Summary
Kinase Translocation Reporters (KTRs) are genetically encoded fluorescent activity sensors that convert kinase activity into a nucleocytoplasmic shuttling equilibrium for visualizing single cell signaling dynamics. Here, we adapt the first generation KTR for extracellular signal-regulated kinase (ERK) to allow easy implementation in vivo. This sensor, “ERK-nKTR,” allows quantitative and qualitative assessment of ERK activity by analysis of individual nuclei, and faithfully reports ERK activity during development and neural function in diverse cell contexts in C. elegans. Analysis of ERK activity over time in the Vulval Precursor Cells (VPCs), a well-characterized paradigm of EGFR-Ras-ERK signaling, has identified dynamic features not evident from analysis of developmental endpoints alone, including pulsatile, frequency-modulated signaling associated with proximity to the EGF source. The toolkit described here will facilitate studies of ERK signaling in other C. elegans contexts, and the design features will enable implementation of this technology in other multicellular organisms.
eTOC blurb
A genetically-encoded biosensor for ERK activity allows qualitative or quantitative assessment by analysis of individual nuclei during development and neural function in diverse cell contexts in C. elegans. In-depth analysis of ERK activity over time in the Vulval Precursor Cells has identified dynamic features including pulsatile, frequency-modulated signaling.
Introduction
Studies of Extracellular signal-regulated kinase (ERK) function have revealed its profound and pervasive roles in development, physiology and disease (Arur et al., 2009; Kolch et al., 2015). ERK is the most downstream protein kinase of the canonical Ras-Raf-MEK-ERK cascade that is activated by a diverse set of stimuli and transduced by growth factor or G protein-coupled receptors. Upon stimulation, positive and negative feedback mechanisms act on multiple levels of the pathway, operating in a stimulus-specific and cell type-specific manner. As a consequence, a variety of ERK activity kinetics are observed in a range of cell types or in response to different stimuli; these include transient, sustained, and pulsatile ERK responses (Albeck et al., 2013; Aoki et al., 2013; Hiratsuka et al., 2015; Lim et al., 2015; Mattingly et al., 2015; Santos et al., 2007; Shankaran et al., 2009). It has been proposed that such dynamic characteristics of signaling may in themselves account for the diversity of ERK-dependent outcomes, such as its ability to trigger graded or switch-like events at the molecular level, and its potential to drive a number of different biological processes at the cellular level (Avraham and Yarden, 2011). In a classic example, rat PC12 cells employ different positive or negative feedback mechanisms to regulate the Nerve Growth Factor receptor (TrkA) and Epidermal Growth Factor Receptor (EGFR), resulting in either sustained ERK activity and differentiation in response to TrkA activation, or transient ERK activity and proliferation in response to EGFR activation (Marshall, 1995; Ryu et al., 2015; Santos et al., 2007).
Our understanding of how protein kinases transform stimuli into cellular responses has been enhanced by the development of genetically encoded fluorescent activity sensors, the first generation of which were based on Förster resonance energy transfer (FRET) (Harvey et al., 2008; Ting et al., 2001). Assayed in living cells, genetically encoded sensors allow monitoring of ERK activity in a longitudinal fashion, in the same cells over time, and reveal additional levels of ERK signaling complexity. For example, in steady-state conditions in cultured mammalian cells, ERK activity oscillates with a frequency modulated by EGF dose as a result of mechanisms acting at the level of the receptor (Albeck et al., 2013; Aoki et al., 2013; Sparta et al., 2015). Intriguingly, within intact living mouse skin, pulses of ERK activity spontaneously arise and spread to neighboring cells (Hiratsuka et al., 2015). Together, these observations highlight the relevance of visualizing ERK signaling activity in real time in a variety of cellular contexts in vitro and in vivo.
Kinase translocation reporters (KTRs) offer a powerful approach to studying kinase activity and dynamics in vivo that is not based on FRET technology (Regot et al., 2014). KTRs are genetically encoded fluorescent sensors that convert phosphorylation state into a nucleocytoplasmic localization within the cell. KTRs are constructed by juxtaposing a docking site for a kinase to a module containing a phospho-inhibited nuclear localization signal (NLS), a phospho-enhanced nuclear export signal (NES), and a fluorescent protein. When the cognate kinase is inactive, the KTR is unphosphorylated and mostly nuclear; when the cognate kinase is active, phosphorylation of the KTR results in cytoplasmic translocation (Fig. 1A). The modularity of KTRs enabled the development of ERK-KTR, a sensor for ERK kinase activity, utilizing conserved ERK docking sites from the ERK substrate Elk1 (Fig. 1B) (Regot et al., 2014). Furthermore, this sensor was shown to be a rapid, specific, and quantitative reporter for ERK kinase activity in human cultured cells, revealing fast pulsatile dynamics in individual cells in response to growth factors; the kinetics are similar to a FRET-based reporter but has the advantage of a higher dynamic range and responding more rapidly to downregulation of kinase activity (Regot et al., 2014; Sparta et al., 2015). The success of this approach in cultured cells suggested that, for developmental biology questions, the versatility of the ERK-KTR method would be especially valuable compared to other methods, as it can be observed by eye without need for image processing, making it appropriate for rapid visual screens, or alternatively, used for image capture and quantification of ERK activity dynamics during development.
Figure 1. The nuclear ratio ERK-nKTR design.
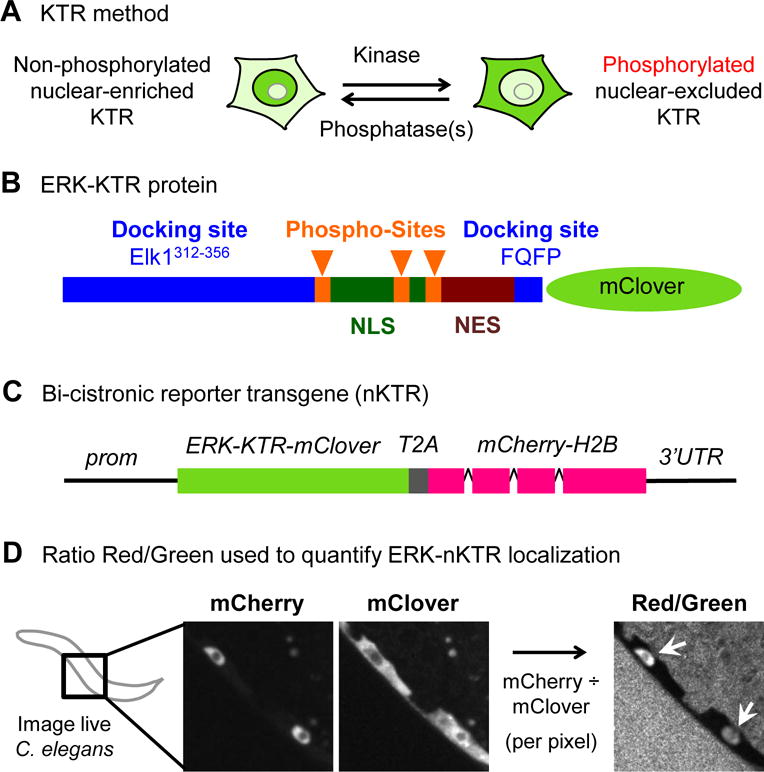
(A) The KTR method (Regot et al., 2014). When kinase activity is low, the KTR sensor protein is not phosphorylated and is nuclear-enriched. When kinase activity is high, the KTR sensor is phosphorylated and excluded from the nucleus. (The KTR is always largely excluded from the nucleolus.) (B) The ERK-KTR sensor is composed of peptides from Elk1 (Elk1312–356 and FQFP), which promote ERK docking to substrates, followed by a module containing NLS and NES sequences, S/T-P phospho-acceptor residues for phosphorylation by ERK (Phospho-Sites), and the green fluorescent protein mClover. An annotated protein sequence of ERK-nKTR-mClover is shown in Table S2. (C) Bicistronic transgene design of ERK-nKTR: the coding region for a viral 2A peptide (T2A) is inserted between the coding regions for ERK-KTR-mClover and mCherry-H2B. The T2A peptide triggers “ribosomal pausing” efficiently in C. elegans (Ahier and Jarriault, 2014), such that ERK-nKTR-mClover and mCherry-H2B are encoded by a single transcript but produced as two separate, functional proteins in an equimolar ratio. Tissue-specific expression of the bicistronic transcript is achieved using appropriate promoter (prom) and 3′ UTR sequences. (C) After imaging of live C. elegans, the ratio of mCherry to mClover intensity is calculated per pixel, producing a new image (Red/Green), a pictorial representation of the the Red/Green ratio within nuclei (arrows), as quantified in Figs. 2 and 3.
Here, we report that we have successfully adapted this technology for visualizing ERK activity in vivo, and show that it has the anticipated versatility for both visual inspection and quantitative analysis. We term the adapted design a “nuclear KTR” (“nKTR”), because it allows for quantitation of kinase activity based solely on the level of the sensor in nuclei. This adaptation facilitates the use of the KTR approach when cells are closely packed together, a situation often encountered in vivo. We then showed that the ERK-nKTR sensor faithfully reports activity of the ERK ortholog MPK-1 in several cell types and different developmental processes in C. elegans: Vulval Precursor Cell (VPC) fate specification, Sex Myoblast migration, AWC and ASE sensory neurons, and germ line development. Finally, we analyzed ERK activity dynamics in VPCs, where the role of EGFR-Ras-ERK signaling promotes VPC “competence” and vulval fate induction. Our observations indicate that sensors have the potential to reveal insights into spatial, temporal and dynamic aspects of signaling in vivo.
Design
A bicistronic transgene strategy for quantifiable KTR activity in nuclei (“nKTR”)
An ERK-KTR sensor developed in mammalian cells utilized a conserved ERK docking site from the mammalian ERK substrate Elk1, and kinase activity was quantified by measuring the ratio of cytoplasmic to nuclear fluorescent signal (Fig. 1A, B)(Regot et al., 2014). Within whole organisms, however, most cells do not reside in flat monolayers; instead, they may be packed tightly against other cells in three dimensions, have complex cytoplasmic shapes, and change in size, shape, or form syncytia even while the nucleus remains discrete and stable (except during cell division). These situations can make quantitation of the cytoplasm unreliable (Fig. S1A); we therefore sought to develop a method that would allow us to quantify KTR localization based solely on fluorescence level in the nucleus, and furthermore would not rely on absolute values of fluorescence signal and thus would be relatively insensitive to cell size and transgene vagaries such as transcriptional noise and promoter strength.
Our solution was to create reporters in which equimolar expression of a KTR-mClover biosensor and the nuclear marker mCherry-H2B is achieved from a single transcript using the 2A peptide. This virus-derived structural motif causes “ribosomal pausing,” leading to the production of distinct proteins during translation, and works efficiently in C. elegans (Ahier and Jarriault, 2014) (Fig. 1C). We designate such reporters “nKTRs,” for nuclear ratio Kinase Translocation Reporters. To quantify kinase activity using the nKTR form of the ERK-KTR, we (i) acquire Z-stacks of nKTR-mClover and mCherry-H2B fluorescence, (ii) generate a ratiometric image by dividing the mCherry (“Red”) by the mClover (“Green”) intensities, pixel-by-pixel, resulting in a new image called “Red/Green” (Fig. 1D) (STAR Methods), and (iii) use the mCherry-H2B image to segment and track the nuclei of all cells over the entire Z stack. This design innovation will facilitate analysis in any other context, including cultured cells, and should be able to be incorporated into any KTR-type reporter, because the docking site is what governs kinase specificity (Regot et al., 2014).
We also incorporated other features and tests into the reporter design, as will be described further below. First, to simplify image collection and cell identification as we developed and tested the design, we used tissue-specific regulatory sequences to drive ERK-nKTR expression in defined cells. Second, to avoid expression level vagaries typical of multicopy transgenes in C. elegans and to minimize protein overexpression that might overwhelm the equilibrium of nuclear import and export, we made single-copy integrated transgenes for all tissue-specific ERK-nKTR reporters. Third, we made variant ERK-nKTRs in an attempt to improve the dynamic range of the reporter. Finally, we performed genetic tests to validate ERK-nKTR as a faithful reporter of ERK activity in different cell contexts.
Results
Vulval Precursor Cells (VPCs): optimizing and validating the ERK-nKTR reporter
Studies of VPC fate specification have illuminated many fundamental and conserved features of canonical EGFR-Ras-ERK signaling, including the initial discovery of how ERK docks with its substrates (Jacobs et al., 1998; 1999). The tools and knowledge accumulated through these studies have made it an attractive paradigm for developing and validating the ERK-nKTR approach.
The VPCs are six polarized epithelial cells, numbered P3.p-P8.p, that in the L2 stage are multipotent and have the potential to generate vulval cells. In the L3 stage, the VPCs adopt different fates in a defined pattern via cell-cell interactions (Fig. 2A; reviewed in (Sternberg, 2005). The pattern is established by an EGF-like signal produced by the anchor cell of the gonad, which activates a canonical EGFR-Ras-Raf-MEK-ERK cascade in VPCs. There are different roles for EGFR-Ras-ERK signaling over the course of the lifetime of the VPCs, and different responses to the EGF-like signal. We will consider these properties in more detail below, but in this section, we focus our analysis on the mid-L2 stage. At this time, the LIN-3/EGF signal evokes a graded ERK response in which activity is highest in P6.p, lower in the neighboring VPCs, P5.p and P7.p, and not detactable in other VPCs, based on a transcriptional reporter (Yoo et al., 2004). We first demonstrate that the pattern of nuclear accumulation of the “nKTR” form of the mammalian ERK-KTR reflects the inferred pattern of MPK-1/ERK activity. Then, to validate the new nKTR and nuclear quantitation method, we show that the observed pattern of ERK-nKTR localization requires phosphorylation of the reporter protein and that its localization is correlated with MPK-1/ERK activity levels. Later, we examine ERK-nKTR in a broader range of times to examine signaling dynamics.
Figure 2. The ERK-nKTR reports MPK-1/ERK activity in a phosphorylation-dependent manner.
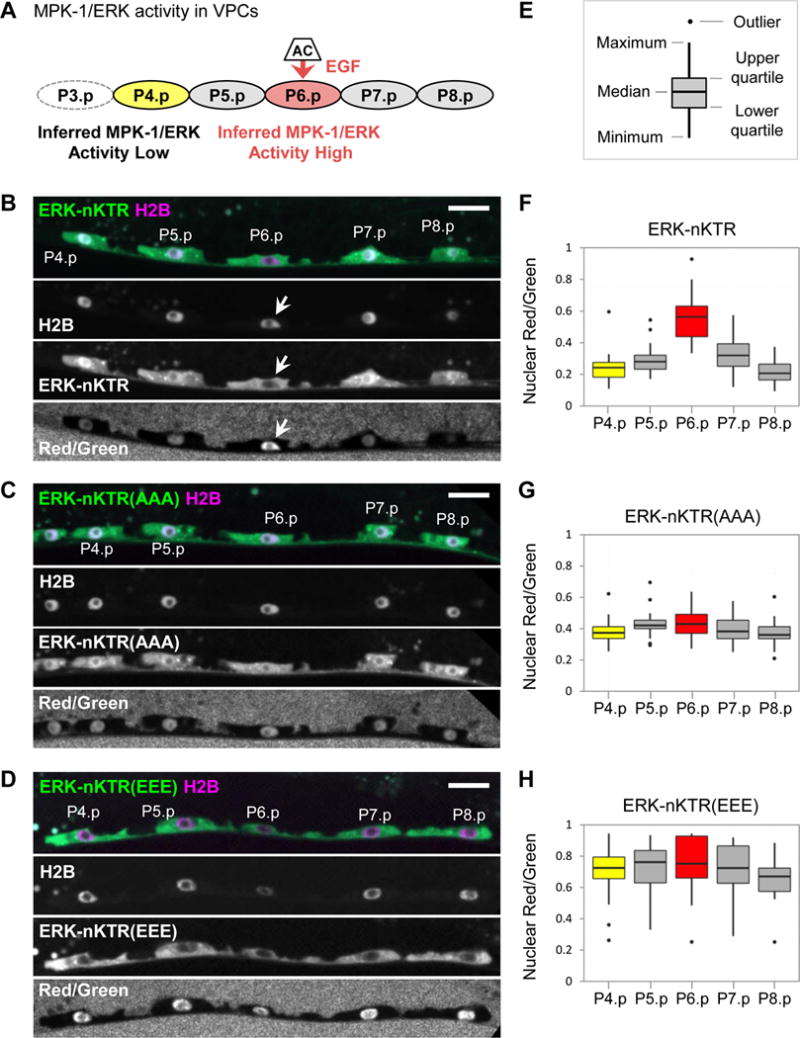
(A) The six VPCs, P3.p-P8.p, receive an EGF signal from the Anchor Cell (AC). Genetic and transcriptional reporter analyses indicate that the EGF signal results in high MPK-1/ERK activity in P6.p (red), the VPC closest to the AC. In the L2 stage, there is low activation of the EGFR-Ras-ERK pathway in P5.p and P7.p (Yoo et al., 2004); VPCs further from the AC, such as P4.p (yellow) have no MPK-1 activity. This drawing and all images of VPCs are oriented with anterior at the left and dorsal at the top. In half of wild-type hermaphrodites, P3.p often fuses with the hypodermal syncytium in the L2 stage instead of remaining as a VPC, so it is omitted from our analysis. (B-D) Here, and in all following images of ERK-nKTR expression (Figs. 3–5), the ERK-nKTR-mClover reporter (ERK-nKTR) is shown in green and mCherry-H2B (H2B) is in magenta. ERK-nKTR, H2B, and the calculated Red/Green ratio (Red/Green) are also shown separately in grayscale. Scale bars are 10 μm. (B) The transgene arTi85 expresses ERK-KTR(NLS3), which we call “ERK-nKTR”, in VPCs. ERK-nKTR protein is excluded from the nucleus of P6.p (arrows). (C) Mutation of ERK-nKTR phospho-sites to alanines in the ERK-nKTR(AAA) transgene arTi101 prevents key phosphorylations that occur upon ERK activation; thus, this mutant form is not responsive to ERK activity and accumulates in the nucleus of P6.p. (D) Mutation of the phospho-sites to glutamic acids, mimicking constitutive phosphorylation, in the ERK-nKTR(EEE) transgene arTi100 also makes the mutant form independent of ERK activity, and is excluded from nuclei in all the VPCs. (E) Each boxplot graph indicates the median, upper quartile, lower quartile, maximum, minimum values, and outliers (values more than 1.5X the interquartile range from either the upper or lower quartile). (F-H) Red/Green ratios for VPC nuclei expressing (F) ERK-nKTR (n=23), (G) ERK-nKTR(AAA) (n=19), and (H) ERK-nKTR(EEE) (n=19).
To assess whether the localization of each reporter is patterned in VPCs, a one-way ANOVA was performed to compare the Red/Green ratios in all VPCs. For ERK-nKTR, there is a highly significant effect on Red/Green ratios when different VPCs are compared (at the p<0.01 level, F(4,109)=32.1, p-value=1.16×10−17). Post hoc pairwise comparisons find significant differences between Red/Green ratios for the following: P6.p vs. P4.p, P5.p, P7.p, or P8.p (all p-values<0.01), P5.p vs. P4.p or P8.p (p-values<0.01), and P7.p vs. P8.p (p-value<0.05) (see statistical tests in Supplemental Materials). In contrast to ERK-nKTR, neither phospho-site mutant is patterned; there are not significant differences in Red/Green ratios between VPCs expressing ERK-nKTR(AAA) (at the p<0.01 level, F(4,90)=2.36, p-value=0.060) or ERK-nKTR(EEE) (at the p<0.01 level, F(4,89)=0.661, p-value=0.621).
The mammalian ERK-KTR sensor is excluded from the nucleus specifically in P6.p
We fused the original ERK-KTR-mClover sensor sequence characterized in mammalian cells (Regot et al., 2014) to T2A-mCherry-H2B and expressed it specifically in VPCs. All details about this transgene and further variants are described in STAR Methods and Table S1. We refer to this reporter as ERK-nKTR(NLS0) to denote the original nuclear localization sequence (NLS), as subsequently we mutated the NLS in attempts to achieve a greater dynamic range. We found that localization of the ERK-nKTR(NLS0) reporter was indeed correlated with the inferred pattern of MPK-1 activity in VPCs: it was visibly excluded from the nucleus of P6.p, consistent with high activity of MPK-1, whereas it was present in both the cytoplasm and nucleus of all other VPCs, where MPK-1 is less active (Fig. S1B).
Analysis of variants to improve the dynamic range
Our initial results with the original ERK-nKTR(NLS0) were promising, because quantification indicated a significant Red/Green signal in P6.p (Fig. S1C). To see if it would be possible to achieve a greater dynamic range, we tested if substituting known C. elegans bipartite NLS sequences for the original NLS would improve its nuclear localization and dynamic range (Fig. S2A–C). We found that, although the NLS sequence from EGL-13 or OCR-2 is sufficient to produce nuclear localization of GFP (Ezak and Ferkey, 2011; Lyssenko et al., 2007), neither NLS was as effective as the original NLS0 when introduced into the sensor (Fig. S2D). Most of the smaller engineered changes to NLS0, to alter the number or spacing of basic residues known to be important for NLS function (Conti et al., 1998; Fontes et al., 2000), also did not result in a significant improvement (Fig. S2D). However, one variant, ERK-nKTR(NLS3), seemed to be as effective as NLS0, as it was visibly excluded from the nucleus of P6.p where MPK-1 activity is high, and present in the nucleus of other VPCs where MPK-1 activity is low (Fig. 2B). When quantified, ERK-nKTR(NLS3) allowed detection of significant differences not only between P6.p and other VPCs, but also between the VPCs flanking P6.p, which also receive the EGF-like signal (P5.p and P7.p) and the outermost VPCs (P4.p and P8.p) which do not (Fig. 2F). These data suggest that the dynamic range of the ERK-nKTR(NLS3) has an improved linear correlation with the levels of ERK activity found in VPCs. Therefore, we have used the NLS3 variant as the canonical reporter for all subsequent analysis, and designate it ERK-nKTR for simplicity.
Mutation of ERK-nKTR indicates that the sensor subcellular localization is phosphorylation-dependent
The translocation of KTRs upon phosphorylation depends on the negative charge introduced into the NLS segment by kinase-specific phosphorylation. Thus, non-phosphorylatable and phospho-mimetic mutants should represent the full dynamic range of the sensor, and would be expected to have a similar Red/Green ratio in all VPCs even though ERK activity state differs among them. We therefore created variants in which three phospho-acceptor sites (see ERK-nKTR protein sequence in Table S2) were mutated to either alanine (AAA) or glutamatic acid (EEE). Localization of ERK-nKTR(AAA) should be the most nuclear possible for ERK-nKTR in VPCs; conversely, localization of ERK-KTR(EEE) should be the most cytoplasmic possible. Indeed, unlike wild-type ERK-nKTR, the non-phosphorylatable ERK-nKTR(AAA) form was not excluded from the nucleus in P6.p (Fig. 2C), whereas the phospho-mimetic ERK-nKTR(EEE) form was excluded from the nucleus in all VPCs (Fig. 2D).
We quantified the intensity of the Red/Green ratio image within VPC nuclei in worms expressing ERK-nKTR, ERK-nKTR(AAA) and ERK-nKTR(EEE) reporters (Fig. 2F–H). Our analysis of ERK-nKTR indicates that Red/Green in P6.p is on average two-fold higher than the other VPCs. Interestingly, the phospho-mimetic ERK-nKTR(EEE) mutant also shows a two-fold increase in all VPCs with respect to the non-phosphorylatable ERK-nKTR(AAA) (compare Fig. 2G and 2H), similar to what is observed in P6.p for the wild-type sensor. These data indicate that the dynamic range of the ERK-nKTR sensor captures the MPK-1 activity level in P6.p.
The subcellular localization of ERK-nKTR in VPCs in mutants correlates with ERK activity
In order to assess if ERK-nKTR localization reflects EGFR-Ras-ERK signaling, we examined it in genetic backgrounds in which ERK activity is lost or constitutively active. We found that ERK-nKTR accumulates uniformly in all VPC nuclei in an mpk-1 null mutant background [hereafter mpk-1(0)] (Fig. 3A, C), similar to that of the non-phosphorylatable ERK-nKTR(AAA) in a wild-type background (Fig 2C, G). Conversely, ERK-nKTR is excluded from the nuclei of all VPCs when MPK-1/ERK is activated by uniform expression of a highly active and stable form of LIN-45/Braf in all VPCs (transgene arTi31, see STAR Methods), (Fig. 3B, D), similar to the phospho-mimetic ERK-nKTR(EEE) (Fig. 2D, H). Together, the results of the cis-mutations in ERK-nKTR and manipulation of MPK-1/ERK activity in trans indicate that ERK-nKTR functions as a faithful reporter for ERK activity.
Figure 3. The ERK-nKTR is sensitive to levels of MPK-1/ERK activity.
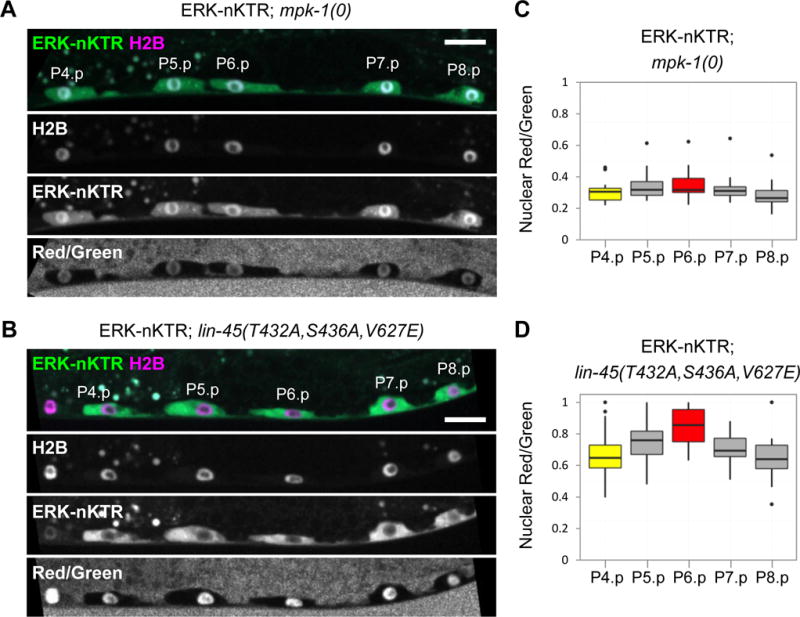
Images of ERK-nKTR transgene arTi85 in genetic backgrounds with altered MPK-1/ERK activity show ERK-nKTR-mClover (ERK-nKTR), mCherry-H2B (H2B), and the calculated Red/Green ratio (Red/Green). (A) Loss of MPK-1/ERK activity in mpk-1(0) causes nuclear accumulation of ERK-nKTR in P6.p, similar to the pattern seen with the non-phosphorylatable ERK-nKTR(AAA) (see Fig. 2D). (B) Ectopic activation of MPK-1 in all VPCs, achieved by expression of a constitutively active and stable form of LIN-45/Raf (see STAR Methods), results in ectopic exclusion of ERK-nKTR from the nuclei of P4.p, P5.p, P7.p, and P8.p, similar to the result seen with the phospho-mimetic ERK-KTR(EEE) (see Fig. 2E). (C–D) Red/Green ratios for VPC nuclei expressing ERK-nKTR in (C) mpk-1(0) (n=19), or in (D) the presence of hyperactive LIN-45/Raf (n=16).
To assess whether the localization of ERK-nKTR is patterned in VPCs of each genotype, one-way ANOVAs were performed to compare the Red/Green ratios in all VPCs. ERK-nKTR is not patterned in mpk-1(0); there are not significant differences in Red/Green ratios between VPCs (at the p<0.01 level, F(4,90)=1.66, p-value=0.165). However, in the presence of hyperactive LIN-45/Raf, there is a significant effect on Red/Green ratios when different VPCs are compared (at the p<0.01 level, F(4,79)=4.52, p-value=0.002). In this case, post hoc pairwise comparisons indicate that the Red/Green ratio of P6.p is significantly different from those of P8.p (p-value<0.01) and P4.p (p-value<0.05) (see statistical tests in Supplemental Materials).
ERK-nKTR accurately reflects MPK-1/ERK activity in different cell contexts
To establish the generality of the ERK-nKTR approach, we examined three additional cell contexts. These contexts differ from VPCs in the upstream receptors that lead to activation, and that mpk-1/ERK activity performs a biological function other than cell fate specification (Sundaram, 2013). In all three cases, nuclear exclusion of the sensor is seen, and such exclusion depends on mpk-1/ERK activity, indicating that ERK-nKTR is an accurate reporter of ERK activity in diverse cell types (Fig. 4). All details about the transgenes for these additional contexts are described in STAR Methods and Table S1.
Figure 4. The ERK-nKTR reports MPK-1/ERK activity in multiple cell contexts.
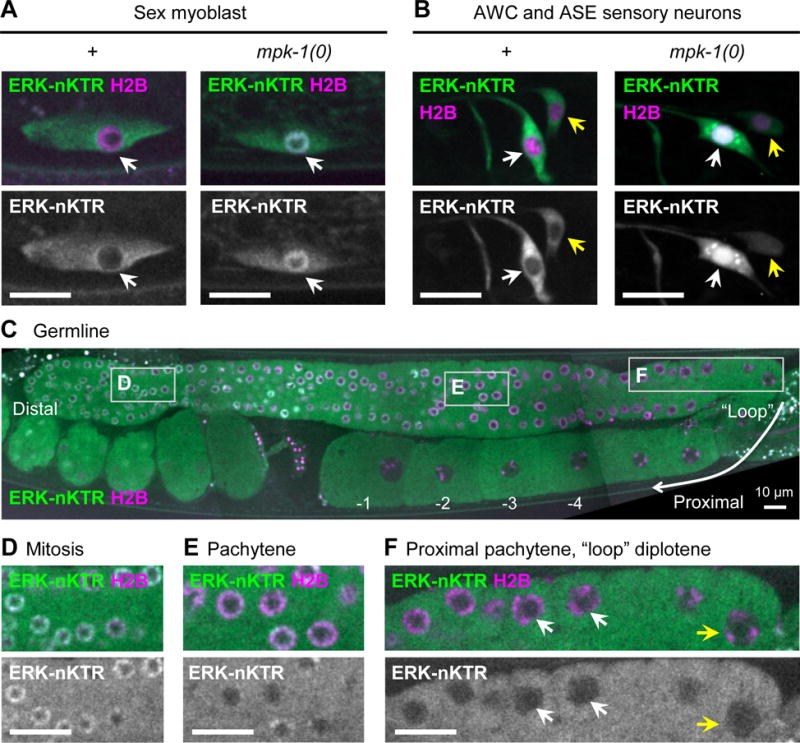
(A) ERK-nKTR is excluded from nuclei of migrating Sex Myoblast (SM) cells in wild type, but not in mpk-1(0). SM-specific ERK-nKTR transgene arTi133, showing ERK-nKTR-mClover (ERK-nKTR) and mCherry-H2B (H2B) in wild type (+) and mpk-1(0). (B) ERK-nKTR is excluded from the nuclei of AWC and ASE sensory neurons in wild type, but not in mpk-1(0). AWC and ASE-specific ERK-nKTR transgene arTi137, showing ERK-nKTR-mClover (ERK-nKTR) and mCherry-H2B (H2B) in wild type (+) and mpk-1(0) AWC (white arrows) and ASE (yellow arrows) neurons. (C) ERK-nKTR is localized in a pattern correlated with MPK-1 activity in the hermaphrodite germline. Germline-expressed ERK-nKTR transgene arSi4, showing ERK-nKTR-mClover (ERK-nKTR) and mCherry-H2B (H2B). The most distal germline nuclei (oriented to left in this micrograph) are in mitosis. More proximally, nuclei enter meiosis and most of the length of germline remains in pachytene of Meiosis I. Proceeding proximally, the germline “loops”, and in this region nuclei enter diplotene, which is followed by diakinesis and oocyte maturation. (D-F) Images from insets indicated by boxes in (C). (D) ERK-nKTR accumulates in nuclei within the distal mitotic region, consistent with low MPK-1 activity detected in fixed samples (Lee et al., 2007). (E,F) ERK-nKTR levels are low in some nuclei in pachytene (E), but becomes highly excluded from nuclei in the proximal pachytene (white arrows) and loop region (yellow arrows) (F). In the most proximal region, ERK-nKTR is excluded from the nuclei undergoing diakinesis and oocyte maturation (C, labeled −1, −2, −3, −4). Nuclear exclusion of ERK-nKTR in proximal pachytene, diplotene, and diakinesis regions is consistent with regions where high levels of phosphorylated substrates are detected in fixed samples (Arur et al., 2011; Drake et al., 2014). Scale bars in all images are 10 μm.
Migrating Sex Myoblasts
The sex muscles that control egg-laying are derived from two precursors, the sex myoblasts (SMs), which are born near the posterior of the larva at the first larval (L1) stage and migrate towards the midbody of the larva in the L2 stage, attracted by an FGF-like signal that emanates from the somatic gonad (Burdine et al., 1998). The precise positioning of the SM at the proper location is mediated by the EGL-15/FGF receptor via the canonical Ras-Raf-MEK-ERK cascade, implying that MPK-1 should be active in migrating SMs (DeVore et al., 1995; Sundaram et al., 1996). Indeed, ERK-nKTR is visibly excluded from nuclei in migrating SMs in a wild-type background, demonstrating that MPK-1/ERK is active. Importantly, ERK-nKTR is no longer excluded from the nucleus in mpk-1(0), indicating that ERK-nKTR is indeed reporting mpk-1 activity in SMs (Fig. 4A).
Sensory neurons
The two AWC neurons mediate chemotaxis to volatile odorants, and the two ASE neurons mediate chemotaxis to water-soluble molecules (Bargmann, 2006). G-protein coupled receptor stimulation of MPK-1/ERK activity has been implicated in the function of these sensory neurons (Chen et al., 2011; Hirotsu and Iino, 2005; Tomida et al., 2012). We examined ERK-nKTR in well-fed worms, in which sensory neurons should be well stimulated by compounds produced by bacteria and chemicals in the growth media, and thus ERK should be active. Indeed, we find that ERK-nKTR is excluded from nuclei of AWC and ASE, and that nuclear exclusion is abrogated in mpk-1(0), indicating that ERK-nKTR is reporting MPK-1/ERK activity in these sensory neurons (Fig. 4B).
Germline
The developing germline displays a spatial pattern along a proximo-distal axis: at the distal end, germline progenitors proliferate mitotically; as they move more proximally, away from the proliferative signal, they enter meiosis. Continuing proximally, germ cells progress through meiosis and become oocytes, undergoing maturation during ovulation. mpk-1 acts autonomously in the germ line for multiple, genetically separable roles including meiotic progression and oocyte maturation (Arur et al., 2009; Church et al., 1995; Kim et al., 2012; Lee et al., 2007; Miller et al., 2001; Sundaram, 2013). Staining of fixed worms with an anti-phospho-ERK antibody revealed activity of this pathway in a corresponding spatial pattern: while MPK-1/ERK protein is present throughout the germ line, phospho-ERK immunostaining is highest in two zones: the proximal half of the pachytene zone and in the oocytes undergoing diakinesis (Lee et al., 2007). Phosphorylation of MPK-1 substrate proteins is also patterned, with high levels of phospho-substrates in the proximal pachytene, loop, and diakinesis (Arur et al., 2011; Drake et al., 2014). The germline therefore provides an opportunity for assessing the performance of the nKTR in a syncytium with spatially graded ERK activity.
Using a germline-specific promoter we expressed ERK-nKTR throughout the germ line and found that ERK-nKTR localization in vivo correlates with independent, direct measurements of active MPK-1/ERK and its phosphorylated substrates seen with immunostaining in extruded and fixed germ-lines described above. ERK-nKTR is enriched in nuclei in the mitotic zone where genetic analysis and phospho-ERK staining has indicated that MPK-1/ERK activity is low (Fig. 4C, D). ERK-nKTR is excluded from the nuclei of germ cells in the proximal pachytene region, the loop, and in diakinesis (Fig. 4C, E, F), a pattern that closely resembles phosphorylated MPK-1 substrates.
Quantitative imaging of ERK-nKTR in VPCs over time
The VPCs are born in the mid-L1 stage. During the L2 stage, Wnt produced from different cellular sources and EGF from the gonad maintains VPC competence for later vulval induction (Myers and Greenwald, 2007; Sternberg, 2005). At that time, graded expression of the egl-17 transcriptional reporter suggests graded activation of EGFR-Ras-ERK signal transduction in VPCs, with a peak in P6.p and lower activity in two neighboring VPCs, P5.p and P7.p (Burdine et al., 1998; Yoo et al., 2004). As development proceeds, the role of EGF changes, specifying P6.p to adopt the 1° fate. The fate specification process appears to begin in the L2 molt, based on the temperature-sensitive period of the let-23/EGFR gene (Ferguson et al., 1987) and the activation of lag-2, a second ERK-dependent transcriptional reporter that is directly regulated by the E RK substrate LIN-1/Elk1 (Zhang and Greenwald, 2011). P6.p becomes committed to the 1° fate early in the L3 phase, prior to entry into S phase of the cell cycle (Ambros, 1999). Fate commitment of P6.p is associated with positive and negative feedback mechanisms on the EGFR-Ras-ERK pathway (Berset et al., 2005; de la Cova and Greenwald, 2012). The flanking VPCs, P5.p and P7.p, are specified to adopt the 2° fate by activation of LIN-12/Notch, which negatively regulates EGFR-Ras-ERK signaling in the prospective 2° fate cells (Berset et al., 2001; Yoo et al., 2004).
Although useful, reliance on transcriptional reporters for precise inferences about ERK activation status and its quantitation is problematic because: (i) there may be a significant lag between the time transcription is activated and fluorescence is visualized, (ii) the available evidence suggests that egl-17 is not a direct transcriptional target of the EGFR-Ras-ERK pathway and responds to other inputs (Cui and Han, 2003; Tiensuu et al., 2005) and (iii) the restricted regulatory regions, multicopy nature and random insertion sites of “classic” reporter transgenes may affect their expression dynamics. The ERK-nKTR sensor we developed here can overcome these problems as it more directly reports ERK activation status.
We examined ERK activity in VPCs during the course of development by generating time-lapse movies of individual larvae. We used two different imaging protocols. In one protocol, we imaged at two-minute intervals within an hour time period in the late L2 stage, so as to capture potential ERK activation dynamics and resolve the frequency and amplitude of pulsatile activity in wild type (Fig. 5A–D). In the second protocol, we imaged at ten-minute intervals within 3–1/2 hour windows, limited by the efficacy of the anesthetic and the inability of larvae to survive the molting period on a slide. This protocol allowed us to span the entire L2 phase and early L3 stage, exclusive of the molt, and revealed lower-resolution information on ERK activity dynamics, including the earliest time of ERK activation in P6.p (Fig. 5E).
Figure 5. Live imaging of ERK-nKTR during VPC development.
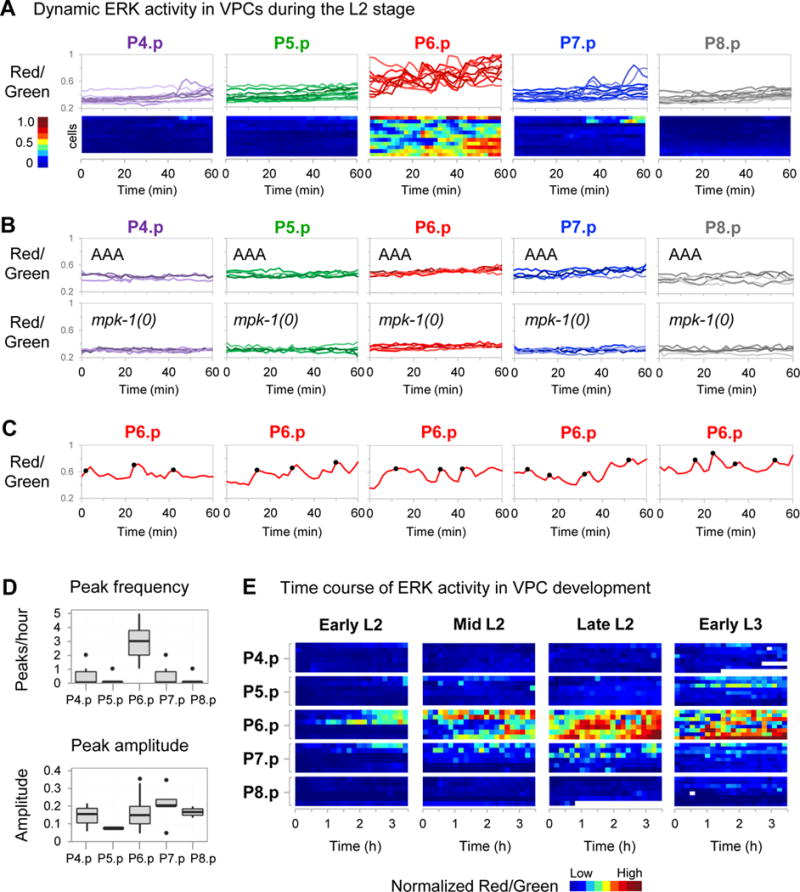
Unless otherwise indicated, movies were made of strains containing the transgene arTi85, which expresses ERK-nKTR in VPCs. (A) To examine ERK activity dynamics in wild-type late L2 larvae, time-lapse movies were captured by imaging at 2 minute intervals over a 1 hour time period (see STAR Methods). The Red/Green ratio of individual VPC nuclei over time is shown in two ways: all individual traces overlaid for each VPC (top), and a heatmap (bottom) where each row represents a cell, and Red/Green ratio is represented by color (n=11 larvae). (B) Time-lapse movies were captured as described in (A) to examine arTi101 larvae that express non-phosphorylatable ERK-nKTR(AAA) in an otherwise wild-type background (top, n=5 larvae), and mpk-1(0) larvae that express ERK-nKTR (bottom, n=5 larvae). Shown are all individual traces overlaid for each VPC. (C) Time-lapse Red/Green data for all individual nuclei were analyzed for peaks (STAR Methods). Shown are five representative traces for P6.p cells, where Red/Green of individual nuclei is shown over time, and the position of activity peaks are indicated by black circles. (D) Peak frequency (peaks/hour) is shown for each VPC (top). A two-sample k-test was performed to make pairwise comparisons of peak number (per hour) between VPCs. The peak frequency found in P6.p differed significantly from peak frequency in all other VPCs (p-value<1×10−6). The relative peak amplitude (difference between Red/Green ratio at peak and nearest trough) is shown for each VPC (bottom). No significant differences in relative peak amplitude were found in pairwise comparisons of different VPCs using a two-sample k-test. (E) Time-lapse movies were captured by imaging at 10 minute intervals over 4 overlapping 3.5-hour time periods after egg lay to examine ERK activity in wild-type VPC development: 20–23.5 hours (Early L2), 23–26.5 hours (Mid L2), 26–29.5 hours (Late L2), and 30–33.5 hours (Early L3) (see also STAR Methods, Fig. S3A). Heat maps where each row represents a cell, and Red/Green ratio of nuclei is represented by color. To correct for variation between animals, the average Red/Green value for the P4.p nucleus of a given larva was used as an internal baseline for normalization of other VPCs in that individual, since P4.p experiences minimal EGF signal (see Discussion and STAR Methods). Occasional missing values in heatmaps (white) result when the nucleus is out of focus for the time point.
Frequency modulated signaling in P6.p
In P6.p, ERK activity is consistently elevated compared to other VPCs, and displays pulse-like changes over time when sampled at high resolution (Fig. 5A, C); in addition, we observed infrequent activity pulses in other VPCs (Fig. 5A). The ERK activity dynamics we observe are the result of meaningful biochemical activity and not an artifact of our experimental system because ERK activity pulses are absent in the ERK-nKTR(AAA) reporter in an otherwise wild-type background, and when the wild-type ERK-nKTR reporter is examined in the mpk-1(0) mutant background (Fig. 5B).
Analysis of the frequency and amplitude of ERK activity pulses in VPCs shows that P6.p sustains significantly more peaks over time, but the relative peak amplitude does not differ from other VPCs (Fig. 5D). These and similar observations in cultured cells have been modeled as a frequency-modulated response where frequency of ERK activity pulses is proportional to EGF concentration, and where very high EGF concentration produces highly frequent pulses manifested as a steady, elevated response. In sum, our observations of signaling dynamics during VPC development show frequency-modulated signaling in a developmental context, underscoring the utility of the ERK-nKTR as a reliable reporter for single cell and real-time characterization of ERK signaling in vivo.
LIN-3/EGF-dependent ERK activity in early L2 stage
In movies covering 3.5-hour intervals across the L2 and L3 stages, we were surprised to find that the earliest ERK activity begins in P6.p an hour after the L1–L2 molt (Fig. 5E), before the anchor cell has been specified and prior to expression of the egl-17 transcriptional reporter used to assess EGFR-Ras-ERK activation (Yoo et al., 2004). At this time, however, the four somatic gonad cells with anchor cell potential transcribe the lin-3/EGF gene (Hwang and Sternberg, 2004), suggesting that ERK-nKTR activity may still be responding to gonadal lin-3/EGF. Indeed, ERK activity is absent in a mutant that lacks lin-3/EGF expression in the anchor cell (Fig. S3C), indicating that EGF, and not just the transcript, is produced at this early stage and is responsible for ERK activation.
During early and mid-L2, the amplitude of ERK activity in P6.p steadily rises and exhibits fluctuations (Fig. 5E). By late L2, activity in P6.p continues to fluctuate and appears to reach a plateau in amplitude. We also detect less frequent ERK activity pulses in other VPCs, notably P7.p, in mid and late L2 stages. In the early L3 stage, ERK remains active in P6.p, in the same time frame that it becomes committed to the 1° fate (Ambros, 1999). The infrequent ERK activity pulses observed in P7.p in the L2 stage appear to be diminished in early L3. We verified that the ERK activity we detect at all stages is absent in the ERK-nKTR(AAA) reporter and in the wild-type ERK-nKTR reporter in the mpk-1(0) background (Fig. S3A, B).
Finally, we note that we used two-minute interval movies to assess the premise that the Red/Green nuclear ratio is an accurate method. First, we examined the intensity of ERK-nKTR and H2B over time and find that the observed pulsatile changes in Red/Green are due to fluctuations in nuclear ERK-KTR, while nuclear H2B is constant (Fig. S1D). Second, we compared Red/Green to the ratio of Cytoplasmic/Nuclear Green as used for the original KTR (Regot et al., 2014), and 1/Green, using the nucleus only as a mask (Fig. S1E). The dynamics look similar regardless of the ratiometric paradigm used. However, the Red/Green nucleus-only design facilitates segmentation and hence remains our preferred method of data analysis.
Discussion
We have established that the ERK-nKTR biosensor technology is readily implemented through standard transgenesis and can be used for monitoring ERK activity in individual cells in C. elegans, at single time-points or over prolonged periods of time. We validated the sensor as responsive to ERK in developing polarized epithelial cells (VPCs), migrating muscle cell precursors (SMs), sensory neurons (AWC and ASE), and in the germ line. Each of these cases has been paradigmatic for developmental or neural processes in mammals. Furthermore, the bicistronic sensor design described here and the approach to image analysis, and the various considerations we describe in developing it, will be useful in extending ERK-nKTR to other experimental systems, as well as for adapting KTR sensors for other kinases (Regot et al., 2014) for use in vivo.
Limitations of fluorescent sensor technology and potential solutions in vivo
A limitation of KTR technology (and other fluorescent sensors of kinase activity) is that it is necessary to correct for kinase-independent parameters such as expression level, protein stability, and nuclear import/export rates that might otherwise affect the reporter baseline. Reporter baseline must be established by including controls to measure reporter activity in the absence of phosphorylation and kinase activity.
In our experiments, we determined the baseline Red/Green value in two ways. First, we used expression of a non-phosphorylatable derivative, ERK-nKTR(AAA), to measure basal variation in each VPC in the absence of phosphorylation. Similarly, we used mpk-1(0) to assess basal variation in each VPC in the absence of ERK activity. As a general approach for normalization in long-term imaging in other cellular paradigms, we suggest that ERK-nKTR(AAA) could be co-expressed with the native ERK-nKTR using the T2A linker; this strategy would be similar to that used for the “in-cell” control in the study of JNK-KTR (Regot et al., 2014). For in vivo analysis, coexpression of the two forms may best be done by incorporating the non-phosphorylatable KTR form into a bicistronic transcript with a native KTR; an additional nuclear marker would be necessary for segmentation.
We note also that the KTR approach measures the phosphorylation state of the ERK substrate, which reflects both the forward phosphorylation of the substrate and dephosphorylation by unknown phosphatases. This same limitation applies to other approaches to measuring ERK activity in live individuals, such as FRET sensors, as well as genetic assays such as suppression or enhancement of phenotypes or expression of transcriptional reporters, which are furthermore likely to have additional inputs other than ERK activity per se that affect the output. Of these various options, we think the ERK-nKTR approach is the most generally useful and easily implemented technology.
Signaling dynamics in VPCs in wild-type hermaphrodites
ERK-nKTR has allowed us to quantify and compare ERK signaling activity and signaling dynamics in VPCs in wild-type hermaphrodites, yielding a more complete and finer-grained picture of ERK activity in this cell fate specification paradigm. Our analysis revealed frequency-modulated dynamics in VPCs in response to EGF. Previously, ERK activity sensors revealed frequency modulation in human cells provided different concentrations of EGF (Albeck et al. 2013), similar to what we have observed in VPCs. In contrast, amplitude modulation was observed in intact mouse skin epithelia mediated by EGFR (Hiratsuka et al., 2015). These observations suggest that there are different modes of regulating ERK activity dynamics.
Data collected from time-lapse movies indicate that ERK activity in VPCs is dynamic and pulsatile, and P6.p is distinguished from all other VPCs in sustaining more frequent ERK activity peaks. At this time, all VPCs have the potential to adopt the 1° fate (Greenwald et al., 1983; Sternberg and Horvitz, 1986; Sulston and White, 1980); however, P6.p is closest to the EGF-producing anchor cell. Thus, we infer EGF level differences found in physiological gradients in vivo are capable of producing different frequencies of pulsatile ERK activity. Interestingly, while the frequency of ERK activity pulses differed in P6.p compared to other VPCs, the relative amplitude of pulses did not, a behavior comparable to the response of human cells provided different concentrations of EGF (Albeck et al. 2013). Although our short time-period movies did not capture a significant difference in the ERK pulse frequency of P5.p and P7.p compared to more distant VPCs, we were able to observe a graded pattern of ERK activity when quantified in a large number of animals at a single time point, consistent with graded expression of the egl-17 transcriptional reporter at this stage (Yoo et al., 2004). Future genetic manipulations of LIN-3/EGF and LET-23/EGFR signaling will help illuminate if different frequencies of pulsatile ERK activity relates to different ERK-triggered outcomes.
In our survey of ERK activity across VPC developmental stages (Fig. 5E), we find that the earliest detectable ERK activity in VPCs is evident in P6.p in the first hour of the L2 stage, an earlier time than activation of known ERK-dependent transcriptional outcomes. This activity is LIN-3/EGF-dependent, reflecting the immediacy of KTR technology for assessing signaling compared to transcriptional reporters, which require transcription, translation, and maturation of the fluorescent protein. This early time frame is before the anchor cell has been specified, and suggests that LIN-3/EGF is produced, not just transcribed, in the four somatic gonad cells with anchor cell potential (Hwang and Sternberg, 2004; Sallee and Greenwald, 2015; Seydoux and Greenwald, 1989). This early L2 stage ERK activity is interesting in view of the redundant role of EGF with Wnt signaling to maintain P6.p in an uncommitted, competent state (Myers and Greenwald, 2007). Interestingly, the number of cells with vulval competence is a variable feature in nematode evolution and there has been divergence in the quantitative contribution of EGF-like signal to vulval induction (Félix, 2007; Kiontke et al., 2007); furthermore, the robustness of VPC fate is influenced by signal strength (Barkoulas et al., 2013; Braendle and Félix, 2008).
The analysis described here for VPCs in wild-type development provides a road map for future examination of such dynamics in different cell types in vivo. This description of in vivo ERK signaling dynamics will also provide the baseline for interpreting experimental perturbations of Ras-Raf-MEK-ERK regulation mechanistically, and to understand the contributions of individual conserved regulators such as SEL-10/Fbw7 (de la Cova and Greenwald, 2012), GAP-1/RasGTPase-activating protein (Hajnal et al., 1997), and others to signaling dynamics in development.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Iva Greenwald (isg4@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains
Strain names and full genotypes are listed in the Key Resources Table. Caenorhabditis elegans strain N2 was the parent strain in which transgenes were generated. The following alleles were obtained from the Caenorhabditis Genetics Center: LGIII: mpk-1(ga117), oxTi615[eft-3∷tdTomato-H2B∷unc-54 3′UTR], LGIV: lin-3(e1417).
Because mpk-1(ga117) (null) adults are sterile, strains carrying mpk-1(ga117) were maintained as heterozygotes over the visible marker oxTi615. Homozygous mpk-1(ga117) segregants—designated mpk-1(0) in text—were picked for analysis. We note that in all cases where we analyzed ERK-nKTR in mpk-1(0), ERK-nKTR was not excluded from the nucleus, indicating that potential maternal contribution was negligible and consistent with the completely penetrant Vulvaless phenotype of such hermaphrodites.
jccTi1, which marks all VPCs, is a single-copy insertion containing lin-31p∷mCherry-his-58∷unc-54 3′UTR, and was a gift of Julie Canman.
arTi22, which marks the four proximal somatic gonad cells with anchor cell (AC) potential in the L2 stage, is a single-copy transgene containing hlh-2prox∷gfp-H2B∷unc-54 3′UTR made by Michelle Attner, based on the promoter hlh-2prox (Sallee and Greenwald, 2015).
The transgene arTi31 encodes a mutant LIN-45 protein that is constitutively active and stable. It is a single-copy insertion containing lin-31p∷lin-45(T432A,S436A,V627E)::unc-54 3′UTR and was modeled after transgenes described in de la Cova and Greenwald (2012).
METHOD DETAILS
Plasmids and transgenes used for expression in C. elegans
All plasmids used are listed in the Key Resources Table and new plasmids and transgenes generated in this work are described in Table S1. The ERK-nKTR cDNA was derived from pENTR-ERKKTRClover (Addgene, #59138) and encodes ERK-KTR fused to mClover (Regot et al., 2014) followed by a T2A peptide (Ahier and Jarriault, 2014) fused to mCherry and HIS-11. Established regulatory sequences were used for tissue-specific expression.
VPCs
The VPC-specific transgene lin-31p::ERK-KTR-mClover-T2A-mCherry-his-11::unc-54 3′UTR contains 5′ and intronic regulatory sequences derived from the lin-31 gene (Tan et al., 1998), designated “lin-31p.” VPC-specific transgenes that express “AAA” and “EEE” mutant derivatives were identical other than the phosphorylation site mutations.
SM
The SM-specific transgene hlh-8p::ERK-KTR-mClover-T2A-mCherry-his-11::unc-54 3′UTR contains regulatory sequences from −1791 to +1 upstream of the hlh-8 gene (Harfe et al., 1998), designated “hlh-8p.”
AWC and ASE
The sensory neuron-specific transgene ceh-36p::ERK-KTR-mClover-T2A-mCherry-his-11::unc-54 3′UTR contains regulatory sequences named “prom2” derived from the ceh-36 gene (Etchberger et al., 2007).
Germline
The germline-specific transgene mex-5p::ERK-KTR-mClover-T2A-mCherry-his-11::tbb-2 3′UTR contains regulatory sequences from the mex-5 and tbb-2 genes that allow uniform germline expression throughout larval and adult development (Merritt et al., 2008).
All somatically-expressed transgenes (driven by regulatory sequences from the lin-31, hlh-8, or ceh-36 genes) contain the unc-54 3′UTR, and were cloned in the miniMos transgenesis vector pCFJ910 (Frøkjær-Jensen et al., 2014) (Addgene, #44481). Random, single-copy miniMos transgenic strains were generated in the N2 strain by germline injection of hermaphrodites with 10 ng/μL transgene plasmid with a co-injection mixture of pCFJ601[eft-3::mos1 transposase::tbb-2 3′UTR], pCFJ90[myo-3::mCherry::unc-54 3′UTR], pGH8[rab-3::mCherry::unc-54 3′UTR], and pMA122[hsp-16.41::peel-1::tbb-2 3′UTR]. Strains carrying integrated transgenes were isolated as described by (Frøkjær-Jensen et al., 2014).
The germline-expressed mex-5-derived transgene contains the tbb-2 3′ UTR, and was cloned in pWZ111, a vector designed for Crispr/Cas9-mediated single-copy insertion at the germline-permissive ttTi4348 locus (Frøkjær-Jensen et al., 2012). Crispr/Cas9-mediated single-copy transgenic strains were generated in N2 by germline injection of 10 ng/μL transgene plasmid with a co-injection mixture of pAP082[eft-3::Cas9::tbb-2 3′UTR, U6::sgRNA targeting the ttTi4348 locus], pCFJ90[myo-3::mCherry::unc-54 3′UTR] and pGH8[rab-3::mCherry::unc-54 3′UTR]. Strains carrying integrated transgenes were isolated as described (Dickinson and Goldstein, 2016; Dickinson et al., 2015). We gratefully acknowledge that the reagents used for insertion at this locus were made by Ari Pani and Bob Goldstein, and gifted to us by David Matus.
Image acquisition
To image ERK-KTR in VPCs and SMs in the L2 stage, larvae were grown at 25°C, mounted on an agarose pad at 24 hours after an egg lay, and anesthetized using 0.5 μM Levamisole in M9. Similarly, the ERK-KTR was imaged in sensory neurons and germline of young adults mounted on agarose pads at 48 hours after an egg lay. To image development in real-time, L2 larvae were anesthetized for 20 minutes using a mixture of 0.1% Tricaine/0.01% Tetramisole, then mounted on an agarose pad in M9 without anesthetic as described in (McCarter et al., 1999). We used different imaging protocols to generate 1) short, high-resolution data, and 2) longer, low-resolution data. To capture ERK dynamics with high resolution in time, we imaged at two-minute intervals within a 1 hour time period beginning at 26 hours after an egg lay. To image ERK activity over longer developmental periods, we imaged at ten-minute intervals over four developmental periods (time in hours after egg lay): early L2 (20–23.5 hr), mid L2 (23–26.6 h), late L2 (26–29.5 hr), early L3 (30–33.5 hr). The duration times of our movies were limited to 3–4 hours because the anesthetic typically became ineffective after 4 hours. Additionally, developing larvae did not survive the molting period on slides even in the absence of anesthetic.
Z-stacks of mClover and mCherry fluorescence were captured simultaneously using a spinning disk confocal, dual camera system (Carl Zeiss). For all images, exposure times used for mClover and mCherry were equal: 500 ms was used for “still” images of a single time point, and 150 ms was used for movie images. For all experiments, blank images were collected using the same parameters.
Image Quantification
A workflow of the image quantitation methods is provided in Table S4. Matlab and Cellprofiler scripts used in this study are provided in https://github.com/GreenwaldLab/de-la-Cova-2017. Illumination correction was done using blank images obtained for every experiment. A ratiometric image was calculated by dividing mCherry intensity values by mClover intensity values for each pixel. The resulting image (Red/Green) was normalized between the percentiles 0.1 and 99.9% to avoid artifacts from hot and cold pixels in the camera. Segmentation and object quantification was performed with CellProfiler (Kamentsky et al., 2011), treating each Z stack image independently. Nuclei were segmented using the mCherry-H2B signal and the upper quartile intensity of the ratiometric Red/Green image was obtained. Tracking of cells within the Z-stack and manual annotation of the VPC identities were performed using custom software. For each VPC nucleus, the mean Red/Green of the five most equatorial Z sections is presented as the measurement “Red/Green”.
To compare Red/Green values acquired at different developmental stages, data from VPCs at each time interval was normalized using the mean Red/Green value of P4.p in the same larva. P4.p was utilized as an internal control because Red/Green values in this cell were well correlated with Red/Green values of ERK-nKTR in mpk-1(0) (Fig. 2F, Fig. 3C).
To detect pulsatile peaks in ERK activity, time-lapse ERK-nKTR Red/Green data was analyzed using a custom peak finding algorithm in Matlab. Specifically, peaks where identified using slope and relative amplitude thresholds that were manually set to maximize agreement with visually distinct peaks. The relative amplitude threshold (0.05 in Red/Green units) was greater than the average difference between time points observed in ERK-nKTR(AAA) at each developmental stage (mean difference ± 95% confidence interval was 0.015 ±0.024 in early L2; 0.020 ± 0.029 in mid-L2; 0.019 ± 0.016 in late L2; 0.027 ± 0.022 in early L3). To measure the relative amplitude of individual ERK activity peaks, the difference between Red/Green ratio at peak and nearest trough was calculated.
QUANTIFICATION AND STATISTICAL ANALYSIS
When comparing all VPCs for differences in mean Red/Green ratio (Fig. 2, 3, Tables S5, S6, and S7), we performed one-way ANOVA tests followed by pairwise comparisons using the Tukey HSD test. In pairwise comparisons of VPCs for differences in peak number and relative peak amplitude (Fig. 5D), we performed two-sample k-tests. For all significant test results, p-values are provided within Figure legends.
DATA AND SOFTWARE AVAILABILITY
Software and algorithms used in this work are listed in the Key Resources Table and Table S4.
Supplementary Material
Highlights.
ERK-nKTR is a genetically-encoded biosensor for extracellular signal-regulated kinase
Qualitative or quantitative assessment is possible by analyzing individual nuclei
ERK-nKTR is a faithful reporter for ERK activity in several different cell types
In the Vulval Precursor Cells, there is pulsatile, frequency-modulated signaling
Acknowledgments
We gratefully acknowledge Markus Covert, Jake Hughey and Sajia Akhter for assisting with custom software development; Ari Pani, Bob Goldstein and David Matus for generously sharing reagents and advice for CRISPR/Cas9-mediated single copy transgene insertion; Julie Canman for the gift of a VPC marker; and Swathi Arur and Tim Schedl for helpful discussion about the germ line. We are also indebted to Daniel Shaye and Claudia Tenen for helpful comments on the manuscript. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center, which is supported by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by Senior Scholar Award AG-SS-2951-12 from the Ellison Medical Foundation and grant R01 GM114140 from the NIH (to I.G.), and a 2016 Kimmel Scholar Award (to S.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.D., R.T., S.R., and I.G. designed the experiments. C.D. and R.T. conducted the experiments. S.R. designed and supervised the image quantification methods. C.D., R.T., S.R., and I.G. wrote the paper.
References
- Ahier A, Jarriault S. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics. 2014;196:605–613. doi: 10.1534/genetics.113.160846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Molecular Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development. 1999;126:1947–1956. doi: 10.1242/dev.126.9.1947. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Molecular Cell. 2013 doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Berkseth M, Nayak S, Hansen D, Zarkower D, Schedl T. MPK-1 ERK controls membrane organization in C. elegans oogenesis via a sex-determination module. Developmental Cell. 2011;20:677–688. doi: 10.1016/j.devcel.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc Natl Acad Sci USA. 2009;106:4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nature Publishing Group. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. In: Chemosensation in C. elegans. WormBook, editor. The C. elegans Research Community, WormBook; 2006. (October 25, 2006) [Google Scholar]
- Barkoulas M, van Zon JS, Milloz J, Van Oudenaarden A, Félix MA. Robustness and Epistasis in the C. elegans Vulval Signaling Network Revealed by Pathway Dosage Modulation. Developmental Cell. 2013;24:64–75. doi: 10.1016/j.devcel.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science. 2001;291:1055–1058. doi: 10.1126/science.1055642. [DOI] [PubMed] [Google Scholar]
- Berset TA, Hoier EF, Hajnal A. The C. elegans homolog of the mammalian tumor suppressor Dep-1/Scc1 inhibits EGFR signaling to regulate binary cell fate decisions. Genes Dev. 2005;19:1328–1340. doi: 10.1101/gad.333505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C, Félix MA. Plasticity and Errors of a Robust Developmental System in Different Environments. Developmental Cell. 2008;15:714–724. doi: 10.1016/j.devcel.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- Chen L, Fu Y, Ren M, Xiao B, Rubin CS. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron. 2011;70:51–65. doi: 10.1016/j.neuron.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- de la Cova C, Greenwald I. SEL-10/Fbw7-dependent negative feedback regulation of LIN-45/Braf signaling in C. elegans via a conserved phosphodegron. Genes Dev. 2012;26:2524–2535. doi: 10.1101/gad.203703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore DL, Horvitz HR, Stern MJ. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Goldstein B. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics. 2016;202:885–901. doi: 10.1534/genetics.115.182162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015;200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M, Furuta T, Suen KM, Gonzalez G, Liu Bin, Kalia A, Ladbury JE, Fire AZ, Skeath JB, Arur S. A Requirement for ERK-DependentDicer Phosphorylation in Coordinating Oocyte-to-Embryo Transition in C. elegans. Developmental Cell. 2014;31:614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezak MJ, Ferkey DM. A functional nuclear localization sequence in the C. elegans TRPV channel OCR-2. PLoS ONE. 2011;6:e25047. doi: 10.1371/journal.pone.0025047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature. 1987;326:259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Félix MA. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr Biol. 2007;17:103–114. doi: 10.1016/j.cub.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Meth. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, Jorgensen EM. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Meth. 2014;11:529–534. doi: 10.1038/nmeth.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hajnal A, Whitfield CW, Kim SK. Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev. 1997;11:2715–2728. doi: 10.1101/gad.11.20.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci USA. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T, Fujita Y, Naoki H, Aoki K, Kamioka Y, Matsuda M. Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. Elife. 2015;4:e05178. doi: 10.7554/eLife.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu T, Iino Y. Neural circuit-dependent odor adaptation in C. elegans is regulated by the Ras-MAPK pathway. Genes Cells. 2005;10:517–530. doi: 10.1111/j.1365-2443.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Sternberg PW. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development. 2004;131:143–151. doi: 10.1242/dev.00924. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Beitel GJ, Clark SG, Horvitz HR, Kornfeld K. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics. 1998;149:1809–1822. doi: 10.1093/genetics/149.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Spike C, Greenstein D. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv Exp Med Biol. 2012;757:277–320. doi: 10.1007/978-1-4614-4015-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Barrière A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DHA, Félix MA. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer. 2015;15:515–527. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple Functions and Dynamic Activation of MPK-1 Extracellular Signal-Regulated Kinase Signaling in Caenorhabditis elegans Germline Development. Genetics. 2007;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Dsilva CJ, Levario TJ, Lu H, Schüpbach T, Kevrekidis IG, Shvartsman SY. Dynamics of Inductive ERK Signaling in the Drosophila Embryo. Curr Biol. 2015;25:1784–1790. doi: 10.1016/j.cub.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko NN, Hanna-Rose W, Schlegel RA. Cognate putative nuclear localization signal effects strong nuclear localization of a GFP reporter and facilitates gene expression studies in Caenorhabditis elegans. BioTechniques. 2007;43:596–598. 560. doi: 10.2144/000112615. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Mattingly HH, Chen JJ, Arur S, Shvartsman SY. A Transport Model for Estimating the Time Course of ERK Activation in the C. elegans Germline. Biophys J. 2015;109:2436–2445. doi: 10.1016/j.bpj.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Developmental Biology. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs Are the Primary Regulators of Gene Expression in the C. elegans Germline. Current Biology. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Myers TR, Greenwald I. Wnt signal from multiple tissues and lin-3/EGF signal from the gonad maintain vulval precursor cell competence in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:20368–20373. doi: 10.1073/pnas.0709989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Chung M, Dobrzyński M, Fey D, Blum Y, Lee SS, Peter M, Kholodenko BN, Jeon NL, Pertz O. Frequency modulation of ERK activation dynamics rewires cell fate. Molecular Systems Biology. 2015;11:838. doi: 10.15252/msb.20156458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee MD, Greenwald I. Dimerization-driven degradation of C. elegans and human E proteins. Genes Dev. 2015;29:1356–1361. doi: 10.1101/gad.261917.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SDM, Verveer PJ, Bastiaens PIH. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57:1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Molecular Systems Biology. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparta B, Pargett M, Minguet M, Distor K, Bell G, Albeck JG. Receptor Level Mechanisms Are Required for Epidermal Growth Factor (EGF)-stimulated Extracellular Signal-regulated Kinase (ERK) Activity Pulses. Journal of Biological Chemistry. 2015;290:24784–24792. doi: 10.1074/jbc.M115.662247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW. In: Vulval development. WormBook, editor. The C. elegans Research Community, WormBook; 2005. Jun 25, p. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. Pattern formation during vulval development in C. elegans. Cell. 1986;44:761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Developmental Biology. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Yochem J, Han M. A Ras-mediated signal transduction pathway is involved in the control of sex myoblast migration in Caenorhabditis elegans. Development. 1996;122:2823–2833. doi: 10.1242/dev.122.9.2823. [DOI] [PubMed] [Google Scholar]
- Sundaram MV. In: Canonical RTK-Ras-ERK signaling and related alternative pathways. WormBook, editor. The C. elegans Research Community, WormBook; 2013. (July 1, 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci USA. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida T, Oda S, Takekawa M, Iino Y, Saito H. The Temporal Pattern of Stimulation Determines the Extent and Duration of MAPK Activation in a Caenorhabditis elegans Sensory Neuron. Science Signaling. 2012;5:ra76–ra76. doi: 10.1126/scisignal.2002983. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- Zhang X, Greenwald I. Spatial regulation of lag-2 transcription during vulval precursor cell fate patterning in Caenorhabditis elegans. Genetics. 2011;188:847–858. doi: 10.1534/genetics.111.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


