Summary
In C. elegans, the skn‐1 gene encodes a transcription factor that resembles mammalian Nrf2 and activates a detoxification response. skn‐1 promotes resistance to oxidative stress (Oxr) and also increases lifespan, and it has been suggested that the former causes the latter, consistent with the theory that oxidative damage causes aging. Here, we report that effects of SKN‐1 on Oxr and longevity can be dissociated. We also establish that skn‐1 expression can be activated by the DAF‐16/FoxO transcription factor, another central regulator of growth, metabolism, and aging. Notably, skn‐1 is required for Oxr but not increased lifespan resulting from over‐expression of DAF‐16; concomitantly, DAF‐16 over‐expression rescues the short lifespan of skn‐1 mutants but not their hypersensitivity to oxidative stress. These results suggest that SKN‐1 promotes longevity by a mechanism other than protection against oxidative damage.
Keywords: aging, C. elegans, oxidative stress, transcription regulation
Introduction, Results, Discussion
SKN‐1 is the C. elegans functional ortholog of the mammalian Nrf transcription factors. It protects against stress such that deletion or over‐expression of skn‐1 results in animals that are hypersensitive or resistant, respectively, to stress (Blackwell et al., 2015). skn‐1 also protects against aging: loss of skn‐1 shortens lifespan and skn‐1 over‐expression or gain‐of‐function usually increases lifespan (Blackwell et al., 2015; Tang & Choe, 2015). As stress resistance and increased lifespan (Age) are often correlated, one possibility is that protection against stress causes longer life (Ristow & Schmeisser, 2011).
Correlated stress resistance and longevity is also observed in worms with reduced insulin/IGF‐1 signaling (rIIS), dependent upon the transcription factor DAF‐16/FoxO (Kenyon, 2010). Results from combined mRNA and chromatin profiling suggest that DAF‐16 acts as a central regulator within a gene network (Schuster et al., 2010; Tullet, 2014). Notably, although these predicted direct DAF‐16 targets include few effectors of stress resistance, one of them is skn‐1 (Schuster et al., 2010). Like daf‐16, skn‐1 contributes to rIIS Age and stress resistance (Blackwell et al., 2015).
This raises the possibility that activation of skn‐1 expression by DAF‐16 promotes stress resistance and, consequently, increased lifespan.
If skn‐1 expression is activated by DAF‐16, then SKN‐1 could mediate the phenotypic effects of DAF‐16 activation. daf‐16 over‐expression (oe) using zIs356, a multicopy transgene array, increases resistance to stress (Henderson & Johnson, 2001) and extends lifespan (Qi et al., 2012). We therefore assessed whether SKN‐1 is required for daf‐16(oe) stress resistance and Age. First, we used zIs356 to compare the resistance of daf‐16(oe) and daf‐16(oe); skn‐1(zu135) worms to oxidative stress. daf‐16(oe) animals proved to be resistant to tert‐butyl hydroperoxide (t‐BOOH) and sodium arsenite, and this resistance was dependent on skn‐1 (Fig. 1A and 1B). Similar results were also obtained with respect to paraquat resistance, although here skn‐1 only partially suppressed the resistance of daf‐16(oe) (Fig. 1C). However, SKN‐1 was dispensable for daf‐16(oe) resistance to heat (Fig. S1). Thus, daf‐16(oe) wholly or partially requires SKN‐1 to promote Oxr but not thermotolerance.
Figure 1.
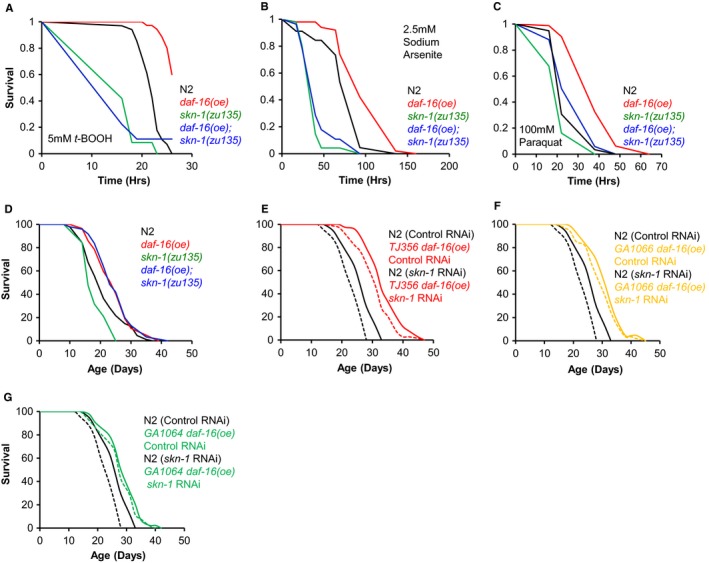
SKN‐1 is required for daf‐16(oe) Oxr but not Age. (A–C) skn‐1 is required for daf‐16(oe) Oxr. One representative trial shown in each case. (A) 5 mm t‐BOOH. daf‐16(oe) increases survival by +14.1%, P < 0.0001 (log rank test; combined data from 3 trials). (B) 2.5 mm sodium arsenite. daf‐16(oe) increases survival by +34.1%, P < 0.0001 (log rank test; representative data from 2 trials). (C) 100 mm Paraquat. daf‐16(oe) increases survival by +34.2%, P < 0.0001 (log rank test; combined data from 3 trials). (D) daf‐16(oe) (zIs356) Age does not require SKN‐1 (trial 1 in Table S1). (E,F) daf‐16(oe) Age is not suppressed by skn‐1 RNAi (both trial 1 in Table S1). (G) Age resulting from intestine‐specific daf‐16(oe) is not suppressed by skn‐1 RNAi (trial 1 in Table S1). Assays performed at 20 °C with 40 μm (D) or 80 μm FUDR (E–G).
Next we tested whether SKN‐1 was required for daf‐16(oe) Age. skn‐1(zu135) alone reduced lifespan but, strikingly, not in daf‐16(oe) animals (3 trials; Fig. 1D, Table S1). Thus, the effects of skn‐1 on daf‐16(oe) Age and Oxr are separable. It is striking here that the life‐shortening effects of skn‐1 are suppressed by daf‐16(oe), even though the worms remain sensitive to oxidative stress. This indicates that the life‐shortening effect of loss of function of skn‐1 is not due to the concomitant increase in sensitivity to oxidative stress.
We then sought to verify this unexpected conclusion using skn‐1(RNAi), and daf‐16(oe) achieved by three means: zIs356, muEx176 (Pdaf‐16::daf‐16a::gfp) or muEx227 (Pges‐1::daf‐16a::gfp, intestine‐limited over‐expression) (Libina et al., 2003; Alic et al., 2014). In most trials, skn‐1(RNAi) either did not reduce daf‐16(oe) Age (4/7 trials), or it reduced lifespan to a similar extent in N2 and daf‐16(oe) populations (2/7 trials; P = 0.46, 0.88; Cox proportional hazard analysis [CPHA]) (Fig. 1E–G, Table S1). In only one trial did skn‐1(RNAi) reduce lifespan marginally more in the daf‐16(oe) populations (P = 0.04, CPHA). By contrast, skn‐1(RNAi) significantly reduced N2 lifespan in all trials (Table S1). Thus, in all RNAi trials, daf‐16(oe) either fully or partially suppressed the short lifespan resulting from skn‐1 RNAi, consistent with results with skn‐1(zu135).
Effects of daf‐16(oe) on aging can be masked by premature death associated with daf‐16(oe)‐induced germline hyperplasia, but treatment with an inhibitor of DNA replication 5‐fluoro‐2‐deoxyuridine (FUDR) prevents this, unmasking the effect of daf‐16(oe) on lifespan (Qi et al., 2012). One possibility is that skn‐1 does not suppress daf‐16(oe) longevity because it also rescues the daf‐16(oe) germline abnormality. However, skn‐1 did not alter the frequency of germ cells outside the basal gonad membrane (Fig. S2) arguing against this. Another possibility is that skn‐1 does not reduce daf‐16(oe) lifespan because FUDR suppresses effects of skn‐1 on lifespan. However, the short lifespan on FUDR of three different skn‐1 mutants argues against this (Fig. S3A, Table S1).
It is notable that in daf‐2 mutants, where DAF‐16 is activated, longevity is skn‐1 dependent, but in daf‐2(+); daf‐16(oe) worms it is not. This could imply that daf‐16(oe) Age is only SKN‐1 dependent given rIIS. To test this, we compared the life spans of daf‐16(oe) and daf‐16(oe); skn‐1 worms subjected to daf‐2 RNAi. As expected, daf‐2 RNAi greatly extended the lifespan of WT worms and this was partially suppressed by skn‐1 (P < 0.0001) (Tullet et al., 2008). daf‐2 RNAi also increased the lifespan of daf‐16(oe) worms but notably this was not suppressed by skn‐1 (Fig. S3B, Table S1). Unexpectedly, daf‐16(oe) reduced the lifespan of worms subjected to daf‐2 RNAi (P < 0.0001 in each of 2 trials) (Fig. S3B, Table S1), perhaps reflecting excessive DAF‐16 activity. In summary, it is not the case that longevity induced by increased DAF‐16 activity is only SKN‐1 dependent given rIIS. Moreover, daf‐16(oe) suppresses the SKN‐1 dependence of rIIS longevity.
A long‐standing theory in the aging field is that aging is caused by accumulated oxidative damage, but some C. elegans studies have argued against this (reviewed in Gems & Partridge, 2013). However, SKN‐1 not only promotes longevity but also resistance to pro‐oxidants. If protection against molecular damage promotes daf‐16(oe) Age, then our lifespan results could be explained by daf‐16(oe) compensating for loss of skn‐1 by inducing other antioxidant defences. If correct, this predicts that elevation of protein oxidation levels in skn‐1 mutants should be suppressed by daf‐16(oe). We tested this by measuring protein carbonyl levels in worm protein extracts. Although results were variable, there was a trend toward skn‐1 worms having increased levels of protein oxidation compared to wild‐type (WT), as seen previously (Rea et al., 2007); this was also seen in daf‐16(oe) animals (Fig. S4), which is consistent with a previous study (Cabreiro et al., 2011). The latter trend was not worsened by skn‐1(zu135), but lessened (Fig. S4). Similar results were also observed in trials using skn‐1 RNAi (Fig. S4). Thus, daf‐16(oe) does not reduce overall levels of protein oxidation in skn‐1 mutants.
We had previously identified skn‐1 as potentially bound and transcriptionally activated by DAF‐16 (Schuster et al., 2010). To test this further, we verified binding of DAF‐16 to the skn‐1 promoter, comparing daf‐2 and daf‐16; daf‐2 adults using chromatin immunoprecipitation (ChIP) and PCR. Our previous chromatin profiles suggested two DAF‐16 binding sites at the skn‐1 locus (Fig. 2A). Re‐examining this confirmed DAF‐16 binding to the skn‐1b/c promoter (Fig. 2B).
Figure 2.
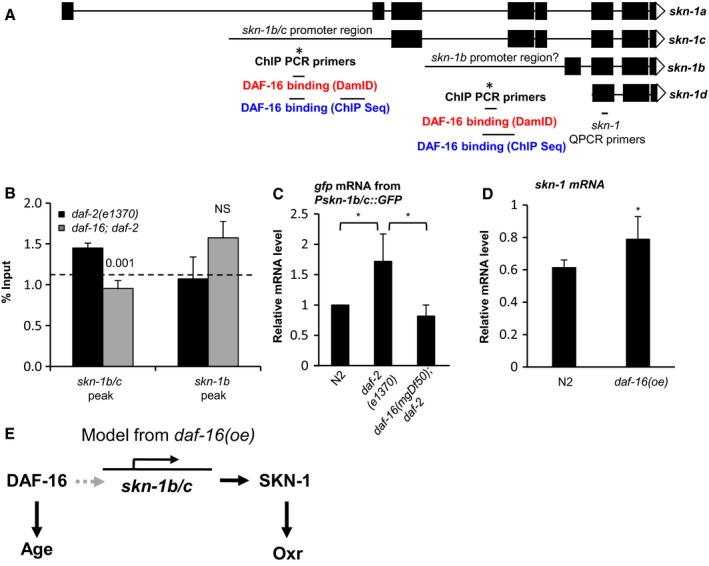
Pskn‐1b/c has the capacity for transcriptional activation by DAF‐16/FoxO. (A) Schematic representation of the skn‐1 locus. This shows the location of putative DAF‐16 binding sites identified by DamID (Schuster et al., 2010) and ChIP Seq (Niu et al., 2011), and of ChIP PCR primers. (B) DAF‐16 binds to Pskn‐1b/c but not Pskn‐1b. A DAF‐16‐specific antibody (Santa Cruz) was used for ChIP. The horizontal dotted line indicates % input from a region 5′ of Pskn‐1b/c not predicted to bind DAF‐16 (Schuster et al., 2010). One representative experiment (of three) is shown which contained 3 IP replicates from the same chromatin preparation (mean ± SD). (C) daf‐16‐dependent increase in gfp mRNA levels in Pskn‐1b/c::gfp daf‐2 animals. *P < 0.05, mean ± SD, 3 independent trials. Prior to transgene expression analysis animals were maintained at 15 °C until the L4 stage and then shifted to 25 °C for 24 h. (D) skn‐1 mRNA level is increased by daf‐16(oe) (zIs356). *P < 0.05, mean ± SD, 3 independent trials. (E) Scheme showing the DAF‐16/SKN‐1 portion of the DAF‐16 gene‐regulatory network, based on the daf‐16(oe) context where SKN‐1 promotes Oxr but not Age. Dashed arrow denotes context dependent capacity for transcriptional activation from Pskn‐1b/c by DAF‐16. Refer to supplement for methods to these and subsequent experiments.
mRNA profile data (microarrays) showed a 2.4‐fold increase in skn‐1 mRNA (q = 0.09) in glp‐4(bn2); daf‐2(m577) relative to daf‐16(mgDf50) glp‐4; daf‐2 in young adult hermaphrodites (McElwee et al., 2007). However, comparison of skn‐1 mRNA levels in daf‐2(e1370) vs daf‐16; daf‐2 animals using RT–QPCR did not confirm this (Fig. S5A). As an additional test of DAF‐16 regulation of skn‐1 expression, we created transgenic worm lines containing a Pskn‐1b/c::gfp transcriptional reporter and crossed them into daf‐2(e1370) and daf‐16; daf‐2 backgrounds. In a WT genetic background Pskn‐1b/c::GFP was expressed in mesendodermal tissues (Fig. S5B), consistent with the established role of SKN‐1 in development (Blackwell et al., 2015) and was broadly similar at all developmental stages. daf‐2(e1370) increased expression from this reporter (gfp mRNA levels), dependent upon daf‐16 (Fig. 2C) consistent with direct activation of the skn‐1b/c promoter by DAF‐16. GFP fluorescence was not changed by rIIS (Fig. S5C), but this could reflect the global reduction of protein synthesis caused by daf‐2(e1370) (Depuydt et al., 2013).
To understand these results, we reasoned that skn‐1 may possess a latent capacity to be up‐regulated by DAF‐16 that becomes detectable using the skn‐1::GFP transgene array. This could reflect the increased gene copy number in the array and/or greater stability of gfp mRNA (Fig. 2C). Consistent with this interpretation, we detected an increase in skn‐1 mRNA levels in daf‐16(oe) worms relative to WT using RT–PCR (Fig. 2D). Together, these results suggest that by virtue of its DAF‐16 binding site (Schuster et al., 2010) skn‐1 has the capacity to be upregulated by DAF‐16 via binding to the skn‐1b/c promoter. However, this capacity remains latent in some contexts in which DAF‐16 activity is increased, and it is possible that it is only manifested under artificial conditions such as daf‐16 over‐expression.
AMP‐activated protein kinase (AMPK) is, like SKN‐1, required for daf‐2 longevity and, like skn‐1, it is directly up‐regulated by DAF‐16 (Tullet, 2014). AMPK also acts upstream of DAF‐16 (Greer et al., 2007). We therefore wondered whether daf‐16(oe) Age might also be AMPK independent. Mutation of the AMPK α subunit gene aak‐2 fully suppresses daf‐2 longevity (Apfeld et al., 2004). However, aak‐2 did not suppress daf‐16(oe) Age (Fig. S3C). That neither SKN‐1 nor AMPK are required for daf‐16(oe) Age could imply that DAF‐16 effectors that extend lifespan vary according to whether DAF‐16 activation results from rIIS or daf‐16(oe) or, in the case of AMPK, may signify that DAF‐16 activation circumvents the need for AMPK upstream.
To conclude, this study reveals that the effects of SKN‐1 on Oxr and Age can be separated, implying that promotion of longevity by SKN‐1 can act by mechanisms other than oxidative stress resistance (Fig. 2E). This is consistent with the general conclusion that oxidative damage does not play a significant role in aging in C. elegans. SKN‐1 also transcriptionally regulates genes involved with other processes, for example, autophagy and collagen synthesis and, like IIS, plays a role in early development (Blackwell et al., 2015). Identification of the actual effector mechanisms by which SKN‐1 protects against aging is an important future challenge, particularly given the evolutionary conserved role of skn‐1/Nrfs in the control of aging (Sykiotis & Bohmann, 2008).
Funding
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a Wellcome Trust Strategic Award (CA, AFG, DG and JMAT) and a Royal Society Research grant (JMAT).
Conflict of interest
None declared.
Supporting information
Fig. S1 Resistance to heat stress measured in liquid.
Fig. S2 skn‐1 mutation does not affect the germline hyperplasia and basal membrane disruption of the germline of daf‐16(oe) animals.
Fig. S3 Tests for interactions between factors affecting lifespan.
Fig. S4 No difference in protein damage detected in response to skn‐1 mutation or skn‐1 RNAi in N2 or daf‐16(oe) animals.
Fig. S5 skn‐1 mRNA and Pskn‐1b/c::GFP fluorescence levels.
Table S1 Statistics for lifespan measurements.
Data S1 Supplemental material.
Acknowledgments
We thank Ralf Baumeister for sharing findings relating to daf‐16(oe) Age prior to publication. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Alic N, Tullet JM, Niccoli T, Broughton S, Hoddinott MP, Slack C, Gems D, Partridge L (2014) Cell‐nonautonomous effects of dFOXO/DAF‐16 in aging. Cell Rep. 6, 608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R (2004) The AMP‐activated protein kinase AAK‐2 links energy levels and insulin‐like signals to lifespan in C. elegans . Genes Dev. 18, 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M (2015) SKN‐1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman BP, Gems D (2011) Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic. Biol. Med. 51, 1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt G, Xie F, Petyuk VA, Shanmugam N, Smolders A, Dhondt I, Brewer HM, Camp DG, Smith RD, Braeckman BP (2013) Reduced insulin/IGF‐1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol. Cell Proteomics 12, 3624–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644. [DOI] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A (2007) The energy sensor AMP‐activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE (2001) daf‐16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C (2003) Tissue‐specific activities of C. elegans DAF‐16 in the regulation of lifespan. Cell 115, 489–502. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 8, R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, Slightham C, Jiang L, Hyman AA, Kim SK, Waterston RH, Gerstein M, Snyder M, Reinke V (2011) Diverse transcription factor binding features revealed by genome‐wide ChIP‐seq in C. elegans . Genome Res. 21, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Huang X, Neumann‐Haefelin E, Schulze E, Baumeister R (2012) Cell‐nonautonomous signaling of FOXO/DAF‐16 to the stem cells of Caenorhabditis elegans. PLoS Genet. 8, e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE (2007) Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5, e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S (2011) Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 51, 327–336. [DOI] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece‐Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D (2010) DamID in C. elegans reveals longevity‐associated targets of DAF‐16/FoxO. Mol. Syst. Biol. 6, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Choe KP (2015) Characterization of skn‐1/wdr‐23 phenotypes in Caenorhabditis elegans; pleitrophy, aging, glutathione, and interactions with other longevity pathways. Mech. Ageing Dev. 149, 88–98. [DOI] [PubMed] [Google Scholar]
- Tullet JMA (2014) DAF‐16 target identification in C. elegans: past, present and future. Biogerontology 16, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK (2008) Direct inhibition of the longevity‐promoting factor SKN‐1 by insulin‐like signaling in C. elegans . Cell 132, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Resistance to heat stress measured in liquid.
Fig. S2 skn‐1 mutation does not affect the germline hyperplasia and basal membrane disruption of the germline of daf‐16(oe) animals.
Fig. S3 Tests for interactions between factors affecting lifespan.
Fig. S4 No difference in protein damage detected in response to skn‐1 mutation or skn‐1 RNAi in N2 or daf‐16(oe) animals.
Fig. S5 skn‐1 mRNA and Pskn‐1b/c::GFP fluorescence levels.
Table S1 Statistics for lifespan measurements.
Data S1 Supplemental material.


