Abstract
Lipid droplets are cytoplasmic organelles that store neutral lipids and are critically important for energy metabolism. Their function in energy storage is firmly established and increasingly well characterized. However, emerging evidence indicates that lipid droplets also play important and diverse roles in the cellular handling of lipids and proteins that may not be directly related to energy homeostasis. Lipid handling roles of droplets include the storage of hydrophobic vitamin and signaling precursors, and the management of endoplasmic reticulum and oxidative stress. Roles of lipid droplets in protein handling encompass functions in the maturation, storage, and turnover of cellular and viral polypeptides. Other potential roles of lipid droplets may be connected with their intracellular motility and, in some cases, their nuclear localization. This diversity highlights that lipid droplets are very adaptable organelles, performing different functions in different biological contexts.
1.0 Introduction
Lipid droplets (LDs) are intracellular organelles specialized for the storage of energy in the form of neutral lipids such as triglycerides and sterol esters. They are ubiquitous organelles, present in animals, plants, fungi, and even bacteria [1, 2]. LDs comprise a core of neutral lipids surrounded by a polar lipid monolayer containing many different proteins, some of which are involved in lipid metabolism [1-4]. Synthesis of neutral lipids in the endoplasmic reticulum (ER) membrane by enzymes such as diglyceride acyltransferase 1 (DGAT1) leads to the formation of nascent LDs, which emerge as organelles partially or completely distinct from the ER [5, 6]. After formation, LDs can continue to grow by local synthesis of neutral lipids or – in certain specialized tissues like adipocytes – via fusion [5, 7]. In times of need, the neutral lipids stored in LDs can be broken down by either of two pathways, lipolysis or lipophagy [8]. Lipolysis is mediated via lipid droplet-associated lipases such as adipose triglyceride lipase (ATGL) whereas lipophagy involves lysosomal acid lipase (LAL) acting upon LDs delivered to autolysosomes via autophagy. Fatty acids (FA) liberated from neutral lipid breakdown can then be used for energy production via mitochondrial or peroxisomal beta-oxidation [9, 10]. LDs therefore respond to the cellular balance of lipogenesis and lipolysis and so play a key role in energy homeostasis. This homeostatic role is critical for normal cellular and organismal function and becomes impaired in many human pathologies including obesity, diabetes, cardiovascular disease and fatty liver disease [11].
The unique cell-biological properties of LDs and their important physiological roles have resulted in an explosion of research in recent years. This has led to a better understanding of the basic mechanisms by which LDs form, grow, and are turned over to regulate energy homeostasis. These aspects of LDs have been reviewed extensively (e.g., [1-7, 11-21]); see also the reviews in this issue by Olzmann, Mashek, Zechner and Kratky, and Yen and Reue. Yet the importance of LDs is not restricted to energy storage. Emerging evidence suggests that LDs also function in the storage of a wide range of lipids with diverse functions as well as protecting against some forms of cellular stress. In addition, LDs have important roles in protein homeostasis, assisting the maturation, storage, and turn-over of many different proteins. Here we review these non-energy storage roles of LDs and identify important recent developments in this emerging field.
2.0 Lipid droplets and lipid handling
Lipidomics reveals that the core of an LD can contain over 100 different species of neutral lipids [22-26]. This repertoire is sure to expand over the next few years with the development of increasingly sophisticated lipidomics methods as well as imaging techniques based on Raman and mass spectrometry [27-34]. In many cell types, including adipocytes and hepatocytes, the majority of lipids in the droplet core correspond to triglycerides and sterol esters with a range of different FA side chains. These play important functions in energy storage and also as a major source of phospholipid precursors and cholesterol for cell membranes. Such precursors can be released from LDs for use in maintaining the homeostasis of the ER and other membranes, particularly during nutrient stress [35-38]. LDs are also part of the machinery that allows cells to adjust the balance of lipid flux that goes to membranes (for growth) or to storage (for starvation/stress survival). A key metabolic branchpoint here involves regulating how much phosphatidic acid (PA) in the ER membrane is used to synthesize membrane phospholipids versus LD triglycerides. In yeast, phosphatidate phosphatase (Pah1) converts PA into diglycerides, the precursors of triglycerides [39]. Nutrient depletion leads to relocalization of Pah1 from the cytoplasm to the nuclear envelope, which is contiguous with the ER, at a subdomain that contacts growing LDs. Lack of nutrients in yeast leads to cytoplasmic acidification and this is thought to activate Pah1, via the Nem1-Spo7 complex [39]. Hence, activation of the Pah1 pathway during starvation may be an important part of the cellular survival response that diverts fatty acyl chains from PA away from membrane biogenesis and into storage in LDs.
2.1.1 What's inside a lipid droplet?
LDs can store more unusual cargo than triglycerides and sterol esters. These lipophilic molecules play diverse functions not directly related to energy storage. Neutral ether lipids of the monoalk(en)yl diacylglycerol (MADAG or MDG) family account for ∼20% of the droplet lipids isolated from mammalian cell lines grown in the presence of oleate [22]. MADAG is also present in droplets isolated from liver but not from white or brown fat, indicating that it is a cell-type specific component of the droplet core [22]. As with other ether lipids, the first steps of MADAG biosynthesis occur in peroxisomes and, correspondingly, it is absent in cells from Zellweger patients defective in peroxisomal biosynthesis [22]. It has therefore been proposed that MADAG precursors are transferred from peroxisomes to LDs via direct contacts that have been observed between these two organelles [40, 41] See also review in this issue by Maya Schuldiner. MADAG in droplets may then function primarily as a precursor supply depot for the synthesis of ether phospholipids contributing to the phospholipid monolayer surrounding LDs and also to the bilayer of other cell membranes [22, 41].
Numerous plants, insects and microbes synthesize substantial quantities of hydrocarbons such as alkanes, alkenes, and isoprenoids [42-45]. These neutral lipids are likely to partition into the droplet core, although this has only been shown thus far in a few cases [42, 46]. For the hydrocarbon precursor of cholesterol, squalene, it has been shown in yeast that its sequestration in LDs is essential for preventing toxicity [47]. Natural rubber (cis-1,4-polyisoprene) is a commercially important hydrocarbon produced by laticifer cells of the rubber tree Hevea brasiliensis, but also made in varying amounts by thousands of other plant species. It is stored in specialized LDs called rubber particles, which lack structural proteins of the oleosin family common on other types of plant LDs [42]. Rubber is synthesized by the sequential condensation of isopentenyl diphosphate subunits [48]. The enzyme responsible, rubber transferase, likely corresponds to one or more cis-prenyltransferases localized to the ER and/or to the phospholipid monolayer of the rubber particle [42, 48]. The polyisoprene latex cargo of rubber particle LDs plays important roles in the protection and defense of plants against wounding, insect herbivory, and environmental stresses [49].
Another emerging function for LDs is in the cellular detoxification of endogenous or exogenous lipophilic molecules. A fungus residing asymptomatically inside lichens, Phaeosphaeria, can kill competing fungal species by producing toxic perylenequinones (PQs)[50]. PQs are toxic because light irradiation stimulates them to generate lethal levels of reactive oxygen species (ROS). PQ resistance of the producer fungus and also that the intended target species requires triglyceride and LD biosynthesis genes such as a diglyceride acyltransferase (DGAT). Storage of PQs in LDs is thought to provide a “safe haven” by somehow decreasing the ability of PQs to generate ROS. This and related detoxification functions for LDs may also have therapeutic implications. For example, some antifungal agents are more potent against yeast strains with defective LD biosynthesis [50]. Moreover, several drugs and prodrugs are known to accumulate in LDs [51, 52]. The hydrophobic anticancer prodrug CHR2863 localizes to LDs before being converted by esterase(s) into its active hydrophilic form, a cytotoxic aminopeptidase inhibitor [52]. Interestingly, myelomonocytic cell sublines selected for resistance to CHR2863 display increased LD abundance [52]. This raises the possibility that sequestration in LDs could be a therapeutically important mode of drug resistance and that inhibitors of droplet biogenesis might therefore be useful adjuncts for improving drug efficacy.
Many man-made environmental toxicants partition selectively into triglycerides rather than phospholipids and so would be expected to be concentrated in LDs [53]. Confirmed LD-associated toxicants include polycyclic aromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), and dioxins [54-57]. Hence, the ability of LDs to accumulate selectively several types of toxicants makes them a sensitive indicator of exposure to environmental pollutants. In the case of plants, it has even been proposed that detoxification of hydrophobic pollutants by LDs could be developed as a “biomimetic” strategy for environmental remediation [57].
2.1.2 Storage of vitamins and signaling precursors
For some fat-soluble vitamins and their metabolites, the LD core is known to provide a physiologically relevant storage site, but for others this is less clear. Vitamin E (tocopherol) is an antioxidant that accumulates in the LDs of plant chloroplasts and cyanobacteria [58, 59]. It is also present in the LDs of human adipocytes [60], although the functions of tocopherol storage here are not yet clear. More is known about vitamin A (retinol), a precursor of the nuclear receptor ligand retinoic acid. Vitamin A is esterified with FAs and accumulates in the core of LDs as retinyl esters [61-64]. The droplets of hepatic stellate cells are a major storage site for 70-80% of vitamin A in the body. These hepatic droplet stores ensure that there is a constant supply of vitamin A during periods of dietary insufficiency. Smaller quantities of retinyl esters are also stored in the LDs of extrahepatic stellate cells in the intestine, lung, and kidney as well as in hepatocytes and adipocytes [61, 62, 64-66]. Retinol is released from retinyl-esters by the action of retinyl-ester hydrolases [63, 67]. This enzymatic reaction is broadly analogous to FA mobilization from TAG by adipocyte triglyceride lipase (ATGL, see review in this issue by Zechner and Kratky). There appears to be no consensus yet on which enzymes are the most physiologically relevant hydrolases for retinyl-esters but it may be that ATGL itself contributes to this function in hepatic stellate cells, with hormone sensitive lipase (HSL) performing it in adipocytes [63, 68, 69]. It is also not yet clear how retinol is transferred from its retinyl-ester storage site in the LDs of hepatic stellate cells to its major site of release, complexed with retinol-binding protein 4, in hepatocytes. What is clear, however, is that hepatic LDs are an important source of vitamin A and they are critical for its homeostatic regulation.
The LD core can accumulate the precursors of molecules important for intercellular communication, such as steroid hormones and FA signals [70, 71]. In specialized endocrine cells of the gonads and adrenals, cholesteryl esters stored in LDs provide an important source of cholesterol for the mitochondrial biosynthesis of several different steroid hormones [72]. Proteomic analyses of LDs from several types of steroid-producing cells indicate that they copurify with some steroidogenic enzymes [71, 73]. This has led to the idea that, in addition to cholesterol storage, LDs may also function as a site for steroidogenic enzymes and/or the transfer of steroid intermediates [71, 73]. Cholesteryl ester-rich LDs may utilize different perilipins from their triglyceride-rich counterparts as it was observed that adrenocortical cells cultured with extra cholesterol upregulate PLIN1c and PLIN4, whereas those supplemented with oleic acid upregulate PLIN1a and PLIN5 [74]. Interestingly, inside a single adrenocortical cell, there is coexistence of separate cholesteryl ester-rich and triglyceride-rich LDs with distinct PLIN profiles [74].
LDs can also store the precursors of eicosanoids, a large family of signaling lipids that include prostaglandins, thromboxanes, and leukotriene [75]. They are ultimately derived from arachidonic acid (AA) and related 20-carbon polyunsaturated fatty acids (PUFAs). Free AA can be derived from enzymatic breakdown of membrane phospholipids but, in some cells, it is released from AA-rich triglycerides stored in LDs [75]. This is the case for mast cells, immune cells involved in inflammation, which have a considerable pool of AA triglycerides [76]. The lipase ATGL can release AA from these triglycerides and thus promote the production of eicosanoids [77, 78]. Inhibition of ATGL, genetically or pharmacologically, also impairs eicosanoid release in other types of immune cells, such as neutrophils [79]. In the intracellular parasites Leishmania and Trypanosoma, LDs can also act as a source of immunomodulatory prostaglandins implicated in improving the survival of the parasite within its host [80, 81]. The available evidence suggests that the enzymatically released AA is locally metabolized, as both eicosanoid precursors and eicosanoid producing enzymes have been found associated with LDs [75]. These observations suggest that LDs are not just a passive source of AA, but are intimately involved in the regulation of eicosanoid production. Newly developed immunofluorescence-based assays may now make it possible to visualize the site(s) of eicosanoid production within the cell and to explore the contribution of LDs in this process [82].
2.2 Roles for lipid droplets in managing cell stress
LDs increase in abundance in many diseases involving disrupted or rewired lipid metabolism, including non-alcoholic fatty liver disease and cancer [11, 83-86]. In murine hepatocytes, the FA composition and degree of saturation in the triglyceride fraction of purified LDs are both very sensitive to nutritional and genetic perturbations [24, 87, 88]. A common denominator associated with LD induction in non-adipose tissues is cell stress, which can be triggered by excess free FAs, nutrient deprivation and/or redox imbalances. In these contexts, LDs often appear to function in stress mitigation: either sequestering unwanted/harmful lipids or maintaining lipid homeostasis in membranes. Nevertheless, unambiguous assignment of cellular roles to LDs requires careful genetic analysis, and the outcomes are likely to depend upon which step of their biogenesis, maintenance or cargo turnover is being experimentally manipulated.
2.2.1 ER stress
Environmental insults or elevated protein synthesis can result in protein misfolding in the endoplasmic reticulum (ER). An imbalance between unfolded proteins and their chaperones, known as ER stress, then triggers the unfolded protein response (UPR) in order to try to restore protein homeostasis [89-91]. Excess FAs can also trigger the UPR, with saturated species acting as a more potent trigger than unsaturated ones [91-94]. High levels of saturated FAs appear to trigger the UPR by a process that does not require the unfolded-protein sensing domains of IRE1 and PERK but may involve sensing of perturbed ER membrane composition [95]. LDs increase in abundance in many contexts where the UPR is activated, raising the question of whether they function to ameliorate or to exacerbate this stress response. Yeast cells deficient in phosphatidylcholine synthesis activate the UPR and develop regions of abnormally fragmented ER [96]. These are called ER aggregation sites and they are associated with LDs as well as with a major component of the protein refolding machinery: the ER chaperone Kar2p/BiP. LDs are important during phosphatidylcholine deficiency as the triglyceride and LD biosynthesis enzymes Dga1p and Lro1p are needed for survival. LDs appear to act by stimulating the removal of unfolded ER proteins, which can then be degraded by ESM1-dependent microlipophagy in the yeast vacuole [96]. In another stress context, nitrogen starvation, yeast mutants deficient in triglyceride and sterol ester synthesis, and thus lacking LDs, displayed disrupted ER homeostasis and also a defect in a key step in autophagy - autophagosome biogenesis [38]. Together, these yeast studies suggest that LDs may regulate autophagy via a function in ER quality control, although it has also been proposed that LDs act by providing a lipid source for autophagosome membranes [36, 38, 97].
ER stress mitigation by increased fatty acid storage in LD triglycerides has also been reported in mammalian cells and tissues. Knockout of adipocyte triglyceride lipase (ATGL) in mice (see review by Zechner and Kratky) increases hepatic triglyceride storage in LDs and protects against tunicamycin-induced ER stress, decreasing stress markers such as GRP78 and spliced XBP1 [98]. Similarly, in human cardiomyocyte-derived cells, palmitate-induced ER stress can be inhibited by increasing triglycerides via the overexpression of PPARγ or Acyl-CoA synthetase 1 [99]. Conversely, ER stress markers can be increased by boosting the release of free FAs from LD triglycerides via the overexpression of ATGL [99]. In clear-cell renal cell carcinoma (ccRCC), constitutive Hypoxia Inducible Factor-2 alpha (HIF-2α) signaling upregulates the lipid droplet protein PLIN2 and induces the abundant LDs characteristic of this type of cancer [100]. PLIN2 knockdown abrogates LDs, dilates the ER, and activates PERK, IRE-1α and other UPR target genes. In the reverse experiment, providing exogenous PLIN2 is sufficient to restore LDs and to suppress the cell death of ccRCC cells lacking HIF-2α activity [100]. In another cancer context, Hela cells, overexpression of the autophagy protein ATG14 stimulates lipophagy, the selective autophagy of lipid droplets, leading to an increase in free FAs and concomitant ER stress [101]. In all four of the above mammalian contexts, the storage of triglycerides in LDs appears to play a critical role in mitigating ER stress. Although the underlying mechanisms are not yet clear, data in yeast are broadly consistent with an “escape hatch” model [96, 102], whereby LDs formed from the ER provide a vehicle for removing misfolded ER proteins as well as for rebalancing ER lipid homeostasis. This LD-mediated function for ER quality control likely functions alongside the ER-associated protein degradation (ERAD) pathway [103, 104].
2.2.2 Oxidative stress
Oxidative stress is associated with the accumulation of LDs in pathologies such as cancer and non-alcoholic fatty liver disease [83, 105-107]. It can be initiated by many different triggers, including hypoxia, chemical pro-oxidants, mitochondrial dysfunction, and ER stress. In each case, oxidative stress occurs when reactive oxygen species (ROS) increase to harmful levels that overwhelm cellular defense enzymes such as glutathione peroxidase (GPx), superoxide dismutase (SOD) and catalase. ER stress and oxidative stress are closely intertwined in multiple ways, and the former can trigger the latter by several UPR-dependent mechanisms [108, 109]. These include ROS induction by misfolded proteins, inhibition of NFE2-related factor 2 (Nrf2) and its antioxidant enzyme targets, as well as disruption of the mitochondrial electron transport chain via calcium leakage from the ER. Hypoxia is a common source of oxidative stress experienced by poorly vascularized cancer cells. It is sensed by Hypoxia Inducible factors (HIF-1α and/or HIF-2α) but also by other mechanisms including phospholipid mediators and lactate-dependent signaling [110-113]. HIF-1α upregulates the reductive carboxylation pathway, reducing glutamine to citrate in order to fuel FA synthesis during hypoxia [114, 115]. Metabolic tracer studies support the idea that de novo FA synthesis tends to decrease during hypoxia, although in some cases there is an increase in fatty acid synthase (FASN) expression and sterol regulatory element-binding protein 1 (SREBP1) activity [113, 116-120]. To compensate for an overall decrease in FA synthesis, hypoxic cancer cells can increase the scavenging of FAs from the extracellular medium [119-121]. In particular, scavenging of unsaturated FA chains from extracellular lysophospholipids can partially compensate for a decrease in lipid desaturation (lower oleate/stearate ratio), thought to result from hypoxic inhibition of the oxygen-consuming enzyme stearoyl-CoA desaturase 1 (SCD1) [119, 121]. The chemotherapy drug cisplatin also induces oxidative stress and, in medulloblastoma cells, this is concomitant with the accumulation of LDs [122, 123]. As with hypoxia, cisplatin decreases overall FA desaturation in whole cell extracts. Interestingly, however, unsaturated FAs such as the polyunsaturated fatty acid (PUFA) linoleate are enriched in LDs [123]. In the hypoxic state, HIF-1α not only modulates FA synthesis but it also regulates the conversion of FAs into LD triglycerides. This effect appears to be cell-type specific as HIF-1α promotes LD accumulation in hepatoblastoma cells, yet it dampens down triglyceride accumulation in colorectal cancer cells [124] [113].
Although LDs correlate with oxidative stress in many cell types, their functions during this process have only been pinpointed in a few cases. In the eye of the fruit fly Drosophila melanogaster, mitochondrial dysfunction in neurons leads to increased ROS and the accumulation of LDs in glial-like pigment cells, which presages neurodegeneration [107]. LD induction depends upon c-Jun-N-terminal Kinase (JNK) signaling and SREBP, and can be inhibited by overexpression of Brummer (ATGL) or Lipase 4, which is related to human acid lipases. Expression of either of these lipases also decreases lipid peroxidation and partially alleviates the neurodegeneration phenotype due to mitochondrial dysfunction. This study provided evidence that ROS and LDs function together in a harmful manner to promote neurodegeneration [107]. It would be interesting to know whether or not the same pro-neurodegenerative contribution of LDs is apparent if they are inhibited at the level of decreased biogenesis, not enhanced lipolysis. Evidence that LD biogenesis is used as a protective mechanism to lower rather than to increase ROS levels comes from two diverse contexts: human cancer cells and Drosophila neural stem cells. Glioblastoma and breast cancer cell lines accumulate LDs during hypoxia in a HIF-1α dependent manner [120]. The lipid-droplet protein PLIN2, as well as two proteins involved in FA uptake (Fabp3 and Fabp7) are required for droplet induction as well as for proliferation during hypoxia in cell culture, and for growth in tumor xenografts [120]. The loss of Fabp3, Fabp7 or PLIN2 leads to an increase during hypoxia of the NADP/NADPH ratio and of ROS levels. This study suggests that the uptake and storage of FAs in LDs somehow help to protect cancer cells from ROS toxicity during hypoxia [120]. In Drosophila, the divisions of neural stem cells (neuroblasts) during development are highly resistant to hypoxia and pro-oxidant chemicals [27]. The ability of neuroblasts to grow and divide during oxidative stress correlates with the induction of LDs in neighboring glial cells. The droplet induction mechanism in this context is not yet clear but it may be independent of Drosophila HIF1 (Sima). Both the LD protein PLIN2 (Lsd-2) and the last enzyme in triglyceride biosynthesis, DGAT1, are required specifically in glial cells for LDs and also to dampen ROS levels and mitigate oxidative damage in glia and in neuroblasts [27]. The notion that only a subpopulation of cells may need to accumulate LDs in order to protect the entire tissue has also been proposed for non-alcoholic fatty liver disease, where LD abundance is very variable from one hepatocyte to another [125]. In Drosophila, one of the beneficial roles of glial LDs is to store the abundant PUFA linoleate as triglyceride, thus sequestering it away from the membrane phospholipid pool [27]. Evidence from lipidomics and lipid peroxidation sensors suggests that PUFAs in LDs may be less vulnerable to peroxidation chain reactions than in membranes, and that this effect could help to limit ROS levels in the nervous system during oxidative stress [27] (Figure 1). The chemical and biophysical mechanisms underlying apparent PUFA protection in the neutral lipid core of droplets are not yet clear. Moreover, computational modeling suggests that, once oxidized, triglycerides are thermodynamically unstable in the neutral lipid core and so may move into the phospholipid monolayer [126]. Curiously, studies of oil-and-water emulsions indicate that TAGs in small LDs are less mobile and better protected from peroxidation chain reactions than those in larger LDs [127]. One explanation for this could be that free diffusion is restricted by long-range ordering of triglycerides and/or sterol esters in the vicinity of the phospholipid interface [128].
Figure 1. Lipid droplets protect against ROS damage in the neural stem cell niche of Drosophila.
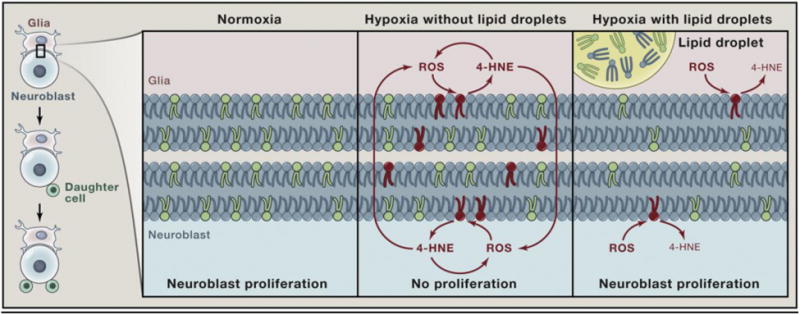
Left: During Drosophila CNS development, neural stem cells (neuroblasts) divide to generate daughter cells that will give rise to neurons. Adjacent glia constitute the neural stem cell niche, a microenvironment that promotes neuroblast divisions.
Right: Magnified view of glia and neuroblast membranes. Neuroblasts are able to sustain their divisions during normoxia or during hypoxia if adjacent glia can synthesize triglycerides stored in lipid droplets. During hypoxia without glial lipid droplets, ROS accumulate and induce the peroxidation of membrane lipids containing PUFAs (green) leading to their damage (red). In the case of the PUFA linoleic acid, this gives rise to a reactive aldehyde (4-hydroxynonenal, 4-HNE), which forms damaging adducts with macromolecules. 4-HNE also triggers more ROS, resulting in an escalating feedback loop. The accumulating damage then inhibits neuroblast divisions. During hypoxia with glial lipid droplets, a proportion of the total membrane fatty acids (including those containing PUFAs) are diverted into lipid droplets where they are protected from ROS-induced damage and thus from triggering the feedback loop between ROS and 4-HNE. This antioxidant role of lipid droplets helps to minimize cellular damage during hypoxia, enabling neuroblasts to continue dividing.
Reprinted from Cell, Vol. 163, Michael A. Welte, How brain fat conquers stress, pp. 269-270, Copyright (2015) [232], with permission from Elsevier.
3.0 Lipid droplets and protein handling
LDs have also been implicated in various steps of the life cycle of specific proteins. LDs can assist with proper folding and assembly of higher order complexes and mediate temporary sequestration of proteins from other cellular compartments, with the best studied examples involving nuclear proteins. For some proteins, association with LDs promotes their degradation. Although these roles in protein homeostasis likely make an important contribution to LD function in health and disease, they remain much less well understood than the roles of LDs in the handling of lipids for energy storage. In particular, it has yet to be established whether protein handling functions of LDs are integrated with their roles in lipid metabolism or whether they represent a second, independent role for LDs.
3.1 Protein maturation
Viruses have evolved to subvert a myriad of functions in their host cells, including those involving LDs. For example, dengue virus upregulates the autophagic breakdown of LDs, ultimately resulting in increased beta-oxidation and thus energy production to sustain viral replication [129]. LDs are also exploited as a crucial platform in the assembly of the next generation of certain viruses: viral proteins transiently associate with the surface of LDs before being incorporated into newly forming viral particles. This LD localization appears to be a critical maturation step as, without it, production of mature viruses can be severely curtailed.
This phenomenon is best understood for Hepatitis C virus. This enveloped, positive strand RNA virus has been intensely studied because it infects well over 100 million people worldwide and can lead to severe liver disease, including liver cirrhosis and liver cancer [130]. After infection, the viral RNA genome is translated into a single polyprotein, which is proteolytically processed into 10 HCV proteins, most of which are associated with the ER [131, 132]. Two of those products, the nucleocapsid core and the non-structural protein NS5A, subsequently associate with LDs. In both cases, LD as well as ER localization requires amphipathic helices within the proteins, and these are also necessary for proper viral assembly [132]. Delivery to LDs specifically depends upon physical interactions with DGAT1 [133, 134]. Since triglycerides make up the bulk of the LD core in most cells, the presence of DGAT1 presumably marks those sites in the ER where nascent LDs are produced and thus where binding of the viral proteins to DGAT1 channels them towards newly formed LDs. When DGAT1 is either absent or enzymatically inhibited, cells can still form LDs via another triglyceride-synthesis enzyme, DGAT2, but these do not accumulate core and NS5A and assembly of new infectious viral particles is severely compromised [133, 134]. NS5A accumulates not only on LDs, but also at ER-derived virus-induced structures, so called double-membrane vesicles (DMVs) [131]. DMVs likely represent sites of viral RNA replication, and they require NS5A for their formation [135, 136]. It has been hypothesized that NS5A may facilitate viral assembly by bringing together LDs and sites of RNA replication [132], such that core can encapsulate newly made viral RNA. Virion formation is then mediated by another set of viral proteins and occurs next to LDs, at ER membranes enriched in viral envelope proteins [137, 138].
Like HCV, dengue is also a positive-strand RNA virus of the family Flaviviridae. Transmitted to humans via mosquitos, it can cause severe and widespread disease, including hemorrhagic fever [139]. The capsid of dengue virus is initially synthesized on the ER and then transiently accumulates on LDs [140, 141]. When this interaction is impaired (either through mutations that prevent capsid targeting to LDs or by reducing LDs via a FA synthase inhibitor), production of infectious viral particles is severely curtailed [140]. Because capsid binding to LDs is impaired after limited proteolysis of LDs, it is presumably mediated by protein-protein interactions with LD proteins [142, 143]. Rotaviruses are double-stranded RNA viruses and a leading cause of diarrhea in children. They replicate using cytoplasmic inclusion bodies named viroplasms [144]. Viroplasms often colocalize with LDs, and a close physical contact between NSP5, a non-structural viral protein, and the LD protein Perilipin A was demonstrated by fluorescent resonance energy transfer and cofractionation [145]. Evidence that this localization is functionally important comes from the observation that compounds that lead to fewer LDs in cells interfere with the production of new viruses [145]. But for exactly which step of virus production LDs are important remains to be discerned: rotavirus infection induces widespread changes in many lipid classes, possibly indicating a broader metabolic role for LDs in infection [146], and treatments to interfere with LDs lead to reduction in viral dsRNA, implicating a role in viral replication [145]. There are indications that a range of other viruses may also exploit LDs for virus maturation. LD-association has been reported for non-structural proteins of orbiviruses [147], bovine viral diarrhea virus [148], and bunyavirus [149, 150], but the functional significance of this LD localization remains to be investigated.
Exactly how LDs promote viral assembly has not been resolved. Assembly may be facilitated because viral proteins are present at high local concentrations, both because LDs protect them from degradation and because they accumulate on a specific surface rather than throughout the ER [132]. Assembly may further be enhanced because LDs are motile: Hepatitis C core protein induces an intracellular redistribution of LDs to regions rich in viral replication sites, in a microtubule and dynein dependent manner [151]. Whether LDs promote viral assembly in addition to providing a platform and a vehicle has not been addressed.
Recent studies in budding yeast raise the intriguing possibility that LDs can play an active role in protein maturation, modulating the protein's folding state [152]. Misfolded cytoplasmic proteins are recognized by the protein quality control machinery and are either degraded, gathered in aggregates, or refolded [153]. Proteins destined for refolding transiently accumulate in structures rich in chaperones, called stress foci or inclusion bodies (IBs). The uncharacterized, yet broadly conserved protein Iml2 localizes to IBs and is important for clearing of misfolded proteins from IBs. Intriguingly, Iml2 physically interacts with known LD proteins, and IBs and LDs are in very close contact during misfolding stress. When LDs are genetically ablated, clearing of misfolded proteins is impaired. These observations suggest that IBs and LDs are brought into physical contact via interactions between Iml2 and LD proteins and that LDs provide an activity necessary for refolding [152]. To identify this activity, the contribution of the neutral lipids stored in the LD core was examined. Lack of triglycerides left refolding activity intact, while lack of sterol esters abolished it. It was proposed that LDs release a sterol-derived molecule that acts as a chemical chaperone to promote refolding of proteins in close contact with LDs [152].
3.2 Protein storage – the case of histones
Like the eggs of many other animals, those of the fruit fly Drosophila contain abundant LDs [154]. These LDs are generated by the mother during oogenesis and provide a crucial energy source for the embryo, as the developing animal cannot feed until it hatches from the egg, almost a day after fertilization. The mother also provisions the eggs with large amounts of proteins, including enough histones to package the chromatin of thousands of nuclei [155]. A subset of these histones is present on LDs: the core histones H2A and H2B as well as the histone variant H2Av colocalize with LDs as assessed by imaging and are detected biochemically in purified droplet fractions [155, 156]. Curiously, the core histones H3 and H4 are similarly overabundant, but present in a different, as yet uncharacterized location. LD localization of histones is mediated by the novel protein Jabba, which likely acts as histone anchor: Jabba physically interacts with these histones, histones are absent from embryonic LDs in Jabba mutants, and the expression of Jabba in cultured Drosophila cells is sufficient to induce recruitment of histones to LDs [156, 157].
In Jabba mutant embryos, the large maternal histone stores of H2A, H2B, and H2Av are absent, not just from the LDs, but from the entire embryo. Thus, Jabba allows long-term storage of maternally produced histones [156]. Because in other cells, excess histone proteins are known to be turned over via the proteasome [158], it was proposed that supernumerary histones, not bound to LDs, are degraded [156]. Surprisingly, lack of Jabba results in excess nuclear accumulation of the histone variant H2Av in very early embryos [159], suggests that degradation of excess histones is not entirely efficient. It remains possible that degradation is more stringent during oogenesis, when the histone stores are produced, or that Jabba plays a different, oogenesis-specific role in ensuring proper histone accumulation.
LD-bound histones are not restricted to Drosophila embryos. The presence of histones on LDs has been directly demonstrated in the embryos of house flies [155], in mouse oocytes and early embryos [160], and in rat sebocytes [161]. In addition, by proteomic analysis, histones have been found to be associated with LDs purified from many different sources, ranging from unicellular eukaryotes to a diverse group of mammalian cell types (for a recent review, see [154]). Because histones are abundant cellular proteins, it is conceivable that, in some studies, their presence on LDs represents biochemical contamination. For example, histones were initially found as potential components of the budding yeast LD proteome, but not in more stringent follow-up studies [40, 162]. A comprehensive understanding of where and when histones can be found on LDs remains an important challenge for the future.
Why are specific histones present on LDs at all? Ongoing research suggests three different biological functions in Drosophila embryos: long-term storage of maternal histones to aid rapid chromatin assembly during early development, short-term sequestration of newly synthesized histones to buffer imbalanced histone production, and a reservoir for histones as antibacterial agents to fight intracellular pathogens [156, 159, 163]. The relative importance of these roles for survival under natural conditions remains to be determined.
Jabba mutant embryos lack the maternal supply of H2A, H2B, and H2Av almost entirely [156], yet early embryogenesis is only slightly impaired [156, 159]. Such mild impairment is made much more severe if embryonic synthesis of new histones is attenuated [156]. Thus, histones stored on LDs support early embryonic development, but – at least under laboratory conditions – serve mainly as a back-up system.
As development proceeds, Jabba mutant embryos also overaccumulate the histone variant H2Av in their nuclei [159], an overaccumulation tied to reduced mitotic fidelity and increased embryo lethality. As new synthesis of histones H2A and H2Av is imbalanced in Jabba mutants, it was proposed that in the wild type excess H2Av can be transiently sequestered on LDs, preventing an incorrect ratio of H2A and H2Av in the nucleus. Indeed, transplantation and ectopic expression experiments showed that histones can be loaded onto droplets in the embryo. These observations suggest that short-term sequestration of histones on LDs has the potential to buffer the histone supply, thereby controlling levels of histones in the nucleus.
When various bacteria are injected into the cytoplasm of Drosophila embryos, the outcome is very different between the wild type and Jabba mutants [163]. At appropriate titers, bacterial growth is restricted in wild-type embryos; and the embryos hatch successfully. Under the same conditions, bacteria proliferate massively in Jabba mutant embryos, causing their death. Since histones have potent anti-microbial properties in vitro [164], it was proposed that histones stored on LDs provide antibacterial agents to fight invaders. In support of this notion, LDs purified from embryos can readily kill bacteria in vitro, a phenomenon dependent upon Jabba and histones [163]. Furthermore, bacterial cell wall components trigger release of histones from LDs in vitro [163]. Histone-packed LDs may thus provide a previously unrecognized innate immune strategy to fight intracellular pathogens. This strategy may be broadly conserved since hepatic LDs from mice are associated with histones, and LD histone levels go up upon mock infection [163].
Although it is clear that histones on LDs are not a fly-specific phenomenon, research in other systems is currently handicapped because the histone anchor on LDs has so far been identified only in Drosophila. Because Jabba's primary sequence is not conserved beyond insects, there exist no obvious vertebrate candidates for such anchors. Identifying the mechanism that targets histones to LDs in mammals will be crucial for further progress, such as for determining if LD histones contribute to embryonic development and innate immunity in mammals.
3.3 Other proteins transiently stored on LDs
Although protein storage on LDs is best characterized for histones, it is by no means restricted to this class of proteins. An exciting recent finding is that in mice the LD protein PLIN5 can, under certain conditions, translocate from the droplet surface to the nucleoplasm and stimulate gene expression [165]. PLIN5 is a member of the Perilipin family of proteins, the most heavily studied family of LD proteins [17]. PLIN5 is preferentially expressed in tissues with high oxidative capacity, such as the heart, liver, brown adipose tissue, and skeletal muscle [166]. It controls lipolysis by regulating access of cytoplasmic lipases to the neutral lipids stored in LDs, and also mediates LD-mitochondrial interactions. In starved animals and in cells after stimulation of the protein kinase A (PKA) pathway, a substantial fraction of PLIN5 was found to relocalize to the nucleus [165]. Phosphorylation of PLIN5 by PKA drives relocalization and assembly into a protein complex with PGC-1α and SIRT-1. PGC-1 α (Peroxisome proliferator-activated receptor gamma coactivator 1-alpha) is a transcriptional regulator important for mitochondrial biogenesis and function; SIRT-1 (Sirtuin 1) is a deacetylase that controls PGC-1 α's acetylation status and activity. PLIN5 mediates PKA-dependent activation of PGC-1α, presumably by disinhibiting SIRT-1, and thus activates PGC-1α target genes [165]. PLIN5 thus serves two functions in two different cellular locations: on LDs it regulates the activity of lipases and, in the nucleus it acts as a transcriptional activator. The two functions are linked because in the absence of PKA-mediated activation, lipases are inactive and PLIN5 is restricted to the LD surface, keeping it out of the nucleus. Upon PKA activation, lipolysis is activated and a portion of PLIN5 travels to the nucleus to upregulate transcription and ultimately to promote mitochondrial biogenesis. The dual role of PLIN-5 at the LD surface and in the nucleus was proposed to coordinate release of FAs during lipolysis and their efficient usage by mitochondria [165].
Another transcriptional regulator, NFAT5 (nuclear factor of activated T cells 5) may also be sequestered on LDs [167]. NFAT5 controls the expression of osmoprotective genes in response to hypotonic stress [168]. NFAT5 can physically interact with the LD protein Fsp27/CIDEC, prominently expressed in adipose tissue. When ectopically expressed in a heterologous system, Fsp27 keeps some NFAT5 out of the nucleus and reduces NFAT5-dependent gene expression. Although the relevance of these findings for adipocyte function remains to be determined, they raise the possibility that Fsp27 can regulate NFAT5's transcriptional activity by controlling its entry into the nucleus. Finally, in the bacterium Rhodococcus jostii, LDs sequester a transcriptional repressor and thus promote expression of a major LD protein [169].
The anti-viral protein viperin (Virus inhibitory, ER associated, Interferon inducible) can interfere with a broad range of RNA and DNA viruses (from influenza and HIV to dengue and hepatitis C viruses), targeting various steps in the viral life cycle, including replication and viral budding [170]. In uninfected cells, it accumulates both on ER membranes and on LDs, and viral infection can induce its relocalization to various other compartments. The function of viperin's LD localization is not clear. Possibilities include that viperin on LDs enhances targeting of those viruses that use LDs as assembly platforms, such as hepatitis C and dengue viruses, or that LDs are a convenient reservoir for excess viperin until needed elsewhere. The mapping of an amphipathic helix in viperin that is necessary and sufficient for LD recruitment provides a tool for addressing this question, although the same sequence also mediates ER localization [171]. Localization of viperin to LDs/ER is necessary for its activity against HCV, and viperin interacts with HCV proteins at the LD surface, suggesting that viperin indeed targets the LD step of viral assembly [172]. In contrast, LD/ER localization of viperin is not necessary for its action against dengue virus, even though this virus also assembles on LDs [173].
3.4 Protein turnover
A number of proteins accumulate on LDs before they undergo degradation. These observations led to the proposal that LDs might serve as an obligatory platform for the turnover of specific proteins [174]. This intermediary LD step may assist the degradation of certain highly hydrophobic proteins that would otherwise form hard-to-clear aggregates in the cytoplasm [175] or it may facilitate the wholesale delivery of a large set of damaged proteins to the lysosome via autophagy [96]. For some proteins, the involvement of LDs in turnover is well established, but it is not yet clear how widespread this strategy is and what properties of proteins target them to this degradation pathway.
Proteasome-mediated protein turnover
VLDL or very low-density lipoprotein is a lipoprotein secreted by the liver that serves to transport lipids through the bloodstream. Its primary protein component, Apolipoprotein B (ApoB), is cotranslationally imported into the ER lumen, where it associates with lipid; further assembly and maturation steps then lead to the nascent VLDL particles ready for secretion [176]. When lipid supply is limiting and VLDL assembly cannot occur correctly, ApoB is transferred into the cytoplasm and turned over via proteasome mediated degradation [177]. LDs play a crucial role in this process. ApoB in the ER destined for turnover transiently accumulates in so called ApoB crescents, located within the ER, but also closely associated with LDs; these structures may represent LDs not fully budded from the ER [175, 178]. ApoB is then transferred to the cytoplasmic surface of LDs, a process involving the ER transmembrane protein Derlin-1 [179]. ApoB is ubiquitinated and eventually degraded by the proteasome system. Loss of ubiquitinated ApoB requires the ubiquitin-like (UBX)-domain containing protein UBXD8, which also recruits the AAA-ATPase p97/VCP to the LD surface [179]. p97/VCP is thought to mediate extraction of proteins from membranes before they are degraded [180], and thus may similarly deliver ubiquitinated ApoB from the LD surface to the proteasome. Cofractionation and immunofluorescence experiments indeed suggest that proteasomes are associated with LDs [179]. This degradation pathway is not restricted to ApoB as UBXD-8 knockdown or proteasome inhibition leads to accumulation of many other ubiquitinated proteins on LDs [178, 179]. In particular, levels of the triglyceride lipase ATGL are controlled by UBXD-8 dependent recruitment of p97/VCP to LDs [181]. Which of the proteins turned over by this pathway are bona-fide LD proteins, like ATGL, and which come originally from the ER or even other compartments, like ApoB, remains to be established.
LDs also play a role in the turnover of HMG-CoA reductase (3-hydroxy-3-methyl-glutaryl-coenzyme A reductase), the rate-limiting enzyme in the cholesterol biosynthetic pathway. HMG-CoA reductase is an ER membrane protein whose degradation is induced when sterols are abundant. Under these conditions, HMG-CoA reductase relocates to ER regions closely associated with LDs [182], and is ubiquitinated, extracted from the membrane by the AAA-ATPase p97/VCP, and then degraded by the proteasome. Ubiquitination of HMG-CoA reductase depends upon the LD protein AUP-1 (ancient ubiquitous protein-1) [183]. AUP-1 recruits the ubiquitin-conjugating enzyme Ubc7 to LDs [183, 184] and promotes its binding to two ubiquitin ligases (gp78 and Trc8), which - in turn - are responsible for HMG-CoA reductase ubiquitination. Indeed, when AUP-1 is knocked down, ubiquitination of HMG-CoA reductase is reduced [183]. It was proposed that a close association of ER and LDs allows the formation of a multi-protein complex containing HMG-CoA reductase, the ligases, Ubc7, and AUP-1, thus promoting ubiquitination [183].
A number of the molecules involved in the turnover of HMG-CoA reductase and ApoB are also components of ERAD, raising the possibility that LDs play a general role in the ERAD pathway. However, this is not the case. Budding yeast cells unable to generate any LDs still turn over several well-characterized ERAD substrates [185, 186], and when LD formation is abolished in mammalian cells by knockout or inhibition of the two enzymes that synthesize triglycerides, the one ERAD substrate tested was still degraded normally [187]. Thus, either only a subset of ERAD substrates requires LDs, or part of the ERAD machinery is also used in a separate, LD-associated pathway.
One of the regulatory strategies by which proteasome activity is controlled is O-GlcNAc modification [188]. This post-translational modification involves the addition of a single β-N-acetylglucosamine (GlcNAc) moiety to a protein, via the enzyme O-GlcNAc transferase. In the case of the proteasome, this modification inhibits its activity. Curiously, the enzyme that removes O-GlcNAc (O-GlcNAcase or OGA) has an isoform that is specifically targeted to LDs [189]. Knockdown of this isoform results in increased proteasome activity and higher cellular abundance of poly-ubiquitinated proteins. Local activation of the proteasome at the LD surface might provide a mechanism to coordinately control protein turnover LD wide. Consistent with this, knockdown of the LD specific OGA isoform increased levels of two LD associated proteins [189]. Such coordination may also underlie the observation that in budding yeast a LD-localized phospholipase has a role in proteasome mediated protein degradation, though the exact mechanism remains to be worked out [190].
Lipophagy
There are a few indications that lipophagy may also contribute to the turnover of proteins from other cellular compartments that are transiently associated with LDs. ApoB crescents are associated with LDs and represent an intermediate in ApoB degradation [175]. Proteasome inhibition leads to accumulation of ApoB crescents because their normal turnover is abolished. However, over the long term, crescents decrease again and ApoB is found in lysosomes. These changes are prevented in the presence of an autophagy inhibitor, implying that ApoB associated with LDs can also be cleared by lipophagy [175]. Lipophagy has also been implicated in combating ER stress [96] and the removal of unwanted mitochondrial proteins [191]. As elaborated in section 2.2.1, lipid imbalance in budding yeast results in protein misfolding in the ER [192] and the formation of fragmented ER structures that frequently colocalize with LDs [96]. LDs purified under these conditions are enriched for polyubiquitinated proteins and an ER luminal chaperone, suggesting that misfolded ER proteins are associating with LDs. Intriguingly, these LDs are taken up into the vacuole, the yeast equivalent of the lysosome, via a specialized type of autophagy (microlipophagy). These observations led to the proposal that damaged ER proteins are sequestered by LDs and subsequently cleared via lipophagy [96]. A similar pathway may also exist for mitochondria. Several proteins normally present in the outer mitochondrial membrane can relocalize to the surface of LDs under certain physiological conditions [191]. In yeast, LDs carrying mitochondrial proteins were observed in the vacuole, hence it was hypothesized that LDs serve as a temporary holding place for such proteins before they, along with the entire LD, are degraded by lipophagy.
Chaperone-mediated autophagy
Some cytosolic proteins are turned over by a specialized form of autophagy: chaperone-mediated autophagy (CMA). In this system, a chaperone from the Hsp70 family recognizes target proteins in the cytosol and delivers them to the lysosome surface [193]. The proteins are then translocated into the lysosome lumen and degraded. CMA has recently been shown to also promote the turnover of LD proteins PLIN2 and PLIN3 [194, 195]. Given this precedent, future studies of the role of LDs in the turnover of proteins from other compartments should also probe the contribution of CMA.
4.0 Other roles of lipid droplets
As LD research expands, new connections of LDs to various biological processes are being discovered. In these cases, it is often not yet clear if energy storage or some other role of LDs is relevant. Below we discuss two such cases: nuclear lipid droplets and droplet motility.
Several groups have recently shown that LDs are not only cytoplasmic but can also be found inside the nucleus [196-198]. LDs in the nucleus are smaller than their cytoplasmic counterparts (∼0.5 μm or less), and their abundance varies drastically between cell types, e.g., they are readily detectable in cells of hepatic origin. Nuclear LDs grow and shrink depending on the metabolic state of the cells [199] and are associated with some, but not all, of the proteins characteristic of cytoplasmic LDs [197]. Because of their relatively small size and low abundance, they contribute only modestly to the overall lipid storage capacity of cells. Their physiological role remains to be characterized, but it has been speculated that they may modulate transcriptional regulation mediated by lipidic signals, control nuclear membrane composition, or act as nucleus-specific protein storage sites [200]. Recent work in the oleaginous bacterium Rhodococcus jostii even raises the possibility that nuclear LDs might directly modulate DNA-based processes, since here LDs bind to genomic DNA via a major LD protein, possibly protecting the DNA from damage under stressful conditions [169]. A functional analysis of nuclear LDs is within reach because components of a pathway controlling the frequency of nuclear LDs has been identified [197]. Nuclear LDs are frequently associated with two intranuclear structures: type I nucleoplasmic reticulum, which represents extensions of the inner nuclear membrane, and PML (premyelocytic leukemia) nuclear bodies. The colocalization with PML bodies appear to be relevant since knockdown or overexpression of a specific constituent of these bodies, PML-II, results - respectively - in fewer or greater numbers of nuclear LDs. Knockdown of either the inner nuclear envelope protein SUN2 or the ER proteins REEP3/4 results in increased numbers of nuclear LDs. Both SUN2 and REEP3/4 have been implicated in the removal of membranes from chromosomes during mitosis [201, 202], leading to the hypothesis that nuclear LDs are similarly cleared from chromosomes and thus tend to be lost to the cytoplasm when the nuclear envelope reforms after mitosis. PML-II is proposed to counteract this mitotic clearance by tethering the nuclear LDs to chromatin [197].
In many types of cells, LDs undergo active, directed motion [203]. For example, in the cells of the mammary epithelium, LDs originate from basally located ER and then move to apical regions, often growing and fusing during this process [204]; the apical LDs are then secreted and give rise to the fat globules present in milk. An analogous situation occurs during insect oogenesis where LDs produced in nurse cells, the sister cells of the oocyte, are moved, via cytoplasmic bridges, to the oocyte and ultimately serve as an energy supply for the developing embryo [154]. Certain intracellular pathogens, like Chlamydia [205] and Mycobacterium [206], induce intracellular relocation of LDs to the vesicle within which the pathogen resides, thus ensuring a sufficient lipid supply for the pathogen. In budding yeast, directed movement of LDs to the growing bud mediates proper allocation of fat stores to the daughter cell [207].
In addition to this highly directed LD transport, LDs can also display active, seemingly random, motion over short distances in various directions. A dramatic example occurs in early Drosophila embryos, where essentially all LDs move incessantly back and forth along microtubules (reviewed in [154]). Similar “chaotic” LD motion is observed in many cells (reviewed in [203]), though the pattern of motion and the fraction of LDs that display motility varies considerably. Although seemingly chaotic, this motion can still be under tight biological control. For example, in fly embryos, the parameters of motion change reproducibly with the developmental stage [154, 208], and in cultured Vero cells, LD motion is regulated by the nutrient supply [209]. The biological role of such motion has long been debated, with proposals ranging from a means to sweep the cytoplasm free of toxic molecules to mediating dispersal of lipids to membrane-bound organelles [203]. Recent studies in cultured mammalian cells have demonstrated that LD motion can facilitate the transfer of FAs from LDs to mitochondria and thus contribute to effective energy production when cells have to tap into their neutral lipid stores. Thus, when Vero cells are nutrient starved, LDs disperse throughout the cytoplasm. This dispersal increases interactions with mitochondria and thus promotes channeling of FAs from LDs to mitochondria for beta-oxidation [209]. FAs are transferred to mitochondria only at a limited number of LD-mitochondria contact sites, yet they can still undergo broad intramitochondrial dispersal because mitochondria fuse into an interconnected network during starvation [210].
Mechanisms driving LD motion vary substantially between cells. In a number of cases, LD distribution is mediated by the actin-myosin system. In budding yeast, for example, the actin-based motor Myo6 promotes vectorial transport of LDs from the mother cell into the developing bud [207]; during fission yeast spore formation, actin polymerization promotes, directly or indirectly, LD clustering around nuclei; and in early zebrafish embryos, drugs that disrupt or promote actin polymerization or inhibit non-muscle myosin interfere with the developmental redistribution of LDs [211]. In most systems studied to date, however, LD motility is largely or exclusively due to transport along microtubules [203], for examples in various cultured mammalian cells [209, 212] or in Drosophila embryos [154]. For most of these cases, LDs are presumably directly pulled by microtubule motors, such as kinesin and dynein [154, 212], but in the fungus Ustilago maydis, LDs themselves do not move but are actively spread out along microtubules via transient interactions with motor-propelled early endosomes [213, 214].
LDs are ideally suited for optical trapping experiments [215], and this property makes it possible to measure the forces with which LDs move, both in vitro and in vivo [154, 216, 217]. Such force measurements can be used, for example, to determine the number of motor molecules moving a single droplet [218, 219], to determine how opposing motors interact [220], and how the activity of motors and their force production is regulated [221-224].
Regulated droplet motility can control the dispersal status of LDs within the cell. In Drosophila embryos, LDs are either dispersed throughout the peripheral cytoplasm or concentrated around the central yolk, depending upon the developmental stage. The overall LD distribution is controlled by a multi-component transport system that includes the motors kinesin-1 and cytoplasmic dynein, various dynein co-factors (BicD, dynactin), the Perilipin LSD-2/PLIN2, the kinase GSK-3, and the nesprin Klar (for a recent review, see [154]). Here, a key factor mediating temporal regulation of LD motion is the novel protein Halo that acts as rate-limiting kinesin-1 cofactor and increases travel lengths in kinesin-1 direction [208, 225]. In Vero cells, LDs are typically clustered near the nucleus; starvation-induced LD dispersal towards the periphery involves upregulated LD motility and preferential recruitment of LDs, as well as mitochondria, to a specific subset of microtubules that are detyrosinated [209]. Restriction of both organelles to the same 1D tracks may promote enhanced interactions between them. Clustering of LDs is also frequently observed when certain droplet-localized proteins are up- or downregulated or posttranslationally modified, such as PLIN1 [212, 226], ancient ubiquitous protein 1 (AUP1 [227]), and CG9186 [157, 228]. To what extent such clustering involves altered droplet motility has yet to be examined but, for U2OS cells, it was recently shown that dissociation of clustered droplets involves actin and myosin [229].
5.0 Conclusions
In contrast to the energy storage roles of LDs, other functions have so far been studied in much less detail. The literature is littered with documented examples of proteins from other cellular compartments present on LDs, including enzymes involved in nucleotide metabolism, cytosolic chaperones, and splicing factors (for a review, see [230]). In addition, many published LD proteomes include a long list of candidate proteins from other cellular compartments. Some of these candidates will, of course, just represent biochemical contamination, but given that other unexpected interactions have been verified and proven to be biologically important, it is important not to dismiss those candidates without further study. In the long run, what is needed is more stringent methods to judge LD localization on a proteomic scale (e.g., [231]). Similarly, advances in lipidomics have recently made the analysis of lipids more sensitive and comprehensive, which sets the stage for discovering exactly which lipids are handled by LDs.
The discovery that the functions of LDs extend well beyond energy storage to diverse and important roles in lipid and protein handling is an exciting development. As evidenced by other articles in this special issue, LD research is booming, revealing that these organelles make important contributions to many more cellular and physiological processes than previously appreciated. Although most of these studies focus on how management of energy storage impinges on these novel biological functions, it is important to consider whether the capacity of LDs as general lipid and protein handling platforms also plays a role. More broadly, many diseases are associated with LD dysfunction and are accompanied by over- or understorage of neutral lipids [2]. Aberrant LD numbers have the potential to disrupt various aspects of lipid and protein homeostasis, potentially causing disease on its own or modulating the severity and outcome of maladies due to aberrant lipid storage (Figure 2).
Figure 2. Potential intersection between lipid and protein homeostasis.
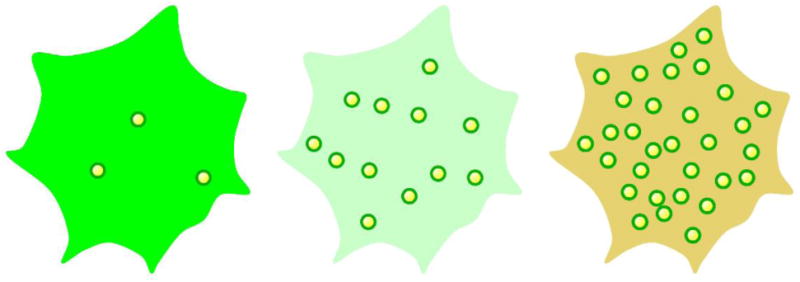
Droplet number per cell might modulate the effective concentration of associated proteins: Too many droplets (right) might tie up too much of the protein. Too few droplets (left) might allow a toxic protein to reach a free concentration that is detrimental.
The diverse roles of LDs in lipid and protein handling are all mediated by the ability of LDs to store or sequester proteins and lipophilic molecules. Molecules stored in LDs for use at other sites in the cell include vitamins, signaling precursors, histones, and transcription factors. LDs also sequester molecules that would otherwise be toxic to the cell or prone to damage and degradation. The former includes free fatty acids, environmental toxins, and aggregation-prone proteins like ApoB; the latter encompasses PUFAs and viral components. In many cases LDs are also an important node for regulating the metabolism and homeostasis of their lipid or protein cargo. LDs thus promote eicosanoid synthesis as well as protein maturation, folding, and turnover. LDs are important for maintaining homeostasis during cellular stress and, in the case of ER stress, lipid and protein handling seem to work together to mitigate it. In many cases, these functions of LDs have yet to be clarified at the molecular level. What seems much clearer, however, is that new roles for LDs will continue to be discovered.
Highlights.
Lipid droplets have important functions beyond energy homeostasis
They store vitamins, signaling precursors, and other hydrophobic molecules
They mitigate some harmful effects of ER and oxidative stress
They function in protein maturation, storage, and turnover
They are motile and can exist in the nucleus
Acknowledgments
This work was supported by the National Institutes of Health grant 1R01 GM102155 (MAW), the University of Rochester (MAW), an Investigator Award 104566/Z/14/Z from The Wellcome Trust (APG), and by the Francis Crick Institute (APG), which receives its core funding from Cancer Research UK (FC001088, the UK Medical Research Council (FC001088), and the Wellcome Trust (FC001088).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 2012;249:541–585. doi: 10.1007/s00709-011-0329-7. [DOI] [PubMed] [Google Scholar]
- 2.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kory N, Farese RV, Jr, Walther TC. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 2016;26:535–546. doi: 10.1016/j.tcb.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilfling F, Haas JT, Walther TC, Farese RV., Jr Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Li P. The size matters: regulation of lipid storage by lipid droplet dynamics. Sci China Life Sci. 2017;60:46–56. doi: 10.1007/s11427-016-0322-x. [DOI] [PubMed] [Google Scholar]
- 8.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerner J, Hoppel C. Fatty acid import into mitochondria. Biochim Biophys Acta. 2000;1486:1–17. doi: 10.1016/s1388-1981(00)00044-5. [DOI] [PubMed] [Google Scholar]
- 10.Eaton S. Control of mitochondrial beta-oxidation flux. Prog Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- 11.Krahmer N, Farese RV, Jr, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:973–983. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrese EL, Saudale FZ, Soulages JL. Lipid Droplets as Signaling Platforms Linking Metabolic and Cellular Functions. Lipid Insights. 2014;7:7–16. doi: 10.4137/LPI.S11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashemi HF, Goodman JM. The life cycle of lipid droplets. Curr Opin Cell Biol. 2015;33:119–124. doi: 10.1016/j.ceb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Andrea S. Lipid droplet mobilization: The different ways to loosen the purse strings. Biochimie. 2016;120:17–27. doi: 10.1016/j.biochi.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Kimmel AR, Sztalryd C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu Rev Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- 18.Schneider MR. Beyond the adipocyte paradigm: Heterogeneity of lipid droplets and associated proteins. Exp Cell Res. 2016;340:171. doi: 10.1016/j.yexcr.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Shatz O, Holland P, Elazar Z, Simonsen A. Complex Relations Between Phospholipids, Autophagy, and Neutral Lipids. Trends Biochem Sci. 2016;41:907–923. doi: 10.1016/j.tibs.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Wang CW. Lipid droplets, lipophagy, and beyond. Biochim Biophys Acta. 2016;1861:793–805. doi: 10.1016/j.bbalip.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 21.Thiam AR, Beller M. The why, when and how of lipid droplet diversity. J Cell Sci. 2017;130:315–324. doi: 10.1242/jcs.192021. [DOI] [PubMed] [Google Scholar]
- 22.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta. 2011;1811:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chitraju C, Trotzmuller M, Hartler J, Wolinski H, Thallinger GG, Lass A, Zechner R, Zimmermann R, Kofeler HC, Spener F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J Lipid Res. 2012;53:2141–2152. doi: 10.1194/jlr.M028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt C, Ploier B, Koch B, Daum G. Analysis of yeast lipid droplet proteome and lipidome. Methods Cell Biol. 2013;116:15–37. doi: 10.1016/B978-0-12-408051-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 26.Vrablik TL, Petyuk VA, Larson EM, Smith RD, Watts JL. Lipidomic and proteomic analysis of Caenorhabditis elegans lipid droplets and identification of ACS-4 as a lipid droplet-associated protein. Biochim Biophys Acta. 2015;1851:1337–1345. doi: 10.1016/j.bbalip.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey AP, Koster G, Guillermier C, Hirst EM, MacRae JI, Lechene CP, Postle AD, Gould AP. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell. 2015;163:340–353. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H, Goulbourne CN, Tatar A, Turlo K, Wu D, Beigneux AP, Grovenor CR, Fong LG, Young SG. High-resolution imaging of dietary lipids in cells and tissues by NanoSIMS analysis. J Lipid Res. 2014;55:2156–2166. doi: 10.1194/jlr.M053363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czamara K, Majzner K, Selmi A, Baranska M, Ozaki Y, Kaczor A. Unsaturated lipid bodies as a hallmark of inflammation studied by Raman 2D and 3D microscopy. Sci Rep. 2017;7:40889. doi: 10.1038/srep40889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bautista G, Pfisterer SG, Huttunen MJ, Ranjan S, Kanerva K, Ikonen E, Kauranen M. Polarized THG microscopy identifies compositionally different lipid droplets in mammalian cells. Biophys J. 2014;107:2230–2236. doi: 10.1016/j.bpj.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta R, Alfonso-Garcia A, Cinco R, Gratton E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Sci Rep. 2015;5:9848. doi: 10.1038/srep09848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Napoli C, Pope I, Masia F, Langbein W, Watson P, Borri P. Quantitative Spatiotemporal Chemical Profiling of Individual Lipid Droplets by Hyperspectral CARS Microscopy in Living Human Adipose-Derived Stem Cells. Anal Chem. 2016;88:3677–3685. doi: 10.1021/acs.analchem.5b04468. [DOI] [PubMed] [Google Scholar]
- 34.Stiebing C, Matthaus C, Krafft C, Keller AA, Weber K, Lorkowski S, Popp J. Complexity of fatty acid distribution inside human macrophages on single cell level using Raman micro-spectroscopy. Anal Bioanal Chem. 2014;406:7037–7046. doi: 10.1007/s00216-014-7927-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang PL, Hsu TH, Wang CW, Chen RH. Lipid droplets maintain lipid homeostasis during anaphase for efficient cell separation in budding yeast. Mol Biol Cell. 2016;27:2368–2380. doi: 10.1091/mbc.E16-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velazquez AP, Graef M. Autophagy regulation depends on ER homeostasis controlled by lipid droplets. Autophagy. 2016;12:1409–1410. doi: 10.1080/15548627.2016.1190074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbosa AD, Sembongi H, Su WM, Abreu S, Reggiori F, Carman GM, Siniossoglou S. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Biol Cell. 2015;26:3641–3657. doi: 10.1091/mbc.E15-03-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laibach N, Post J, Twyman RM, Gronover CS, Prufer D. The characteristics and potential applications of structural lipid droplet proteins in plants. J Biotechnol. 2015;201:15–27. doi: 10.1016/j.jbiotec.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Blomquist GJ, Bagneres AG. Insect hydrocarbons: biology, biochemistry, and chemical ecology. Cambridge University Press; Cambridge: 2010. [Google Scholar]
- 44.Makki R, Cinnamon E, Gould AP. The development and functions of oenocytes. Annu Rev Entomol. 2014;59:405–425. doi: 10.1146/annurev-ento-011613-162056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herman NA, Zhang W. Enzymes for fatty acid-based hydrocarbon biosynthesis. Curr Opin Chem Biol. 2016;35:22–28. doi: 10.1016/j.cbpa.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Peramuna A, Morton R, Summers ML. Enhancing alkane production in cyanobacterial lipid droplets: a model platform for industrially relevant compound production. Life (Basel) 2015;5:1111–1126. doi: 10.3390/life5021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valachovic M, Garaiova M, Holic R, Hapala I. Squalene is lipotoxic to yeast cells defective in lipid droplet biogenesis. Biochem Biophys Res Commun. 2016;469:1123–1128. doi: 10.1016/j.bbrc.2015.12.050. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita S, Yamaguchi H, Waki T, Aoki Y, Mizuno M, Yanbe F, Ishii T, Funaki A, Tozawa Y, Miyagi-Inoue Y, Fushihara K, Nakayama T, Takahashi S. Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. Elife. 2016;5 doi: 10.7554/eLife.19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer G, Gorb SN, Klein MC, Nellesen A, von Tapavicza M, Speck T. Comparative study on plant latex particles and latex coagulation in Ficus benjamina, Campanula glomerata and three Euphorbia species. PLoS One. 2014;9:e113336. doi: 10.1371/journal.pone.0113336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang W, Zhang M, Zheng S, Li Y, Li X, Li W, Li G, Lin Z, Xie Z, Zhao Z, Lou H. Trapping toxins within lipid droplets is a resistance mechanism in fungi. Sci Rep. 2015;5:15133. doi: 10.1038/srep15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandoz KM, Valiant WG, Eriksen SG, Hruby DE, Allen RD, 3rd, Rockey DD. The broad-spectrum antiviral compound ST-669 restricts chlamydial inclusion development and bacterial growth and localizes to host cell lipid droplets within treated cells. Antimicrob Agents Chemother. 2014;58:3860–3866. doi: 10.1128/AAC.02064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verbrugge SE, Al M, Assaraf YG, Kammerer S, Chandrupatla DM, Honeywell R, Musters RP, Giovannetti E, O'Toole T, Scheffer GL, Krige D, de Gruijl TD, Niessen HW, Lems WF, Kramer PA, Scheper RJ, Cloos J, Ossenkoppele GJ, Peters GJ, Jansen G. Multifactorial resistance to aminopeptidase inhibitor prodrug CHR2863 in myeloid leukemia cells: down-regulation of carboxylesterase 1, drug sequestration in lipid droplets and pro-survival activation ERK/Akt/mTOR. Oncotarget. 2016;7:5240–5257. doi: 10.18632/oncotarget.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandermann H., Jr Differential lipid affinity of xenobiotics and natural compounds. FEBS Lett. 2003;554:165–168. doi: 10.1016/s0014-5793(03)01143-8. [DOI] [PubMed] [Google Scholar]
- 54.Murphy G, Jr, Rouse RL, Polk WW, Henk WG, Barker SA, Boudreaux MJ, Floyd ZE, Penn AL. Combustion-derived hydrocarbons localize to lipid droplets in respiratory cells. Am J Respir Cell Mol Biol. 2008;38:532–540. doi: 10.1165/rcmb.2007-0204OC. [DOI] [PubMed] [Google Scholar]
- 55.Bourez S, Le Lay S, Van den Daelen C, Louis C, Larondelle Y, Thome JP, Schneider YJ, Dugail I, Debier C. Accumulation of polychlorinated biphenyls in adipocytes: selective targeting to lipid droplets and role of caveolin-1. PLoS One. 2012;7:e31834. doi: 10.1371/journal.pone.0031834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bui A, Xiao R, Perveen Z, Kleinow K, Penn A. Zebrafish embryos sequester and retain petrochemical combustion products: developmental and transcriptome consequences. Aquat Toxicol. 2012;108:23–32. doi: 10.1016/j.aquatox.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 57.Hanano A, Almousally I, Shaban M, Rahman F, Blee E, Murphy DJ. Biochemical, Transcriptional, and Bioinformatic Analysis of Lipid Droplets from Seeds of Date Palm (Phoenix dactylifera L. and Their Use as Potent Sequestration Agents against the Toxic Pollutant, 2,3,7,8-Tetrachlorinated Dibenzo-p-Dioxin. Front Plant Sci. 2016;7:836. doi: 10.3389/fpls.2016.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spicher L, Kessler F. Unexpected roles of plastoglobules (plastid lipid droplets) in vitamin K1 and E metabolism. Curr Opin Plant Biol. 2015;25:123–129. doi: 10.1016/j.pbi.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Peramuna A, Summers ML. Composition and occurrence of lipid droplets in the cyanobacterium Nostoc punctiforme. Arch Microbiol. 2014;196:881–890. doi: 10.1007/s00203-014-1027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Traber MG, Kayden HJ. Tocopherol distribution and intracellular localization in human adipose tissue. Am J Clin Nutr. 1987;46:488–495. doi: 10.1093/ajcn/46.3.488. [DOI] [PubMed] [Google Scholar]
- 61.Senoo H, Kojima N, Sato M. Vitamin A-storing cells (stellate cells) Vitam Horm. 2007;75:131–159. doi: 10.1016/S0083-6729(06)75006-3. [DOI] [PubMed] [Google Scholar]
- 62.Schreiber R, Taschler U, Preiss-Landl K, Wongsiriroj N, Zimmermann R, Lass A. Retinyl ester hydrolases and their roles in vitamin A homeostasis. Biochim Biophys Acta. 2012;1821:113–123. doi: 10.1016/j.bbalip.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grumet L, Taschler U, Lass A. Hepatic Retinyl Ester Hydrolases and the Mobilization of Retinyl Ester Stores. Nutrients. 2016;9 doi: 10.3390/nu9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blaner WS, Li Y, Brun PJ, Yuen JJ, Lee SA, Clugston RD. Vitamin A Absorption, Storage and Mobilization. Subcell Biochem. 2016;81:95–125. doi: 10.1007/978-94-024-0945-1_4. [DOI] [PubMed] [Google Scholar]
- 65.Senoo H, Mezaki Y, Morii M, Hebiguchi T, Miura M, Imai K. Uptake and storage of vitamin A as lipid droplets in the cytoplasm of cells in the lamina propria mucosae of the rat intestine. Cell Biol Int. 2013;37:1171–1180. doi: 10.1002/cbin.10140. [DOI] [PubMed] [Google Scholar]
- 66.Nagy NE, Holven KB, Roos N, Senoo H, Kojima N, Norum KR, Blomhoff R. Storage of vitamin A in extrahepatic stellate cells in normal rats. J Lipid Res. 1997;38:645–658. [PubMed] [Google Scholar]
- 67.O'Byrne SM, Blaner WS. Retinol and retinyl esters: biochemistry and physiology. J Lipid Res. 2013;54:1731–1743. doi: 10.1194/jlr.R037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taschler U, Schreiber R, Chitraju C, Grabner GF, Romauch M, Wolinski H, Haemmerle G, Breinbauer R, Zechner R, Lass A, Zimmermann R. Adipose triglyceride lipase is involved in the mobilization of triglyceride and retinoid stores of hepatic stellate cells. Biochim Biophys Acta. 2015;1851:937–945. doi: 10.1016/j.bbalip.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tuohetahuntila M, Molenaar MR, Spee B, Brouwers JF, Houweling M, Vaandrager AB, Helms JB. ATGL and DGAT1 are involved in the turnover of newly synthesized triacylglycerols in hepatic stellate cells. J Lipid Res. 2016;57:1162–1174. doi: 10.1194/jlr.M066415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papackova Z, Cahova M. Fatty acid signaling: the new function of intracellular lipases. Int J Mol Sci. 2015;16:3831–3855. doi: 10.3390/ijms16023831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen WJ, Azhar S, Kraemer FB. Lipid droplets and steroidogenic cells. Exp Cell Res. 2016;340:209–214. doi: 10.1016/j.yexcr.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kraemer FB, Khor VK, Shen WJ, Azhar S. Cholesterol ester droplets and steroidogenesis. Mol Cell Endocrinol. 2013;371:15–19. doi: 10.1016/j.mce.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamaguchi T, Fujikawa N, Nimura S, Tokuoka Y, Tsuda S, Aiuchi T, Kato R, Obama T, Itabe H. Characterization of lipid droplets in steroidogenic MLTC-1 Leydig cells: Protein profiles and the morphological change induced by hormone stimulation. Biochim Biophys Acta. 2015;1851:1285–1295. doi: 10.1016/j.bbalip.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 74.Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bozza PT, Bakker-Abreu I, Navarro-Xavier RA, Bandeira-Melo C. Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot Essent Fatty Acids. 2011;85:205–213. doi: 10.1016/j.plefa.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 76.Dichlberger A, Schlager S, Lappalainen J, Kakela R, Hattula K, Butcher SJ, Schneider WJ, Kovanen PT. Lipid body formation during maturation of human mast cells. J Lipid Res. 2011;52:2198–2208. doi: 10.1194/jlr.M019737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dichlberger A, Schlager S, Maaninka K, Schneider WJ, Kovanen PT. Adipose triglyceride lipase regulates eicosanoid production in activated human mast cells. J Lipid Res. 2014;55:2471–2478. doi: 10.1194/jlr.M048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schreiber R, Zechner R. Lipolysis meets inflammation: arachidonic acid mobilization from fat. J Lipid Res. 2014;55:2447–2449. doi: 10.1194/jlr.C055673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlager S, Goeritzer M, Jandl K, Frei R, Vujic N, Kolb D, Strohmaier H, Dorow J, Eichmann TO, Rosenberger A, Wolfler A, Lass A, Kershaw EE, Ceglarek U, Dichlberger A, Heinemann A, Kratky D. Adipose triglyceride lipase acts on neutrophil lipid droplets to regulate substrate availability for lipid mediator synthesis. J Leukoc Biol. 2015;98:837–850. doi: 10.1189/jlb.3A0515-206R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Araujo-Santos T, Rodriguez NE, Moura-Pontes S, Dixt UG, Abanades DR, Bozza PT, Wilson ME, Borges VM. Role of prostaglandin F2alpha production in lipid bodies from Leishmania infantum chagasi: insights on virulence. J Infect Dis. 2014;210:1951–1961. doi: 10.1093/infdis/jiu299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toledo A, Roque NR, Teixeira L, Milan-Garces EA, Carneiro AB, Almeida MR, Andrade GF, Martins JS, Pinho RR, Freire-de-Lima CG, Bozza PT, D'Avila H, Melo RC. Lipid Body Organelles within the Parasite Trypanosoma cruzi: A Role for Intracellular Arachidonic Acid Metabolism. PLoS One. 2016;11:e0160433. doi: 10.1371/journal.pone.0160433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bandeira-Melo C, Paiva LA, Amorim NR, Weller PF, Bozza PT. EicosaCell:An Imaging-Based Assay to Identify Spatiotemporal Eicosanoid Synthesis. Methods Mol Biol. 2017;1554:127–141. doi: 10.1007/978-1-4939-6759-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koizume S, Miyagi Y. Lipid Droplets: A Key Cellular Organelle Associated with Cancer Cell Survival under Normoxia and Hypoxia. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gluchowski NL, Becuwe M, Walther TC, Farese RV., Jr Lipid droplets and liver disease: from basic biology to clinical implications. Nat Rev Gastroenterol Hepatol. 2017 doi: 10.1038/nrgastro.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carr RM, Ahima RS. Pathophysiology of lipid droplet proteins in liver diseases. Exp Cell Res. 2016;340:187–192. doi: 10.1016/j.yexcr.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chitraju C, Trotzmuller M, Hartler J, Wolinski H, Thallinger GG, Haemmerle G, Zechner R, Zimmermann R, Kofeler HC, Spener F. The impact of genetic stress by ATGL deficiency on the lipidome of lipid droplets from murine hepatocytes. J Lipid Res. 2013;54:2185–2194. doi: 10.1194/jlr.M037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hartler J, Kofeler HC, Trotzmuller M, Thallinger GG, Spener F. Assessment of lipidomic species in hepatocyte lipid droplets from stressed mouse models. Sci Data. 2014;1:140051. doi: 10.1038/sdata.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 91.Volmer R, Ron D. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol. 2015;33:67–73. doi: 10.1016/j.ceb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pineau L, Ferreira T. Lipid-induced ER stress in yeast and beta cells: parallel trails to a common fate. FEMS Yeast Res. 2010;10:1035–1045. doi: 10.1111/j.1567-1364.2010.00674.x. [DOI] [PubMed] [Google Scholar]
- 93.Baiceanu A, Mesdom P, Lagouge M, Foufelle F. Endoplasmic reticulum proteostasis in hepatic steatosis. Nat Rev Endocrinol. 2016;12:710–722. doi: 10.1038/nrendo.2016.124. [DOI] [PubMed] [Google Scholar]
- 94.Han J, Kaufman RJ. The role of ER stress in lipid metabolism and lipotoxicity. J Lipid Res. 2016;57:1329–1338. doi: 10.1194/jlr.R067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, Pon LA. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell. 2015;35:584–599. doi: 10.1016/j.devcel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fuchs CD, Claudel T, Kumari P, Haemmerle G, Pollheimer MJ, Stojakovic T, Scharnagl H, Halilbasic E, Gumhold J, Silbert D, Koefeler H, Trauner M. Absence of adipose triglyceride lipase protects from hepatic endoplasmic reticulum stress in mice. Hepatology. 2012;56:270–280. doi: 10.1002/hep.25601. [DOI] [PubMed] [Google Scholar]
- 99.Bosma M, Dapito DH, Drosatos-Tampakaki Z, Huiping-Son N, Huang LS, Kersten S, Drosatos K, Goldberg IJ. Sequestration of fatty acids in triglycerides prevents endoplasmic reticulum stress in an in vitro model of cardiomyocyte lipotoxicity. Biochim Biophys Acta. 2014;1841:1648–1655. doi: 10.1016/j.bbalip.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, Bobrovnikova-Marjon E, Diehl JA, Keith B, Simon MC. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mukhopadhyay S, Schlaepfer IR, Bergman BC, Panda PK, Praharaj PP, Naik PP, Agarwal R, Bhutia SK. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic Biol Med. 2017;104:199–213. doi: 10.1016/j.freeradbiomed.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 102.Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- 103.Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ito J, Ishii N, Akihara R, Lee J, Kurahashi T, Homma T, Kawasaki R, Fujii J. A high-fat diet temporarily renders Sod1-deficient mice resistant to an oxidative insult. J Nutr Biochem. 2016;40:44–52. doi: 10.1016/j.jnutbio.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 106.Ashraf NU, Sheikh TA. Endoplasmic reticulum stress and Oxidative stress in the pathogenesis of Non-alcoholic fatty liver disease. Free Radic Res. 2015;49:1405–1418. doi: 10.3109/10715762.2015.1078461. [DOI] [PubMed] [Google Scholar]
- 107.Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, Bellen HJ. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tariq Z, Green CJ, Hodson L. Are oxidative stress mechanisms the common denominator in the progression from hepatic steatosis towards non-alcoholic steatohepatitis (NASH)? Liver Int. 2014;34:e180–190. doi: 10.1111/liv.12523. [DOI] [PubMed] [Google Scholar]
- 109.Hasnain SZ, Prins JB, McGuckin MA. Oxidative and endoplasmic reticulum stress in beta-cell dysfunction in diabetes. J Mol Endocrinol. 2016;56:R33–54. doi: 10.1530/JME-15-0232. [DOI] [PubMed] [Google Scholar]
- 110.Bishop T, Ratcliffe PJ. HIF hydroxylase pathways in cardiovascular physiology and medicine. Circ Res. 2015;117:65–79. doi: 10.1161/CIRCRESAHA.117.305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee DC, Sohn HA, Park ZY, Oh S, Kang YK, Lee KM, Kang M, Jang YJ, Yang SJ, Hong YK, Noh H, Kim JA, Kim DJ, Bae KH, Kim DM, Chung SJ, Yoo HS, Yu DY, Park KC, Yeom YI. A lactate-induced response to hypoxia. Cell. 2015;161:595–609. doi: 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 113.Valli A, Rodriguez M, Moutsianas L, Fischer R, Fedele V, Huang HL, Van Stiphout R, Jones D, McCarthy M, Vinaxia M, Igarashi K, Sato M, Soga T, Buffa F, McCullagh J, Yanes O, Harris A, Kessler B. Hypoxia induces a lipogenic cancer cell phenotype via HIF1alpha-dependent and -independent pathways. Oncotarget. 2015;6:1920–1941. doi: 10.18632/oncotarget.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 117.Furuta E, Pai SK, Zhan R, Bandyopadhyay S, Watabe M, Mo YY, Hirota S, Hosobe S, Tsukada T, Miura K, Kamada S, Saito K, Iiizumi M, Liu W, Ericsson J, Watabe K. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008;68:1003–1011. doi: 10.1158/0008-5472.CAN-07-2489. [DOI] [PubMed] [Google Scholar]
- 118.Lee J, Homma T, Kurahashi T, Kang ES, Fujii J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem Biophys Res Commun. 2015;464:229–235. doi: 10.1016/j.bbrc.2015.06.121. [DOI] [PubMed] [Google Scholar]
- 119.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, Zhang Q, Wakelam MJ, Karpe F, Schulze A, Harris AL. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 121.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, Giannoukos DN, Bobrovnikova-Marjon E, Diehl JA, Keith B, Simon MC. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27:1115–1131. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deavall DG, Martin EA, Horner JM, Roberts R. Drug-induced oxidative stress and toxicity. J Toxicol. 2012;2012:645460. doi: 10.1155/2012/645460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pan X, Wilson M, McConville C, Arvanitis TN, Griffin JL, Kauppinen RA, Peet AC. Increased unsaturation of lipids in cytoplasmic lipid droplets in DAOY cancer cells in response to cisplatin treatment. Metabolomics. 2013;9:722–729. doi: 10.1007/s11306-012-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mylonis I, Sembongi H, Befani C, Liakos P, Siniossoglou S, Simos G. Hypoxia causes triglyceride accumulation by HIF-1-mediated stimulation of lipin 1 expression. J Cell Sci. 2012;125:3485–3493. doi: 10.1242/jcs.106682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Herms A, Bosch M, Ariotti N, Reddy BJ, Fajardo A, Fernandez-Vidal A, Alvarez-Guaita A, Fernandez-Rojo MA, Rentero C, Tebar F, Enrich C, Geli MI, Parton RG, Gross SP, Pol A. Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr Biol. 2013;23:1489–1496. doi: 10.1016/j.cub.2013.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mohammadyani D, Tyurin VA, O'Brien M, Sadovsky Y, Gabrilovich DI, Klein-Seetharaman J, Kagan VE. Molecular speciation and dynamics of oxidized triacylglycerols in lipid droplets: Mass spectrometry and coarse-grained simulations. Free Radic Biol Med. 2014;76:53–60. doi: 10.1016/j.freeradbiomed.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nakaya K, Ushio H, Matsukawa S, Shimizu M, Ohshima T. Effects of droplet size on the oxidative stability of oil-in-water emulsions. Lipids. 2005;40:501–507. doi: 10.1007/s11745-005-1410-4. [DOI] [PubMed] [Google Scholar]
- 128.Chaban VV, Khandelia H. Distribution of neutral lipids in the lipid droplet core. J Phys Chem B. 2014;118:11145–11151. doi: 10.1021/jp506693d. [DOI] [PubMed] [Google Scholar]
- 129.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 131.Meyers NL, Fontaine KA, Kumar GR, Ott M. Entangled in a membranous web: ER and lipid droplet reorganization during hepatitis C virus infection. Curr Opin Cell Biol. 2016;41:117–124. doi: 10.1016/j.ceb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med. 2015;21:34–42. doi: 10.1016/j.molmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 133.Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Jr, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Camus G, Herker E, Modi AA, Haas JT, Ramage HR, Farese RV, Jr, Ott M. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem. 2013;288:9915–9923. doi: 10.1074/jbc.M112.434910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Berger C, Romero-Brey I, Radujkovic D, Terreux R, Zayas M, Paul D, Harak C, Hoppe S, Gao M, Penin F, Lohmann V, Bartenschlager R. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology. 2014;147:1094–1105 e1025. doi: 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 136.Chatterji U, Bobardt M, Tai A, Wood M, Gallay PA. Cyclophilin and NS5A inhibitors, but not other anti-hepatitis C virus (HCV) agents, preclude HCV-mediated formation of double-membrane-vesicle viral factories. Antimicrob Agents Chemother. 2015;59:2496–2507. doi: 10.1128/AAC.04958-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boson B, Granio O, Bartenschlager R, Cosset FL. A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 2011;7:e1002144. doi: 10.1371/journal.ppat.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stapleford KA, Lindenbach BD. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J Virol. 2011;85:1706–1717. doi: 10.1128/JVI.02268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jain B, Chaturvedi UC, Jain A. Role of intracellular events in the pathogenesis of dengue; an overview. Microb Pathog. 2014;69–70:45–52. doi: 10.1016/j.micpath.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 140.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Iglesias NG, Mondotte JA, Byk LA, De Maio FA, Samsa MM, Alvarez C, Gamarnik AV. Dengue Virus Uses a Non-Canonical Function of the Host GBF1-Arf-COPI System for Capsid Protein Accumulation on Lipid Droplets. Traffic. 2015;16:962–977. doi: 10.1111/tra.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Carvalho FA, Carneiro FA, Martins IC, Assuncao-Miranda I, Faustino AF, Pereira RM, Bozza PT, Castanho MA, Mohana-Borges R, Da Poian AT, Santos NC. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J Virol. 2012;86:2096–2108. doi: 10.1128/JVI.06796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faustino AF, Martins IC, Carvalho FA, Castanho MA, Maurer-Stroh S, Santos NC. Understanding Dengue Virus Capsid Protein Interaction with Key Biological Targets. Sci Rep. 2015;5:10592. doi: 10.1038/srep10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lever A, Desselberger U. Rotavirus replication and the role of cellular lipid droplets: New therapeutic targets? J Formos Med Assoc. 2016;115:389–394. doi: 10.1016/j.jfma.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 145.Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gaunt ER, Zhang Q, Cheung W, Wakelam MJ, Lever AM, Desselberger U. Lipidome analysis of rotavirus-infected cells confirms the close interaction of lipid droplets with viroplasms. J Gen Virol. 2013;94:1576–1586. doi: 10.1099/vir.0.049635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Belhouchet M, Mohd Jaafar F, Firth AE, Grimes JM, Mertens PP, Attoui H. Detection of a fourth orbivirus non-structural protein. PLoS One. 2011;6:e25697. doi: 10.1371/journal.pone.0025697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Isken O, Langerwisch U, Schonherr R, Lamp B, Schroder K, Duden R, Rumenapf TH, Tautz N. Functional characterization of bovine viral diarrhea virus nonstructural protein 5A by reverse genetic analysis and live cell imaging. J Virol. 2014;88:82–98. doi: 10.1128/JVI.01957-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu X, Qi X, Liang M, Li C, Cardona CJ, Li D, Xing Z. Roles of viroplasm-like structures formed by nonstructural protein NSs in infection with severe fever with thrombocytopenia syndrome virus. FASEB J. 2014;28:2504–2516. doi: 10.1096/fj.13-243857. [DOI] [PubMed] [Google Scholar]
- 150.Sun Q, Qi X, Zhang Y, Wu X, Liang M, Li C, Li D, Cardona CJ, Xing Z. Synaptogyrin-2 Promotes Replication of a Novel Tick-borne Bunyavirus through Interacting with Viral Nonstructural Protein NSs. J Biol Chem. 2016;291:16138–16149. doi: 10.1074/jbc.M116.715599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 152.Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M. Lipid Droplets Are Essential for Efficient Clearance of Cytosolic Inclusion Bodies. Dev Cell. 2015;33:603–610. doi: 10.1016/j.devcel.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 153.Amen T, Kaganovich D. Dynamic droplets: the role of cytoplasmic inclusions in stress, function, and disease. Cell Mol Life Sci. 2015;72:401–415. doi: 10.1007/s00018-014-1740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Welte MA. As the fat flies: The dynamic lipid droplets of Drosophila embryos. Biochim Biophys Acta. 2015;1851:1156–1185. doi: 10.1016/j.bbalip.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 156.Li Z, Thiel K, Thul PJ, Beller M, Kuhnlein RP, Welte MA. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol. 2012;22:2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kolkhof P, Werthebach M, van de Venn A, Poschmann G, Chen L, Welte M, Stuhler K, Beller M. A luciferase-fragment complementation assay to detect lipid droplet-associated protein-protein interactions. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M116.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr Opin Genet Dev. 2006;16:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 159.Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol. 2014;24:1485–1491. doi: 10.1016/j.cub.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kan R, Jin M, Subramanian V, Causey CP, Thompson PR, Coonrod SA. Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev Biol. 2012;12:19. doi: 10.1186/1471-213X-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nagai A, Sato T, Akimoto N, Ito A, Sumida M. Isolation and identification of histone H3 protein enriched in microvesicles secreted from cultured sebocytes. Endocrinology. 2005;146:2593–2601. doi: 10.1210/en.2004-1478. [DOI] [PubMed] [Google Scholar]
- 162.Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV., Jr High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res. 2014;55:1465–1477. doi: 10.1194/jlr.M050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Anand P, Cermelli S, Li Z, Kassan A, Bosch M, Sigua R, Huang L, Ouellette AJ, Pol A, Welte MA, Gross SP. A novel role for lipid droplets in the organismal antibacterial response. Elife. 2012;1:e00003. doi: 10.7554/eLife.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gallardo-Montejano VI, Saxena G, Kusminski CM, Yang C, McAfee JL, Hahner L, Hoch K, Dubinsky W, Narkar VA, Bickel PE. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1alpha/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat Commun. 2016;7:12723. doi: 10.1038/ncomms12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kimmel AR, Sztalryd C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr Opin Lipidol. 2014;25:110–117. doi: 10.1097/MOL.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ueno M, Shen WJ, Patel S, Greenberg AS, Azhar S, Kraemer FB. Fat-specific protein 27 modulates nuclear factor of activated T cells 5 and the cellular response to stress. J Lipid Res. 2013;54:734–743. doi: 10.1194/jlr.M033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Aramburu J, Drews-Elger K, Estrada-Gelonch A, Minguillon J, Morancho B, Santiago V, Lopez-Rodriguez C. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem Pharmacol. 2006;72:1597–1604. doi: 10.1016/j.bcp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 169.Zhang C, Yang L, Ding Y, Wang Y, Lan L, Ma Q, Chi X, Wei P, Zhao Y, Steinbuchel A, Zhang H, Liu P. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat Commun. 2017;8:15979. doi: 10.1038/ncomms15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Helbig KJ, Beard MR. The role of viperin in the innate antiviral response. J Mol Biol. 2014;426:1210–1219. doi: 10.1016/j.jmb.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 171.Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, McCartney EM, Jangra RK, Lemon SM, Beard MR. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, Beard MR. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl Trop Dis. 2013;7:e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fujimoto T, Ohsaki Y. Proteasomal and autophagic pathways converge on lipid droplets. Autophagy. 2006;2:299–301. doi: 10.4161/auto.2904. [DOI] [PubMed] [Google Scholar]
- 175.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tiwari S, Siddiqi SA. Intracellular trafficking and secretion of VLDL. Arterioscler Thromb Vasc Biol. 2012;32:1079–1086. doi: 10.1161/ATVBAHA.111.241471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhou M, Fisher EA, Ginsberg HN. Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 178.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 179.Suzuki M, Otsuka T, Ohsaki Y, Cheng J, Taniguchi T, Hashimoto H, Taniguchi H, Fujimoto T. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell. 2012;23:800–810. doi: 10.1091/mbc.E11-11-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 181.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci U S A. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285:19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jo Y, Hartman IZ, DeBose-Boyd RA. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol Biol Cell. 2013;24:169–183. doi: 10.1091/mbc.E12-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Olzmann JA, Kopito RR. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J Biol Chem. 2011;286:27872–27874. doi: 10.1074/jbc.C111.266452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nakatsukasa K, Kamura T. Subcellular Fractionation Analysis of the Extraction of Ubiquitinated Polytopic Membrane Substrate during ER-Associated Degradation. PLoS One. 2016;11:e0148327. doi: 10.1371/journal.pone.0148327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.To M, Peterson CW, Roberts MA, Counihan JL, Wu TT, Forster MS, Nomura DK, Olzmann JA. Lipid disequilibrium disrupts ER proteostasis by impairing ERAD substrate glycan trimming and dislocation. Mol Biol Cell. 2017;28:270–284. doi: 10.1091/mbc.E16-07-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 189.Keembiyehetty CN, Krzeslak A, Love DC, Hanover JA. A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J Cell Sci. 2011;124:2851–2860. doi: 10.1242/jcs.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Weisshaar N, Welsch H, Guerra-Moreno A, Hanna J. Phospholipase Lpl1 Links Lipid Droplet Function with Quality Control Protein Degradation. Mol Biol Cell. 2017 doi: 10.1091/mbc.E16-10-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bischof J, Salzmann M, Streubel MK, Hasek J, Geltinger F, Duschl J, Bresgen N, Briza P, Haskova D, Lejskova R, Sopjani M, Richter K, Rinnerthaler M. Clearing the outer mitochondrial membrane from harmful proteins via lipid droplets. Cell Death Discov. 2017;3:17016. doi: 10.1038/cddiscovery.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DT. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–438. doi: 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Layerenza JP, Gonzalez P, Garcia de Bravo MM, Polo MP, Sisti MS, Ves-Losada A. Nuclear lipid droplets: a novel nuclear domain. Biochim Biophys Acta. 2013;1831:327–340. doi: 10.1016/j.bbalip.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 197.Ohsaki Y, Kawai T, Yoshikawa Y, Cheng J, Jokitalo E, Fujimoto T. PML isoform II plays a critical role in nuclear lipid droplet formation. J Cell Biol. 2016;212:29–38. doi: 10.1083/jcb.201507122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Uzbekov R, Roingeard P. Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res Notes. 2013;6:386. doi: 10.1186/1756-0500-6-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lagrutta LC, Montero-Villegas S, Layerenza JP, Sisti MS, Garcia de Bravo MM, Ves-Losada A. Reversible Nuclear-Lipid-Droplet Morphology Induced by Oleic Acid: A Link to Cellular-Lipid Metabolism. PLoS One. 2017;12:e0170608. doi: 10.1371/journal.pone.0170608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Farese RV, Jr, Walther TC. Lipid droplets go nuclear. J Cell Biol. 2016;212:7–8. doi: 10.1083/jcb.201512056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schlaitz AL, Thompson J, Wong CC, Yates JR, 3rd, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Turgay Y, Champion L, Balazs C, Held M, Toso A, Gerlich DW, Meraldi P, Kutay U. SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J Cell Biol. 2014;204:1099–1109. doi: 10.1083/jcb.201310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Welte MA. Fat on the move: intracellular motion of lipid droplets. Biochem Soc Trans. 2009;37:991–996. doi: 10.1042/BST0370991. [DOI] [PubMed] [Google Scholar]
- 204.Masedunskas A, Chen Y, Stussman R, Weigert R, Mather IH. Kinetics of milk lipid droplet (LD) transport, growth and secretion revealed by intravital imaging: LD release is intermittently stimulated by oxytocin. Mol Biol Cell. 2017 doi: 10.1091/mbc.E16-11-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kumar Y, Cocchiaro J, Valdivia RH. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 2006;16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 206.Barisch C, Paschke P, Hagedorn M, Maniak M, Soldati T. Lipid droplet dynamics at early stages of Mycobacterium marinum infection in Dictyostelium. Cell Microbiol. 2015;17:1332–1349. doi: 10.1111/cmi.12437. [DOI] [PubMed] [Google Scholar]
- 207.Knoblach B, Rachubinski RA. Transport and retention mechanisms govern lipid droplet inheritance in Saccharomyces cerevisiae. Traffic. 2015;16:298–309. doi: 10.1111/tra.12247. [DOI] [PubMed] [Google Scholar]
- 208.Arora GK, Tran SL, Rizzo N, Jain A, Welte MA. Temporal control of bidirectional lipid-droplet motion in Drosophila depends on the ratio of kinesin-1 and its co-factor Halo. J Cell Sci. 2016;129:1416–1428. doi: 10.1242/jcs.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Herms A, Bosch M, Reddy BJ, Schieber NL, Fajardo A, Ruperez C, Fernandez-Vidal A, Ferguson C, Rentero C, Tebar F, Enrich C, Parton RG, Gross SP, Pol A. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat Commun. 2015;6:7176. doi: 10.1038/ncomms8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dutta A, Kumar Sinha D. Turnover of the actomyosin complex in zebrafish embryos directs geometric remodelling and the recruitment of lipid droplets. Sci Rep. 2015;5:13915. doi: 10.1038/srep13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Orlicky DJ, Monks J, Stefanski AL, McManaman JL. Dynamics and molecular determinants of cytoplasmic lipid droplet clustering and dispersion. PLoS One. 2013;8:e66837. doi: 10.1371/journal.pone.0066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Guimaraes SC, Schuster M, Bielska E, Dagdas G, Kilaru S, Meadows BR, Schrader M, Steinberg G. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J Cell Biol. 2015;211:945–954. doi: 10.1083/jcb.201505086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lin C, Schuster M, Guimaraes SC, Ashwin P, Schrader M, Metz J, Hacker C, Gurr SJ, Steinberg G. Active diffusion and microtubule-based transport oppose myosin forces to position organelles in cells. Nat Commun. 2016;7:11814. doi: 10.1038/ncomms11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gross SP. Application of optical traps in vivo. Methods Enzymol. 2003;361:162–174. doi: 10.1016/s0076-6879(03)61010-4. [DOI] [PubMed] [Google Scholar]
- 216.Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- 217.Barak P, Rai A, Dubey AK, Rai P, Mallik R. Reconstitution of microtubule-dependent organelle transport. Methods Enzymol. 2014;540:231–248. doi: 10.1016/B978-0-12-397924-7.00013-3. [DOI] [PubMed] [Google Scholar]
- 218.Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, Welte MA, Gross SP. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Barak P, Rai A, Rai P, Mallik R. Quantitative optical trapping on single organelles in cell extract. Nat Methods. 2013;10:68–70. doi: 10.1038/nmeth.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Leidel C, Longoria RA, Gutierrez FM, Shubeita GT. Measuring molecular motor forces in vivo: implications for tug-of-war models of bidirectional transport. Biophys J. 2012;103:492–500. doi: 10.1016/j.bpj.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bartsch TF, Longoria RA, Florin EL, Shubeita GT. Lipid droplets purified from Drosophila embryos as an endogenous handle for precise motor transport measurements. Biophys J. 2013;105:1182–1191. doi: 10.1016/j.bpj.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Reddy BJ, Mattson M, Wynne CL, Vadpey O, Durra A, Chapman D, Vallee RB, Gross SP. Load-induced enhancement of Dynein force production by LIS1-NudE in vivo and in vitro. Nat Commun. 2016;7:12259. doi: 10.1038/ncomms12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Weaver C, Leidel C, Szpankowski L, Farley NM, Shubeita GT, Goldstein LS. Endogenous GSK-3/shaggy regulates bidirectional axonal transport of the amyloid precursor protein. Traffic. 2013;14:295–308. doi: 10.1111/tra.12037. [DOI] [PubMed] [Google Scholar]
- 224.Xu J, Reddy BJ, Anand P, Shu Z, Cermelli S, Mattson MK, Tripathy SK, Hoss MT, James NS, King SJ, Huang L, Bardwell L, Gross SP. Casein kinase 2 reverses tail-independent inactivation of kinesin-1. Nat Commun. 2012;3:754. doi: 10.1038/ncomms1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gross SP, Guo Y, Martinez JE, Welte MA. A determinant for directionality of organelle transport in Drosophila embryos. Curr Biol. 2003;13:1660–1668. doi: 10.1016/j.cub.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 226.Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 227.Lohmann D, Spandl J, Stevanovic A, Schoene M, Philippou-Massier J, Thiele C. Monoubiquitination of ancient ubiquitous protein 1 promotes lipid droplet clustering. PLoS One. 2013;8:e72453. doi: 10.1371/journal.pone.0072453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jackle H, Beller M. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Sci. 2013;126:2198–2212. doi: 10.1242/jcs.120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pfisterer SG, Gateva G, Horvath P, Pirhonen J, Salo VT, Karhinen L, Varjosalo M, Ryhanen SJ, Lappalainen P, Ikonen E. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat Commun. 2017;8:14858. doi: 10.1038/ncomms14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 231.Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Welte MA. How Brain Fat Conquers Stress. Cell. 2015;163:269–270. doi: 10.1016/j.cell.2015.09.046. [DOI] [PubMed] [Google Scholar]


