Abstract
Bronchiolitis is one of the most severe diseases affecting infants worldwide. An imbalanced oropharynx (OP) microbiota has been reported in infants hospitalized with bronchiolitis; however, the microbiota dynamics in the OP and faeces during therapy remain unexplored. In total, 27 infants who were hospitalized with bronchiolitis were selected for this study, and sampling was conducted before therapy and after clinical recovery. We also recruited 22 age-matched healthy infants for this study. The faecal and OP microbiota diversity in the patients was lower than that in the healthy children. The faecal microbiota (FM) in the diseased children significantly differed from that in the healthy subjects and contained accumulated Bacteroides and Streptococcus. The OP microbiota in both the healthy and diseased infants was dominated by Streptococcus. After the treatment, the FM and OP microbiota in the patients was comparable to that before the treatment. This study may serve as an additional reference for future bronchiolitis studies, and the “risk microbiota model” of clinically recovered infants suggests an increased susceptibility to pathogen intrusion.
Introduction
Bronchiolitis is the leading cause of hospitalization of infants worldwide and is characterized by rapid spreading of upper respiratory tract (URT) infections to the lower respiratory tract (LRT)1. Respiratory syncytial virus (RSV) is the main agent causing bronchiolitis in infants2, 3. Rhinovirus, bocavirus, and human metapneumovirus infections are also commonly observed in paediatric bronchiolitis2, 3.
Common pathogens, including viruses and bacteria, can be identified through conventional culturing or PCR amplification4. However, these pathogens represent a small fraction of the microbial organisms living in the gastrointestinal and respiratory tracts5, 6. Ubiquitous opportunistic pathogens are present in the URT, but not all infants develop respiratory diseases7–10. Opportunistic pathogens are also commonly present in the intestine during infancy. These pathogens foster stable homeostasis with other microbial commensals11–13. Therefore, a stable microbial structure may be important for the prevention of infection and disease.
Several studies have explored imbalances in the faecal and nasopharyngeal microbiota during the onset of bronchiolitis, which has improved the understanding of the micro-ecology of bronchiolitis14–17. Carlos A. Camargo Jr. et al. performed a retrospective study involving 1,005 infants hospitalized for bronchiolitis14, 15. In total, 4 nasopharyngeal microbiota profiles were identified; of these profiles, the Haemophilus-dominated profile was closely associated with intensive care use (ICU) and a prolonged hospitalization time14, 15. This finding was also confirmed in an additional 307 infants hospitalized for bronchiolitis14. The microbiota profile was associated with the disease severity only in infants with low serum cathelicidin (LL-37 ≤46 ng/ml)15. Five additional nasopharyngeal microbiota clusters enriched with Haemophilus influenzae, Streptococcus, Corynebacterium, Moraxella, and Staphylococcus aureus have been identified16. The H. influenzae- and Streptococcus-dominant profiles were positively associated with RSV infections, host expression of toll-like receptors, and neutrophil and macrophage activation and signalling16. Carlos A. Camargo Jr. et al. also conducted a study investigating the faecal microbiota (FM) in 40 infants hospitalized for bronchiolitis and 115 healthy infants17. The infants with the Enterobacter/Veillonella-dominant microbiota cluster exhibited the lowest incidence of bronchiolitis, and the Bacteroides-dominant microbiota profile was associated with the greatest incidence of bronchiolitis onset17. The Escherichia- and Bifidobacterium-enriched microbiota clusters were associated with a medium incidence17. A longitudinal study involving 265 neonates with healthy records was performed until the participants were 3 years of age18. The infants who harboured positive cultures of Streptococcus pneumoniae, H. influenzae, and Moraxella catarrhalis in their hypopharynx at four weeks of age had a higher incidence of pneumonia and bronchiolitis18.
The oropharynx (OP) microbiota is more analogous to the LRT microbiota than to the NP microbiota19, 20. However, few reports have evaluated the OP microbiota in patients with bronchiolitis. Furthermore, the dynamics of the OP microbiota and FM during clinical therapy remain unclear, and such knowledge could improve the understanding of repeated infections in paediatric bronchiolitis. In this study, 27 infants who were hospitalized for bronchiolitis and 22 age-matched healthy infants were recruited from Shenzhen Children’s Hospital, and a comparative analysis of the FM and OP microbiota was performed. We explored the following two issues in this study: 1. whether the FM and OP microbiota in the diseased children differed from those in the healthy infants and 2. whether the FM and OP microbiota structures changed after clinical therapy.
Results
Participants and data output
We selected 27 infants who were hospitalized for mild bronchiolitis from Shenzhen Children’s Hospital (Table 1, Supplementary Table); all infants were diagnosed with a human RSV infection. The primary medical treatment during hospitalization was nebulized budesonide combined with salbutamol (Supplementary Table). Two bacterial pathogens, i.e., Streptococcus pneumonia and Haemophilus parainfluenzae, were identified in bacterial cultures of sputum from 5 hospitalized infants (Supplementary Table). Based on clinical experience or diagnoses of bacterial pathogens, 11 diseased infants received antibiotic treatment (Supplementary Table). No inpatient was admitted to the paediatric intensive care unit (PICU) or given mechanical ventilation during hospitalization. Most hospitalized infants remained in the hospital for 3–10 days, except for the following two infants: one infant stayed in the hospital for 13 days, and another infant stayed in the hospital for 26 days (Supplementary Table). The longer hospital stays of these two inpatients were due to secondary infections with rotavirus after clinical remission of bronchiolitis. In addition, 22 age-matched healthy infants were recruited in Shenzhen, China (Table 1, Supplementary Table). In total, 4,898,610 tags were obtained using 16S rDNA amplicon sequencing, ranging from 10,617 to 55,487 tags per sample.
Table 1.
Sample information.
| Patients (n = 27) | Healthy (n = 22) | |
|---|---|---|
| Demographic characteristics | ||
| Age (months) | 4.3 (1.3–11) | 6.1 (1.4–10.7) |
| Male | 18 (66.7%) | 7 (31.8%) |
| Breast feeding | 16 (59.3%) | 12 (54.5%) |
| Premature | 2 (0.07%) | 5 (22.7%) |
| C-section | 11 (40.7%) | 11 (50%) |
| History of eczema | 6 (22.2%) | 1(4.5%) |
| Maternal asthma | 4 (14%) | 3(13.6%) |
| Maternal smoking | 14 (51.9%) | 4(18.2%) |
| Disease situation | ||
| Hospitalization time (days) | 7 (3–26) | NA |
| Fever | 14 (51.9%) | NA |
| Wheezing | 18 (66.7%) | NA |
| Dyspnea | 3 (11.1%) | NA |
| Three concave sign | 5 (18.5%) | NA |
| Anhelation | 13 (48.1%) | NA |
| Cyanosis | 5 (18.5%) | NA |
| Moist rales | 17 (63%) | NA |
| Increase of lung markings | 21 (77.8%) | NA |
| Patch shadow | 12 (44.4%) | NA |
| Eosnophils (0.5–5%) | 20 (74.1%) | NA |
| CRP (<0.499) | 13 (48.1%) | NA |
Confounder analysis
Bronchiolitis onset, age, body weight, gender, delivery mode, feeding pattern, history of eczema, maternal asthma and smoking status were selected for the analysis of the main contributing factors to the inter-group discrepancies. According to the association analysis, the bronchiolitis onset significantly contributed to the differences in the FM/OP microbiota between the healthy and diseased children (q-value < 0.001, Supplementary Table).
FM and OP microbiota of the infants with bronchiolitis differed from that of the healthy infants
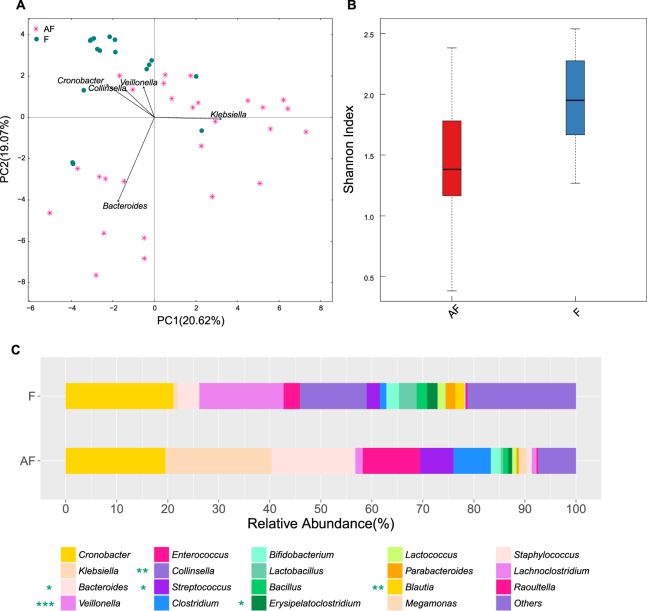
The FM structure in the hospitalized infants differed from that in the age-matched healthy infants (Fig. 1-A). The FM in the diseased infants exhibited a lower diversity than that in the healthy infants (Fig. 1-B). Firmicutes Bacteroidetes, Proteobacteria and Actinobacteria accounted for >99% of the FM in both the hospitalized and healthy infants (Supplementary Table), and Proteobacteria (42.14%) and Bacteroidetes (17.31%) were enriched in the FM in the hospitalized infants (Supplementary Table). At the genus level, the diseased infants harboured more Bacteroides (16.44% vs 4.26% in the healthy infants, q-value < 0.05) and Streptococcus (6.54% vs 2.60% in the healthy infants, q-value < 0.05) in the FM (Fig. 1-C, Supplementary Table). Klebsiella (20.80% vs 0.86% in the healthy infants), Clostridium (7.32% vs 1.27% in the healthy infants) and Enterococcus (11.20% vs 3.19% in the healthy infants) were also more prevalent in the FM in the children with bronchiolitis, but this difference was not statistically significant (Fig. 1-C, Supplementary Table). By contrast, Collinsella (0.03% vs 13.09% in the healthy infants, q-value < 0.01), Veillonella (1.46% vs 16.51% in the healthy infants, q-value < 0.001), Blautia (0.08% vs 1.78% in the healthy infants, q-value < 0.01), and Erysipelatoclostridium (0.77% vs 2.04% in the healthy infants, q-value < 0.05) accumulated in the FM in the hospitalized infants (Fig. 1-C, Supplementary Table).
Figure 1.
Comparison of the FM in the patients and healthy infants. AF: faecal samples collected from diseased children within 24 h of hospitalization. F: faecal samples collected from the healthy infants. (A) Pink plots represent the patients, and green circles represent the healthy infants. (B) Boxplot of the alpha diversity in the FM. (C) Stacked bar of the relative abundance at the genus level. *, ** and *** noted next to the genus represent q-values ≤0.05, ≤0.01 and ≤0.001, respectively.
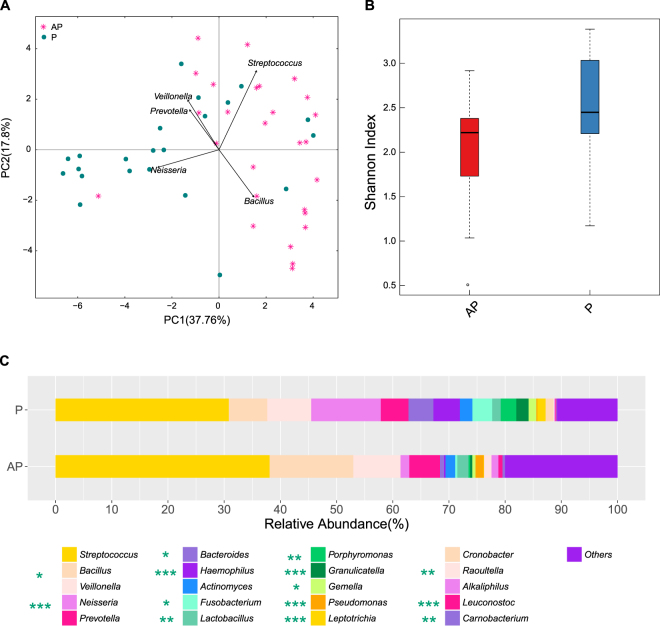
Similar to the FM, the OP microbial structure also differed between the healthy and diseased infants (Fig. 2-A), and the OP microbiota diversity in the hospitalized infants was lower than that in the healthy infants (Fig. 2-B). Firmicutes (84.79%, q-value < 0.01) was dominant in the diseased infants, while Bacteroidetes (13.94%, q-value < 0.01) and Proteobacteria (20.46%, q-value < 0.05) accumulated dramatically in the healthy children (Supplementary Table). The OP microbiota in both the healthy and hospitalized infants was dominated by Streptococcus (Fig. 2-C, Supplementary Table). Several genera accounted for the lowered abundance of the OP microbiota in the hospitalized infants, including Neisseria (1.57% vs 12.40% in the healthy infants, q-value < 0.001), Bacteroides (0.71% vs 4.43% in the healthy infants, q-value < 0.05), Haemophilus (0.28% vs 4.72% in the healthy infants, q-value < 0.001), Granulicatella (0.40% vs 2.19% in the healthy infants, q-value < 0.001), Leptotrichia (0.07% vs 1.46% in the healthy infants, q-value < 0.001), and Porphyromonas (0.29% vs 2.78% in the healthy infants, q-value < 0.01) (Fig. 2-C, Supplementary Table). By contrast, Bacillus (14.90% vs 6.86% in the healthy infants, q-value < 0.05), Pseudomonas (1.44% vs 0.19% in the healthy infants, q-value < 0.001), and Raoultella (1.28% vs 0.16% in the healthy infants, q-value < 0.01) accumulated in the OP microbiota in the diseased infants (Fig. 2-C, Supplementary Table).
Figure 2.
Comparison of OP microbiota in the patients and healthy controls. AP: oropharyngeal swabs sampled from diseased children over the course of 24 h of hospitalization. P: oropharyngeal swabs sampled from healthy infants. (A) Pink plots represent the patients, and green circles represent the healthy infants. (B) Boxplot of the alpha diversity in the OP microbiota. (C) Stacked bar of the relative abundance at the genus level. *, ** and *** noted next to the genus represent q-values ≤ 0.05, ≤ 0.01 and ≤ 0.001, respectively.
Treatment-induced changes in the FM/OP microbiota were few and individual-specific
The FM and OP microbiota structures of clinically recovered patients were similar to that before therapy(Figs 3–4). A comparative analysis of each case was also conducted to understand the microbiota changes in each hospitalized infant (Supplementary Figures 1–2).
Figure 3.
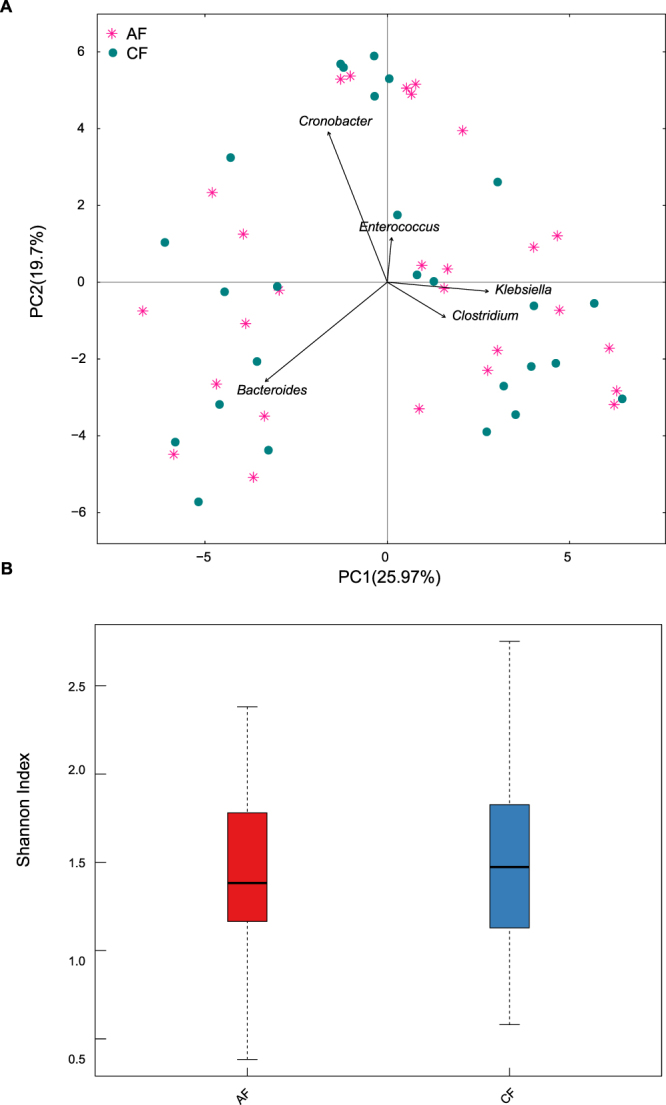
Comparison of the FM at two sampling time points in the patients. AF: within 24 h of hospitalization. CF: clinical recovery. (A) Pink plots represent the AF, and green circles represent the CF. (B) Boxplot of the alpha diversity in the FM.
Figure 4.
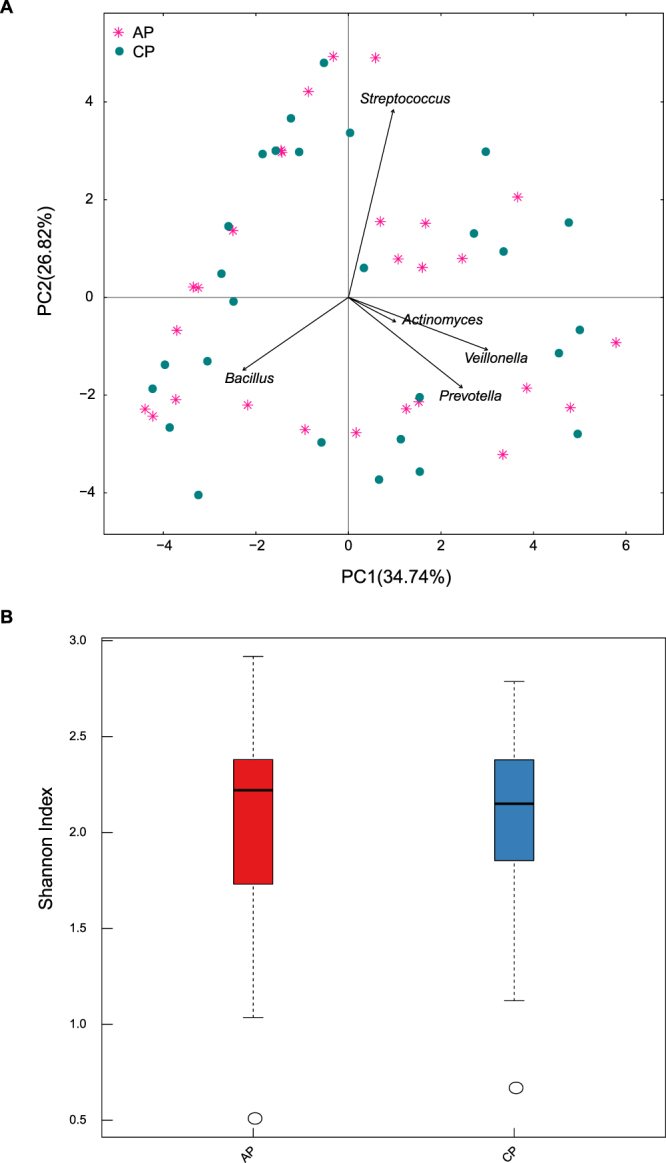
Comparison of the OP microbiota at two sampling time points in the patients. AP: within 24 h of hospitalization. CP: clinical recovery. (A) Pink plots represent the AP, and green circles represent the CP. (B) Boxplot of the alpha diversity in the OP microbiota.
Haemophilus influenzae or Streptococcus pneumoniae were identified in sputum cultures from 5 patients, whose FM diversity was similar to that of the other patients (Supplementary Figure 1A, Supplementary Table). Only two diseased children (patients 6 and 7) received oral antibiotics (Supplementary Table), and their FM diversity significantly decreased after therapy (Supplementary Figure 1B). By contrast, a comparable or higher FM diversity was identified in the other patients (Supplementary Figure 1A) who received intravenous or no antibiotic therapy (Supplementary Table). Klebsiella, Enterococcus, Bacteroides, Cronobacter and/or Clostridium dominated the FM in the diseased infants and did not change after the different therapies (Supplementary Figure 1B). After clinical therapy, the OP microbiota diversity in the hospitalized infants increased or remained the same, even in patients who received oral antibiotics (Supplementary Figure 2A). Streptococcus remained dominant in the OP microbiota in the diseased infants during therapy (Supplementary Figure 2B). Bacillus, Veillonella, Prevotella and Raoultella also dominated after treatment (Supplementary Figure 2B).
Discussion
Bronchiolitis is mainly caused by viral infections and co-infections with bacterial pathogens2, 3. Viral and bacterial infections have been shown to induce an immune response and microbiota imbalance21. This study revealed an imbalanced FM/OP microbiota in children with viral bronchiolitis, which is consistent with that identified in other respiratory diseases, such as asthma and cystic fibrosis22, 23.
The distal effect of the gut microbiota (GM) on respiratory health is profoundly affected by the regulation of the host immune system23. The GM plays a crucial role in the immune response to and protection from pulmonary infection24–28. Bifidobacterium and Lactobacillus tend to accumulate in healthy infants but not in significant amounts. Bifidobacterium spp. was found to be protective against both bacterial and viral infections of the respiratory tract27, 29, 30. Probiotics composed of Bifidobacterium and Lactobacillus are also promising for the control of respiratory infections31–34. In the healthy children, enriched Veillonella was positively associated with Th17-mediated immunity in the lungs35 and negatively associated with the risk of asthma36. Bacteroides and Streptococcus accumulated in significant amounts and represented a high proportion of FM in diseased children. Several members of Streptococcus spp. could induce inflammation in the epithelial mucus37, including the insignificantly enriched Klebsiella in diseased infants38. By contrast, various Bacteroides spp. have the potential to relieve inflammation by expanding the Treg cell population and suppressing inflammatory responses23. The aforementioned prior study could partially explain the GM imbalance observed in the diseased children. In all patients, except for patients 6 and 7, who took oral antibiotics, the FM diversity either remained unchanged or decreased after the treatment. This finding indicated the need to decrease exposure to oral antibiotics, which could block GM recovery for several months39.
Numerous reports have described the OP microbiota in healthy children, which was primarily dominated by Streptococcus, Rothia, Prevotella, Gemella, Veillonella, Fusobacteria, Haemophilus, and Neisseria 4. Nader Shaikh et al. also reported prevalent streptococcal carriage in children without pharyngitis symptoms40. We found microbial carriage in the OP in the healthy infants that was identical to that described in these reports4, 40. However, the OP microbial structures in the hospitalized infants significantly differed from those in the healthy infants, which potentially indicates transmission to the lung19, 20. Several reports have identified four microbiota profiles in the NP in infants hospitalized for bronchiolitis, and the Haemophilus-dominated profile was associated with the highest clinical severity14, 15. We established a Streptococcus-dominant OP microbiota profile in both the hospitalized and healthy infants, which could be attributed to the differing microbiota components between the NP and OP19, 20. Moreover, the OP microbiota remained unchanged in most patients who recovered clinically. This finding may explain the widespread repeated respiratory infections in children after therapy because high incidences of respiratory diseases have been reported in the presence of “unstable” URT microbiota8, 41. Therefore, the OP microbiota has promising potential in bronchiolitis prognosis, therapy optimization, and evaluation of recovery.
This study also had some limitations. A total of 47 infants was not sufficient for partitioning information according to discrepant clinical symptoms, such as that performed by Carlos A and Camargo Jr. et al.14, 15. The detailed clinical symptoms and host responses, including immune cytokines, should be considered to assess the contributions of microbiota. Long-term investigations exploring the incidence of acute respiratory infection in infants who recovered clinically should also be performed.
In conclusion, this study reviewed the imbalanced FM and OP microbiota in Chinese children with bronchiolitis and provided additional reference data for associated studies. More importantly, we identified comparable OP and FM microbiota among patients before and after treatment. This finding suggested a risk model of OP microbiota in children who recover clinically42.
Materials and Methods
Ethics statement
This study was approved by the Ethical Committee of Shenzhen Children’s Hospital under the registration number 2015020. All procedures were performed in accordance with the relevant guidelines and regulations stipulated by the Ethical Committee of Shenzhen Children’s Hospital. We obtained written informed consent from the parents of all participants, who approved their children’s participation in the study.
Sample preparation
In total, 27 inpatients (aged ≤1 year of age) were sampled within 24 h of hospitalization (before therapy) in Shenzhen Children Hospital, and the second sampling was conducted after clinical recovery from bronchiolitis (average 7–10 days after treatment). In addition, 22 age-matched healthy infants were recruited according to the following inclusion criteria: no wheezing, fever, cough, or other respiratory/allergic symptoms at the time of sampling and for 2 weeks prior to the study and no respiratory symptoms for 1 week after sampling. None of the infants was exposed to antibiotics for two weeks before sampling. Sterile oropharyngeal swabs (155 C, COPAN, Murrieta, California, USA) were used for sampling the OP and faeces. For the diseased infants, we performed the sample collection during the following two time points: 24 hours after hospitalization and upon clinical recovery (average 7–10 days after treatment). The collected samples were immediately stored at −80 °C, and the DNA extraction was performed within one week. The sputum samples were collected by performing endotracheal suctioning (AARC (American Association for Respiratory Care) Clinical Practice Guidelines)43 and cultured for several bacterial pathogens.
DNA extraction, sequencing, and analysis
DNA was extracted using a PowerSoil® DNA Isolation Kit (Mo Bio Laboratories) according to the manufacturer’s protocol. The hypervariable V3–V4 region of the 16S rRNA gene was amplified as previously reported44 and was sequenced using an Illumina MiSeq platform. The paired-end reads were filtered using Mothur’s Miseq SOP45 and connected to tags using FLASH (v1.2.11, http://ccb.jhu.edu/software/FLASH/index.shtml). The qualified tags were clustered into operational taxonomic units (OTUs), and OTUs from chimaeras were removed using USEARCH (v7.0.1090). The OTUs were assigned a taxonomic classification by aligning to the RDP 16S rRNA database (201408).
The multivariable analysis was conducted via a PERMANOVA (permutations = 9999, P-value ≤0.05) using the R package “vegan” (version 2.4-3)46 to identify the important factors that could be associated with the microbiota structure. The Wilcoxon rank-sum test was used to compare the inpatients to the healthy infants, and a time-series comparison of the hospitalized infants was performed using the Wilcoxon signed-rank test. P-values were adjusted according to the false discovery rate for multiple tests. *, ** and *** represent q-values ≤0.05, ≤0.01 and ≤0.001, respectively. Increases/decreases in the Shannon index of FM and OP microbiota diversity >50% were considered significant. R (version 3.2.3) and SVG (version 1.1) software packages were used for visualization.
Accession number
Clean reads were deposited in the GenBank database under accession number PRJNA362484.
Electronic supplementary material
Acknowledgements
This work was supported by the Guangdong Provincial Nature Science Foundation (2015A030313759).
Author Contributions
F.W. and Q.H. designed the project. Y.Z., Q.H., D.F., D.D., Q.Y., S.L., and W.W. selected the inpatients, collected the samples, and performed the clinical detection. Q.H. and G.L. recruited the age-matched healthy infants and recorded the relative individual information. D.L. and Z.Y. extracted the DNA and processed the sequencing data. W.D., Q.Z. and Y.L. performed the bioinformatics analysis. Q.H. and W.D. interpreted the results and prepared the paper. S.L. guided the analysis. All authors reviewed this manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Qian Hu, Wenkui Dai and Qian Zhou contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11311-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oymar K, Skjerven HO, Mikalsen IB. Acute bronchiolitis in infants, a review. Scand J Trauma Resusc Emerg Med. 2014;22:23. doi: 10.1186/1757-7241-22-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florin TA, Plint AC, Zorc JJ. Viral bronchiolitis. Lancet. 2017;389:211–224. doi: 10.1016/S0140-6736(16)30951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meissner HC. Viral Bronchiolitis in Children. NEJM. 2016;374:1793–1794. doi: 10.1056/NEJMra1413456. [DOI] [PubMed] [Google Scholar]
- 4.Stearns JC, et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 2015;9:1246–1259. doi: 10.1038/ismej.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14:R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch AA, et al. Development of Upper Respiratory Tract Microbiota in Infancy is Affected by Mode of Delivery. EBioMedicine. 2016;9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biesbroek G, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 9.Biesbroek G, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190:298–308. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 10.Teo SM, et al. The infant airway microbiome in health and disease impacts later asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuang YS, et al. Composition of gut microbiota in infants in China and global comparison. Sci Rep. 2016;6:36666. doi: 10.1038/srep36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Backhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa K, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J. 2016;48:1329–1339. doi: 10.1183/13993003.00152-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa, K. et al. The Fecal Microbiota Profile and Bronchiolitis in Infants. Pediatrics. 138, doi:10.1542/peds.2016-0218 (2016). [DOI] [PMC free article] [PubMed]
- 16.de Steenhuijsen Piters WA, et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa K, et al. Serum cathelicidin, nasopharyngeal microbiota, and disease severity among infants hospitalized with bronchiolitis. J Allergy Clin Immunol. 2016;139:1383–1386. doi: 10.1016/j.jaci.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188:1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 19.Bassis CM, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ES, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brealey, J. C., Sly, P. D., Young, P. R. & Chappell, K. J. Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett. 362, doi:10.1093/femsle/fnv062 (2015). [DOI] [PubMed]
- 22.Marsland BJ, Trompette A, Gollwitzer ES. The Gut-Lung Axis in Respiratory Disease. Ann Am Thorac Soc. 2015;12(Suppl 2):S150–156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 23.Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. 2015;6:1085. doi: 10.3389/fmicb.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LW, Chen PH, Hsu CM. Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock. 2011;36:67–75. doi: 10.1097/SHK.0b013e3182184ee7. [DOI] [PubMed] [Google Scholar]
- 25.Schuijt TJ, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65:575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. PNAS. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, et al. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr Microbiol. 2013;67:414–422. doi: 10.1007/s00284-013-0380-z. [DOI] [PubMed] [Google Scholar]
- 28.Fagundes CT, et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. 2012;188:1411–1420. doi: 10.4049/jimmunol.1101682. [DOI] [PubMed] [Google Scholar]
- 29.Kawahara T, et al. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol Immunol. 2015;59:1–12. doi: 10.1111/1348-0421.12210. [DOI] [PubMed] [Google Scholar]
- 30.Vieira AT, et al. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A) Microbes Infect. 2016;18:180–189. doi: 10.1016/j.micinf.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Jespersen L, et al. Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: a randomized, double-blind, placebo-controlled, parallel-group study. Am J Clin Nutr. 2015;101:1188–1196. doi: 10.3945/ajcn.114.103531. [DOI] [PubMed] [Google Scholar]
- 32.King S, Glanville J, Sanders ME, Fitzgerald A, Varley D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis. Br J Nutr. 2014;112:41–54. doi: 10.1017/S0007114514000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luoto R, et al. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J Allergy Clin Immunol. 2014;133:405–413. doi: 10.1016/j.jaci.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West NP, et al. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clin Nutr. 2014;33:581–587. doi: 10.1016/j.clnu.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Segal LN, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arrieta MC, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 37.Yoo IH, et al. Role of pneumococcal pneumolysin in the induction of an inflammatory response in human epithelial cells. FEMS Immunol Med Microbiol. 2010;60:28–35. doi: 10.1111/j.1574-695X.2010.00699.x. [DOI] [PubMed] [Google Scholar]
- 38.Lu, B. et al. Molecular characterization of Klebsiella pneumoniae isolates from stool specimens of outpatients in sentinel hospitals Beijing, China, 2010–2015 Gut Pathog. 9, 39, doi:10.1186/s13099-017-0188-7 (2017). [DOI] [PMC free article] [PubMed]
- 39.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaikh N, Leonard E, Martin JM. Prevalence of streptococcal pharyngitis and streptococcal carriage in children: a meta-analysis. Pediatrics. 2010;126:e557–564. doi: 10.1542/peds.2009-2648. [DOI] [PubMed] [Google Scholar]
- 41.Bosch, A. A. T. M. et al. Maturation of the Infant Respiratory Microbiota, Environmental Drivers and Health Consequences: A Prospective Cohort Study. Am J Respir Crit Care Med, doi:10.1164/rccm.201703-0554OC. [Epub ahead of print] (2017). [DOI] [PubMed]
- 42.Hasegawa K, Camargo CA., Jr. Airway microbiota and acute respiratory infection in children. Expert Rev Clin Immunol. 2015;11:789–792. doi: 10.1586/1744666X.2015.1045417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.American Association for Respiratory, C. AARC Clinical Practice Guidelines. Endotracheal suctioning of mechanically ventilated patients with artificial airways 2010. Respir Care. 55, 758–764 (2010). [PubMed]
- 44.Teo SM, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warton DI, Wright TW, Wang Y. Distance-based multivariate analyses confound location and dispersion effects. Methods in Ecology and Evolution. 2012;3:89–101. doi: 10.1111/j.2041-210X.2011.00127.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.