chains A–F, Figure 4A) of which five (chains A–E) contributed to the 6-coordinated pentamer and one (chain F) to the 5-coordinated neighboring pentamer located at the icosahedral five-fold axis (Buck and Trus, 2012; Wolf et al., 2010). A pseudo-atomic model of the HPV59 VLP was obtained starting from a rigid-body fit of the crystal structure of one L1 monomer independently into each subunit of seven adjacent pentamers, segmented out from the density map (see Experimental Procedures and Movie S1). A good correlation coefficient value (average ~0.56 for all 35 monomers) of this initial fitting indicated that the structure of pentamers was consistent both in crystals and in assembled VLPs. The β-sheet core structure of L1 monomer had been well resolved in the density map (Figure 4B). Next, we computed a difference map by subtracting the density attributed to the fitted L1 crystal structures from the cryo-EM density map (Figures 4C and 4E). The densities observed in this difference map were attributed mainly to three missing segments in the crystal structure: N-terminal (aa 10–19), trypsin-cleaved loop (aa 406–438) (C-terminal arm), and C-terminal tail (aa 474–508) (Figure 1A). This difference map enabled us to trace most of the missing segments: N-terminal regions in only chains B and F; and trypsin-cleaved loops (referred to as C-terminal arm) in all six chains of the asymmetric unit (Figures 4C–4F). Densities attributed to the “suspended bridge” (exposed surface part of the C-terminal arm) were clearly visible in our cryo-EM map, which allowed us to confidently build the model in this region (Figures 4E and 4F). Therefore, the pseudo-atomic model of HPV59 VLP demonstrated that it also adopts an “invading and reinsertion” assembly mechanism similar to that ascribed to HPV16 and BPV (Cardone et al., 2014; Wolf et al., 2010). However, we were unable to assign with confidence the C-terminal tails, since the relevant densities were too weak or too disconnected to be traced unambiguously.
It is important to note that, consistent with the pseudo-atomic model of the T = 7 HPV16 capsid (Cardone et al., 2014), we also built a random coil instead of an α helix at the C-terminal arm, since the density in that region is much weaker than in other well-resolved α helices in the map. Biochemical evidence has shown the existence of an inter-pentamer disulfide bond between the cysteine at the C-terminal arm and the cysteine at the jellyroll β barrel (Modis et al., 2002; Sapp et al., 1998). The Cys175 in HPV59 L1 protein was mutated into alanine (C175A) and serine (C175S), respectively. Similarly, Cys429 was mutated into C429A and C429S, respectively. The TEM inspection of these four mutants showed that they mainly stayed in the form of capsomers but were unable to correctly assemble into spherical forms of VLP capsid (Figure S1A). Moreover, the amount of disulfide bonds in these four mutants was significantly lower than that of “native” HPV59 L1 VLPs (Figure S1B). Hence, both Cys175 and Cys429 play critical roles in bonding neighboring capsomers to form an intact HPV capsid. Therefore, we positioned Cys429 close to the Cys175 in the neighboring pentamer in our model (Figure 4F). In brief the C-terminal arm, which mediates the interaction between adjacent pentamers, extends from the bottom of the β barrel and then loops toward the crevice (the space between loop BC of one monomer and loop EF of the other) between two monomers from the neighboring pentamers (aa 413–418). Then the “wall region” wedges as a random coil (aa 419–428) into the crevice. After this, the link between neighboring pentamers is further strengthened by the disulfide bond between Cys429 in the C-terminal arm and Cys175 in the EF loop of its target pentamer. The arm then loops back toward the midsection of the β barrel from which it originated and forms the “suspended bridge” (aa 430–437), and goes down toward its base (Figure 4E).
The N-terminal arm (aa 1–20) at the base of the protein shell has also been shown to contribute contacts between pentamers in human and bovine papillomaviruses (Chen et al., 2000; Wolf et al., 2010). In our cryo-EM map, only the N-terminal arms (aa 10–19) in chains B and F could be traced while building the pseudo-atomic model (Figures 4C and 4D). In this model, the elbow-like N-terminal arm of chain F (located close to the icosahedral five-fold axis) lies adjacent to that arm of chain B in the neighboring 6-coordinated pentamer (Figure 4D). The association of these two chains contributes to the pairwise interactions between 5- and 6-coordinated pentamers. However, the other four chains showed no clearly identifiable densities at their N-terminal regions. This may be a consequence of chains B and F having more stabilizing interactions with one another compared with any others (see Discussion).
Following the initial rigid-body docking of the L1 crystal structures into the HPV59 cryo-EM map and the manual building of missing segments, a pseudo-atomic model of the asymmetric unit of the T = 7 VLP structure was refined by means of MDFF procedures (Figures 4A and S4A). This refinement led to an improvement in the local cross-correlation coefficient between model and map from ~0.5 to ~0.7 (Figures S4B–S4E and Table S2), and the RMSD between the models before and after MDFF of Cα is 2.38 Å (Figure S5 and Table S2). A reliable pseudo-atomic model of HPV59 allowed us to carefully compare the HPV59 structure with that of BPV (Figures 4G and S6). Both papillomaviruses have major capsid proteins with very similar, β-barrel core structures. The prominent structural differences occur in the surface loops and extended C-terminal loops (Figures 4G and S6).
C-Terminal Arm of HPV59 Is Involved in Virus-Host Interaction
Although HPV59 and BPV share strikingly similar L1 core structures, the relative arrangement of the BC, DE, EF, FG, and HI surface loops and the C-terminal arm are quite distinct (Figures 4G and S6; Table S3). Not surprisingly, based on the sequence alignments of 36 HPV serotypes (Figure 5A), the five loops and structural domains represent regions of high variability (Figure 5B). In a VLP-based cell binding assay, the HPV59 L1 VLPs attached to but did not penetrate into the cellular membrane of fixed HaCaT cells (Figure 6A). We designed and synthesized eight peptides to probe how these surface loops might be involved in virus-host interactions. These peptides included the BC (aa 50–66), DE (aa 120–147), EF (aa 170–187), FG (aa 263–290), and HI (aa 348–360) loops, the C-terminal arm (CTA; aa 414–438), 19CTA (aa 420–438, which includes the “suspended bridge” and wall region), and 8CTA (aa 431–438, which includes the “suspended bridge”). The N termini of all eight peptides were labeled with fluorescein isothiocyanate (FITC) for the cell binding experiments. The CTA loop and a truncated version of it, 19CTA, bound to the cellular membrane, but none of the other loops showed any detectable binding activity. It is noteworthy that the other truncated CTA peptide, 8CTA, did not bind to cells, which suggests that a specific local conformation might be necessary to maintain binding activity (Figure 6B).
Figure 5. Amino Acid Sequence Alignments of Different HPV L1 Proteins and Location of Variable Regions on the Monomer Surface.
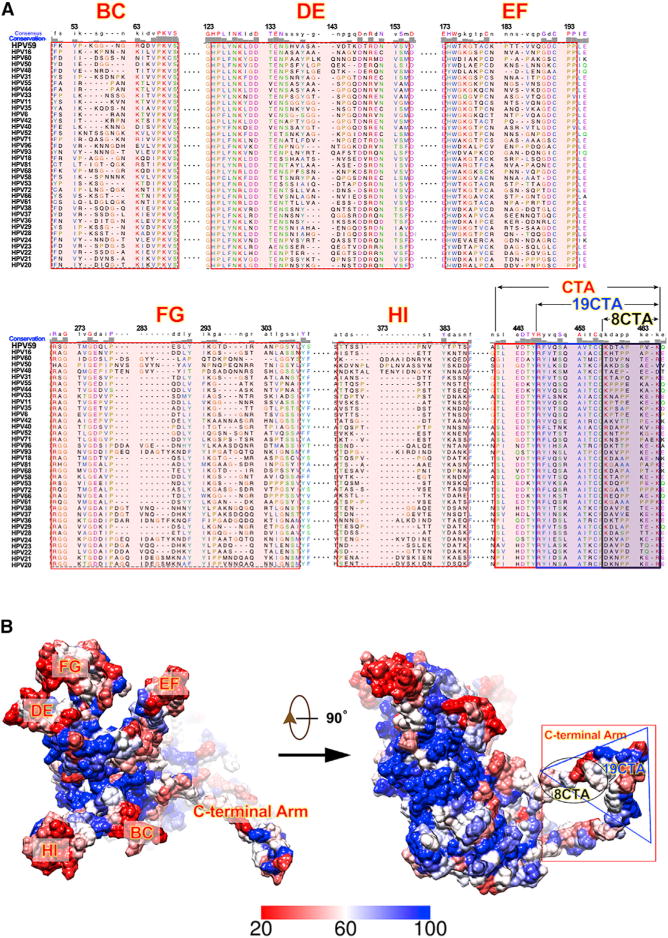
(A) Sequence alignments of the L1 proteins from 36 different HPV serotypes. The six variation regions (BC, DE, EF, FG, HI, and C-terminal arm) are highlighted in red. Alignments were determined and plotted using Chimera (Pettersen et al., 2004).
(B) Locations of regions with high sequence variation on the HPV L1 surface. The magnitude of sequence conservation based on the sequence alignments in (A) are mapped onto the surface of the HPV59 L1 monomer (left: top view; right: side view). Highly conserved regions (60%–100% conservation) are color-coded from white to blue, whereas less conserved regions (from 20% to 60%) are represented coded from red to white.
Figure 6. Variation C-Terminal Arm of HPV59 L1.
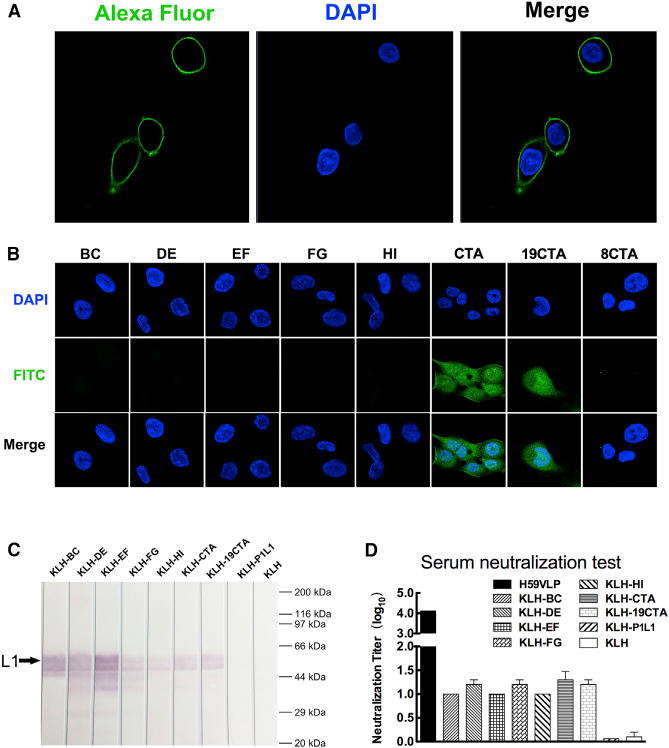
(A) Immunofluorescence assay for detecting the cell binding activity of HPV59 VLPs. The DAPI blue fluorescent stain identifies nuclei, and the green Alexa Fluor-labeled anti-mouse antibodies identify mAb 26D3 that binds specifically to HPV59.
(B) Binding assay of HaCaT cells for eight peptides derived from exposed regions of L1. Three of these peptides include portions of the C-terminal invading-arm.
(C) Western blotting assay of HPV59 L1 by the antisera. The black arrow points to the L1 band.
(D) Neutralization assay of antisera of HPV59 VLP, KLH-BC, KLH-DE, KLH-EF, KLH-FG, KLH-HI, KLH-CTA, KLH-19CTA, KLH-P1L1, and KLH. The P1L1, a peptide representing the extracellular domain the latent membrane protein of human herpesviruses (Delbende et al., 2009), was used as a negative control for the assay. Every experiment was repeated independently three times, and the error bar above the mean value denotes the SD of individual tests.
In contrast to VLPs, which remain localized on the cellular membrane, the peptides CTA and 19CTA are able to attach to and penetrate into host cells. The seven peptides (all aforementioned peptides but not 8CTA) were fused respectively to an adjuvant protein, keyhole limpet hemocyanin (KLH), to generate antiserum in mice. The specificity of antiserum reacting with HPV59 L1 was verified by ELISA and western blotting (Figures 6C, S7A, and S7B). In the ELISA test, KLH-CTA elicited antibody titer one log higher than KLA-19CTA. In addition, we also measured the neutralization titer of the antiserum using a pseudo-virion-based cell neutralization assay. The mean neutralization titer of CTA and 19CTA was about 20, with statistical significance (p < 0.05) compared with KLH alone, although the titer was lower than that of HPV59 VLPs (Figure 6D). The titers of CTA and 19CTA are similar to that of the surface loops (BC, DE, EF, FG, and HI) (Figure 6D), which revealed the C-terminal arm to be equally important to other known loop regions as a key neutralization site (Christensen et al., 1996; Guan et al., 2015b; Lee et al., 2014; Roden et al., 1997). Furthermore, a panel of such neutralizing monoclonal antibodies (mAbs), generated by immunizing HPV59 VLPs, demonstrated much lower binding ability with the assembled HPV59 pentamers (without the C-terminal arms) (Figure S7C). This result indicated that the neutralization sites defined by those mAbs were related to the C-terminal arm. This finding is also consistent with that of a previous study, i.e., an HPV16 neutralizing antibody, H16.U4, recognizes the “suspended bridge” and can preclude HPV16 capsids from attaching to the HaCaT cells (Guan et al., 2015a). Overall, the C-terminal arm, which links neighboring pentamers together, is at least one primary site of virus-host interaction, and some of the antibodies that recognize this arm can neutralize the virus.
DISCUSSION
Maturation is a common process by which virus capsids gain stability. Studies of HPV16 capsid assembly revealed that heat treatment increases the formation of disulfide crosslinks between neighboring capsomers (Buck et al., 2005; Cardone et al., 2014; Zhao et al., 2002). For the HPV59 VLPs used in this study, we also found that particles incubated for 24 hr or more were much more homogeneous and their average size was larger than the particles not going through the maturation process. We also found that the amount of disulfide bond components in mVLPs was higher than that in imVLPs (Figure S1B), which indicated that disulfide bond formation is part of the maturation process. The disulfide bonds associate the neighboring pentamers and stabilize the whole capsid; therefore, the mVLP particles present more homogeneity than imVLPs.
We contend that only after we improved the homogeneity of the HPV59 VLPs by maturation treatment was it possible to reconstruct a 3D density map, albeit at medium resolution. Even after the maturation procedure, there are still many percentages of the particles not fully matured and the full complement of intermolecular disulfide bonds were not attained, as occurs in HPV16 and BPV virions (Buck et al., 2005; Cardone et al., 2014; Wolf et al., 2010). Therefore, building a high-quality density map necessitates an effective method to pick out those “fully matured and shiny” particles. The program Relion allowed us to perform such a task with both 2D and 3D classification. Finally, ~3,100 were selected from more than 20,000 particle images and were included in computing the cryo-EM density map of HPV59 L1-only capsid. The core domain of L1 is clearly strong in our density map (Figures 3E and 4B) and the density of the “suspended bridge” was very strong (Figures 3C, 3E, and 4E). It is possible to trace both the N-terminal (chains B and F) (Figure 4D) and C-terminal regions (all chains) that link adjacent pentamers.
It is noteworthy that the N-terminal arms of the other four chains in the asymmetric unit cannot be modeled because there are no identifiable densities to assign to these regions. This difference might be due to the fact that the first nine aa are missing in our construction of L1 sequence in HPV59 VLP. In the BPV atomic model (PDB: 3IYJ; Wolf et al., 2010), the segment corresponding to the first 14 residues of chains B and F was not identified owing to density features being very weak or absent in these regions, although these same segments in the other four chains were clearly resolved. This distinction indicates that the stable and consistent interactions involving the N-terminal arms might be mainly located beyond the first 14 aa in chain B and F, whereas they might involve the whole leading segments in the case of the other four chains (Wolf et al., 2010). Based on high sequence and tertiary structural similarities between BPV and HPV, it is reasonable to postulate that the N-terminal arms of HPV59 and BPV interact in similar fashion. Therefore, the missing initial nine residues in the recombinant HPV59 L1 protein might selectively disrupt otherwise stable interactions and induce flexibility at the N-terminal regions of chains A, C, D, and E while not affecting chains B or F. Such flexibility could be one factor that has prevented us from achieving higher resolution in our current cryo-reconstruction.
The C terminus (~30 aa) of the HPV16 L1 protein is rich in basic residues as well as serines and threonines, and was shown to contribute to the association between the minichromosome and capsid (Touze et al., 2000). In genome-less VLPs, the C-terminal tail of L1 would likely be quite flexible and might explain why the quality of the density map is too poor to build a reliable atomic model of this portion of L1. However, in some studies the C-terminal tail had little impact on the overall structure of L1 (Bishop et al., 2007; Cardone et al., 2014; Chen et al., 2000). Therefore, such absence would not prevent us from successfully building a relatively precise and confident pseudo-atomic model of HPV L1 VLP.
In the HaCaT cell binding assay, we found that peptides derived from the C-terminal arm and 19CTA can attach to cells whereas the stretch of residues corresponding to the “suspended bridge” (aa 431–438, 8CTA), which is the most exposed region of the VLP, did not bind to cells (Figure 6B). This implies that binding activity might depend significantly on the local conformation of the exposed, inter-pentameric link, and the “suspended bridge” on its own may be too short to maintain a required conformation. Peptides that exhibit positive cell binding activities are able to elicit neutralizing antibody responses. Therefore, identification of epitopes involved in interactions between L1 and neutralizing mAbs or those amino acids involved in HPV and host-cell interaction offer a promising means to locate HPV cell receptors and elucidate the mechanism of HPV infection.
EXPERIMENTAL PROCEDURES
Preparation and Maturation of L1 VLPs
The full-length L1 gene of HPV59 was synthesized (Invitrogen) according to the deposited sequence (GenBank: CAA54856.1). A series of progressively longer, N-terminal truncations were tested to yield highly efficient expression in E. coli. This led to a nine-residue truncation mutant (aa 10–507) that was chosen as the best construct for this study. The genes amplified by PCR were first cloned into the pMD 18-T vector using a TA cloning kit (Takara) and subsequently subcloned into the non-fusion pTO-T7 vector. E. coli strain ER2566, used for protein expression, was induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside for 10 hr at 25°C. After harvesting, cells were re-suspended in buffer (50 mM Tris-HCl [pH 7.2], 10 mM EDTA, 300 mM NaCl) and disrupted by sonication. The supernatant was purified by two chromatographies using SP Sepharose (GE Healthcare) and CHT II resin (Bio-Rad). Following chromatography, the L1 proteins were dialyzed into the above buffer without reducing agent to allow VLP self-assembly. The maturation of HPV59 L1-only VLPs was performed immediately after 24 hr dialysis at room temperature in the absence of reducing agent. The immature VLPs were moved into the same buffer at 37°C and incubated for another 24 hr, then chilled at 4°C for 15 min. Both immature and mature L1-only capsids were purified by size-exclusion chromatography. The C175A, C175S, C429A, and C429S point mutations were constructed from the clone of HPV59 L1DN-9 with site-directed PCR reaction. The mutants of L1 were purified and put through the self-assembly process in the same manner as the preparation of VLPs. For characterization by TEM, the L1 protein was spotted on glow-discharged, carbon-coated grids (EM Science) and stained with 2% (w/v) uranyl acetate, and viewed under a JEM2100 electron microscope. Other characterizations of HPV pentamer and VLP were performed by SDS-PAGE, western blotting, AUC, DLS, MALDI-TOF mass spectrometry, disulfide bonding quantification, and ELISA. For further details, refer to Supplemental Experimental Procedures.
Preparation and Crystallization of HPV59 L1 Pentamer
To prepare pentamers for crystallization, we first purified the HPV59 L1 C175S mutant through a series of chromatography columns as described above for the preparation of VLPs. The mutant protein was then subjected to limited trypsin digestion (trypsin to L1 ratio of 1:1,000) at room temperature overnight, purified by Superdex 200 (GE Healthcare), and concentrated to ~5 mg/ml for crystallization trials. We used the hanging-drop vapor diffusion method to obtain brick-shaped crystals of the HPV59 L1 pentamer at 20°C in the presence of 0.2 M magnesium acetate, 0.1 M sodium cacodylate (pH 6.5), and 10% polyethylene glycol 8000. Crystals grew to a maximum size of 0.3 × 0.1 × 0.05 mm over a 1-week period.
X-Ray Diffraction Data Collection, Processing, and Structure Determination
Crystals were cryoprotected in the reservoir solution, which was supplemented with 30% glycerol, and flash-cooled at 100 K. Diffraction data from the HPV59 L1 pentamer crystals were collected at the Shanghai Synchrotron Radiation Facility beamline BL17U using a Quantum-315r CCD Area Detector. All of the datasets were processed using the HKL-2000 program package (http://www.hkl-xray.com). The structures were solved by molecular replacement with PHASER using chain A of PDB: 2R5I as the search model. The models were built manually in Coot (Emsley and Cowtan, 2004), refined using Phenix (Adams et al., 2010), and analyzed with MolProbity (Chen et al., 2010). In brief, one round of rigid-body refinement was performed after molecular replacement. After manual modification of the refined model in Coot, coordinates and individual B factors were refined in reciprocal space with NCS restraints and reference-model restraints to avoid overfitting. The reference model was the 2.8-Å HPV16 L1 structure (PDB: 3OAE). TLS refinement was performed in the later stages with each monomer as a TLS group. Statistics for the data collection, processing, and structure refinement for the HPV59 L1 pentamer are summarized in Table 1. Ramachandran plots generated by MolProbity also stereochemically validate the side chains of amino acids (Figure S2C).
Cryo-EM and 3D Reconstruction of HPV59 L1 VLPs
Purified, mature, L1-only HPV59 VLP samples at a concentration of ~3 mg/ml were vitrified on Quantifoil holey carbon grids in an FEI Vitrobot. Images were recorded on an FEI Falcon I direct detector camera at 93,000 nominal magnification in an FEI TF30 FEG microscope at 300 kV, with underfocus settings estimated to be between 1.0 and 3.0 μm, and with an electron dose of 25 e/Å2. A total of ~20,000 particle images out of 1,800 micrographs were manually boxed and extracted with the program Robem (Yan et al., 2007b). The origin and orientation parameters for each of these particle images were estimated by means of model-based procedures (Yan et al., 2007b) and an initial model was generated with the random model method (Yan et al., 2007a). After several rounds of reference-free 2D and thereafter 3D classifications using Relion (version 1.4), 3,100 good particles were selected for further 3D refinement. The final 3D density map was computed to a resolution limit of 6Å further sharpened with an inverse B factor of 1/(250 Å 2) with the program BFACTOR (Zhang et al., 2008). Local-resolution variations were estimated using ResMap (Kucukelbir et al., 2014).
Manual Building of Missing Residues and Molecular Dynamics Flexible Fitting
A density map comprising seven pentamers representing all unique inter-pentamer interaction patterns (see Movie S1) was segmented out of the complete T = 7 VLP map using the color zone and split map tools in Chimera (Pettersen et al., 2004). We then fitted the crystal structure of one L1 monomer (extracted from the crystal structure of the pentamer determined in this study) independently into the above segmented map (tool fit in map). A difference density map was obtained by deleting those densities corresponding to the fitted seven-pentamer crystal structures (tools e2proc3d.py in EMAN2 and vop resample and vop subtract in Chimera). This difference map was used as a guide to “patch” the three missing segments onto the crystal structure: N terminus (aa 10–19), trypsin-cleaved loop (aa 406–438), and C-terminal tail (aa 474–508). This “patching” procedure was performed manually with the program Coot (Emsley and Cowtan, 2004). To ensure the model obeyed the reasonable stereochemical constraints, we used Regularize Zone with planar peptide restraints (refinement weight of weight matrix is 60.00) instead of Real Space Refine Zone based on the cryo-EM map. The preliminary pseudo-atomic model of the HPV59 L1 VLP was then obtained by imposing icosahedral symmetry on one asymmetric unit (five monomers in a 6-coordinated pentamer plus one monomer in the 5-coordinated pentamer at the icosahedral fivefold axis). These same seven pentamers were re-extracted and further refined using MDFF procedures (Trabuco et al., 2008, 2011): first, the model was loaded into cryo-EM map with rigid-body docking; then it was solvated in a box of water molecules with 150 mM NaCl and 10 Å of padding in all directions using the visual molecular dynamics program (Humphrey et al., 1996); counterions were added to neutralize the simulated system, which was bound by a 310-Å wide cubic box and a total of 253,252 atoms; simulation was performed with NAMD 2.10 (Phillips et al., 2005) using the CHARMM27 force field (Mackerell et al., 2004); finally, the asymmetric unit was extracted from the seven-pentamer model and was used to create the final pseudo-atomic model of HPV59 VLP with icosahedral symmetry operation. Electron density maps of the atomic models were computed using the e2pdb2mrc.py program in EMAN2 (Tang et al., 2007), and the FSC curve between our final model and the cryo-EM map of HPV59 L1 were computed using the e2proc3d.py program in EMAN2 (Tang et al., 2007).
Immunofluorescence
For VLP-cell binding assays, HaCaT cells (1.56 × 105 cells per well) were seeded on sterile glass coverslips in 24-well plates. The wells were rinsed on the following day with PBS and the cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Coverslips were then washed three times in PBS. The mAb H59.26D3 was used to detect the bound VLP. Fluorophore-labeled secondary antibodies were anti-mouse Alexa Fluor 488 immunoglobulin G (Thermo Fisher Scientific). All immunofluorescence images were recorded in a Zeiss LSM780 spectral confocal microscope.
Peptide Synthesis
Eight peptides, BC (F50KVPKGGNGRQDVPKVS66), DE (H120PLYNKLDD TENSHVASAVDTKDTRDNV147), EF (T170KGTACKPTTVVQGDCPP187), FG (R263SGTMGDQLPESLYIKGTDIRANPGSYL290), HI (T348TSSIPNVYTPTS360), CTA (S414LVDTYRFVQSAAVTCQKDTAPPVK438), 19CTA (R420FVQSAAVTC QKDTAPPVK438), and 8CTA (K431DTAPPVK438), according to the L1 sequence of HPV type 59 (GenBank: CAA54856.1), were synthesized with FITC labeling. Meanwhile, all eight peptides were synthesized and conjugated to KLH for immunogenicity analysis (Sangon Biotech). Another peptide, P1L1 (MSDWTGGALCLWNLHGQ), according to the LMP1 sequence of human herpes 4 (GenBank: NC_007605.1), was also synthesized and conjugated to KLH, which was used as the negative control of the immunogenicity assay.
Peptide Binding Assays
HaCaT cells were seeded and fixed on glass coverslips as described in the aforementioned immunofluorescence experiment. Each FITC-labeled peptide was firstly re-suspended in PBS containing 10% fetal bovine serum and 3% BSA at a final concentration of 50 μg/ml. An aliquot of 200 μl was pipetted into the wells to immerse the coverslips. The 24-well plates were incubated for 1.5 hr at room temperature and then washed three times with PBS containing 0.3% BSA and 0.1% Triton X-100. The HaCaT cells binding with FITC-labeled peptide were visualized by a Zeiss LSM780 spectral confocal microscope.
Ethics Statement
Animal experiments were approved by Xiamen University Laboratory Animal Center (XMULAC). All procedures were conducted in accordance with animal ethics guidelines and approved protocols. The Animal Ethics Committee approval number was XMULAC20130200.
Animals, Immunizations, and Serological Analysis
1 mg of KLH-CTA and KLH-19CTA were mixed with Freund’s complete adjuvant at final concentration of 5 mg/ml. 200 μl of the mixture was subcutaneously immunized to BALB/c female mice, 4–6 weeks old, four mice per group. Further vaccinations were boosted on days 14 and 21 using 1 mg of each antigen mixed otherwise with Freund’s incomplete adjuvant. Either 1 mg/mouse KLH or 5 μg/mouse HPV59 L1 VLP were immunized on the same schedule, serving as negative and positive control, respectively. Serum samples collected at 1 month after three immunizations were used for immunization analysis. Detailed analysis of the immune sera is provided in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
T = 7 HPV59 L1-only capsid solved to 6 Å resolution by cryo-EM
Pseudo-atomic structure of HPV59 capsid built from 72 pentamer crystal structures
The C-terminal arm exposed on the capsid mediates the virus-host interaction
The C-terminal arm elicits neutralizing antibody in mice when conjugated with KLH
In Brief.
The structural information of HPV capsid is limited. Using crystallography and cryo-EM, Li et al. present a pseudoatomic structure of T = 7 HPV59 L1 capsid and find that the C-terminal arms are involved in virus-host interaction and elicit neutralizing antibodies in mice while conjugating to KLH.
Acknowledgments
The authors would like to acknowledge the use of beamline 17U of the Shanghai Synchrotron Radiation Facility for data collection, and also thank Dr. Jason S. McLellan (Dartmouth Medical School) for assistance with the X-ray structure determination. S.L., H.Y., Q. Zheng, Y.G., and Q. Zhao would like to acknowledge funding support from the Chinese government: National Natural Science Foundation (grant nos. 81172885, 81471934, 81371818, 81401669), National Key Projects in Science and Technology (grant nos. 2012ZX09101316, 2014AA021302), and Fujian Provincial Science Fund (grant no. 2013J05053). Work performed at UCSD was supported in part by funding to T.S.B. from the NIH (grant R37-GM33050) and from the University of California San Diego and the Agouron Foundation.
Footnotes
ACCESSION NUMBERS
Atom coordinates and structure factors for the HPV59 L1 pentamer and atom coordinates for the final pseudo-atomic model of HPV59 VLP have been deposited in the PDB (PDB: 5J6R and 5JB1, respectively). The EM density map for HPV59 VLP has been deposited in the Electron Microscopy DataBank (EMDB: EMD-8147).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, three tables, one movie, and two pdb models and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.04.008.
AUTHOR CONTRIBUTIONS
S.L. and N.X. Conceived and designed the experiments. Z.L., H.Y., D.W., S.S., Y.L., M.H., Q.H., Q. Zheng, M.E.W.J., G.C., and N.H.O. performed the experiments. Z.L., X.Y., H.Y., Q. Zhao, Y.G., J.Z., T.S.B., S.L., and N.X. analyzed the data. T.S.B., S.L., and N.X. contributed reagents, materials, and analysis tools. Z.L., X.Y., and S.L. wrote the paper.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- Broutian TR, Brendle SA, Christensen ND. Differential binding patterns to host cells associated with particles of several human alphapapillomavirus types. J Gen Virol. 2010;91:531–540. doi: 10.1099/vir.0.012732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Trus BL. The papillomavirus virion: a machine built to hide molecular Achilles’ heels. Adv Exp Med Biol. 2012;726:403–422. doi: 10.1007/978-1-4614-0980-9_18. [DOI] [PubMed] [Google Scholar]
- Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. Maturation of the human papillomavirus 16 capsid. mBio. 2014;5:e01104–01114. doi: 10.1128/mBio.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. v5-Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- Culp TD, Budgeon LR, Christensen ND. Human papillomaviruses bind a basal extracellular matrix component secreted by keratinocytes which is distinct from a membrane-associated receptor. Virology. 2006a;347:147–159. doi: 10.1016/j.virol.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol. 2006b;80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta J, Bienkowska-Haba M, Ortega ME, Patel HD, Bodevin S, Spillmann D, Bishop B, Sapp M, Chen XS. Structural basis of oligosaccharide receptor recognition by human papillomavirus. J Biol Chem. 2011;286:2617–2624. doi: 10.1074/jbc.M110.160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbende C, Verwaerde C, Mougel A, Tranchand Bunel D. Induction of therapeutic antibodies by vaccination against external loops of tumor-associated viral latent membrane protein. J Virol. 2009;83:11734–11745. doi: 10.1128/JVI.00578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccines. 2010;6:407–419. doi: 10.4161/hv.6.5.11023. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S. The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM. J Virol. 2015a;89:12108–12117. doi: 10.1128/JVI.02020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology. 2015b;483:253–263. doi: 10.1016/j.virol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Tanaka K, Kanda T. Mutational analysis of human papillomavirus type 16 major capsid protein L1: the cysteines affecting the intermolecular bonding and structure of L1-capsids. Virology. 2003;308:128–136. doi: 10.1016/s0042-6822(02)00099-5. [DOI] [PubMed] [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. A CryoEM study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol. 2014;89:1428–1438. doi: 10.1128/JVI.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cripe TP, Estes PA, Lyon MK, Rose RC, Garcea RL. Expression of the human papillomavirus type 11 L1 capsid protein in Escherichia coli: characterization of protein domains involved in DNA binding and capsid assembly. J Virol. 1997;71:2988–2995. doi: 10.1128/jvi.71.4.2988-2995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ, International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol. 2013;87:11426–11437. doi: 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden RB, Kirnbauer R, Jenson AB, Lowy DR, Schiller JT. Interaction of papillomaviruses with the cell surface. J Virol. 1994;68:7260–7266. doi: 10.1128/jvi.68.11.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden RB, Armstrong A, Haderer P, Christensen ND, Hubbert NL, Lowy DR, Schiller JT, Kirnbauer R. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol. 1997;71:6247–6252. doi: 10.1128/jvi.71.8.6247-6252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- Sadeyen JR, Tourne S, Shkreli M, Sizaret PY, Coursaget P. Insertion of a foreign sequence on capsid surface loops of human papillomavirus type 16 virus-like particles reduces their capacity to induce neutralizing antibodies and delineates a conformational neutralizing epitope. Virology. 2003;309:32–40. doi: 10.1016/s0042-6822(02)00134-4. [DOI] [PubMed] [Google Scholar]
- Salunke DM, Caspar DL, Garcea RL. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp M, Fligge C, Petzak I, Harris JR, Streeck RE. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J Virol. 1998;72:6186–6189. doi: 10.1128/jvi.72.7.6186-6189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa T, Pushko P, Steers G, Gschmeissner SE, Hajibagheri MA, Finch J, Crawford L, Tommasino M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 1995;206:126–135. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Touze A, Mahe D, El Mehdaoui S, Dupuy C, Combita-Rojas AL, Bousarghin L, Sizaret PY, Coursaget P. The nine C-terminal amino acids of the major capsid protein of the human papillomavirus type 16 are essential for DNA binding and gene transfer capacity. FEMS Microbiol Lett. 2000;189:121–127. doi: 10.1111/j.1574-6968.2000.tb09217.x. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Schreiner E, Gumbart J, Hsin J, Villa E, Schulten K. Applications of the molecular dynamics flexible fitting method. J Struct Biol. 2011;173:420–427. doi: 10.1016/j.jsb.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, Booy FP. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci USA. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stehle T, Liddington RC, Zhao H, Harrison SC. Structure determination of simian virus 40 and murine polyomavirus by a combination of 30-fold and 5-fold electron-density averaging. Structure. 1996;4:157–164. doi: 10.1016/s0969-2126(96)00019-6. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wu S, Manger W, Gadam SM. (US Patent No.: US 6436402).Process for Making Human Papillomavirus Virus-like Particles with Improved Properties. 2002:B1. [Google Scholar]
- Zhao Q, Guo HH, Wang Y, Washabaugh MW, Sitrin RD. Visualization of discrete L1 oligomers in human papillomavirus 16 virus-like particles by gel electrophoresis with Coomassie staining. J Virol Methods. 2005;127:133–140. doi: 10.1016/j.jviromet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Potter CS, Carragher B, Lander G, Sworen J, Towne V, Abraham D, Duncan P, Washabaugh MW, Sitrin RD. Characterization of virus-like particles in GARDASIL(R) by cryo transmission electron microscopy. Hum Vaccin Immunother. 2014;10:734–739. doi: 10.4161/hv.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


