Abstract
Lipophagy is defined as the autophagic degradation of intracellular lipid droplets (LDs). While the field of lipophagy research is relatively young, an expansion of research in this area over the past several years has greatly advanced our understanding of lipophagy. Since its original characterization in fasted liver, the contribution of lipophagy is now recognized in various organisms, cell types, metabolic states and disease models. Moreover, recent studies provide exciting new insights into the underlying mechanisms of lipophagy induction as well as the consequences of lipophagy on cell metabolism and signaling. This review summarizes recent work focusing on LDs and lipophagy as well as highlighting challenges and future directions of research as our understanding of lipophagy continues to grow and evolve.
Keywords: lipophagy, autophagy, lipid droplets
1. Introduction
LDs represent the most energetically dense and, often, primary form of energy storage in most cell types. These dynamic organelles form and expand or shrink and dissolve in response to changes in energy status of cells. In times of energy demand, fatty acids (FAs) liberated from LD-containing triacylglycerol (TAG) degradation are substrates for β-oxidation and ultimately the generation of ATP needed for cell survival. On an organism level, LD degradation in white adipose tissue (WAT) is critical to increase circulating FAs that provide fuel in non-adipose tissues during nutrient insufficiency. Although LDs have historically been recognized for their importance in energy storage, a growing body of literature has more recently identified important roles in cell signaling and function that link LD accumulation to the etiology of numerous diseases [1]. Thus, the LD represents a dynamic organelle that serves a multitude of functions.
Historically, the degradation of TAG and cholesterol ester (CE) stored within LDs was attributed to the actions of hormone-sensitive lipase (HSL) as first discovered over a half-century ago [2,3]. Subsequent work has revealed that HSL primarily hydrolyzes diacylglycerol, in addition to CE, which led to additional studies that ultimately identified adipose triglyceride lipase (ATGL) as the primary cytosolic lipase in numerous metabolically active tissues including adipose, heart, liver and intestine [4–6]. In response to lipolytic stimuli, ATGL is recruited to LDs to facilitate the initial step in TAG catabolism followed by subsequent reactions catalyzed by HSL and monoacylglycerol lipase. Until the discovery of lipophagy, this pathway was thought to be the primary mechanism through which TAG contained within LDs was degraded.
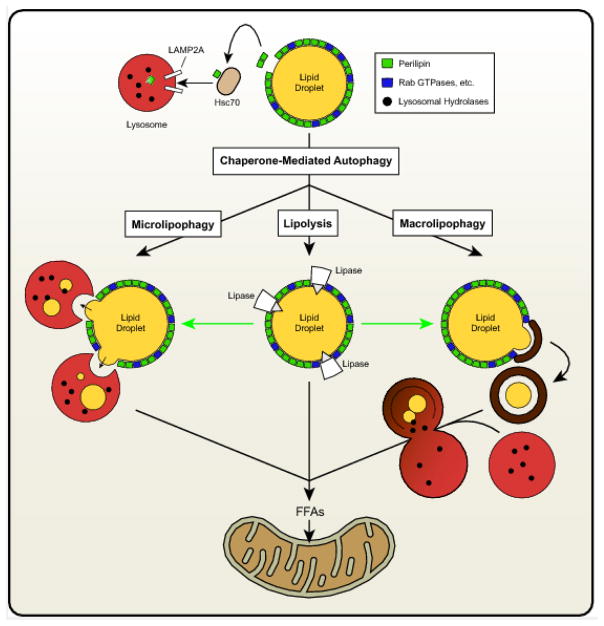
Although the effects of autophagy on degradation of various organelles has been known since the early 1960s [7,8], only recently has the contribution of autophagy to LD degradation been identified. Putative links between autophagy and LDs arose following the observation that mutations in lysosomal acid lipase (LAL), which is responsible for lysosomal lipid degradation, leads to the accumulation of LDs in various organs [9–11]. Indeed, LAL enzymatic insufficiency is the cause of Wolman Disease and Cholesterol Ester Storage Disease (CESD). However, because of the concomitant lipodystrophy and reduced hepatic catabolism of endocytosed lipoproteins in subjects lacking functional LAL, the contribution of lipophagy to LD accumulation was not known. A groundbreaking study by Singh et al. in 2009 clearly demonstrated in hepatocytes that autophagy contributes to the degradation of LDs, leading to the origin of the term “lipophagy” [12]. Similar to non-specific canonical autophagy, the selective process of lipophagy can occur via both macro- and micro- based mechanisms. Macrolipophagy involves the classical autophagosome-mediated pathway of budding off and sequestering LDs for their subsequent delivery to autolysosomes. Microlipohagy reflects the direct and transient interactions of lysosomes with LDs as a means to degrade LD-derived lipids. Chaperone-mediated autophagy (CMA), another arm of autophagy involving targeted protein degradation, is not directly responsible for LD degradation, but may influence lipophagy indirectly as discussed below.
2. Proteins Involved in Lipophagy Induction
2.1 PLINs and lipases
Although LDs differ in lipid composition, size, and cellular location, perhaps the characteristic that best highlights their dynamic nature is their proteome. The surface of the LDs comprises hundreds of resident and transient proteins that influence LD metabolism and signaling. An emerging role of these proteins is to regulate LD-specific functions including lipophagy. The perilipin family members are the best characterized LD proteins and play numerous roles in LD biology [13]. This family of proteins (PLIN1-5) differ in regards to tissue expression profiles and biological functions and share varying degrees of homology. Numerous PLINs also influence LD catabolism through their ability to modulate access of lipases to the LD surface [13]. A role for PLIN proteins in linking CMA to LD catabolism has recently been identified [14]. These studies reveal that ablation of lysosomal-associated membrane protein 2 (LAMP2A), which is required for CMA, leads to LD accumulation. The authors also show that the heat shock cognate protein of 70 kDa (Hsc70) binds a CMA recognition motif (KFERQ) within PLIN2, thereby targeting it for CMA-mediated degradation in the lysosome; similar effects with PLIN3 were also shown. Moreover, blocking CMA reduced cytosolic lipase-mediated lipolysis and lipophagy, suggesting that the degradation of PLIN2 (and perhaps PLIN3) is required to allow access of ATGL and autophagic proteins to the surface of LDs to promote LD catabolism. Additional work has identified AMP-activated protein kinase as a critical protein involved in phosphorylation of PLIN2, an event that is required for CMA-mediated degradation of PLIN2 [15]. Thus, these studies place CMA-mediated degradation of PLIN proteins as an upstream event critical for initiating lipophagy.
The above studies highlight an important role for ATGL in promoting TAG catabolism in response to PLIN2 degradation. Additional work has extended the links between ATGL and autophagy/lipophagy. For example, Martinez-Lopez et al. showed that LC3, a classical marker of the autophagosome, was able to directly interact with ATGL at the surface of LDs [16]; interactions between LC3 and HSL were also identified. LC3 binds ATGL via an LC3 interacting region (LIR) at residues 145–150 (STFIPV). Ablation of this LIR reduces basal ATGL localization to LDs and prevents the translocation of ATGL to the LD surface in response to serum starvation. Therefore, these studies suggest that LC3 is critical for translocation of ATGL to the surface of LDs to facilitate TAG hydrolysis. However, the biological rationale for why ATGL would require LC3 for LD localization remains to be elucidated.
The prevailing model of LD catabolism, including the above studies, is that both ATGL and lipophagy directly contribute to LD degradation. A recent study from our laboratory has explored if there is linearity in the relationship between ATGL and autophagy/lipophagy [17]. In this work, ATGL was shown to be both necessary and sufficient to promote the expression of autophagy genes in the liver. Moreover, ATGL promotes LD turnover, positively regulates autophagic flux and increase interactions of lysosomes and LC3 with LDs in hepatocytes suggesting increased lipophagy. Importantly, genetic or chemical inhibition of autophagy or lysosomal lipid degradation (e.g. LAL knockdown or inhibition) blocks the effects of ATGL overexpression on LD turnover and the subsequent oxidation of liberated FAs. Based on previous work showing that ATGL promotes the activity of SIRT1 [18], a major driver of autophagy [19], additional studies show that SIRT1 mediates the effects of ATGL on promoting autophagy/lipophagy. This work is also supported by studies showing that PNPLA5 and PNPLA8, members of the patatin-like phospholipase domain-containing protein family that also contain ATGL (PNPLA2), promote autophagy/lipophagy in a variety of cell types [20,21]. Regarding PNPLA5, Dupont et al. suggest that diacylglycerol generated by PNPLA5 is critical for autophagosomal membrane synthesis and may influence membrane curvature and, thus, protein trafficking. Taken together, this work suggests that ATGL, and potentially other lipases, act as upstream regulators of autophagy/lipophagy and that lipophagy (rather than ATGL itself or related lipases) is responsible for the bulk breakdown of LDs (see Figure 1 for an overview of these pathways). It should be noted that macrophages that lack ATGL also have impaired lipophagy but normal macroautophagy, suggesting that unique cell type-specific effects may exist [22]. Since lipolysis is a highly-regulated process influenced by numerous signaling networks (cAMP/PKA, AMPK, etc.) and a host of proteins known to directly interact with ATGL or other LD proteins that indirectly influence ATGL activity (or presumably other PNPLA lipases), much remains to be learned about how ATGL-mediated signaling integrates these various inputs to coordinate lipophagy.
Figure 1. Overview of the major arms of LD degradation.
CMA promotes LD catabolism via the degradation of PLIN LD proteins, thus, allowing access for lipases and lipophagic organelles. Additionally, lipases such as ATGL promote both macro- and micro-lipophagy to facilitate LD catabolism leading to the generation of free FAs that are largely channeled towards oxidative pathways.
2.2 Rab GTPases
Numerous Rab proteins are now recognized to influence LD biology and metabolism following their initial identification on LDs via proteomic studies [23–26]. The Rab family of approximately 70 small GTPases are historically considered to be important mediators of endosomal trafficking events, conferring a discrete “identity” upon individual trafficking organelles [27]. Rabs function as molecular switches, cycling between active and inactive GTP- and GDP-bound states, respectively. Interconversion between these states regulates associations with cytoskeletal motor proteins and membrane fusion complexes, thus promoting an intricate vesicular trafficking network within the cell. The nearly 30 Rab GTPases found on the surface of LDs to date [28] have led to numerous investigations geared toward the identification of their potential biological functions in the context of LD metabolism. Importantly, recent reports have revealed that perturbations to some members of this family of small GTPases have detrimental effects on LD turnover in response to classical lipophagy-inducing cues.
One of the most predominant Rabs populating the surface of the LD is Rab7, a well characterized marker of the late endocytic pathway [29] and a known participant in the process of autophagosomal maturation [30,31]. Rab7 assists in the regulation of key late endocytic and autophagic membrane fusion events (e.g. lysosome-autophagosome interactions) by coordinating interactions between various SNARE proteins and members of the HOPS tethering complex [32,33]. Furthermore, interactions of Rab7 together with downstream effector proteins such as FYVE and coiled-coil domain containing protein 1 (FYCO1) or Rab7-interacting-like protein (RILP) promote plus- and minus-end directed transport of organelles along microtubules in association with kinesin and dynein-dynactin motors, respectively [34–36]. Rab7 can therefore be considered to occupy a central role in catalyzing interactions between various cellular compartments and facilitating dynamic processes such as autophagy. Indeed, recent studies have demonstrated important contributions for Rab7 in the process of mitophagy, where it appears to assist in the encapsulation of mitochondria within autophagic membranes together with the GTPase-activating proteins TBC1D15 and TBC1D17 [37].
A potential function for Rab7 in LD homeostasis was first identified because β-adrenergic stimulation of numerous cell types resulted in an increase in the co-localization of this small GTPase with LDs as well as lysosomes and autophagic membranes [38]. siRNA-mediated knockdown of endogenous Rab7 or the overexpression of a dominant-negative form of Rab7 results in the aberrant accumulation of LDs in hepatoma cells [39]. Upon nutrient deprivation, this small GTPase becomes activated directly on the LD surface, resulting in its increased affinity for GTP over GDP. This activated state promotes recruitment of degradative structures (i.e. multivesicular bodies and lysosomes) into the vicinity of the LD, potentially “priming” the LD for targeted degradation via lipophagy [40]. This result suggests that Rab7 may be an important LD-localized node for the regulation of hepatic lipophagy. Indeed, a very recent report shows that Rab7 is also a primary target for ethanol-induced hepatic steatosis [41]. The authors found that rats chronically fed an alcohol-containing diet are resistant to starvation-induced lipophagy, which ultimately results in liver steatosis. Importantly, as compared to control hepatocytes, hepatocytes exposed to ethanol had significant reductions in levels of active Rab7, suggestive of defects in the autophagic recognition of LDs for targeted turnover. Further studies are required to determine the exact mechanisms through which ethanol suppresses this regulator of hepatic lipophagy.
Rab10 is a second LD-localized member of the Rab family that has been identified as potentially participating in lipophagy. This GTPase has important roles in the regulation of insulin-stimulated GLUT4 vesicle trafficking [42] and trafficking from the Golgi during epithelial polarization [43]. Recently, Rab10 was also shown to be critical for coordinating tubular extension and fusion during the process of ER morphogenesis [44]. Like Rab7, depletion or genetic perturbations to Rab10 also result in hepatocellular lipid accumulation [45]. Interestingly, nutrient-depleted cells exhibit a significant redistribution of activated Rab10 to the LD surface. Under these conditions, this GTPase co-localizes at the LD together with markers for autophagic membranes, including LC3 and Atg16. Rab10 therefore appears to act downstream of Rab7 as part of a complex, together with its binding partner EH-domain binding protein 1 and the membrane-deforming ATPase EHD2, to promote the envelopment of the LD by an expanding phagophore during lipophagic progression [45].
Other Rabs have clear connections to LD metabolism and conventional lipolysis, but putative roles in the selective autophagy of LDs remain unclear. Rab32 co-localizes with the LD as well as with markers of autophagic or lysosomal membranes in the Drosophila fat body [46]. Moreover, knockdown of Rab32 results in an increase in ATGL expression and reduced LD abundance and size [47]. The mechanisms of ATGL upregulation were not studied, however, and therefore require further investigation. An additional Rab GTPase shown to have a connection with LD homeostasis is Rab18. In adipose tissue, this small GTPase is tightly associated with LDs and exhibits sex- and fat-depot-specific differences that are associated with overall adiposity [48]. The bulk of cellular Rab18 appears to be localized exclusively to the LD surface, especially following β-adrenergic stimulation [49,50]. Rab18 appears to bind to discrete subsets of LDs, suggesting differential recruitment that is dependent on the metabolic status of individual LDs, an enticing concept with respect to the selectivity inherent to lipophagy. A recent study shows that interactions between the transport protein particle (TRAPP-II) and the coatomer protein COP-I function to re-localize TRAPP-II to the LD surface. TRAPP-II is then able to activate Rab18, promoting its subsequent recruitment to the LD to regulate lipolysis [51]. TRAPP complexes are known to be involved in the autophagic process [52]; therefore, future investigations will be useful in determining whether Rab18-TRAPP interactions play any defined role in the selective process of lipophagy. Other Rabs, such as Rab25, have recently been suggested to participate in the autophagic turnover of retinyl ester-enriched LDs from hepatic stellate cells (HSCs) [53]. Generation of ROS during the process of HSC activation was found to result in an increase in Rab25 expression. Genetic depletion of Rab25 using siRNA prevented LD turnover and, as a consequence, inhibited the process of HSC activation. This interesting result suggests a new role for yet another Rab GTPase in lipophagy; however, it is not clear whether this pool of Rab25 functions specifically at the HSC LD surface, nor whether this Rab is also involved in other arms of the autophagic process.
2.3 LD-localized receptors for lipophagy
A great deal of interest has focused on the identification of cargo-specific selective autophagy receptors that may provide structural links between a given organelle and the autophagic machinery (for a recent review, see [54]). Unique membrane-bound receptors that engage the autophagic machinery have been identified for nearly all organelles within the cell aside from the LD. Many of these diverse receptors are known to interact with LC3B, the classical marker of the autophagic membrane, via LC3-interacting region (LIR) motifs, as mentioned above [55]. For example, the identification of clearly defined roles for the mitochondrial cargo receptors NDP52 and optineurin have provided exciting new insights into the selective process of mitophagy [56,57]. In addition to NDP52 [58], which may play roles in other types of autophagy such as xenophagy [59], general candidate receptors for the targeted turnover of other cellular organelles have also been identified, including p62/SQSTM1, NBR1, and Huntingtin [60–62]. Mutations in the latter have been shown to result in significant LD accumulation, thus suggesting its potential function as a LD recognition receptor protein [62,63]. While LC3-binding to ATGL appears important to facilitate LD degradation, as mentioned above, no other LD-resident proteins have been demonstrated to unambiguously mediate lipophagy via an LC3-binding mechanism, suggesting that other proteins (i.e. Rab7, above) may potentially assist in the recruitment of the autophagic machinery through as-of-yet undefined mechanisms. Alternatively, protein modification via polyubiquitination may serve as a lipophagy-promoting signal. For example, interactions between ancient ubiquitous protein 1 (AUP1) and the E2 ubiquitin conjugase G2 at the LD surface may promote the tagging of droplets for degradation [64–66]. Further work will be required to precisely define the signals initiating and regulating the autophagic turnover of LDs.
3. Lysosomal Lipid Degradation
Once internalized into the lysosome, LAL is responsible for the hydrolysis of TAG and CE. LAL is perhaps most recognized for its deficiency, which results in Wolman disease and CESD [67,68]. Mutations in LAL that render the enzyme catalytically dead result in the more severe Wolman Disease, where some residual activity remains in mutations that lead to CESD. Clinical manifestations of Wolman disease include massive accumulation of TG and CE in the liver and spleen, intestinal malabsorption, severe cachexia, and adrenal calcification [69]. Patients with the relatively milder CESD can progressively develop hepatosplenomegaly and dyslipidemia. In 2015, recombinant enzyme replacement therapy with sebelipase alfa was approved and has been shown to normalize symptoms and prolong survival for those with severe Wolman disease [70]. Ablation of LAL in mice results in a phenotype that mirrors CESD in humans [71]. Although the massive accumulation of lipids should be suggestive of a major role of lipophagy in the liver, it is difficult to dissociate the contribution of remnant lipid uptake, which requires LAL, and altered whole-body lipid trafficking that results from progressive loss of adipose tissue in these mice [72]. Interestingly, hematopoietic cell transplantation can normalize symptoms of subjects with LAL deficiency [73]. These data suggest a potential role of immune cells in the development of the phenotype via secretion of LAL and its subsequent uptake and restoration of lysosomal neutral lipid hydrolase activity in hepatocytes. Because LAL is the only known neutral lipid hydrolase in lysosomes, it plays a crucial role in mediating lipid flux through the lysosome and, therefore, likely affects both lipophagy and the subsequent release, trafficking, and signaling of downstream lipid breakdown products.
4. Regulation of Lipophagy
4.1 Transcriptional regulation
The past few years have brought major advances in our understanding into the transcriptional control of autophagy and, to some degree, lipophagy. The most studied transcriptional regulators of autophagy/lipophagy are the members of the microphthalmia-associated/TFE subfamily of basic/helix-loop-helix/leucine zipper transcription factors that include TFEB and TFE3 in mammals and HLH-30 in C. elegans. In addition to activating peroxisome proliferator-activated receptor (PPAR)-γ coactivator 1α (PGC-1α) and PPAR-α target genes involved in fat catabolism, TFEB has also been shown to promote lipophagy [74]. A study performed in C. elegans revealed that coordinated activation of the TFEB homolog HLH-30 promotes lipophagy and lysosomal lipid hydrolysis by increasing the expression of the numerous lysosomal lipases that have been shown to exist in worms [75]. The study also showed that the fasting-mediated induction of LAL expression in mouse liver is dependent on TFEB. TFE3 also regulates lipophagy in the liver. Specifically, liver-specific ablation of TFE3 promotes steatosis, and overexpression reduces steatosis via lipophagy [76]. In contrast, adipose-specific overexpression of TFE3 increases adiposity [77], which may be explained by studies showing that autophagy promotes adipocyte differentiation [78–80]. These studies highlight distinct roles of TFE3 in differentially regulating lipid metabolism between the liver and adipose tissue. Activation of the transcription factor forkhead box O1 (FOXO1) through nutrient depletion has been shown to induce lipophagy and LAL in adipocytes [81]. Moreover, FOXO1/3/4 liver-specific knockout mice develop hepatic steatosis and hypertriglyceridemia with decreased autophagic flux, which can be reversed upon overexpression of the autophagy gene ATG14 [82]. Related work by Seok et al., shows that cAMP response element-binding protein (CREB) promotes lipophagy in fasted states via TFEB activation and in the presence of nutrients, this cascade is suppressed by the nuclear receptor farsenoid X receptor (FXR) [83]. A similar regulatory interplay between FXR and PPAR-α was described by Lee et al., wherein starvation-induced PPARα activation suppresses FXR-mediated inhibition of lipophagy [84]. Taken together, these studies suggest that transcription factors and co-activators induced with fasting (TFEB, TFE3, PPAR-α, PGC-1α, FoxO1 and CREB) promote lipophagy, whereas feeding-induced transcription factors (FXR) suppress lipophagy.
In addition to the above fasting/feeding transcriptional networks, a role of sterol response element binding protein-2 (SREBP-2) in autophagy/lipophagy regulation has also been identified. Knockdown of SREBP-2 activates autophagy genes during sterol depletion and is responsible for autophagosome formation to promote lipid droplet turnover [85]. A subsequent study revealed that SREBP-2 promotes the expression of PNPLA8, which facilitates lipophagy and prevents hepatic steatosis [21]. These studies would suggest that SREBP-2, in response to reduced intracellular cholesterol levels, promotes lipophagy as a means to increase cholesterol levels, in addition to directly promoting the expression of cholesterol synthesis genes. These studies also highlight the importance of lipid signaling in the regulation of lipophagy.
4.2 Hormonal and nutrient-mediated regulation
The mammalian target of rapamycin (mTOR) is well characterized as a major signaling node that responds to nutrients (amino acids, glucose, etc.) and hormones (insulin) to coordinate downstream metabolic pathways amongst a host of other functions. Consistent with its activation in response to nutrients, mTOR is a potent inhibitor of autophagy. Pharmacological approaches to activate autophagy using rapamycin, an inhibitor of mTOR, have been shown to improve lipid oxidation and enhance lipid degradation [86]. Additionally, induction of autophagy through serum deprivation decreases phosphorylation of mTOR, thereby rendering it inactive, and subsequently promotes lipid turnover in hypothalamic neurons [87]. Coordination between autophagy and lipolysis via mTOR inhibition was further supported in an elegant study in C. elegans [88]. This study shows that rapamycin promotes the expression of the lysosomal lipase lipl-4, which increases lifespan. Thus, mTOR appears to regulate lipophagy in a manner similar to autophagy although the mechanisms by which it promotes lipophagy remain to be fully elucidated.
In contrast to anabolic signaling, hormones that typically promote catabolic pathways also promote lipophagy. β-adrenergic signaling promotes lipophagy in adipocytes, a process which is dependent on Rab7 [38] similar to what is observed in hepatocytes [39]. Thyroidine (T3) is well documented to promote lipid catabolism including hepatic mitochondrial β-oxidation. Consistent with this role, a functional thyroid receptor is required for the mobilization of lipids via autophagy [89]. Additional studies are required to address the role of T3 hormonal regulation on lipophagy in extrahepatic tissues.
Lipids themselves are also known to regulate lipophagy. Acute stimulus with a physiological fatty acid such as oleic acid activates lipophagy [12]. This activation is thought to occur as a mechanism to consume excess lipid influx into the cell. Similar induction of lipophagy by oleic acid has also been reported in hypothalamic neurons, suggesting that this could be a ubiquitous effect observed in multiple cell types [87]. In contrast to acute stimulation, over-nutrition or high-fat diet feeding leads to hepatic steatosis coinciding with decreased LC3 in liver LDs, a phenotype that can be reversed by restoring hepatic ATG7 levels [90]. High-fat feeding also reduces LAMP-2A [91], which could result in reduced CMA-mediated degradation of PLIN2 leading to reduced lipophagy.
4.3 Small molecule regulation
Several natural compounds modulate lipophagy. Epigallocatechin-3-gallate (commonly referred to as EGCG), a polyphenolic compound enriched in green tea, has pro-autophagic/lipophagic effects in the liver [92,93]. Additionally, caffeine exerts protective effects on fatty liver disease and enhances fatty acid oxidation in liver cells via lipophagy [94]. Parfati et al. explored the role of a dietary polyphenol bergamot, derived from the peel of the bergamot citrus fruit, in alleviating hepatic steatosis. A 3-month treatment of 50 mg/kg in rats resulted in decreased total hepatic lipid content and enhanced association between LDs and autophagic compartments [95]. Additionally, the red wine bioactive resveratrol alleviates hepatic steatosis via induction of autophagy/lipophagy [96,97]. Contrary to the above molecules that enhance lipid turnover through autophagic signaling, tetrandrine, a bisbenzylisoquinoline alkaloid, impairs autophagy and thereby induces lipid accumulation in hepatic stellate cells [98]. While a broad spectrum of similar compounds have been shown to regulate autophagy, specific effects of other small molecules on lipophagy have not been tested [99].
5. Lipophagy in Disease States
5.1 Fatty liver disease
The finding that autophagy serves a central role in hepatic lipid homeostasis [12] has resulted in numerous studies focused on elucidating the contribution of this catabolic process to the onset of non-alcoholic fatty liver disease (NAFLD). NAFLD is estimated to have a global prevalence of nearly 25% in adults and represents the second leading indication for liver transplant in the United States [100]. A great deal remains to be understood regarding potential connections between human liver disease and autophagy, with uncertainty persisting as to whether defects in hepatic autophagy represent either a cause or result of liver injury [101–103]. In contrast to those reports focused on general bulk macroautophagy, studies specifically examining the role for selective autophagy of LDs in the context of liver disease are relatively few in number.
How might lipophagy be regulated or altered in the steatotic liver? One possibility is that a transcriptional regulator such as TFEB, which controls lipid utilization through the coordination of PPARα and PGC-1α, may itself be dysregulated [103]. Alternatively, a recent study by Zubiete-Franco et al. provided evidence that the decreased expression of a specific enzyme, glycine N-methyltransferase, may result in abnormally high levels of serum methionine and S-adenosylmethionine, resulting in impaired lipophagy within a subset of NAFLD patients [104]. Conversely, an autophagy-inhibiting protein, Rubicon, exhibits slight elevation in liver samples taken from patients with NAFLD [105]. Studies performed using Rubicon-knockout mice confirmed an enhancement in levels of hepatic lipophagy, evidenced by electron micrographs demonstrating the presence of numerous double-membrane structures tightly wrapped along the LD surface. Other factors are also of interest, including the mitochondrial protein IRGM (Immunity-related GTPase family M). This protein is perhaps best characterized for its role in the autophagic clearance of bacteria [106]. Interestingly, in addition to a reduction in xenophagy, knockdown or naturally occurring mutations of this protein result in increased hepatic lipid accumulation [107,108]. Clearly, a number of targets connected broadly to the autophagic pathway have intricate links to the turnover of hepatic LDs.
These findings suggest that the targeted stimulation of lipophagy may prove an attractive route to promote the resolution of fatty liver. As such, various treatments known to upregulate conventional autophagy are currently being investigated as therapeutic avenues toward the treatment of NAFLD [109]. For example, Lin et al. have shown that intraperitoneal injections of carbamazepine or rapamycin alleviated both diet- and alcohol-induced hepatic steatosis in mice [86]. As mentioned above, numerous small molecules (e.g. resveratrol, caffeine) or genetic manipulations (e.g. TFEB overexpression) have been shown in mice to attenuate hepatic steatosis. Moreover, in other disease states associated with hepatic triglyceride accumulation, such as glucose-6-phosphatase deficiency (von Gierke’s disease), genetic or pharmacological stimulation of autophagy was able to reduce liver steatosis [110]. A better understanding of the molecular regulation of lipophagy will undoubtedly result in important insights not only into the lipid accumulation that is characteristic to the early stages of NAFLD, but perhaps also to the early stages of fibrosis relevant to later stages of this disease. Indeed, the autophagic flux and lipophagy-driven catabolism of vitamin A-containing LDs appears to be a hallmark of hepatic stellate cell activation and the subsequent deposition of extracellular matrix in the fibrotic liver [111]. Therefore, lipophagy likely plays an important role at multiple points in the progression of NAFLD.
5.2 Obesity
In contrast to NAFLD, a concrete role for the selective autophagy of LDs in obesity has yet to be identified. However, evidence for a role of generalized autophagy in obesity does exist. A program of lysosomal biogenesis activated by obesity in adipose tissue macrophages was recently identified by Xu et al. [112]. These authors demonstrated that an inhibition of lysosomal function in macrophages not only favored increased lipid retention, but also resulted in reduced levels of lipolysis in the WAT. The membrane curvature-sensing protein endophilin B1, known to be highly expressed in human adipose tissue, may also play a key role in obesity: genetic knockouts of endophilin B1 resulted in mice that displayed an enhanced susceptibility to body weight gain compared to control littermates [113]. In contrast to WAT, lipophagy may play a more important role in lipid homeostasis in the metabolically active brown adipose tissue: indeed, Martinez-Lopez et al. showed that exposure of mice to cold resulted in the co-localization of the autophagosomal marker LC3 with LDs, which appears to assist in the coordination of the downstream recruitment of cytoplasmic lipases to the LD for conventional lipolysis in brown adipocytes [16]. Whether such interactions occur in WAT or are disturbed in the context of obesity remains unclear.
At the cellular level, lipophagy appears to have some connections to the lipid accumulation characteristic to the obese phenotype. As mentioned above, the small GTPase Rab7 contributes to the regulation of autophagy in the adipocyte, playing an important role in the control of both basal and β-adrenergic mediated lipolysis [38]. Additionally, a study of the effects of visible light irradiation on cultured adipocytes revealed that extended exposure to light at a wavelength of 590 nm resulted in LD turnover that was accompanied by an increase in autophagic flux [114]. These results suggest a possible role for lipophagy in the adipocyte; however, it is important to note that other studies have also shown autophagy to be important in the adipocyte differentiation process. For example, MEFs derived from Atg5−/− mice were found to be defective in adipogenesis [115]. Likewise, knockdowns of Atg7 in cultured preadipocytes were unable to effectively differentiate into mature adipocytes [80]. Importantly, this same study also demonstrated that adipose-tissue specific knockouts of Atg7 resulted in mice that exhibit a lean phenotype and elevated insulin sensitivity. The multilevel roles of lipophagy and autophagy at the cellular and organismal levels reinforce a need to urgently define the molecular mechanisms of these processes in order to more completely understand the basis for complex metabolic disease states such as NAFLD and obesity.
5.3 Cancer
While the general field of metabolism has received increasing attention in cancer research over the past decade, we are only beginning to understand the regulation and role of LDs, and specifically lipophagy, in cancer. p53, a well-documented tumor suppressor protein, activates lipid hydrolysis and the subsequent oxidation of FAs through the direct control of genes involved in autophagy/lipophagy and FA β-oxidation [116]. Global genomic profiling revealed that p53 coordinates the expression of several autophagy genes such as ATG7, ATG4 and UVRAG amongst others [117]. Pharmacological inhibition of p53 with pifithrin-α results in lipid accumulation, suggestive of a role of p53 in lipophagy induction [118]. p53 is also required for oleate-mediated induction of lipophagy in Chang liver cancer cells, further supporting a unique role of this tumor suppressor in lipophagy [118]. Knockdown of microtubule-associated protein 1S (MAP1S) in renal cells causes an impairment of autophagic clearance of lipid droplets, whereas overexpression of MAP1S promotes lipophagy [119]. Levels of MAP1S are higher in normal renal cells compared to those with clear cell renal cell carcinoma. Since high levels of MAP1S are associated with a reduced malignancy and metastasis and predict better survival rates in clear cell renal cell carcinoma patients, these data indirectly highlight a potential beneficial role of lipophagy. Additional work in HeLa cells highlights a role of ATG14 and lipophagy in cell viability. ATGL14 overexpression promotes lipophagy and intracellular FA accumulation leading to ER stress, ROS generation, and apoptosis. Inhibition of lipophagy in ATG14-overexpressing HeLa cells enhances cellular viability [120]. In contrast to the anti-proliferative effects of lipophagy listed above, lipophagy may promote cell growth in androgen-sensitive prostate cancer cell lines [121]. Thus, like many metabolic pathways, the role of lipophagy in cancer cell metabolism, signaling and growth is likely very dependent upon the unique aspects of the cancer itself. Undoubtedly, future studies will expand our knowledge into the role and regulation of lipophagy in diverse cancer types and highlight its therapeutic potential.
6. Relevance of Lipophagy to Cellular Signaling
Once internalized in lysosomes, complex lipids such as TAG, phospholipids and CEs are hydrolyzed to simple lipids such as FAs and cholesterol. Fatty acids have diverse signaling roles including ligands for transcription factors, allosteric modulators and substrates for the generation of other signaling molecules (i.e. eicosanoids, sphingolipids, etc.). Cholesterol also has diverse signaling effects, although perhaps the most noted is its negative feedback regulation of SREBP2 to control the cholesterol synthetic pathway. The detailed mechanism through which lysosome-derived cholesterol is trafficked to regulate SREBP2 remains unclear. Given the signaling properties of FAs and cholesterol, the lysosome can therefore be considered as a source of these signaling molecules. A recent study in worms highlights a novel signaling network that links the lysosome to transcriptional regulation. Folick et al. identified that overexpression of LIPL-4, the C. elegans lysosomal acid lipase, increases lifespan [122]. The lipid binding protein LBP-8 was required for the lifespan-extending effects of LIPL-4, suggesting that lipid signals may link LIPL-4 to longevity. Further analysis found that oleolyethanolamide (OEA), a metabolite of oleic acid, was increased in the LIPL-4 transgenic worms and bound both LBP-8 and transcription factor NHR-80 (homolog of mammalian HNF4). Moreover, feeding OEA increased lifespan in wild type worms, but not in those overexpressing LIPL-4 or LBP-8, suggesting it may work via a similar mechanism. Thus, these studies suggest that lipid signaling molecules derived from lysosomal lipid hydrolysis may act to alter gene expression to influence lifespan. To date, this study provides the most direct evidence of a lysosome-derived signaling network involving lipids. These data are consistent with the well-known function of lysosomal-generated amino acids regulating mTORC1 and downstream signaling cascades [123]. Undoubtedly, as we expand our understanding into lipophagy and lysosomal biology, additional signaling networks that link lipophagy to cell function will be identified.
7. Future Directions, Questions and Challenges
7.1 Lipophagic targeting of LDs
The past several decades of autophagy research have largely involved characterizing the core proteins and signaling networks that govern the global autophagy pathway. Given the breadth of substrates that undergo autophagic degradation, a current and future focus of the field will be to identify and characterize specific proteins and signaling networks that facilitate the individual arms of autophagy such as lipophagy. While numerous proteins (Rabs, etc.) involved in lipophagy have been identified, many of these proteins also influence autophagy on a more global scale. Thus, a major challenge to the field is to identify how individual lipid droplets are recognized by the autophagic machinery. Which proteins, metabolites and signaling networks regulate this process? Since the accumulation of LDs is involved in the etiology of numerous diseases, activation of lipophagy is a viable therapeutic target to prevent or treat such diseases. However, global activation of autophagy is likely to cause undesirable effects due to the unnecessary degradation of cellular components tangential to LDs. Therefore, a more in depth characterization of lipophagy is needed to develop drugs that can specifically target autophagic organelles to LDs.
7.2 Measuring lipophagic flux
The methods to measure autophagic flux have evolved significantly over the past several years as static measures of autophagy (LC3 punctae, abundance of autophagic proteins, etc.) have given way to acid-sensitive/resistant fluorescently-tagged proteins that allow true measures of flux into the lysosome [124]. While valid and useful for measuring flux through many arms of autophagy, there are inherent problems with fluorescent microscopy-mediated approaches for the measurement of lipophagy. For example, dual-labeled PLIN2 has been used to indicate lipophagic flux [39] although, as mentioned above, PLIN2 is also degraded by CMA [14]. Thus, a major challenge in the field is to construct bona fide markers of the LD that accurately measure the engulfment and degradation of these organelles through macro- or microlipophagic pathways with high sensitivity. Ideally, a fluorescent lipid that is resistant to lysosomal degradation or quenching of the signal would allow for the optimal measurement of lipophagic flux. Unfortunately, a probe has yet to be identified for such an application. In addition, we must be careful to draw conclusions based on chemical or genetic models of autophagy inhibition. Ablating one autophagy gene is likely not sufficient for making general inferences about autophagy itself. As we learn more about autophagy/lipophagy, it is becoming clear that inhibiting various proteins or steps in the autophagy pathways can elicit very different effects. Thus, multiple approaches to inhibit autophagy/lipophagy are truly required to show the contribution of the more global pathway. Moreover, the conditions in which we measure autophagy and lipophagy are important. A commonly used model of autophagy/lipophagy induction is complete nutrient removal (i.e. incubation with Earle’s Balanced Salt Solution). While this clearly induces autophagy, the physiological relevance of this model is questionable since cells in vivo, even when an organism is fasting, are always exposed to minimal levels of nutrients and in some case, especially with regard to FAs, increased levels. Thus, culturing cells in reduced nutrient levels or reduced growth factor/serum levels is likely to provide a more physiological and, thus, translational model to study lipophagy.
7.3 Lysosomal sensing/lipid trafficking
Lysosomes are now recognized to be a major site of nutrient sensing in cells. The translocation of mTOR to lysosomes in response to nutrients (amino acids, glucose) facilitates the formation of a protein complex involving mTOR, ragulator and the RAG-heterodimer allowing for downstream mTOR signaling [125]. In addition, mTOR activation promotes lysosomal/cytosolic sequestration of TFEB and TFE3 to limit the expression of autophagy-related genes [126,127]. In addition to sensing extra-lysosomal nutrient levels, some of which are influenced by efflux from the lysosome, the lysosomal nutrient sensing machinery is also influenced by the levels of intraluminal nutrients such as amino acids [123]. Exogenous fatty acids regulate lysosomal nutrient sensing depending on their structure - saturated FAs activate and unsaturated FAs inhibit [128,129] - although caution must be noted in interpreting data involving exposure to a high concentration of a single FA as this is unphysiological compared to the diverse mixture of FAs cell are exposed to in vivo. To date, no studies have evaluated the ability of the lysosome to sense intraluminal levels of FAs or other lipids. Undoubtedly, research in this area will greatly advance our understanding of the contribution of lipophagy to nutrient sensing and cell signaling.
8. Conclusions
Despite its relative youth compared to autophagy, the field of lipophagy has seen tremendous growth and major advances in the past several years. The identification of a subset of proteins crucial for the initiation of lipophagy has facilitated a better understanding of the core components that help the autophagic machinery recognized and degrade LDs. In addition, we have made significant strides in our understanding of the transcriptional networks that govern both lipophagy and autophagy. While these advances have pushed the field forward, much remains to be discovered, especially proteins and signaling networks that specifically modulate lipophagy independent of more global effects on autophagy. Identification of such factors could pave the way for novel therapeutics to modulate the turnover of LDs, which are a hallmark and etiological factor in numerous diseases. The importance of lipophagy and the consequential downstream changes in cell signaling networks will undoubtedly be a major focus of research that could provide novel insights into new regulatory nodes that ultimately link LDs and lipophagy to disease development.
Highlights.
Lipophagy contributes to lipid droplet (LD) degradation in numerous cell types
Perilipins, lipases, and Rab GTPases act as key regulators of autophagic/lipophagic initiation
Alterations in lipophagy are common in various diseases
The mechanisms whereby autophagic machinery target LDs is poorly understood
The downstream effects of lipophagy on cell signaling networks are largely unknown
Acknowledgments
Funding
This work was supported by a NIH grants T32DK007352 (R.J.S.), DK108790 (D.G.M) and DK050456 (Minnesota Obesity Center), and a grant from the American Diabetes Association (1-16-IBS-203) to D.G.M.
Abbreviations
- ATGL
adipose triglyceride lipase
- CE
cholesterol ester
- CESD
cholesterol ester storage disease
- CMA
chaperone-mediated autophagy
- CREB
cAMP response element binding protein
- FA
fatty acid
- FOXO
forkhead box O proteins
- FXR
farsenoid X receptor
- HSL
hormone sensitive lipase
- LAL
lysosomal acid lipase
- LD
lipid droplet
- LIR
LC3-interaction region
- MAP1S
microtubule-associated protein 1S
- mTOR
mammalian target of rapamycin
- LD
lipid droplet
- SREBP
sterol response element binding protein
- TAG
triacylglycerol
- TFEB/3
transcription factor EB/3
- TRAPP-II
transport protein particle II
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizack MA. Activation of an epinephrine-sensitive lipolytic activity from adipose tissue by adenosine 3′,5′-phosphate. J Biol Chem. 1964;239:392–395. [PubMed] [Google Scholar]
- 3.Vaughan M, Steinberg D. Effect of hormones on lipolysis and esterification of free fatty acids during incubation of adipose tissue in vitro. J Lipid Res. 1963;4:193–199. [PubMed] [Google Scholar]
- 4.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 5.Obrowsky S, Chandak PG, Patankar JV, Povoden S, Schlager S, Kershaw EE, Bogner-Strauss JG, Hoefler G, Levak-Frank S, Kratky D. Adipose triglyceride lipase is a TG hydrolase of the small intestine and regulates intestinal PPARα signaling. J Lipid Res. 2013;54:425–435. doi: 10.1194/jlr.M031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:116–126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novikoff AB, Essner E. Cytolysomes and mitochondrial degeneration. J Cell Biol. 1962;15:140–146. doi: 10.1083/jcb.15.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke JA, Schubert WK. Deficient activity of hepatic acid lipase in cholesterol ester storage disease. Science. 1972;176:309–310. doi: 10.1126/science.176.4032.309. [DOI] [PubMed] [Google Scholar]
- 10.Patrick AD, Lake BD. Deficiency of an acid lipase in Wolman’s disease. Nature. 1969;222:1067–1068. doi: 10.1038/2221067a0. [DOI] [PubMed] [Google Scholar]
- 11.Sloan HR, Fredrickson DS. Enzyme deficiency in cholesteryl ester storage idisease. J Clin Invest. 1972;51:1923–1926. doi: 10.1172/JCI106997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimmel AR, Sztalryd C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu Rev Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- 14.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–438. doi: 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, Singh R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2015;23:113–27. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathyanarayan A, Mashek MT, Mashek DG. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep. 2017;19:1–9. doi: 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan SA, Sathyanarayan A, Mashek MT, Ong KT, Wollaston-Hayden EE, Mashek DG. ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1α/PPAR-α signaling. Diabetes. 2015;64:418–426. doi: 10.2337/db14-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol CB. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KY, Jang HJ, Yang YR, Park KI, Seo J, Shin IW, Jeon TI, Ahn S, Suh PG, Osborne TF, Seo YK. SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci Rep. 2016;6:35732. doi: 10.1038/srep35732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goeritzer M, Vujic N, Schlager S, Chandak PG, Korbelius M, Gottschalk B, Leopold C, Obrowsky S, Rainer S, Doddapattar P, Aflaki E, Wegscheider M, Sachdev V, Graier WF, Kolb D, Radovic B, Kratky D. Active autophagy but not lipophagy in macrophages with defective lipolysis. Biochim Biophys Acta. 2015;1851:1304–1316. doi: 10.1016/j.bbalip.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cermelli S, Guo Y, Gross SP, Welte MA. The Lipid-Droplet Proteome Reveals that Droplets Are a Protein-Storage Depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Zhang H, Wang W, Hong Y, Wang J, Zhang S, Xu S, Shu Q, Li J, Yang F, Zheng M, Qian Z, Liu P. Comparative proteomics reveals abnormal binding of ATGL and dysferlin on lipid droplets from pressure overload-induced dysfunctional rat hearts. Sci Rep. 2016;6:19782. doi: 10.1038/srep19782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RGW. Chinese Hamster Ovary K2 Cell Lipid Droplets Appear to Be Metabolic Organelles Involved in Membrane Traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 26.Khan SA, Wollaston-Hayden EE, Markowski TW, Higgins L, Mashek DG. Quantitative analysis of the murine lipid droplet-associated proteome during diet-induced hepatic steatosis. J Lipid Res. 2015;56:2260–2272. doi: 10.1194/jlr.M056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 28.Kiss RS, Nilsson T. Rab proteins implicated in lipid storage and mobilization. J Biomed Res. 2014;28:169–177. doi: 10.7555/JBR.28.20140029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vitelli R, Santillo M, Lattero D, Chiariello M, Bifulco M, Bruni CB, Bucci C. Role of the small GTPase Rab7 in the late endocytic pathway. J Biol Chem. 1997;272:4391–4397. doi: 10.1074/jbc.272.7.4391. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 31.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 32.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.kleine Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 34.Pankiv S, Alemu EA, Brech A, Bruun J-A, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol CB. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 36.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lizaso A, Tan K-T, Lee Y-H. β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9:1228–1243. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The Small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmona-Gutierrez D, Zimmermann A, Madeo F. A molecular mechanism for lipophagy regulation in the liver. Hepatol Baltim Md. 2015;61:1781–1783. doi: 10.1002/hep.27738. [DOI] [PubMed] [Google Scholar]
- 41.Schulze RJ, Rasineni K, Weller SG, Schott MB, Schroeder B, Casey CA, McNiven MA. Ethanol Exposure Inhibits Hepatocyte Lipophagy by Inactivating the Small Guanosine Triphosphatase Rab7. Hepatol Commun. n.d doi: 10.1002/hep4.1021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sano H, Roach WG, Peck GR, Fukuda M, Lienhard GE. Rab10 in insulin-stimulated GLUT4 translocation. Biochem J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- 43.Schuck S, Gerl MJ, Ang A, Manninen A, Keller P, Mellman I, Simons K. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic Cph Den. 2007;8:47–60. doi: 10.1111/j.1600-0854.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 44.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15:169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Schulze RJ, Weller SG, Krueger EW, Schott MB, Zhang X, Casey CA, Liu J, Stöckli J, James DE, McNiven MA. A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci Adv. 2016;2:e1601470. doi: 10.1126/sciadv.1601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Liu Z, Huang X. Rab32 Is Important for Autophagy and Lipid Storage in Drosophila. PLoS ONE. 2012;7:e32086. doi: 10.1371/journal.pone.0032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Wang J, Wan Y, Chen D. Depletion of Rab32 decreases intracellular lipid accumulation and induces lipolysis through enhancing ATGL expression in hepatocytes. Biochem Biophys Res Commun. 2016;471:492–496. doi: 10.1016/j.bbrc.2016.02.047. [DOI] [PubMed] [Google Scholar]
- 48.Pulido MR, Diaz-Ruiz A, Jiménez-Gómez Y, Garcia-Navarro S, Gracia-Navarro F, Tinahones F, López-Miranda J, Frühbeck G, Vázquez-Martínez R, Malagón MM. Rab18 Dynamics in Adipocytes in Relation to Lipogenesis, Lipolysis and Obesity. PLoS ONE. 2011;6:e22931. doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 50.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Luo X, Zhao S, Siu GK, Liang Y, Chan HC, Satoh A, Yu SS. COPI-TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J. 2017;36:441–457. doi: 10.15252/embj.201694866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JJ, Lipatova Z, Segev N. TRAPP Complexes in Secretion and Autophagy. Front Cell Dev Biol. 2016;4:20. doi: 10.3389/fcell.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Zhao S, Yao Z, Wang L, Shao J, Chen A, Zhang F, Zheng S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2016;11:322–334. doi: 10.1016/j.redox.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 55.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 56.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen TN, Padman BS, Lazarou M. Deciphering the Molecular Signals of PINK1/Parkin Mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 59.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 61.Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, Brech A, Johansen T, Kim PK. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 62.Rui Y-N, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, Zhang S. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 64.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lohmann D, Spandl J, Stevanovic A, Schoene M, Philippou-Massier J, Thiele C. Monoubiquitination of ancient ubiquitous protein 1 promotes lipid droplet clustering. PloS One. 2013;8:e72453. doi: 10.1371/journal.pone.0072453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward C, Martinez-Lopez N, Otten EG, Carroll B, Maetzel D, Singh R, Sarkar S, Korolchuk VI. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Mayatepek E, Seedorf U, Wiebusch H, Lenhartz H, Assmann G. Fatal genetic defect causing Wolman disease. J Inherit Metab Dis. 1999;22:93–94. doi: 10.1023/a:1005428122457. [DOI] [PubMed] [Google Scholar]
- 68.Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra J. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res. 2001;42:489–500. [PubMed] [Google Scholar]
- 69.Hoffman EP, Barr ML, Giovanni MA, Murray MF. Lysosomal Acid Lipase Deficiency. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, Bird TD, Ledbetter N, Mefford HC, Smith RJ, Stephens K, editors. GeneReviews(®) University of Washington; Seattle, Seattle (WA): 1993. [accessed March 2, 2017]. http://www.ncbi.nlm.nih.gov/books/NBK305870/ [Google Scholar]
- 70.Desai NK, Wilson DP. Lysosomal Acid Lipase Deficiency. In: De Groot LJ, Chrousos G, Dungan K, Feingold KR, Grossman A, Hershman JM, Koch C, Korbonits M, McLachlan R, New M, Purnell J, Rebar R, Singer F, Vinik A, editors. Endotext. MDText.com, Inc; South Dartmouth (MA): 2000. [accessed March 2, 2017]. http://www.ncbi.nlm.nih.gov/books/NBK395569/ [Google Scholar]
- 71.Aqul A, Lopez AM, Posey KS, Taylor AM, Repa JJ, Burns DK, Turley SD. Hepatic entrapment of esterified cholesterol drives continual expansion of whole body sterol pool in lysosomal acid lipase-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2014;307:G836–847. doi: 10.1152/ajpgi.00243.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Radović B, Vujić N, Leopold C, Schlager S, Goeritzer M, Patankar JV, Korbelius M, Kolb D, Reindl J, Wegscheider M, Tomin T, Birner-Gruenberger R, Schittmayer M, Groschner L, Magnes C, Diwoky C, Frank S, Steyrer E, Du H, Graier WF, Madl T, Kratky D. Lysosomal acid lipase regulates VLDL synthesis and insulin sensitivity in mice. Diabetologia. 2016;59:1743–52. doi: 10.1007/s00125-016-3968-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolar J, Petryk A, Khan K, Bjoraker KJ, Jessurun J, Dolan M, Kivisto T, Charnas L, Shapiro EG, Orchard PJ. Long-term metabolic, endocrine, and neuropsychological outcome of hematopoietic cell transplantation for Wolman disease. Bone Marrow Transplant. 2009;43:21–27. doi: 10.1038/bmt.2008.273. [DOI] [PubMed] [Google Scholar]
- 74.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Jürgen Klisch T, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–58. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiong J, Wang K, He J, Zhang G, Zhang D, Chen F. TFE3 Alleviates Hepatic Steatosis through Autophagy-Induced Lipophagy and PGC1α-Mediated Fatty Acid β-Oxidation. Int J Mol Sci. 2016;17:387. doi: 10.3390/ijms17030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujimoto Y, Nakagawa Y, Satoh A, Okuda K, Shingyouchi A, Naka A, Matsuzaka T, Iwasaki H, Kobayashi K, Yahagi N, Shimada M, Yatoh S, Suzuki H, Yogosawa S, Izumi T, Sone H, Urayama O, Yamada N, Shimano H. TFE3 controls lipid metabolism in adipose tissue of male mice by suppressing lipolysis and thermogenesis. Endocrinology. 2013;154:3577–3588. doi: 10.1210/en.2013-1203. [DOI] [PubMed] [Google Scholar]
- 78.Song B, Chi Y, Li X, Du W, Han Z-B, Tian J, Li J, Chen F, Wu H, Han L, Lu S, Zheng Y, Han Z. Inhibition of Notch Signaling Promotes the Adipogenic Differentiation of Mesenchymal Stem Cells Through Autophagy Activation and PTEN-PI3K/AKT/mTOR Pathway. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2015;36:1991–2002. doi: 10.1159/000430167. [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, He Y, Okutsu M, Ong LC, Jin Y, Zheng L, Chow P, Yu S, Zhang M, Yan Z. Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARγ2 degradation. Am J Physiol Endocrinol Metab. 2013;305:E530–539. doi: 10.1152/ajpendo.00640.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lettieri Barbato D, Tatulli G, Aquilano K, Ciriolo MR. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013;4:e861. doi: 10.1038/cddis.2013.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seok S, Fu T, Choi S-E, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seo Y-K, Jeon T-I, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin C-W, Zhang H, Li M, Xiong X, Chen X, Chen X, Dong XC, Yin X-M. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lapierre LR, Gelino S, Meléndez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinha RA, You S-H, Zhou J, Siddique MM, Bay B-H, Zhu X, Privalsky ML, Cheng S-Y, Stevens RD, Summers SA, Newgard CB, Lazar MA, Yen PM. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest. 2012;122:2428–2438. doi: 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodriguez-Navarro JA, Kaushik S, Koga H, Dall’Armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim H-S, Montana V, Jang H-J, Parpura V, Kim J. Epigallocatechin gallate (EGCG) stimulates autophagy in vascular endothelial cells: a potential role for reducing lipid accumulation. J Biol Chem. 2013;288:22693–22705. doi: 10.1074/jbc.M113.477505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou J, Farah BL, Sinha RA, Wu Y, Singh BK, Bay B-H, Yang CS, Yen PM. Epigallocatechin-3-gallate (EGCG), a green tea polyphenol, stimulates hepatic autophagy and lipid clearance. PloS One. 2014;9:e87161. doi: 10.1371/journal.pone.0087161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, Martinez L, Xie S, Bay B-H, Summers SA, Newgard CB, Yen PM. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 2014;59:1366–1380. doi: 10.1002/hep.26667. [DOI] [PubMed] [Google Scholar]
- 95.Parafati M, Lascala A, Morittu VM, Trimboli F, Rizzuto A, Brunelli E, Coscarelli F, Costa N, Britti D, Ehrlich J, Isidoro C, Mollace V, Janda E. Bergamot polyphenol fraction prevents nonalcoholic fatty liver disease via stimulation of lipophagy in cafeteria diet-induced rat model of metabolic syndrome. J Nutr Biochem. 2015;26:938–948. doi: 10.1016/j.jnutbio.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y, Chen M, Zhou Y, Yi L, Gao Y, Ran L, Chen S, Zhang T, Zhou X, Zou D, Wu B, Wu Y, Chang H, Zhu J, Zhang Q, Mi M. Resveratrol improves hepatic steatosis by inducing autophagy through the cAMP signaling pathway. Mol Nutr Food Res. 2015;59:1443–1457. doi: 10.1002/mnfr.201500016. [DOI] [PubMed] [Google Scholar]
- 97.Tang L, Yang F, Fang Z, Hu C. Resveratrol Ameliorates Alcoholic Fatty Liver by Inducing Autophagy. Am J Chin Med. 2016;44:1207–1220. doi: 10.1142/S0192415X16500671. [DOI] [PubMed] [Google Scholar]
- 98.Miyamae Y, Nishito Y, Nakai N, Nagumo Y, Usui T, Masuda S, Kambe T, Nagao M. Tetrandrine induces lipid accumulation through blockade of autophagy in a hepatic stellate cell line. Biochem Biophys Res Commun. 2016;477:40–46. doi: 10.1016/j.bbrc.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 99.Zenkov NK, Chechushkov AV, Kozhin PM, Kandalintseva NV, Martinovich GG, Menshchikova EB. Plant Phenols and Autophagy. Biochem Biokhimiia. 2016;81:297–314. doi: 10.1134/S0006297916040015. [DOI] [PubMed] [Google Scholar]
- 100.Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, Racila A, Hunt S, Beckerman R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatol Baltim Md. 2016;64:1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- 101.Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kwanten WJ, Martinet W, Michielsen PP, Francque SM. Role of autophagy in the pathophysiology of nonalcoholic fatty liver disease: A controversial issue. World J Gastroenterol WJG. 2014;20:7325–7338. doi: 10.3748/wjg.v20.i23.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martinez-Lopez N, Singh R. Autophagy and Lipid Droplets in the Liver. Annu Rev Nutr. 2015;35:215–237. doi: 10.1146/annurev-nutr-071813-105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zubiete-Franco I, García-Rodríguez JL, Martínez-Uña M, Martínez-Lopez N, Woodhoo A, Juan VG-D, Beraza N, Lage-Medina S, Andrade F, Fernandez ML, Aldámiz-Echevarría L, Fernández-Ramos D, Falcon-Perez JM, Lopitz-Otsoa F, Fernandez-Tussy P, Barbier-Torres L, Luka Z, Wagner C, García-Monzón C, Lu SC, Aspichueta P, Mato JM, Martínez-Chantar ML, Varela-Rey M. Methionine and S-adenosylmethionine levels are critical regulators of PP2A activity modulating lipophagy during steatosis. J Hepatol. 2016;64:409–418. doi: 10.1016/j.jhep.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanaka S, Hikita H, Tatsumi T, Sakamori R, Nozaki Y, Sakane S, Shiode Y, Nakabori T, Saito Y, Hiramatsu N, Tabata K, Kawabata T, Hamasaki M, Eguchi H, Nagano H, Yoshimori T, Takehara T. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease. Hepatology. 2016;64:1994–2014. doi: 10.1002/hep.28820. [DOI] [PubMed] [Google Scholar]
- 106.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 107.Lin Y-C, Chang P-F, Lin H-F, Liu K, Chang M-H, Ni Y-H. Variants in the Autophagy Related Gene IRGM Confer Susceptibility to Nonalcoholic Fatty Liver Disease by Modulating Lipophagy. J Hepatol. 2016;64:1994–2014. doi: 10.1016/j.jhep.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 108.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier J-F, Hébuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 109.Ni H-M, Williams JA, Yang H, Shi Y-H, Fan J, Ding W-X. Targeting autophagy for the treatment of liver diseases. Pharmacol Res. 2012;66:463–474. doi: 10.1016/j.phrs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farah BL, Landau DJ, Sinha RA, Brooks ED, Wu Y, Fung SYS, Tanaka T, Hirayama M, Bay B-H, Koeberl DD, Yen PM. Induction of autophagy improves hepatic lipid metabolism in glucose-6-phosphatase deficiency. J Hepatol. 2016;64:370–379. doi: 10.1016/j.jhep.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 111.Thoen LFR, Guimarães ELM, Dollé L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 112.Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Y, Takahashi Y, Desai N, Zhang J, Serfass JM, Shi Y-G, Lynch CJ, Wang H-G. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci Rep. 2016;6:20453. doi: 10.1038/srep20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi MS, Kim H-J, Ham M, Choi D-H, Lee TR, Shin DW. Amber Light (590 nm) Induces the Breakdown of Lipid Droplets through Autophagy-Related Lysosomal Degradation in Differentiated Adipocytes. Sci Rep. 2016;6:28476. doi: 10.1038/srep28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baerga R, Zhang Y, Chen P-H, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goldstein I, Rotter V. Regulation of lipid metabolism by p53 - fighting two villains with one sword. Trends Endocrinol Metab TEM. 2012;23:567–575. doi: 10.1016/j.tem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 117.Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Park E-J, Lee AY, Chang S-H, Yu K-N, Kim J-H, Cho M-H. Role of p53 in the cellular response following oleic acid accumulation in Chang liver cells. Toxicol Lett. 2014;224:114–120. doi: 10.1016/j.toxlet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 119.Xu G, Jiang Y, Xiao Y, Liu XD, Yue F, Li W, Li X, He Y, Jiang X, Huang H, Chen Q, Jonasch E, Liu L. Fast clearance of lipid droplets through MAP1S-activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival. Oncotarget. 2016;7:6255–6265. doi: 10.18632/oncotarget.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mukhopadhyay S, Schlaepfer IR, Bergman BC, Panda PK, Praharaj PP, Naik PP, Agarwal R, Bhutia SK. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic Biol Med. 2017 doi: 10.1016/j.freeradbiomed.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 121.Kaini RR, Sillerud LO, Zhaorigetu S, Hu C-AA. Autophagy regulates lipolysis and cell survival through lipid droplet degradation in androgen-sensitive prostate cancer cells. The Prostate. 2012;72:1412–1422. doi: 10.1002/pros.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science. 2015;347:83–86. doi: 10.1126/science.1258857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N. An Autophagic Flux Probe that Releases an Internal Control. Mol Cell. 2016;64:835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 125.Mony VK, Benjamin S, O’Rourke EJ. A lysosome-centered view of nutrient homeostasis. Autophagy. 2016;12:619–631. doi: 10.1080/15548627.2016.1147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kwon B, Querfurth HW. Palmitate activates mTOR/p70S6K through AMPK inhibition and hypophosphorylation of raptor in skeletal muscle cells: Reversal by oleate is similar to metformin. Biochimie. 2015;118:141–150. doi: 10.1016/j.biochi.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 129.Yasuda M, Tanaka Y, Kume S, Morita Y, Chin-Kanasaki M, Araki H, Isshiki K, Araki S, Koya D, Haneda M, Kashiwagi A, Maegawa H, Uzu T. Fatty acids are novel nutrient factors to regulate mTORC1 lysosomal localization and apoptosis in podocytes. Biochim Biophys Acta. 2014;1842:1097–1108. doi: 10.1016/j.bbadis.2014.04.001. [DOI] [PubMed] [Google Scholar]