Abstract
Escherichia coli is a commensal or pathogenic bacterium that can survive in diverse environments. Adhesion to surfaces is essential for E. coli colonization, and thus it is important to understand the molecular mechanisms that promote this process in different niches. Autotransporter proteins are a class of cell-surface factor used by E. coli for adherence. Here we characterized the regulation and function of YeeJ, a poorly studied but widespread representative from an emerging class of autotransporter proteins, the inverse autotransporters (IAT). We showed that the yeeJ gene is present in ~40% of 96 completely sequenced E. coli genomes and that YeeJ exists as two length variants, albeit with no detectable functional differences. We demonstrated that YeeJ promotes biofilm formation in different settings through exposition at the cell-surface. We also showed that YeeJ contains a LysM domain that interacts with peptidoglycan and thus assists its localization into the outer membrane. Additionally, we identified the Polynucleotide Phosphorylase PNPase as a repressor of yeeJ transcription. Overall, our work provides new insight into YeeJ as a member of the recently defined IAT class, and contributes to our understanding of how commensal and pathogenic E. coli colonise their environments.
Introduction
Escherichia coli is a versatile bacterium comprising both commensal and pathogenic strains found in intra- and extra-intestinal environments1. The capacity for E. coli to adhere to different surfaces contributes to its ability to colonise and persist in specific host sites2. Among the numerous cell surface structures expressed by commensal or pathogenic E. coli multiple autotransporter (AT) proteins have been shown to contribute to their attachment and colonisation capacities3–11.
AT proteins represent a group of afimbrial adhesins that share several common features: an N-terminal domain leader sequence, a passenger domain that determines the functional characteristics of the protein and a C-terminal translocator or β-barrel domain that integrates into the outer membrane and facilitates transport of the passenger domain12, 13. In E. coli one of these autotransporters is the well-characterised phase-variable antigen 43 (Ag43) protein. Ag43 is encoded by the flu (or agn43) gene, and mediates cell-to-cell aggregation, biofilm formation, and long-term persistence in the urinary tract3, 14, 15. AT proteins have traditionally been grouped into four subclasses: Type Va, Vb, Vc and Vd16. However, more recent studies have revealed a new Type Ve subclass, which is similar to the classical monomeric Type Va AT proteins, but reversed in that they possess an N-terminal translocation domain and a C-terminal passenger domain17, 18. Due to this reversed topology, Type Ve AT proteins are also referred to as inverse AT (IAT) proteins. One such IAT is intimin, which mediates intimate binding to host cells and is an important virulence factor of enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli 19. The N-terminal region of some IATs contains a lysin motif domain (LysM domain), which mediates specific binding to peptidoglycan and acts as a dimerization interface for intimin20, 21.
Genomic analysis of the E. coli K-12 strain MG1655 revealed the presence of numerous genes encoding potentially cryptic adhesins, including AT proteins that contribute to attachment and colonization9, 10. In a previous study, we used the RExBAD cassette to place an arabinose inducible promoter upstream of putative adhesin-encoding genes in E. coli MG1655, and identified novel cryptic adhesins involved in biofilm formation that were not expressed under standard laboratory growth conditions9. This analysis identified YeeJ as an intimin-like protein involved in adhesion to different abiotic surfaces. While deletion of the yeeJ gene did not impact adherence to abiotic surfaces, the constitutive expression of YeeJ promoted strong biofilm formation9. The YeeJ protein from MG1655 is described as a 2,358 amino acid protein that belongs to the family of biofilm-associated proteins (Bap)22. Initially identified in Staphylococcus aureus, Bap is a surface adhesin that mediates biofilm formation and cell-to-cell adhesion, and can form amyloid fibers under specific environmental conditions9, 23–25. Bap-related proteins are present in many non-related Gram-negative and Gram-positive bacteria, including Esp from Enterococcus faecalis, LapA from Pseudomonas fluorescens, LapF from Pseudomonas putida, SiiE from Salmonella enterica and Bap from Acinetobacter baumannii 26–30. A common intriguing feature about these proteins is their very large size and ability to mediate a range of phenotypes, including (i) adhesion to abiotic and biotic surfaces, (ii) cell-to-cell interactions, (iii) biofilm formation, (iv) interaction with host epithelial cells and (v) the capacity to mediate invasion22, 31. Several studies have also shown these large extracellular adhesins also represent therapeutic targets, either as potential vaccines or as targets for anti-adhesion strategies32, 33.
In this study, we took advantage of the large number of E. coli genomes available on public databases to analyze the prevalence and conservation of the yeeJ gene. Our in silico analyses revealed the existence of two distinct variants of YeeJ that share similar functional properties. We show that YeeJ is both surface-located and present in the bacterial supernatant and that its LysM domain binds to peptidoglycan and is required for optimal YeeJ cell-surface localization and biofilm formation. At the regulatory level, our results revealed that, in E. coli K12, transcription of the yeeJ gene is increased in absence of the mRNA regulator PNPase. Taken together, this work enhances our understanding of YeeJ and its contribution to E. coli adhesion and biofilm formation.
Material and Methods
Bacterial strains and growth conditions
Strains used in this study are described in Table 1. Experiments were performed in LB medium or in M9 minimal medium with 0.4% glucose (M9-Glc) at 37 °C. Media were supplemented with the following antibiotics as required: ampicillin (100 μg/mL), chloramphenicol (25 μg/mL), kanamycin (50 μg/mL), spectinomycin (50 μg/mL), tetracycline (7.5 μg/mL) and zeocin (50 μg/mL).
Table 1.
Strains and plasmids.
| Strains | Description | Reference |
|---|---|---|
| MG1655 | K-12 reference strain | E. coli genetic stock center |
| MG1655 ΔyeeJ | yeeJ gene replaced by a Zeo cassette, ZeoR | This study |
| MG1655 PcLyeeJ | yeeJ gene placed under the control of the kmPcLrbs cassette λPR promoter | This study |
| MG1655 PcLyeeJΔLysM | strain MG1655 PcLyeeJ lacking LysM domain using λred recombination | This study |
| MG1655 Δpnp | Δpnp moved by P1 vir from the KEIO collection mutant, KmR | This study |
| MG1655 Δpnp (pZE12CFP) | pZE12CFP in MG1655, AmpR | This study |
| MG1655 Δpnp (pPNP2) | pPNP2 in MG1655, AmpR | This study |
| UMN026 | Wild type UPEC strain | 87 |
| MS427 | MG1655flu | 88 |
| MS427 (pSU2718) | pSU2718 in MS427, CmR | 48 |
| MS427 (pYeeJMG1655) | pYeeJMG1655 in MS427, CmR | This study |
| MS427 (pYeeJUMN026) | pYeeJUMN026 in MS427, CmR | This study |
| OS56 | MG1655flu; GFP + ; AmpR | 89 |
| OS56 (pSU2718) | pSU2718 in OS56, AmpR CmR | 48 |
| OS56 (pYeeJMG1655) | pYeeJMG1655 in OS56, AmpR CmR | This study |
| OS56 (pYeeJUMN026) | pYeeJUMN026 in OS56, AmpR CmR | This study |
| BL21 (DE3) | F– ompT hsdSB(rB–, mB–) gal dcm (DE3) | Stratagene |
| MG1655 ΔlacIZ::cat | lacIZ operon replaced by a Cm cassette, CmR | This study |
| MG1655 ΔlacIZ ΔyeeJ::lacZ | yeeJ replaced by the cassette lacZzeo, RBS of the yeeJ gene conserved; ∆lacIZ::cat. ZeoR, CmR | This study |
| MG1655_λATT::Km-GFPmut3 | Source for cassette kmPcL, KmR | 49 |
| Plasmids | ||
| pSU2718 | pACYC184-derived cloning plasmid | 90 |
| pYeeJMG1655 | yeeJ gene from MG1655 in pSU2718 | This study |
| pYeeJUMN026 | yeeJ gene from UMN026 in pSU2718 | This study |
| pZE12CFP | pZE12-MCS2-derived cloning plasmid | 91 |
| pPNP2 | pnp gene from MG1655 in pZE12CFP | This study |
DNA manipulation and genetic techniques
Genomic DNA (gDNA) was extracted and purified using the Wizard Genomic DNA purification kit (Promega). Isolation of plasmid DNA was carried out using the QIAprep Spin Miniprep kit (Qiagen). Gel extraction and purification of PCR products were performed using the QIAquick Gel Extraction kit (Qiagen) and QIAquick PCR Purification kit (Qiagen) or MiniElute PCR Purification kit (Qiagen), respectively. PCR screening assays were performed with Taq polymerase (New England BioLabs), and PCRs requiring proofreading were performed with the Phusion® High-Fidelity DNA Polymerase (New England BioLabs) or KAPA HiFi DNA Polymerase (Kapa Biosystems) as described by the manufacturers. Restriction endonucleases and T4 ligase were used per the manufacturer’s specifications (New England BioLabs). DNA sequencing was carried out using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) by the Australian Equine Genetics Research Centre.
Bioinformatic analysis
The prevalence and sequence conservation of yeeJ was examined using the FASTA36 software package34 to probe 96 complete E. coli genomes on the National Centre for Biotechnology Information (NCBI) database (Figure S1). The prevalence of genes was determined using a cut-off of >75% over a 75% amino acid sequence alignment. The E. coli strains were classified into major phylogroups (A, B1, B2, D, E and F) based on an in silico analysis of the arpA, chuA, yjaA and TSPE4.C2 loci35. Amino acid alignments were performed using ClustalW36. The Conserved Domain Database (CDD)37, Phyre238 and InterPro39 were used to analyze protein structures, and SignalP4.140 was used to predict the presence of signal sequences. The genomic context of genes was analysed with Easyfig.41. The 118 strains collection is a subcollection of the 122-strains collection previously described42.
PCR screening for the yeeJ gene in two E. coli strain collections
The prevalence of the yeeJ gene was assessed among strains from the ECOR collection43 and a subcollection of 118 strains from a previously described 122-strain collection42. Two primer sets were used: 3765 (5′-gatatgaacagcgagcaagc) and 3766 (5′-gtcattttcgccctgttta) targeting a 669 bp fragment at the 5′ end of the gene, and 3767 (5′-cagaagacaaaataatgagcgg) and 3768 (5′-ggtttttataacatgtcgcataagc) targeting a 591 bp fragment at the 3′ end of the gene. Primers 4225 (5′-gatcaaagtactgctgccctg) and 4226 (5′-gctatcgacgccattacctg) were used to further screen for the 906 bp fragment. Genomic DNA extracted from strains from the two collections was used as template DNA in the PCR assays.
β-galactosidase assays
β-galactosidase activity was measured as described previously44. Overnight cultures were washed and concentrated twice in LB medium. The enzyme activity was measured in triplicate for each strain at 37 °C with two technical repeats per sample. Colour development was measured at an optical density of 420 nm (OD420) in 1-cm cuvettes. Data were presented in arbitrary Miller units and calculated using the following formula: (750 * OD420)/(T * V * OD600), where T is time in minutes and V is volume in ml.
Biofilm formation assays
PVC 9-well microtiter plates (BD Falcon) were used to monitor biofilm formation as described previously45. Briefly, M9-Glc minimum media containing 1 mM IPTG was inoculated with a 1/100 dilution from an overnight culture in M9-Glc minimum media. After inoculation, microtitre plates were incubated at 37 °C for 24 h, rinsed and 150 μl of a 0.1% solution of crystal violet was added to each well. The plates were incubated at room temperature for 30 min and rinsed, and biofilm formation was tested as follows: crystal violet was solubilized by addition of 150 μl of ethanol-acetone (80:20), and the OD595 was determined. Results were presented as the mean of four replicate wells in three independent experiments. Flow cell assays were performed as previously described46. Briefly, OD600 standardized cells pre-grown in M9-Glc minimal media containing the appropriate antibiotics and 1 mM IPTG, if required, were inoculated into flow chambers and biofilms were allowed to develop on plastic coverslips (ProSciTech, Kirwan, QLD, Australia). Scanning confocal laser microscopy was performed using a ZEISS LSM 510 META Confocal Microscope to monitor biofilm formation 24 h post-inoculation.
Immunofluorescence microscopy
Overnight cultures supplemented with the appropriate antibiotics and 1 mM of IPTG were fixed to an OD600 of 0.4, spotted onto a glass slide and allowed to dry. The cells were fixed with 4% paraformaldehyde (PFA) and quenched with 50 mM NH4Cl. After three washes with PBS, the slides were blocked with 0.5% BSA, and incubated with a 1:100 dilution of the appropriate primary antibody in PBS for 30 minutes. The cells were washed with PBS and incubated with a secondary goat anti-rabbit antiserum coupled to fluorescein isothiocyanate (FITC), diluted 1:500 in PBS. The slides were washed and air-dried, mounted with ProLong Gold (Invitrogen), and examined under a ZEISS Axioplan 2 epifluorescence microscope.
Protein preparation from outer membrane vesicles (OMVs)
Overnight cultures were diluted 1/100 into 50 ml of LB broth with the appropriate antibiotics and induced with IPTG as required. After four hours of growth, cells were fixed at an OD600 of 1.0 and pelleted at 4 °C. Cell pellets were washed three times with 1 ml of cold PBS, and resuspended in 1 M of ethylenediaminetetraacetic acid (EDTA) buffer. Samples were incubated at 56 °C for 30 min and centrifuged for 15 min at 4 °C. The supernatants were filter sterilized using 0.22-μm filters. Trichloroacetic acid (TCA) was added to the samples to a final concentration of 20% to precipitate proteins. After an overnight incubation at 4 °C, cold acetone was added to each sample and the samples were centrifuged for 1 h. This wash was repeated twice. Pellets were air-dried and resuspended in 50 μl of resuspension buffer (50 mM ammonium bicarbonate, 3 M urea, 5 mM DTT). The samples were mixed with 50 μl of 2xSDS loading buffer, boiled for 5 min and centrifuged. Commercial anti-OmpA antibodies were used as a control (Antibody Research Corporation).
Peptidoglycan binding assays
Peptidoglycan (PG) from E. coli (Invitrogen) was used for pull-down assays. One milligram of lyophylized peptidoglycan was resuspended in 1 ml of 50 mM Tris-HCl, pH 7, sonicated (Misonix Sonicator 2000; microprobe, 30 s) and used as a stock for pull-down assays as described47. Cell pellets were lysed by B-Per and the protein fraction from the soluble phase was recovered. Protein extracts were concentrated 100× by size exclusion centrifugal filtration and dialyzed overnight against 50 mM Tris-HCl, pH 7. 100 μg of total protein extracts were incubated with 150 μl of the 1 mg/ml peptidoglycan stock (0.15 mg PG final concentration) in a total reaction of 300 μl. A control reaction lacking peptidoglycan was included. Reaction mixtures were incubated for 2 h at 4 °C with gentle agitation and then centrifuged at 20 000 × g. The supernatant containing unbound protein (U) was removed and mixed with SDS sample buffer for Western blot analysis. The pellet containing peptidoglycan and bound protein (B) was treated with 300 μl of 4% SDS in 50 mM Tris-HCl, pH 7 for 15 min at 4 °C and centrifuged. The supernatant containing released protein was collected and mixed with SDS sample buffer. Samples were analysed by Western immunoblotting using anti-YeeJ antibody raised against the C-terminal domain.
SDS-PAGE and Western blot analysis
Cell lysates, OMVs extractions and bacterial concentrated supernatants were subjected to SDS-PAGE and were transferred to PVDF microporous membrane filters as previously described48. Culture supernatants were prepared by filtering (pore size 0.22 μm) to remove intact bacterial cells and concentrated 100× by size exclusion centrifugal filtration. Serum raised against different domains of YeeJ (β-barrel and C-terminal domain) was used as primary antibody. The secondary antibody was an alkaline phosphatase-conjugated anti-rabbit immunoglobulin G. 5-Bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) was used as the substrate in the detection process (Healthcare).
Construction of strains and transposon mutagenesis
The strain constitutively expressing the YeeJ protein from E. coli MG1655 was constructed by the insertion of the λPR constitutive promoter upstream of the yeeJ coding region to generate MG1655 PcLyeeJ. The strain constitutively expressing a YeeJ protein deleted for the LysM domain was constructed using lambda-red recombination of a PCR fragment constructed by overlapping PCRs to generate MG1655 PcLyeeJ∆LysM. Strain MG1655 PcLyeeJ was used as a template for PCRs using primers: yeeJ.500-5 (5′-gttcattaattaacaactttg), delta-LysM-3 (5′-tttttttcactaacttgtgccaccgtattggcatttgcaatggca) and delta-LysM-5 (5′-ttgcaaatgccaatacggtggcacaagttagtgaaaaaaaattaa) and LysM-500-3 (5′-caggtgcgctgcgccagttggt). The two fragments obtained were assembled by PCR using yeeJ.500-5 and LysM-500-3. The assembled fragment was used to recombine using lambda red on the MG1655 genome and selection was performed using kanamycin. The insertion of the PcL promoter and the LysM deletion were verified by PCR with LysM-ext-3 (5′-tggccgggcgtgcttcataatc) and LysMjunction-5 (5′-ccaatacggtggcacaagttag) and sequencing using LysM-seq (5′-gacaaatattatgcgggatg). The MG1655∆yeeJ::lacZ strain was constructed using lambda red recombination employing the following primers: yeeJ-lacZzeo-Long.L-5 (5′-aagtggagaagaaataaatgaccgacaaatattatgcgggatgaccatgattacggattcactg), yeeJ-lacZzeo-Long.L-3 (5′-gccccgaatggaacactctttaagaaattataacggaaaatcagtctcctgctacgaa). Verification was performed using primers YeeJ.ext-5 (5′-gcatcacgctaatacagactcg), YeeJ.ext-3 (5′-gtctaccgtttatccttaccac), lacZ.ATG + 100-3 (5′-gggggatgtgctgcattaag) and zeo.verif-5 (5′-caggaccaggtggtgccggacaacaccc). The MG1655Δpnp strain was constructed using P1 transduction to transfer the pnp mutation from the KEIO collection to strain MG1655 using primers PnpFWDmut (5′-gtgccgtaaggtactgtctaag) and PnpREVmut (5′-gcagcgggagggcaatggc). All constructs were finally verified by sequencing. Random mutagenesis using mariner transposon was performed on strain MG1655ΔyeeJ::lacZΔlacZI as described previously49. Transposon insertion sites were determined using arbitrary PCR. Homology searches were performed using Blast 2.0.
Construction of plasmids
The yeeJ gene was amplified from either E. coli K-12 strain MG1655 or UPEC UMN026 with primers 3797 (5′-ccggcgtcgacatgaaggagggtaagcatggctacgaagaagag) and 3764 (5′-ggccggcatgctcagaggtttttataacatgtcg). The PCR products were digested with SalI (forward primer) and SphI (reverse primer), and ligated to a SalI-SphI digested pSU2718 plasmid to generate pYeeJMG1655 and pYeeJUMN026, respectively. Transcription of yeeJ in these constructs was under the control of the IPTG-inducible lac promoter. The pnp gene was amplified from K-12 strain MG1655 with primers PnpFWD (5′-ggtaccttgcttaatccgatcgttcg) and PnpREV (5′-aagcttttactcgccctgttcagcagc). The PCR products were digested with KpnI (forward primer) and HindIII (reverse primer) and ligated to a KpnI-HindIII digested pZE12CFP plasmid to generate pPNP2 plasmid.
Detection of YeeJ by ELISA
100 µl of bacterial culture adjusted to OD600 = 1 in a carbonate coating buffer (0.1 M NaHCO3, 0.1 M Na2CO3; pH to 9.5) were inoculated into 96 well microtiter plate MaxiSorp (Nunc.) and incubated overnight at 4 °C. Bacterial culture was removed by inversion, washed with T-PBS (×3) and incubated with 2% Milk/T-PBS for 1 hour at room temperature. Primary antibodies were diluted 1/100 for anti-YeeJ and 1/1000 for anti-E. coli in 1% Milk/T-PBS and incubated 1 hour at RT and washed with T-PBS (×3). Secondary antibody anti-rabbit IgG HRP was diluted 1/10000 in 1% Milk/T-PBS, incubated 1 hour at RT and washed with T-PBS (×3). Substrate reagent R&D Systems DY999 and Stop solution R&D Systems DY994 were used to develop and absorbance was read at OD420. The results are averages for four replicate wells in three independent experiments.
RNA extraction and qRT-PCR
500 µl of exponentially growing cells (OD600 = 0.6) were stabilized in 1 ml of RNAprotect Bacteria Reagent (Qiagen), and subsequent RNA extraction was performed using the RNeasy Mini Kit (Qiagen). After treatment with rDNase I (Ambion) to remove contaminating gDNA, the RNA was re-purified using the RNeasy Mini Kit RNA cleanup protocol (Qiagen). First-strand cDNA synthesis was performed using the SuperScript® III First-Strand Synthesis System (Invitrogen) as per manufacturer’s recommendation. Real-time PCR was performed using SYBR® Green PCR Master Mix (Applied Biosystems) on the ViiATM 7 Real-Time PCR System (Applied Biosystems) using primers 6965 (5′-tttgcgactgaccaactg) and 6966 (5′-agaccagtttaccgccaa) for yeeJ. Transcript levels of each gene were normalized to gapA as the endogenous gene control using primers 820 (5′-ggtgcgaagaaagtggttatgac) and 821 (5′-ggccagcatatttgtcgaagttag). Gene expression levels were determined with the 2−ΔΔCT method, with relative fold-difference expressed against wild type MG1655. All experiments were performed as three independent replicates.
Purification of YeeJ and generation of antibodies
Two six-histidine tagged, truncated forms of YeeJ were constructed. Primers 3096 (5′-tacttccaatccaatgcgaacctcgagcaacagatagcc) and 3097 (5′- ttatccacttccaatgtcacgttgtgaccactttaccacc) were used to amplify the portion of the gene encoding for the predicted β-barrel domain of the protein (corresponding to amino acids 119–460), and primers 5560 (5′-tacttccaatccaatgcggatgaaaaactgacactcac) and 5561 (5′-ttatccacttccaatgtcagcttgagttgccagtga) were used to amplify a portion of the predicted extracellular passenger domain of the protein (corresponding to amino acids 1838–1938). The PCR products were purified and inserted into the pMCSG7 vector by ligation independent cloning. The plasmids were transformed separately into E. coli BL21(DE3), and expression of the recombinant proteins was induced with 1 mM IPTG. The recombinant proteins were purified by Ni-nitrilotriacetic acid (NTA) superflow columns (Qiagen) under denaturing conditions (according to manufacturer’s instructions). The purified proteins were quantified with the Bicinchoninic Acid Protein Assay Kit (Sigma) and assessed for purity via SDS-PAGE. Rabbit polyclonal antisera were raised against each recombinant protein using four immunizations (400 µg protein/dose) at the Walter and Eliza Hall Institute Antibody Facility. The antisera were adsorbed against a crude protein extract of MG1655 ΔyeeJ prior to use.
Results
Prevalence and conservation of the yeeJ gene
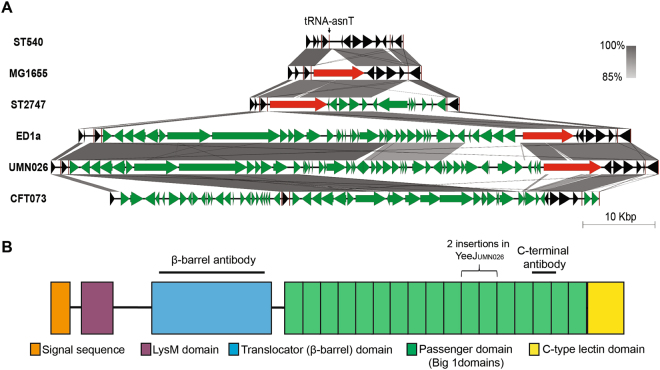
The prevalence of yeeJ was examined in a collection of 96 complete E. coli genome sequences representing all defined phylogroups. An intact gene encoding for a full length YeeJ protein was found in 40% (38/96) of genomes at the same genomic location as yeeJ in MG1655. The genetic context of yeeJ was conserved in the majority of strains (n = 35; represented by MG1655), while differences were observed in three strains (ST2747, ED1a and UMN026; Fig. 1A). Among the 38 yeeJ-positive strains, 25 belong to phylogroup A, three to B1, one to B2, two to D, six to E, and one to F (Figure S1). An additional 15 genomes contained the yeeJ gene at the same position, although further in silico analysis revealed that they had mutations or deletions that disrupt the yeeJ coding sequence.
Figure 1.
Genomic context of yeeJ and domain structure of the YeeJ protein. (A) The genomic context of yeeJ (red) was evaluated using MG1655 as the reference in Easyfig.41. Core (black arrows), variable (green arrows) and tRNA (red lines) genes are compared to the yeeJ-negative strains ST540 and CFT073. (B) Schematic illustration of the YeeJ protein from E. coli MG1655. Indicated are the predicted signal sequence, LysM peptidoglycan-binding domain, the translocator (β-barrel) domain, the passenger domain containing an invasin domain 3, Big 1 domains and the C-type lectin terminal domain. Also shown are the regions used to generate polyclonal antibodies against the β-barrel and passenger domains.
Comparative analysis of the nucleotide sequence of the 38 yeeJ genes revealed two distinct variants, represented by yeeJ from MG1655 (yeeJ MG1655) and yeeJ from the uropathogenic strain UMN026 (yeeJ UMN026). These two variants possessed 86% nucleotide sequence conservation, with the major difference being the presence of a 906 bp fragment in yeeJ UMN026 (but absent in yeeJ MG1655). A total of 11/42 strains possessed the yeeJ UMN026 variant. Of these strains, 2/11 belonged to phylogroup A, 1/11 to B1, 0/11 to B2, 2/11 to D and 6/11 to E (Figure S1).
To extend these findings, we screened two large, well-defined E. coli reference collections for the presence of the yeeJ MG1655 and yeeJ UMN026 variant alleles using a two-stage PCR screening approach. These collections included the E. coli Reference (ECOR) collection of 72 strains43, as well as another previously described collection of 118 strains42, 50–53. Strains from both collections were isolated from an array of hosts and geographical locations, and are representative of the ecological and phylogenetic diversity of the E. coli species. First, the presence of yeeJ was examined using primers designed to amplify two conserved regions of the gene (one region corresponding to the predicted β-barrel domain and the other region corresponding to the predicted passenger domain). The correct sized product for both PCR reactions was found in 36% (26/72) of strains from the ECOR collection, and 39% (44/118) of strains from the 118-strain collection (Tables S1 and S2). Further PCR analysis of these strains using a second primer set designed to identify the 906 bp fragment revealed that the yeeJ UMN026 variant was present in 8/26 strains from the ECOR collection and 17/44 of strains from the 118-strain collection.
Bioinformatic analysis of the YeeJ AT protein
Bioinformatic analysis of YeeJ protein from the strain MG1655 revealed a 2340 amino acid protein (instead of the 2358 amino acid protein indicated in the automated annotation). YeeJMG1655 is predicted to possess a multi-domain structure: (i) an N-terminal signal sequence, (ii) a periplasmic LysM (Lysin Motif) domain (Prosite PS51782), (iii) an inverse autotransporter translocator domain (Pfam PF11924), (iv) a passenger domain containing an invasin domain 3 (Pfam PF09134) and 13 repeated bacterial immunoglobulin-like 1 domains (Pfam PF02369) and (v) a C-type lectin terminal domain (Pfam PF07979) (Fig. 1B).
The LysM domain is found in many bacterial membrane-associated and secreted proteins, and mediates direct interaction with the peptidoglycan layer to stabilize cell-surface associated proteins. It may even contribute to the formation of pores in the peptidoglycan layer that assist protein translocation18, 54. Modeling of the 49 amino acid YeeJ LysM domain using the PHYRE2 program38 indicated that it strongly resembles the domain found in other adhesins including intimin, FsaP of Francisella tularensis and TspA of Neisseria meningitidis (Figure S2)55, 56. The translocator domain of YeeJ revealed a predicted 12-stranded β-barrel structure (Figure S3). This domain also shares 59% amino acid sequence identity with the β-barrel domain of the intimin-like protein FdeC, and is likely to be involved in the insertion of YeeJ into the outer membrane57. The passenger domain is predicted to be extracellular, as described for the intimin and invasin proteins17 and contains 13 bacterial immunoglobulin-like domain (Big) repeats. These repeats are typical of type Ve autotransporter proteins such as intimin and FdeC18, 57. The last 103 amino acids of YeeJ correspond to a C-type lectin domain, a structural domain also found in intimin and some other invasins that bind to carbohydrates58 (Figure S4). Analysis of the YeeJ variant from strain UMN026 revealed a similar overall predicted structure, with the additional 906 bp in the passenger domain. The 906 bp fragment is predicted to encode for three additional Big domains. BLAST analysis of the 906 bp fragment revealed that it is made up of a repeated section of sequence also found in YeeJMG1655.
Cloning and expression of the yeeJ gene from MG1655 and UMN026
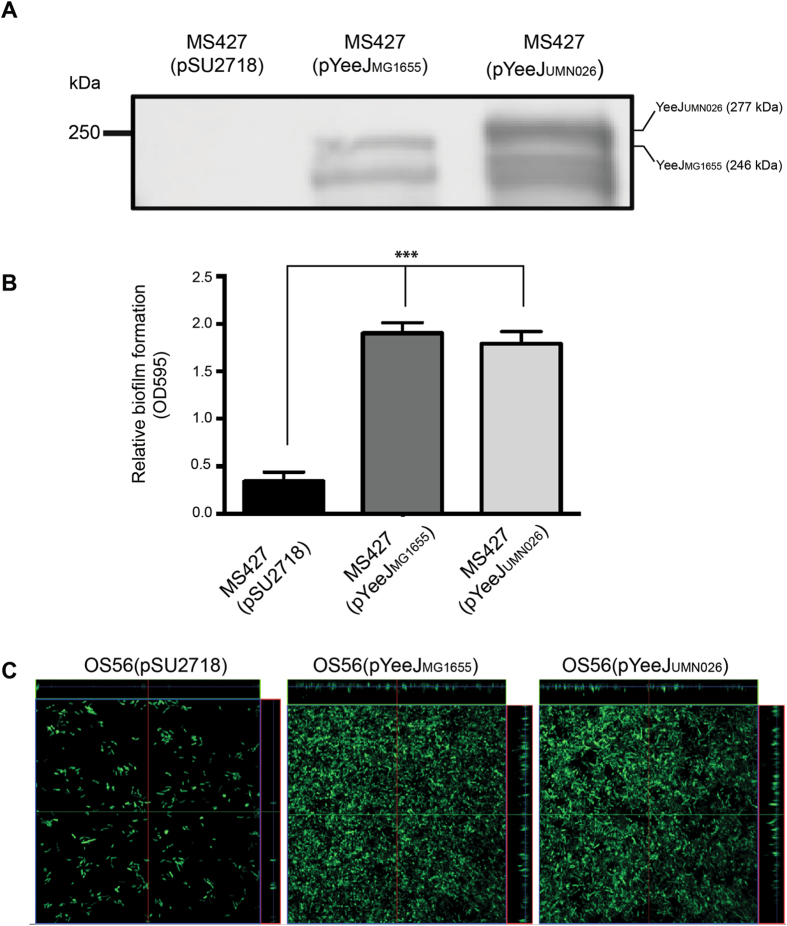
In order to examine the function of YeeJ and the impact of the additional 906 bp region in yeeJ UMN026, the yeeJ MG1655 and yeeJ UMN026 genes were PCR amplified and cloned into the low copy IPTG inducible pSU2718 expression vector to generate the plasmids pYeeJMG1655 and pYeeJUMN026, respectively. To demonstrate the expression of the YeeJ protein, both plasmids were transformed into the E. coli K-12 mutant strain MS427. MS427 contains a mutation in the flu gene (encoding for Ag43), and is unable to form compact cell aggregates and biofilms. Western blot analysis using a YeeJ-specific antiserum raised against the YeeJ C-terminal passenger domain resulted in the detection of a band corresponding to the full-length YeeJMG1655 (246 kDa) and YeeJUMN026 (277 kDa) in whole cell lysates prepared from MS427(pYeeJMG1655) and MS427(pYeeJUMN026), respectively (Fig. 2A). Overexpression of YeeJMG1655 and YeeJUMN026 also resulted in multiple smaller bands presumed to be breakdown products generated during the preparation of these samples. No YeeJ-specific band was detected in whole cell lysates prepared from the MS427(pSU2718) vector control.
Figure 2.
YeeJMG1655 and YeeJUMN026 possess similar functional properties. (A) Western blot analysis of whole cell lysates prepared from MS427(pSU2718) (vector control), MS427(pYeeJMG1655) and MS427(pYeeJUMN026). A band corresponding to full-length YeeJ from MG1655 (246KDa) and YeeJ from UMN026 (277KDa) were detected from MS427(pYeeJMG1655) and MS427(pYeeJUMN026), respectively. The image has been cropped from a larger blot as depicted in Supplementary Information file S7. (B) Biofilm formation by strains MS427 harboring plasmid pSU2718 (vector control), pYeeJMG1655 or pYeeJUMN026. All strains were grown in M9-Glc minimal media in the presence of 1 mM IPTG to induce yeeJ expression. Crystal violet staining was used to quantitate biofilm formation in 96-well microtiter plates. Values represent the average absorbance at 595 nm and error bars show the standard deviation calculated from three separate experiments. One-way ANOVA statistical analysis; ***P < 0.0001. (C) Scanning confocal microscopy images of biofilms formed on plastic coverslips under continuous flow conditions 24 hours post-inoculation with (i) OS56(pSU2718), (ii) OS56(pYeeJMG1655) and (iii) OS56(pYeeJUMN026). The images represent the horizontal sections within each biofilm. Displayed to the top and right of each micrograph are vertical sections representing the xz and yz plane, at the positions indicated by the green and red lines respectively.
Phenotypic properties of YeeJ
Some common features of AT proteins include the ability to mediate cell-to-cell aggregation, adhesion to extracellular matrix (ECM) proteins and epithelial cells, and biofilm formation. However, expression of either YeeJMG1655 or YeeJUMN026 in MS427 did not result in aggregation or adhesion to ECM proteins or to non-polarised and polarised Caco-2 human epithelial colorectal adenocarcinoma cells, T24 human bladder epithelial cells and MDCK dog kidney epithelial cells (data not shown). We then compared the ability of the two YeeJ variants to promote biofilm formation using two distinct systems. First, the two YeeJ proteins were tested for their ability to mediate biofilm formation in a microtiter plate biofilm assay. In this assay, MS427 (pYeeJMG1655) and MS427 (pYeeJUMN026) both formed a significant biofilm compared to the MS427 (pSU2718) control strain following growth in M9-Glc minimal media and induction with IPTG (Fig. 2B). Next, the two YeeJ proteins were tested for their ability to mediate biofilm formation under dynamic conditions using a continuous-flow chamber model. The gfp-tagged OS56 (pYeeJMG1655) and OS56 (pYeeJUMN026) strains were monitored for biofilm formation over 24 h using scanning confocal laser microscopy. In contrast to the OS56 (pSU2718) control strain, both OS56 (pYeeJMG1655) and OS56 (pYeeJUMN026) formed a biofilm with a higher total bio-volume (P < 0.0001), substratum coverage (P < 0.0001) and mean thickness (P < 0.0001) (Fig. 2C). No significant difference was observed between the biofilms formed by strains expressing either YeeJ variant. Taken together, these results demonstrate that YeeJ can promote biofilm formation when expressed in a recombinant E. coli K-12 strain, and that there is no difference between the ability of the two YeeJ variants to mediate biofilm formation under the conditions examined in these experiments.
YeeJ is located in outer membrane vesicles and at the cell surface
Our bioinformatic analysis suggested that yeeJ encodes for a protein with structural similarities to other outer membrane proteins such as FdeC and intimin from E. coli 18, 57. As the YeeJ C-terminal passenger domain is predicted to be extracellular, we investigated the subcellular localization of YeeJ. OMVs are inherently enriched in surface exposed proteins59, and we previously showed that EDTA/heat-induced OMVs of uropathogenic E. coli are strongly enriched with OM and extracellular proteins60, 61. Therefore, using a strain that constitutively expresses YeeJMG1655 (MG1655 PcLyeeJ), we first examined the presence of YeeJ in outer membrane vesicles (OMVs) by Western blot analysis using antibodies raised against the YeeJ C-terminal passenger domain. A band corresponding to the expected full-length YeeJ was detected in MG1655 PcLyeeJ, but not in wild type MG1655 (Fig. 3A).
Figure 3.
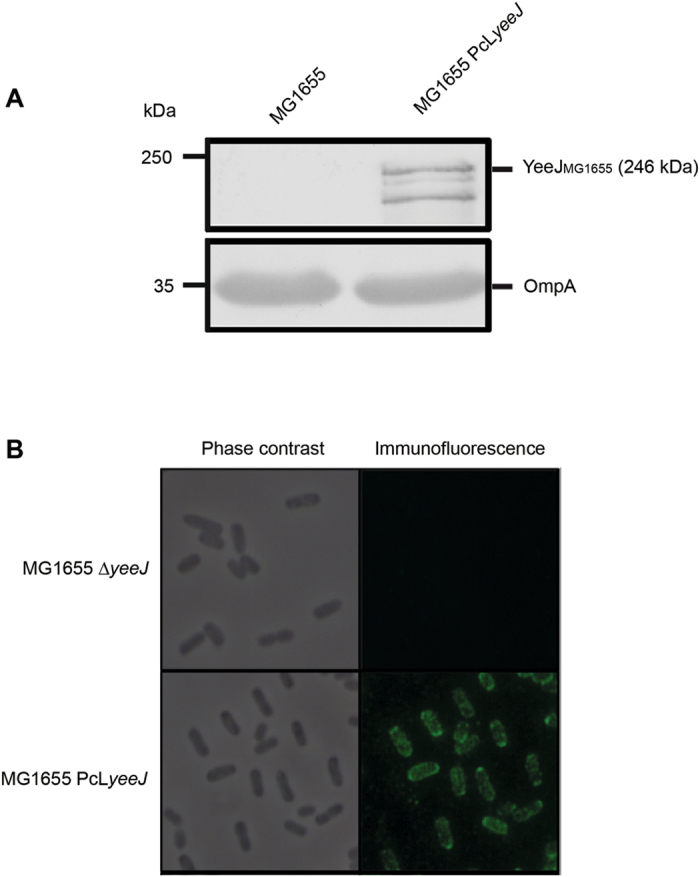
YeeJ is localized to the cell surface of E. coli. (A) Western blot analysis of outer membrane vesicles prepared from strains MG1655 and MG1655 PcLyeeJ using a YeeJ-specific antiserum targeting the C-terminal domain of YeeJ. A band corresponding to full-length YeeJ (246 KDa) was detected in MG1655 PcLyeeJ but not MG1655. The OmpA protein, which is located in the outer membrane92, was added as a control and was detected in both protein preparations. The images have been cropped from larger blots as depicted in Supplementary Information file S7. The same samples have been run in two independent gels, transferred and immunodetected with anti-YeeJ C-terminal or anti-OmpA antibodies. (B) Surface localization of the YeeJ protein. Cells of MG1655 and MG1655 PcLyeeJ were observed by phase contrast (left) or immunofluorescence microscopy (right) using a YeeJ-specific antiserum targeting the C-terminal domain of YeeJ.
In order to validate the OM localization of YeeJ and further assess whether YeeJ is exposed at the cell surface, we performed immunofluorescence microscopy on non-permeabilized cells using two anti-YeeJ antisera – (i) an antiserum against the C-terminus of the potential extracellular passenger domain and (ii) an antiserum against the outer membrane embedded β-barrel domain (Fig. 1B). While the YeeJ antiserum raised against the passenger domain reacted strongly with MG1655 PcLyeeJ (Fig. 3B), no signal was detected using the anti-β-barrel domain (data not shown). Taken together, our results suggest that YeeJ is localised to the outer membrane, and that the passenger domain is exposed at the cell surface.
The mature passenger domain of YeeJ can be released from the cell surface
YeeJ presence was also examined in supernatant fractions prepared from MG1655 PcLyeeJ (Fig. 4). Smaller bands between 125–150 kDa were detected in the TCA-precipitated supernatant using the antisera against the passenger domain. These bands were smaller than the band detected in whole cell lysates, which corresponds to the predicted full-length size of YeeJ (250 kDa) and some degradation products (Fig. 4A). Interestingly, we were not able to detect YeeJ in the supernatant fraction using the antisera against the β-barrel domain (Fig. 4B). The sigma 70 protein was used as a control, and was detected solely in the whole cell extract (pellet) and not in the supernatant (Fig. 4C). Taken together, these data suggest that the mature passenger domain of YeeJ can be processed and released into the supernatant.
Figure 4.
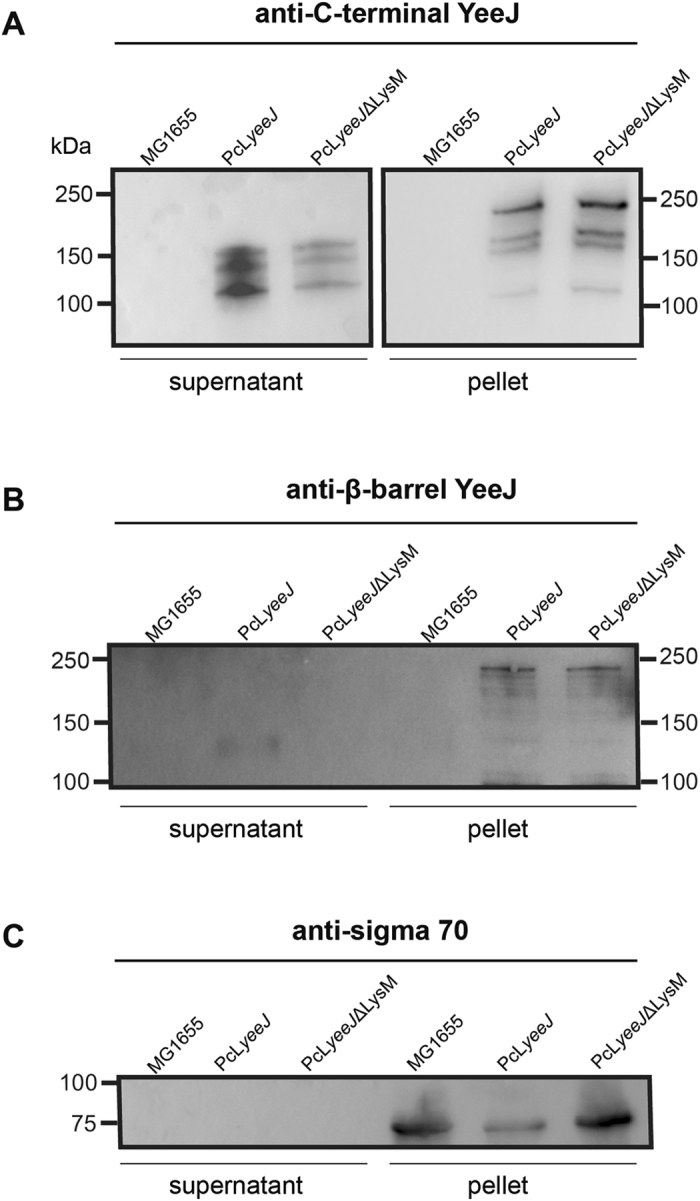
YeeJ can be cleaved from the E. coli cell surface. Western blot analysis of bacterial supernatants and corresponding pellets of strains MG1655, MG1655 PcLyeeJ and MG1655 PcLyeeJΔLysM using a polyclonal antibody targeting (A) the C-terminal region of YeeJ, (B) the β-barrel of YeeJ, and (C) the sigma 70 antibody (control). In (A) the bacterial supernatants and corresponding pellets have been run in two independent gels, transferred and immunodetected with anti-YeeJ C-terminal antibodies. In (B) and (C) the bacterial supernatants and corresponding pellets have been run in the same gel, transferred and immunodetected with YeeJ β-barrel antibodies and anti-Sigma70 antibodies, respectively. In (A–C) the images have been cropped from larger blots as depicted in Supplementary Information file S7.
The LysM domain is involved in YeeJ binding to peptidoglycan and affects YeeJ function
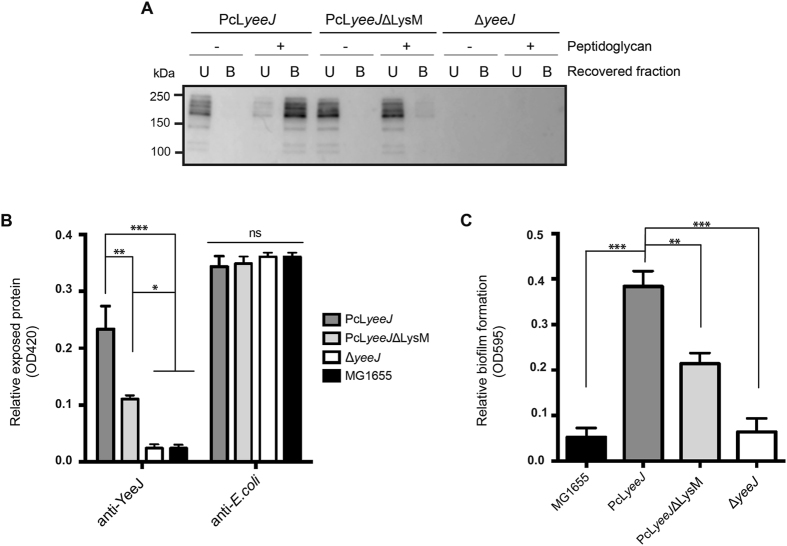
Our in silico analysis identified a 49 amino acid LysM domain located between the signal peptide and the predicted β-barrel domain (Fig. 1B). The LysM domain confers interaction with peptidoglycan, and is present in multiple secreted proteins, outer membrane proteins, lipoproteins and proteins bound in a non-covalent manner to the bacterial cell wall54. We hypothesized that in YeeJ, this domain could also bind to the peptidoglycan layer of the cell and stabilize the protein. Hence, we generated an in-frame deletion of the LysM domain on the chromosome of MG1655 PcLyeeJ, generating the strain MG1655 PcLyeeJΔLysM. Using YeeJ-specific antisera targeting both the passenger and translocator domains we detected a band corresponding to the full length YeeJ in cell pellets of both MG1655 PcLyeeJ and MG1655 PcLyeeJΔLysM strains but not in MG1655 (Figure 4AB). We observed no clear differences in molecular mass of YeeJ vs YeeJ∆LysM, probably because the deletion is too small to be detected by SDS PAGE analysis.
Next, we carried out an in vitro pull-down assay, where we prepared total protein extracts from MG1655 PcLyeeJ, MG1655 PcLyeeJΔLysM and MG1655ΔyeeJ and compared their capacity to bind to commercially purified E. coli peptidoglycan (Fig. 5A). In these assays, we observed that the majority of YeeJ was recovered in the bound fraction (B) when using the PcLyeeJ protein extracts with peptidoglycan compared to the control without peptidoglycan. Conversely, the majority of YeeJΔLysM was recovered in the unbound fraction (U) when using the PcLyeeJΔLysM extracts with or without peptidoglycan. No protein was detected in either fraction prepared with MG1655ΔyeeJ.
Figure 5.
The LysM domain of YeeJ promotes binding to peptidoglycan. (A) Peptidoglycan pull-down assay using total protein extract from strain MG1655 ∆ yeeJ, MG1655 PcLyeeJ and MG1655 PcLyeeJΔLysM obtained as described in the Materials and Methods. Total protein extracts were incubated with ultrapure peptidoglycan (Invivogen). Protein extracts containing PcLyeeJ with LysM motif bound to peptidoglycan (B), while the extracts containing PcLyeeJΔLysM were mainly associated with the unbound fraction (U). Samples were run in the same gel, transferred and immunodetected using antibodies targeted against the YeeJ passenger domain. The image corresponds to the whole blot as depicted in Supplementary Information file S7. (B) Quantification of YeeJ cell surface expression by strains MG1655, MG1655 ΔyeeJ, MG1655 PcLyeeJ and MG1655 PcL-yeeJΔLysM by ELISA using an antibody targeting the passenger domain of YeeJ or E. coli (control). One-way ANOVA statistical analysis; ***P < 0.0001; **P < 0.001; *P < 0.01. (C) Biofilm formation by strains MG1655, MG1655 PcL-yeeJ, MG1655 PcLyeeJΔLysM and MG1655 ΔyeeJ. All strains were grown in M9-Glc minimal medium. Crystal violet staining was used to quantitate biofilm formation in 96-well microtiter plates at 24 h. Values represent the average absorbance at 595 nm and error bars show the standard deviation calculated from three separate experiments. One-way ANOVA statistical analysis was performed using PcLyeeJ strain as reference: ***P < 0,0001; **P < 0,001.
We also examined whether the deletion of the LysM domain would affect YeeJ localization. We could not detect a clear difference in the cell surface localization of YeeJ between MG1655 PcLyeeJ and MG1655 PcLyeeJΔLysM by immunofluorescence microscopy (Figure S5). However, a more quantitative whole cell ELISA showed a reduction in signal for MG1655 PcLyeeJΔLysM compared to MG1655 PcLyeeJ, indicating that there was a lower amount of surface localized YeeJ in the MG1655 PcLyeeJΔLysM strain (Fig. 5B).
Finally, we performed biofilm assays to determine if the absence of the LysM domain would affect YeeJ function. In these assays, we found that despite the lack of the LysM domain, MG1655 PcLyeeJΔLysM formed a stronger biofilm compared to wild type MG1655 and MG1655ΔyeeJ (Fig. 5C). However, consistent with our previous data, biofilm formation by MG1655 PcLyeeJ was enhanced compared to MG1655 PcLyeeJΔLysM. Taken together, our results demonstrate that the LysM domain of YeeJ is able to bind to peptidoglycan, and deletion of this domain results in a reduction in the amount of YeeJ localized to the cell surface, which in turn affects biofilm formation.
The polynucleotide phosphorylase PNP affects yeeJ mRNA levels
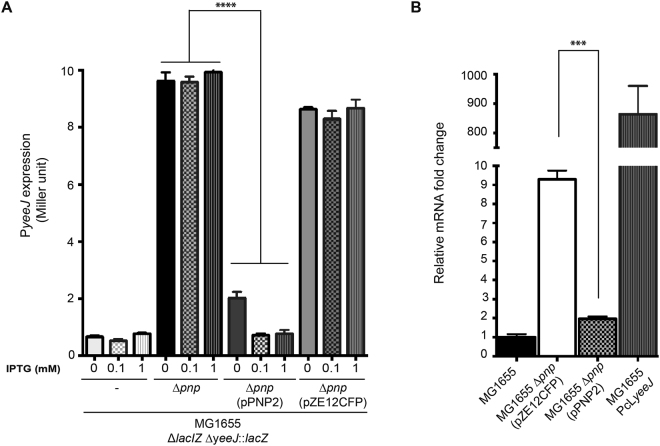
Our western blot and ELISA analyses indicated that YeeJ is either not produced or produced at levels below our limit of detection during growth in LB broth. To investigate the genetic basis of yeeJ regulation, we generated a chromosomal yeeJ promoter-lacZ reporter fusion construct (MG1655 ΔlacIZ ΔyeeJ::lacZ). All MG1655 ΔlacIZ ΔyeeJ::lacZ colonies were white when grown on LB agar supplemented with X-gal at 37 °C, indicating no apparent activity from the yeeJ promoter, a result consistent with the lack of detection of YeeJ in MG1655 (Figs 3 and 4). Since the expression of cell surface adhesins can be modulated by environmental factors, we assessed whether yeeJ would be expressed in MG1655 following growth under different conditions (including different temperatures, static growth, anaerobic conditions and increased osmotic conditions). None of the tested conditions affected yeeJ expression (Figure S6). In order to identify potential transcriptional regulators of yeeJ, the MG1655 ΔlacIZ ∆yeeJ::lacZ reporter strain was subjected to random mariner transposon mutagenesis. The resultant transposon mutants were screened on LB plates supplemented with X-gal to identify blue colonies indicative of an active yeeJ promoter. Three blue transposon mutants were isolated, and the transposon insertion sites in these mutants were determined by arbitrary PCR. All three mutants contained independent insertions within the pnp gene (Accession Number: ECK3152). The pnp gene encodes for a multi-enzyme complex polynucleotide phosphorylase (PNPase), which is involved in RNA metabolism and controls numerous phenotypes such as biofilm formation, motility and bacterial survival62–64. In order to confirm the activity of the yeeJ promoter in the pnp mutant, we generated a specific pnp mutant in the MG1655 ΔlacIZ ∆yeeJ::lacZ strain and performed a ß-galactosidase assay (Fig. 6A). To complement pnp mutation, the pnp gene from strain MG1655 was amplified and cloned into the pZE12CFP plasmid, under the control of an IPTG inducible promoter, to generate plasmid pPNP2. Our results demonstrated that the activity of the yeeJ promoter is induced in the pnp mutant (MG1655 Δpnp ΔlacIZ ΔyeeJ::lacZ), and complementation of the pnp mutation with pPNP2 restores promoter activity to wild type level.
Figure 6.
PNPase is a repressor of yeeJ transcription. (A) Activity of the yeeJ promoter-lacZ fusion assessed by β-galactosidase quantitation in wild type MG1655 (MG1655 ΔlacIZ ΔyeeJ::lacZ), an isogenic pnp mutant, complemented pnp mutant with plasmid pPNP2 and pnp mutant carrying the empty plasmid pZE12CFP. β-galactosidase levels were measured in each individual strain with different IPTG concentrations (0, 0.1 and 1 mM) from an overnight culture. Mutation of pnp gene increased PyeeJ activity and complementation of the pnp mutation restored the PyeeJ basal activity to wild type levels. All experiments were performed in triplicate. One-way ANOVA statistical analysis: ***P < 0.0001; **P < 0.001. (B) Transcription of the yeeJ gene in wild type MG1655, isogenic pnp mutant, complemented mutant with plasmid pPNP2 and the PcLyeeJ strain. The relative fold difference in yeeJ transcript levels relative to MG1655 as determined by qRT-PCR using the 2−ΔΔCT method. Mutation of the pnp gene led to an increase in yeeJ mRNA transcript levels, and complementation of the mutant with a plasmid containing pnp (pPNP) restored yeeJ mRNA transcripts to wild type level. All experiments were performed in triplicate. One-way ANOVA statistical analysis was performed using MG1655 Δpnp as reference: ***P < 0.0001.
To further confirm the effect of PNPase on yeeJ transcription, we deleted pnp in MG1655Δpnp and we evaluated levels of these transcripts by qRT-PCR. The strain MG1655 PcLyeeJ, that constitutively expresses yeeJ was used as a positive control. The level of yeeJ transcript was examined in wild type MG1655, MG1655 Δpnp (pZE12CFP), complemented MG1655 Δpnp (pPNP2) and MG1655 PcLyeeJ strains by qRT-PCR (Fig. 6B). Consistent with our ß-galactosidase assay, inactivation of the pnp gene led to an ~9-fold increase in relative yeeJ transcript level, and complementation of the mutant with pPNP2 plasmid restored relative yeeJ transcripts to wild type level. In contrast to MG1655 Δpnp, the level of yeeJ transcription in MG1655 PcLyeeJ was 900-fold higher than in MG1655. Deletion of the pnp gene in MG1655 did not lead to increased biofilm formation (data not shown), suggesting that a threshold level of YeeJ is required to translate into increased adhesion. Taken together, these results indicate that PNPase negatively regulates the transcription of yeeJ. However, this repression cannot totally explain the very low yeeJ expression observed during laboratory growth.
Discussion
E. coli produces a vast number of factors that contribute to biofilm formation and adhesion to various surfaces, including AT protein adhesins. These adhesion factors are a core component of the type V secretion system that delivers cargo proteins across the outer membrane of Gram-negative bacteria. The Ve subclass of AT proteins are referred to as IATs, due to their similarity with classical monomeric AT proteins, but with the passenger and translocation domain in opposite locations within the primary amino acid sequence. Two well-studied proteins from this subclass include intimin and FdeC of E. coli, both of which have been extensively characterised48. Here, we characterised the YeeJ IAT protein from K-12 strain MG1655; we performed an in silico analysis of the yeeJ gene, determined the prevalence of yeeJ, and demonstrated that it is surface localized and mediates biofilm formation in vitro.
Our analysis revealed that the yeeJ gene in MG1655 is found immediately downstream of the tRNA-asnT gene, a common site for the insertion of horizontally acquired DNA65, 66. Indeed, the tRNA-asnT gene is frequently associated with insertion of the high pathogenicity island that contains genes required for the synthesis of yersiniabactin. This pathogenicity island was originally discovered in Yersinia enterocolitica 67, but has also been identified in multiple pathotypes of E. coli 68–71. Interestingly, this pathogenicity island has been shown to be more frequently associated with pathogenic E. coli isolates72. The genomic location of yeeJ adjacent to the tRNA-asnT, together with the high potential for recombination at this site, may explain why a high number of strains possessed a truncated yeeJ gene, and why a full length yeeJ gene is found more frequently in phylogroup A (non-pathogenic) strains.
Our in silico screen of complete E. coli genomes publicly available on the NCBI database revealed that 38 strains possess an intact yeeJ gene encoding for the full length protein. These strains belong to different pathotypes and phylogroups, suggesting that the yeeJ gene is conserved across a diverse range of strains. An additional 15 strains possess the yeeJ gene, but have frame-shift mutations that result in a truncated YeeJ protein. However, it is possible that these mutations are artifacts of sequencing or post-sequencing genome assembly errors. Indeed, this type of miss-assembly has previously been observed in the highly repetitive upaH AT gene from CFT073, which was initially reported to contain a frame-shift resulting in a truncated protein. Subsequent work determined that the upaH gene was misassembled during genome closure, and in fact it encodes for a full-length surface-expressed protein that mediates biofilm formation73. In addition, we also examined the presence of yeeJ in two large E. coli collections by PCR; the 72-strain ECOR collection and a collection of previously described 118 strains. Both collections correspond to strains isolated from diverse hosts and geographical sites, thus representing the ecological and phylogenetic diversity of the E. coli species. The correct sized PCR products were amplified in roughly 40% of strains, providing further evidence that there is strong selective pressure favouring the conservation of yeeJ gene in the E. coli species.
Although YeeJ has been previously linked to the Bap family of proteins9, 32, our analysis suggests that it is more closely associated with intimin and FdeC, and the IAT family of proteins. Despite having multiple Big domains (like the Bap proteins), YeeJ also contains a N-terminal LysM domain, a translocator β-barrel domain and a passenger domain capped with a C-lectin-like domain, all of which are absent in the Bap proteins but found in other IAT proteins. Moreover, the multiple repeats of Big domains (Big_3_2 (Pfam 12245) and Big_3_4 (Pfam13754)) found in Bap are different from the ones found in YeeJ. Two distinct variants of the yeeJ gene were identified based on the presence/absence of a 906 bp fragment within the passenger-encoding domain. Interestingly, the gene encoding for the longer variant of YeeJ was found in almost all yeeJ-positive pathogenic isolates identified in our bioinformatics analysis, but not in any non-pathogenic isolates. This suggests that the region encoded by this 906 bp fragment may contribute to fitness and/or virulence. Indeed, other adhesins like UpaH and Ag43 exhibit sequence variation that results in altered levels of biofilm formation by different variants14, 74. Hence, one of the aims of this study was to characterize both variants using the genes from E. coli MG1655 and UMN026 as representatives. However, both variants displayed similar phenotypic properties, suggesting that this fragment in the protein does not affect YeeJ function in the assays employed in this study. Additional Big domains found in the longer version of YeeJ might confer differential properties of YeeJ in some context that remains to be elucidated, such as extension of the protein beyond other surface structures that might otherwise mask its function.
The localization of YeeJ was investigated by immunofluorescence microscopy using two different sets of YeeJ antisera. Our results suggest that the C-terminal domain of YeeJ is exposed at the cell surface, whereas the β-barrel domain is likely embedded in the outer membrane, consistent with what has been described for intimin and invasin75. In agreement, immunodetection of outer membrane extractions revealed the presence of YeeJ. Analysis of supernatant fractions showed that the YeeJ passenger domain might be cleaved from the cell surface, yielding a cleavage product of approximately 100 kDa less than full-length YeeJ. Similarly, the passenger domains of other AT proteins may be processed and released into the extracellular surroundings (e.g. Pet and EspP), or cleaved but remain in contact with the cell surface via non-covalent interactions with the β-barrel domain (e.g. AIDA and Ag43)76. Whether the YeeJ cleavage products have a relevant function remains to be determined.
Some ATs like E. coli UpaG, EhaG and Ag43, or meningococcal AutA have been shown to mediate biofilm formation and cell-to-cell aggregation, resulting in the formation of bacterial clumps and flocculation4, 6, 73, 77. Additionally, intimin mediates adhesion of enteropathogenic E. coli strains to the intestinal epithelium, and invasin produced by enteropathogenic strains of Yersinia enterocolitica mediates binding to β1-integins17, 78. The intimin-like FdeC also mediates biofilm formation and colonisation of the bladder and kidney57. However, our results indicate that YeeJ does not mediate cell-to-cell aggregation, or adhesion to ECM proteins and different types of eukaryotic cells. We cannot exclude that the C-type lectin region of YeeJ could recognize a specific, yet unknown, receptor of some eukaryotic cells.
LysM domains are well-conserved domains found in proteins from a large variety of organisms from mammals to bacteria and viruses, and are known to bind different polysaccharides containing N-acetylglucosamine residues. Recently, the molecular mechanism behind LysM-peptidoglycan interaction was described79. A LysM domain is present in the N-terminal region of several adhesins including FsaP from Francisella tularensis, TspA from Neisseria meningitidis and intimin from EHEC and EPEC strains20, 55, 80, and the LysM domain of intimin binds to peptidoglycan20. Consistent with this result, we showed that YeeJ also binds to peptidoglycan, while deletion of the LysM domain from YeeJ results in loss of this interaction. Additionally, the LysM domain of intimin and other similar proteins has been shown to dimerise20, with interaction between the LysM domain and peptidoglycan possibly contributing to translocation. Consistent with this hypothesis, our results suggested that absence of the LysM domain of YeeJ reduced its surface localisation. It is possible that LysM-mediated interaction with peptidoglycan could stabilize YeeJ in the outer membrane. Additionally, LysM mediated dimerisation and potentially higher order oligomerisation could increase its local concentration and enhance its functional activity.
We also attempted to determine the genetic basis for the very low expression of yeeJ during growth under standard laboratory conditions, in order to uncover potential additional layers of its regulatory control. Using a mutagenesis approach, we identified the Polynucleotide phosphorylase (PNPase) as a potential repressor of yeeJ. PNPase is involved in RNA metabolism, for example it degrades various mRNAs and is involved in cold shock regulation. Loss of PNPase leads to an increase in steady-state levels of mRNA81, 82. It has been described that PNPase is involved in the degradation of lac mRNA, rnb mRNA and the RNA-OUT anti-sense molecule62, 83. The yeeJ mRNA can thus be added to the list of mRNA degraded by PNPase. The importance of yeeJ mRNA degradation in the physiology or lifestyle of E. coli remains to be elucidated. Since the deletion of pnp did not lead to increased biofilm formation, we hypothesize that the repression of yeeJ involves additional levels of regulatory control. The regulators and/or environmental conditions that induce yeeJ expression remain to be identified. Interestingly, PNPase has been described to have a pleiotropic effect on other extracellular factors. For instance, PNPase inactivation affects E. coli virulence, particularly increasing Tir protein content and transcription of Type III secretion system components, including intimin, Tir and EspB in E. coli O157:H784. PNPase also negatively affects N. meningitidis aggregation and adhesion mediated by Type IV pili85, as well as transcription of the genes encoding Type IV pili and the Type III secretion system in P. aeruginosa 86. In addition to the control of yeeJ, it is possible that PNPase also regulates the expression of other surface factors in E. coli, although this remains to be demonstrated.
Taken together, our characterisation of YeeJ function and the identification of PNPase as a regulator involved in the control of its expression provides new insight into the potential role of this adhesin in E. coli biofilm formation.
Electronic supplementary material
Acknowledgements
We thank L. Férrières and Laura Grange for the construction of the yeeJ-lacZ reporter fusion. This work was supported by the Institut Pasteur, the French Government Investissement d’Avenir Program Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID), Fondation pour la Recherche Médicale grant “Equipe FRM DEQ. 20140329508” and from the Région Ile-de-France (DIM Malinf), and grants from the Australian National Health and Medical Research Council (NHMRC). MAS is supported by an NHMRC Senior Research Fellowship (GNT1106930). MT is supported by a Queensland University of Technology Vice-Chancellor’s Senior Research Fellowship.
Author Contributions
M.M.G. conducted the experiments for Figures 4, 5 and 6A, K.G.K.G. conducted the experiments for Figures 1, 2, 3 and 6B. E.R. contributed to the the generation of data for Figures 4, 5 and 6A. K.G.K.G. and C.S. purified the YeeJ protein for antibody production. B.A. contributed to the the generation of data for Figure 6A. M.M.G., K.G.K.G., D.G.M., M.T., C.B., M.A.S. and J.M.G. analyzed the data and wrote the manuscript, and all authors read and edited the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Marta Martinez-Gil and Kelvin G.K. Goh contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10902-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mark A. Schembri, Email: m.schembri@uq.edu.au
Christophe Beloin, Email: cbeloin@pasteur.fr.
References
- 1.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Reviews Microbiology. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Antão E-M, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut. Pathogens. 2009;1:22. doi: 10.1186/1757-4749-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjaergaard K, Hasman H, Schembri MA, Klemm P. Antigen 43-Mediated Autotransporter Display, a Versatile Bacterial Cell Surface Presentation System. Journal of bacteriology. 2002;184:4197–4204. doi: 10.1128/JB.184.15.4197-4204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulett GC, Webb RI, Schembri MA. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology. 2006;152:2101–2110. doi: 10.1099/mic.0.28607-0. [DOI] [PubMed] [Google Scholar]
- 5.Klemm P, Vejborg RM, Sherlock O. Self-associating autotransporters, SAATs: Functional and structural similarities. International Journal of Medical Microbiology. 2006;296:187–195. doi: 10.1016/j.ijmm.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Valle J, et al. UpaG, a New Member of the Trimeric Autotransporter Family of Adhesins in Uropathogenic Escherichia coli. Journal of bacteriology. 2008;190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells TJ, Totsika M, Schembri MA. Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology. 2010;156:2459–2469. doi: 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- 8.Korea C-G, Ghigo J-M, Beloin C. The sweet connection: Solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli. BioEssays. 2011;33:300–311. doi: 10.1002/bies.201000121. [DOI] [PubMed] [Google Scholar]
- 9.Roux A, Beloin C, Ghigo JM. Combined Inactivation and Expression Strategy To Study Gene Function under Physiological Conditions: Application to Identification of New Escherichia coli Adhesins. Journal of bacteriology. 2005;187:1001–1013. doi: 10.1128/JB.187.3.1001-1013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korea C-G, Badouraly R, Prevost M-C, Ghigo J-M, Beloin C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environmental microbiology. 2010;12:1957–1977. doi: 10.1111/j.1462-2920.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- 11.Larsonneur F, et al. Functional analysis of Escherichia coli Yad fimbriae reveals their potential role in environmental persistence. Environmental microbiology. 2016;18:5228–5248. doi: 10.1111/1462-2920.13559. [DOI] [PubMed] [Google Scholar]
- 12.Grijpstra J, Arenas J, Rutten L, Tommassen J. Autotransporter secretion: varying on a theme. Research in microbiology. 2013;164:562–582. doi: 10.1016/j.resmic.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Henderson IR, Nataro JP. Virulence Functions of Autotransporter Proteins. Infection and immunity. 2001;69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulett GC, et al. Functional Analysis of Antigen 43 in Uropathogenic Escherichia coli Reveals a Role in Long-Term Persistence in the Urinary Tract. Infection and immunity. 2007;75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luthje P, Brauner A. Ag43 Promotes Persistence of Uropathogenic Escherichia coli Isolates in the Urinary Tract. Journal of Clinical Microbiology. 2010;48:2316–2317. doi: 10.1128/JCM.00611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desvaux M, Parham NJ, Henderson IR. Type V protein secretion: simplicity gone awry? Current issues in molecular biology. 2004;6:111–124. [PubMed] [Google Scholar]
- 17.Oberhettinger P, et al. Intimin and Invasin Export Their C-Terminus to the Bacterial Cell Surface Using an Inverse Mechanism Compared to Classical Autotransport. PLoS ONE. 2012;7:e47069. doi: 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai JC, et al. The Bacterial Intimins and Invasins: A Large and Novel Family of Secreted Proteins. PLoS ONE. 2010;5:e14403. doi: 10.1371/journal.pone.0014403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholls L, Grant TH, Robins-Browne RM. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Molecular microbiology. 2000;35:275–288. doi: 10.1046/j.1365-2958.2000.01690.x. [DOI] [PubMed] [Google Scholar]
- 20.Leo JC, et al. The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan. Molecular microbiology. 2015;95:80–100. doi: 10.1111/mmi.12840. [DOI] [PubMed] [Google Scholar]
- 21.Leo JC, Oberhettinger P, Schütz M, Linke D. The inverse autotransporter family: Intimin, invasin and related proteins. International Journal of Medical Microbiology. 2015;305:276–282. doi: 10.1016/j.ijmm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Latasa C, Solano C, Penadés JR, Lasa I. Biofilm-associated proteins. Comptes Rendus Biologies. 2006;329:849–857. doi: 10.1016/j.crvi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Cucarella C, et al. Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation. Journal of bacteriology. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasa I, Penadés JR. Bap: A family of surface proteins involved in biofilm formation. Research in microbiology. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Taglialegna A, et al. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLOS Pathogens. 2016;12:e1005711. doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toledo-Arana A, et al. The Enterococcal Surface Protein, Esp, Is Involved in Enterococcus faecalis Biofilm Formation. Applied and environmental microbiology. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Molecular microbiology. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Molecular microbiology. 2010;77:549–561. doi: 10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed] [Google Scholar]
- 29.Wagner C, et al. Functional dissection of SiiE, a giant non-fimbrial adhesin of Salmonella enterica. Cellular Microbiology. 2011;13:1286–1301. doi: 10.1111/j.1462-5822.2011.01621.x. [DOI] [PubMed] [Google Scholar]
- 30.Goh HMS, et al. Molecular Analysis of the Acinetobacter baumannii Biofilm-Associated Protein. Applied and environmental microbiology. 2013;79:6535–6543. doi: 10.1128/AEM.01402-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barlag B, Hensel M. The Giant Adhesin SiiE of Salmonella enterica. Molecules. 2015;20:1134–1150. doi: 10.3390/molecules20011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latasa C, et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Molecular microbiology. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- 33.Krachler AM, Orth K. Targeting the bacteria–host interface. Virulence. 2013;4:284–294. doi: 10.4161/viru.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 36.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic acids research. 2014;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell A, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic acids research. 2015;43:D213–221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rendueles, O. et al. Screening of Escherichia coli Species Biodiversity Reveals New Biofilm-Associated Antiadhesion Polysaccharides. mBio2, e00043-00011-e00043-00011, doi:10.1128/mbio.00043-11 (2011). [DOI] [PMC free article] [PubMed]
- 43.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. Journal of bacteriology. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, J. H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria., (Cold Spring Harbor Laboratory Press, 1992).
- 45.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Molecular microbiology. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 46.Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Molecular microbiology. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 47.Wehbi H, et al. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. Journal of bacteriology. 2011;193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Easton DM, et al. The Intimin-Like Protein FdeC Is Regulated by H-NS and Temperature in Enterohemorrhagic Escherichia coli. Applied and environmental microbiology. 2014;80:7337–7347. doi: 10.1128/AEM.02114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Da R, S., Le Quere, B., Ghigo, J. M. & Beloin, C. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Applied and environmental microbiology73, 3391–3403, doi:10.1128/AEM.02625-06 (2007). [DOI] [PMC free article] [PubMed]
- 50.Picard B, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and immunity. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escobar-Paramo P. A Specific Genetic Background Is Required for Acquisition and Expression of Virulence Factors in Escherichia coli. Molecular Biology and Evolution. 2004;21:1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]
- 52.Skurnik D. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. Journal of Antimicrobial Chemotherapy. 2006;57:1215–1219. doi: 10.1093/jac/dkl122. [DOI] [PubMed] [Google Scholar]
- 53.Escobar-Páramo P, et al. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environmental microbiology. 2006;8:1975–1984. doi: 10.1111/j.1462-2920.2006.01077.x. [DOI] [PubMed] [Google Scholar]
- 54.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Molecular microbiology. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 55.Melillo A, et al. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS microbiology letters. 2006;263:102–108. doi: 10.1111/j.1574-6968.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- 56.Oldfield NJ, et al. T-cell stimulating protein A (TspA) of Neisseria meningitidis is required for optimal adhesion to human cells. Cellular Microbiology. 2007;9:463–478. doi: 10.1111/j.1462-5822.2006.00803.x. [DOI] [PubMed] [Google Scholar]
- 57.Nesta, B. et al. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. MBio3, e00010-00012-e00010-00012, doi:10.1128/mBio.00010-12 (2012). [DOI] [PMC free article] [PubMed]
- 58.Sharon, N. & Lis, H. The Structural Basis for Carbohydrate Recognition By Lectins. Advances in Experimental Medicine and Biology, 1–16, doi:10.1007/978-1-4615-1267-7_1 (2001). [DOI] [PubMed]
- 59.Berlanda Scorza F, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli ∆tolR IHE3034 mutant. Molecular & cellular proteomics: MCP. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Wurpel DJ, Moriel DG, Totsika M, Easton DM, Schembri MA. Comparative analysis of the uropathogenic Escherichia coli surface proteome by tandem mass-spectrometry of artificially induced outer membrane vesicles. Journal of proteomics. 2015;115:93–106. doi: 10.1016/j.jprot.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Wurpel DJ, et al. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. Journal of proteomics. 2016;131:177–189. doi: 10.1016/j.jprot.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J, et al. Polynucleotide Phosphorylase Protects Escherichia coli against Oxidative Stress. Biochemistry. 2009;48:2012–2020. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carzaniga T, Antoniani D, Dehò G, Briani F, Landini P. The RNA processing enzyme polynucleotide phosphorylase negatively controls biofilm formation by repressing poly-N-acetylglucosamine (PNAG) production in Escherichia coli C. BMC Microbiology. 2012;12:270. doi: 10.1186/1471-2180-12-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hacker J, Kaper JB. Pathogenicity Islands and the Evolution of Microbes. Annual Review of Microbiology. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt H, Hensel M. Pathogenicity Islands in Bacterial Pathogenesis. Clinical Microbiology Reviews. 2006;19:257–257. doi: 10.1128/CMR.19.1.257.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. Journal of bacteriology. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. Yersinia HPI in septicemic Escherichia coli strains isolated from diverse hosts. FEMS microbiology letters. 2001;196:57–60. doi: 10.1111/j.1574-6968.2001.tb10540.x. [DOI] [PubMed] [Google Scholar]
- 69.Hu J, Kan B, Liu ZH, Yu SY. Enteroaggregative Escherichia coli isolated from Chinese diarrhea patients with high-pathogenicity island of Yersinia is involved in synthesis of siderophore yersiniabactin. World journal of gastroenterology. 2005;11:5816–5820. doi: 10.3748/wjg.v11.i37.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karch H, et al. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infection and immunity. 1999;67:5994–6001. doi: 10.1128/iai.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infection and immunity. 2002;70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infection and immunity. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allsopp LP, et al. UpaH Is a Newly Identified Autotransporter Protein That Contributes to Biofilm Formation and Bladder Colonization by Uropathogenic Escherichia coli CFT073. Infection and immunity. 2010;78:1659–1669. doi: 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allsopp LP, et al. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. Journal of bacteriology. 2012;194:5769–5782. doi: 10.1128/JB.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fairman JW, et al. Crystal Structures of the Outer Membrane Domain of Intimin and Invasin from Enterohemorrhagic E. coli and Enteropathogenic Y. pseudotuberculosis. Structure. 2012;20:1233–1243. doi: 10.1016/j.str.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arenas J, et al. The meningococcal autotransporter AutA is implicated in autoaggregation and biofilm formation. Environmental microbiology. 2015;17:1321–1337. doi: 10.1111/1462-2920.12581. [DOI] [PubMed] [Google Scholar]
- 78.Grassl GA, Bohn E, Müller Y, Bühler OT, Autenrieth IB. Interaction of Yersinia enterocolitica with epithelial cells: invasin beyond invasion. International Journal of Medical Microbiology. 2003;293:41–54. doi: 10.1078/1438-4221-00243. [DOI] [PubMed] [Google Scholar]
- 79.Mesnage S, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun. 2014;5:4269. doi: 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kizil G, et al. Identification and characterization of TspA, a major CD4(+) T-cell- and B-cell-stimulating Neisseria-specific antigen. Infection and immunity. 1999;67:3533–3541. doi: 10.1128/iai.67.7.3533-3541.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kinscherf TG, Apirion D. Polynucleotide phosphorylase can participate in decay of mRNA in Escherichia coli in the absence of ribonuclease II. Mol Gen Genet. 1975;139:357–362. doi: 10.1007/BF00267975. [DOI] [PubMed] [Google Scholar]
- 82.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Molecular microbiology. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 83.Cameron TA, De Lay NR. The Phosphorolytic Exoribonucleases Polynucleotide Phosphorylase and RNase PH Stabilize sRNAs and Facilitate Regulation of Their mRNA Targets. Journal of bacteriology. 2016;198:3309–3317. doi: 10.1128/JB.00624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu J, Zhu MJ. Defects in polynucleotide phosphorylase impairs virulence in Escherichia coli O157:H7. Frontiers in microbiology. 2015;6:806. doi: 10.3389/fmicb.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Engman J, et al. Neisseria meningitidis Polynucleotide Phosphorylase Affects Aggregation, Adhesion, and Virulence. Infection and immunity. 2016;84:1501–1513. doi: 10.1128/IAI.01463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen R, et al. Polynucleotide Phosphorylase Regulates Multiple Virulence Factors and the Stabilities of Small RNAs RsmY/Z in Pseudomonas aeruginosa. Frontiers in microbiology. 2016;7:247. doi: 10.3389/fmicb.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manges AR, et al. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. The New England journal of medicine. 2001;345:1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- 88.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. Development and maturation of Escherichia coli K-12 biofilms. Molecular microbiology. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 89.Sherlock O, Schembri MA, Reisner A, Klemm P. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. Journal of bacteriology. 2004;186:8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 91.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic acids research. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Henning U, Sonntag I, Hindennach I. Mutants (ompA) affecting a major outer membrane protein of Escherichia coli K12. Eur J Biochem. 1978;92:491–498. doi: 10.1111/j.1432-1033.1978.tb12771.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.