Abstract
Background
Interferons (IFNs) are cytokines typically induced upon viral infection but are constitutively expressed also in the absence of acute infection. The physiological role of autocrine and paracrine IFN signaling, however, remains poorly understood, and its function in glioblastoma has not been explored in depth.
Methods
Using RNA interference-mediated gene silencing, we characterized constitutive type I IFN signaling and its role in human glioma cells.
Results
We observed constitutive expression of phosphorylated signal transducer and activator of transcription 1 (pSTAT1) and myxovirus resistance protein A (MxA), a classical IFN-response marker, in the absence of exogenous IFN-β. In vivo, we found higher MxA expression in gliomas than in normal tissue, suggesting that IFN signaling is constitutively active in these tumors. To demonstrate the presence of an autocrine type I IFN signaling loop in glioma cells in vitro, we first confirmed the expression of the type I alpha/beta receptor (IFNAR)1/2, and its ligands, IFN-α and IFN-β. Small interfering RNA–mediated receptor gene silencing resulted in reduced expression of MxA at mRNA and protein levels, as did gene silencing of the ligands, corroborating the hypothesis of an autocrine signaling loop in which type I IFNs induce intracellular signaling through IFNAR1/2. On a functional level, following IFNAR1 or IFNAR2 gene silencing, we observed reduced programmed death ligand 1 (PD-L1) and major histocompatibility complex (MHC) class I and II expression as well as an enhanced susceptibility to natural killer immune cell lysis, suggesting that autocrine IFN signaling contributes to the immune evasion of glioma cells.
Conclusions
Our findings point to an important role of constitutive IFN signaling in glioma cells by modulating their interaction with the microenvironment.
Keywords: glioma, immunogenicity, interferon, PD-L1, STAT1
Importance of the study.
Interferons might be constitutively expressed also in the absence of acute infection, thereby regulating various physiological processes such as cellular proliferation and differentiation and cell viability, as well as immune cell function. However, their role in glioblastoma, especially in glioma-initiating cells, has not been investigated in detail.
Here we show that IFN signaling is constitutively active in gliomas in vitro and in vivo. We demonstrate the presence of an autocrine signaling loop in which the type I IFNs, IFN-α and IFN-β, induce intracellular signaling through the type I IFN receptor, IFNAR1/2. On a functional level, we define the potential impact of constitutive IFN signaling on the immunogenicity of glioma cells. IFNAR1/2 gene silencing reduces PD-L1 and MHC class I and II expression and enhances susceptibility to immune cell lysis, suggesting that constitutive IFN signaling acts as a negative regulator of antitumor immune responses in gliomas.
Gliomas are intrinsic brain tumors which represent a major clinical challenge. Despite intense therapeutic efforts, these tumors typically progress and ultimately result in neurological deterioration and death. This unfavorable prognosis reflects the biological properties of glioma cells, which are paradigmatic for various hallmarks of cancer, such as invasive growth, impaired immunogenicity, and resistance to various apoptotic stimuli.1,2 The underlying mechanisms are only partially understood but it has become clear that various alterations on the genetic and molecular levels contribute to the malignant behavior of glioma cells. Furthermore, the existence of a subpopulation of cells that harbor stem-cell characteristics within gliomas suggests that a rather small percentage of the tumor cells may maintain tumor growth.3 With regard to stem cells, type I interferons (IFNs) such as IFN-α and IFN-β have gained increasing interest within the last years. Constitutive type I IFN signaling may be important for the maintenance and mobilization of hematopoietic stem cells within the niche, since either the absence of constitutive IFN signaling or prolonged elevated IFN signaling deplete the hematopoietic stem cell niche. Upon chronic IFN signaling, an induction of proliferation was observed in dormant hematopoietic stem cells, a finding that might be of relevance also for cancer stem cells.4,5
IFNs are produced by most nucleated cells, and their signaling is mediated through a common cell surface type I IFN receptor complex composed of 2 subunits, IFN alpha/beta receptor 1 (IFNAR1) and IFNAR2.6,7 IFNAR2 is supposed to be responsible for ligand binding, while IFNAR1 holds very weak ligand binding affinity but induces intracellular signaling cascades.8–10 The ligand-mediated association of the 2 subunits promotes a signaling cascade that results in the phosphorylation of IFNAR1 and creation of a docking site for signal transducer and activator of transcription 2 (STAT2). STAT2 phosphorylation creates a docking site for STAT1, which enables phosphorylation of STAT1.7 The STATs form either homodimers of STAT1 or heterodimers of STAT1 and STAT2, which then translocate to the nucleus to induce expression of IFN-stimulated genes. Additionally, type I IFNs can activate other members of the STAT family, such as STAT3, STAT4, STAT5, and STAT6.11
Activity of the IFN-signaling pathway also leads to the induction of myxovirus protein A (MxA) expression, a cytoplasmic GTPase with antiviral activity.12 The genes activated by IFNs play a crucial role not only in cellular processes protecting from viral infections, but also in modulating general immune responses, cell proliferation, and cell survival.13 Moreover, IFN signaling might be of relevance also in different tumor types, since mutations, preventing the production of, or altering the responsiveness to IFNs, have been observed in numerous malignancies like leukemia and melanoma, thus possibly representing a survival advantage for tumor cells.14,15 In addition, the efficacy of type I IFNs as an anticancer treatment is affected by constitutive IFN expression.16 Thus, based on the involvement of type I IFNs in various cellular processes, we aimed at characterizing constitutive IFN signaling and its role in glioma cells, including glioma cells with stem cell properties.
Materials and Methods
Cell Lines and Reagents
The human long-term cell (LTC) lines U87MG and T98G were obtained from the American Type Culture Collection, and LN-18, LN-428, D247MG, LN-319, A172, LN-308, and LN-229 cells were kindly provided by N. de Tribolet (Lausanne, Switzerland). The glioma-initiating cells (GICs) T-269, T-325, S-24, ZH-161, and ZH-305 were isolated from tumors resected in Tübingen, Germany (T-269, T-325), Stuttgart, Germany (S-24), and Zurich, Switzerland (ZH-161, ZH-305) and were used during the first passages.17 Cells were authenticated regularly at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) by short tandem repeat analysis, lastly in 2013 (LTC) and in 2016 (GIC). All LTCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal calf serum (FCS; Invitrogen) and 2 mM glutamine. The GIC cultures were maintained in Neurobasal (NB) medium (Invitrogen) supplemented with B-27 supplement (20 µL/mL), Glutamax (10 µL/mL) (Invitrogen), fibroblast growth factor 2, epidermal growth factor (20 ng/mL each; Peprotech), and heparin (32 IE/mL; Ratiopharm). The natural killer cell line NKL was obtained from M. J. Robertson (Indiana University School of Medicine) and cultured in Roswell Park Memorial Institute 1640 medium (Invitrogen) supplemented with 15% FCS, 2 mM glutamine, and 50 U/mL interleukin-2 (Peprotech). Recombinant IFN-β1b was purchased from AbD Serotec. Antibodies to human CD45 (clone 2B11) was from DakoCytomation, STAT1 and phosphorylated STAT1 (pSTAT1; Tyr701) were purchased from Cell Signaling, phycoerythrin-labeled anti-IFNAR2 from PBL Assay Science, and anti–MHC-I (clone W/32) and anti–human leukocyte antigen (HLA)-DR (clone L243) were kindly provided by S. Stevanović (Tübingen, Germany).18 Antibodies to NKG2D ligands (NKG2DL) were provided by A. Steinle (Frankfurt, Germany), BV421 anti-human programmed death ligand 1 (PD-L1) (clone MIH1) was obtained from BD Biosciences, and actin antibody was purchased from Santa Cruz. The MxA antibody used for immunoblot analysis in Fig. 1B was provided by J. Pavlovic (Zurich, Switzerland). All other immunoblot analyses were performed with a MxA antibody provided by O. Haller (Freiburg, Germany). The MxA antibody for immunohistochemistry and immunofluorescence was purchased from Thermo Fisher Scientific.
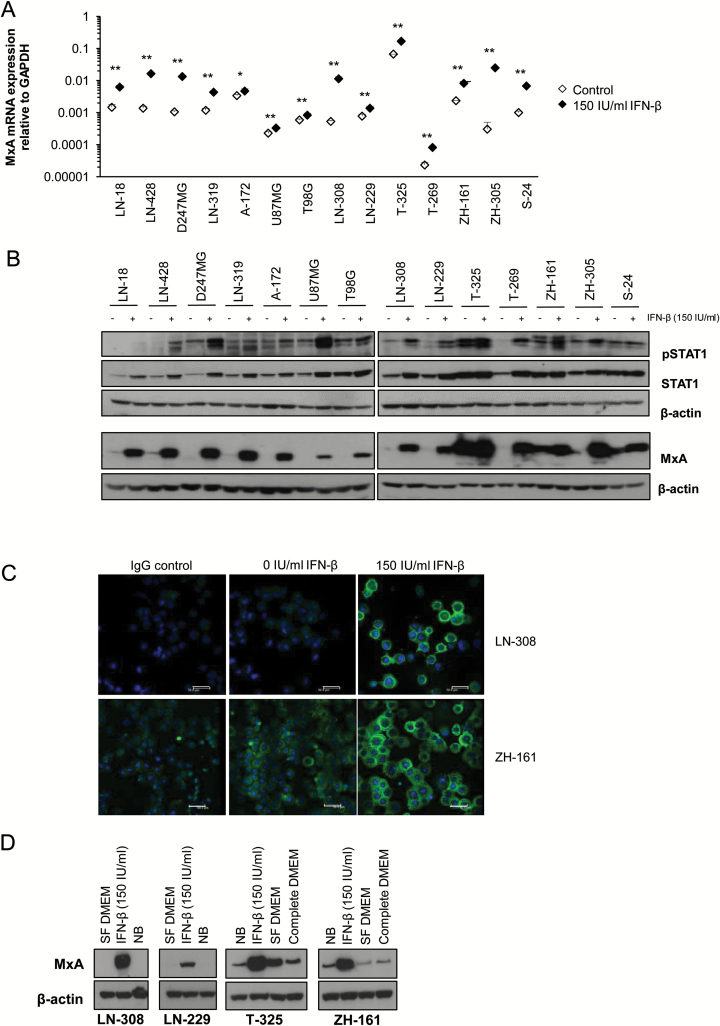
Fig. 1.
IFN-β target genes are expressed in glioma cells. (A) The human LTC LN-18, LN-428, D247MG, LN-319, A172, U87MG, T98G, LN-308 and LN-229, and the GICs T-325, T-269, ZH-161, ZH-305, and S-24 were cultured without or with IFN-β (150 IU/mL) for 48 h and MxA mRNA expression levels were determined using real-time PCR (median expression levels ± SE are shown from 3 independent experiments). (B) The cells were treated as in (A). Whole cell lysates were subsequently analyzed for pSTAT1, STAT1, and MxA protein levels by immunoblot using actin as a loading control (1 out of 2 independent experiments is shown). (C) LN-308 or ZH-161 cells, untreated or exposed to IFN-β (150 IU/mL) for 48 h, were analyzed for MxA protein levels by immunofluorescence (scale bar, 50 µm). (D) LN-308, LN-229, T-325, or ZH-161 cells were cultured in various medium conditions as indicated for 48 h (SF, serum-free DMEM; NB, Neurobasal medium). Whole cell lysates were assessed for MxA expression levels using immunoblot. Actin was used as loading control.
Real-Time PCR
Total RNA was prepared using the NucleoSpin RNA II System (Macherey-Nagel) and transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems. Gene expression was measured in an ABI Prism 7000 Sequence Detection System with SYBR Green Master Mix (Thermo Fisher Scientific). The conditions for the PCR reactions were: 40 cycles, 95°C/15 sec, 60°C/1 min, using the following specific primers (Microsynth): GAPDH forward: 5ʹ-CTCTCTGCTCCTCCTGTTCGAC-3ʹ (NM_002046.4, nt 100–121), GAPDH reverse: 5ʹ-TGAGCG ATGTGGCTCGGC T -3ʹ (NM_002046.4, nt 150–168); IFNAR1 forward: 5ʹ-TATGCTGCGAAAGTCTTCTTGAG-3ʹ (NM_0006 29.2, nt 1510–1532), IFNAR1 reverse: 5ʹ-TCTTGGCTAGTTT GGGAACTGTA-3ʹ (NM_000629.2; nt 1726–1748); IFNAR2 forward: 5ʹ-TCTTGAGGCAAGGTCTCGCTA-3ʹ (NM_20758 4.2, nt 1195–1215), IFNAR2 reverse: 5ʹ-CAGGGATGCACGCTT GTAATC-3ʹ (NM_207584.2, nt 1320–1340); IFN-α14 forward: 5ʹ-TCTTCGGGATTCCCAATGGC-3ʹ (NM_002172.2; nt 30–49), IFN-α14 reverse: 5ʹ-CTTGACTTGCAGCTGAGCAC-3ʹ (NM_002172.2; nt 81–100); IFN-β1 forward: 5ʹ-AGTAGGCGA CACTGTTCGTG-3ʹ (NM_002176.3; nt 67–86), IFN-β1 reverse: 5-AGCCTCCCATTCAATTGCCA-3ʹ (NM_002176.3; nt 221–240); MxA forward: 5`- TGGAGATCAGCTCCCGAGATG-3ʹ (NM_001144925.1, nt 512–532), MxA reverse: 5ʹ- ATTG CCCACAGCCACTCTG-3ʹ (NM_001144925.1, nt 570–588). Data analysis was done using the ΔΔCT method for relative quantification.
RNA Interference-Mediated Gene Silencing
For transient transfections, glioma cells were seeded in a 6-well plate and transfected with 100 nM of specific or scrambled control small interfering (si)RNA by electroporation using the Neon Transfection System (Invitrogen). Control as well as MxA, IFNAR1, and IFNAR2 siRNA oligonucleotides were purchased from Thermo Scientific using siGENOME SMARTpool. SiRNA oligonucleotides targeting IFN-α14, IFN-α17, or IFN-β1 were obtained from Qiagen.
Flow Cytometry
Cells were detached using Accutase (PAA Laboratories), incubated with primary antibody for 30 min at 4°C followed by an R-phycoerythrin-conjugated secondary antibody labeling step for another 30 min in case of a not unlabeled primary antibody. Fluorescence was detected in a CyAn flow cytometer (Beckman Coulter) or a BD FACSVerse flow cytometer. Signal intensity was calculated as the ratio of the median fluorescence of the specific antibody and the isotype control antibody (specific fluorescence index).
Immune Cell Lysis Assay
Immune-mediated glioma cell lysis was determined using a flow cytometry–based cytotoxicity assay.17 Target cells were stained with PKH-26 (Sigma-Aldrich) for 3 min and then co-incubated with NKL effector cells at different effector-to-target (E:T) ratios as indicated for 4 h. Subsequently, live/dead staining was performed with the Zombie Aqua Fixable Viability Kit (BioLegend), followed by assessment of target cell lysis by flow cytometry. Specific cell lysis was expressed as percentage of dead cells within the PKH-26–positive target cells, corrected for spontaneous background lysis. For PD-L1 blocking experiments, T-325 or ZH-161 cells were incubated 1 h prior and during co-incubation with 10 µg/mL anti–PD-L1 (clone 29E.2A3, Biolegend).
Immunoblot Analysis
The cells were treated as indicated and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate) containing phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich) and protease inhibitor (Roche Diagnostics). Thirty micrograms of protein per lane were mixed with Laemmli buffer containing β-mercaptoethanol and analyzed by immunoblot. Visualization of protein bands was accomplished using horseradish peroxidase–coupled secondary antibodies (Santa Cruz) and enhanced chemiluminescence (Thermo Fisher Scientific).
Immunohistochemistry
Immunohistochemistry was carried out on tissue microarrays (TMA) with tissue specimens of normal brain or of gliomas of different World Health Organization (WHO) grades of malignancy that were kindly provided by Karl Frei.19 TMA sections were stained using the rabbit polyclonal primary antibody to MxA (1:200) and visualized using anti-rabbit secondary antibody with the ImmPact DAB kit (Vector Laboratories). The intensity of staining and the percentage of stained tumor cells on the TMA were quantified by H scoring.20,21 For costainings, TMA sections were stained with primary antibodies to MxA (rabbit, 1:200) and CD45 (mouse, 1:50) and visualized using mouse anti-rabbit immunoglobulin (Ig)G–horseradish peroxidase secondary antibody (Santa Cruz) with the ImmPact DAB kit followed by anti-mouse IgG–alkaline phosphatase secondary antibody (Vector Laboratories) with the HighDef green IHC chromogen kit (Enzo).
Immunofluorescence
We cytospun 5 × 105 cells onto a glass slide; they were then dried for 30 min, fixed in ice-cold methanol for 10 min, and permeabilized with 0.5% Triton X100. Subsequently, cells were stained using the rabbit polyclonal primary antibody to MxA (1:100), visualized using an Alexa Fluor 488–conjugated secondary antibody (Southern Biotech) and mounted in Vectashield Mounting Media with 4′,6′-diamidino-2-phenylindole (DAPI).
TCGA Analysis
Kaplan–Meier analysis of survival probability for the glioblastoma 540–MAS5.0–u133a dataset (n = 540) in the database of The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) was done using the R2 microarray analysis and visualization platform (http://r2.amc.nl), and median MxA expression was selected as the scan cutoff mode. Eighty-five samples of this dataset classified according to glioblastoma subtype22 were analyzed for MxA expression using the subtype track mode.23 Correlation analysis for the glioblastoma 540–MAS5.0–u133a dataset (n = 540) or the Glioma–French–284–MAS5.0–u133p2 dataset (n = 284) was done using the module for correlation of 2 genes of the R2 microarray analysis and visualization platform (http://r2.amc.nl). Samples of the Glioma–French–284–MAS5.0–u133p2 dataset (n = 284) were grouped according to the different WHO grades of malignancy and analyzed for MxA expression using the histology subtype track mode.
Data Analysis
Data are representative of at least 2 independent experiments. If not indicated otherwise, analysis of significance was performed using the 2-tailed Student’s t-test (*P < 0.05; **P < 0.01).
Results
STAT1 Phosphorylation and MxA Expression Indicate Autocrine IFN Signaling in Glioma Cells
Binding of type I IFNs to their cognate cell surface receptor induces downstream signaling through STAT1/STAT2 and Jak1/Tyk2 kinases. To demonstrate constitutive IFN signaling as well as responsiveness to exogenous IFN, we assessed expression of MxA, an established marker for activation of the classical IFN-α/β signaling pathway. Highest constitutive MxA levels were seen in T-325 and ZH-161 GICs, and MxA mRNA levels increased in all cell lines upon exposure to IFN-β (Fig. 1A). Several glioma cell lines displayed constitutive MxA protein expression, with T-325 and ZH-161 GICs having the highest constitutive MxA protein levels. Exposure to IFN-β induced MxA expression in all cell lines investigated (Fig. 1B, C). Furthermore, we determined the basal levels of pSTAT1 as well as the induction of STAT1 phosphorylation upon exposure to recombinant IFN-β. We observed constitutive STAT1 phosphorylation to a variable degree in all cell lines and an induction of STAT1 phosphorylation upon exposure to exogenous IFN-β (Fig. 1B).
To rule out an artificial effect of the cell culture medium in the induction of IFN-β target genes such as MxA, we cultured LTCs in NB medium: here, no induction of MxA was observed; further, culturing GICs in serum-free (SF) or complete DMEM had no major consistent effect on MxA levels (Fig. 1D), but interfered with the sphere culture phenotype at longer incubation times (data not shown).
The IFN-β Signaling Pathway Is Activated in Glioma Cells In Vivo
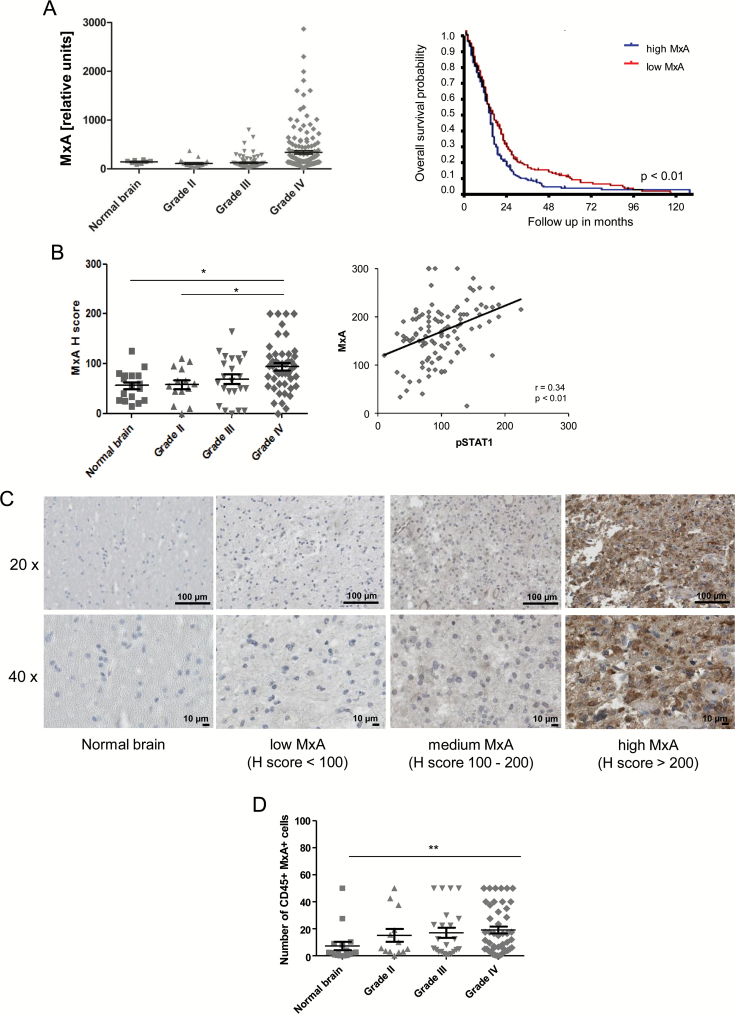
We then explored whether IFN signaling is constitutively active in gliomas in vivo. To this end, we analyzed the expression of the IFN-specific target gene, MxA, using data from glioma patients of different WHO grades deposited in the database of TCGA. There was a trend for increased MxA mRNA levels with increasing WHO grade, with highest levels in glioblastoma (Fig. 2A, left). No differential expression was observed when MxA levels were examined in the molecular subtypes classified by Verhaak et al22 (Supplementary Fig. 1A). The survival of glioblastoma patients with tumors expressing high MxA levels was shorter when the median was used as cutoff (Fig. 2A, right). We observed a strong correlation of MxA expression with the IFN response genes STAT1, STAT2, and STAT3 and IFN regulatory factor 7, but not with IFNAR1, IFNAR2, or type I IFN on the transcriptional level (Supplementary Fig. 1B). To confirm the results obtained by TCGA database interrogation, we analyzed MxA protein levels by immunohistochemistry on a TMA of gliomas of different WHO grades. MxA was mainly found in the cytoplasm. As shown in Fig. 2B and C, MxA protein levels increased with WHO grade. Again, highest MxA levels were observed in glioblastoma. Similarly, pSTAT1 significantly correlated with MxA protein levels (Fig. 2B, right). Since IFNs are also produced by activated immune cells, we looked for the presence of MxA-expressing immune cells in the glioma tissue specimens on the TMA, too. Despite the overall low numbers, we noticed an increase of CD45/MxA double-positive immune cells with histological grade (Fig. 2D).
Fig. 2.
MxA is expressed in gliomas in vivo. (A) MxA mRNA expression levels in gliomas of different WHO grades were analyzed using data from the database of TCGA (left). Overall survival analysis within the TCGA database for glioblastoma patients with high versus low MxA expression was performed by Kaplan–Meier analysis. The median was used as cutoff (right). (B) MxA protein levels were assessed by immunohistochemistry on a glioma tissue microarray and quantified by H scoring (left). Phospho-STAT1 protein levels were analyzed by immunohistochemistry on a TMA and quantified by H scoring. A correlation analysis of pSTAT1 H scores with MxA H scores is shown (right). (C) Representative images of normal brain and glioblastoma specimens with low, intermediate, and high MxA levels are shown (scale bar, 100 µm or 10 µm for 20x or 40x magnification, respectively). (D) MxA/CD45 costaining was performed on a glioma TMA and the number of double-positive cells was counted.
IFN Receptors Are Expressed by Glioma Cells
IFN-α and IFN-β mediate their effects through the type I IFN heterodimer cell surface receptor complex composed of 2 subunits, IFNAR1 and IFNAR2. First, we confirmed that IFNAR1 and IFNAR2 are expressed on a transcriptional level in the panel of glioma cells tested (Fig. 3A, left). Moreover, as observed previously, only IFNAR2, but not IFNAR1, was detectable by flow cytometry using currently available antibodies (Fig. 3A, right).24
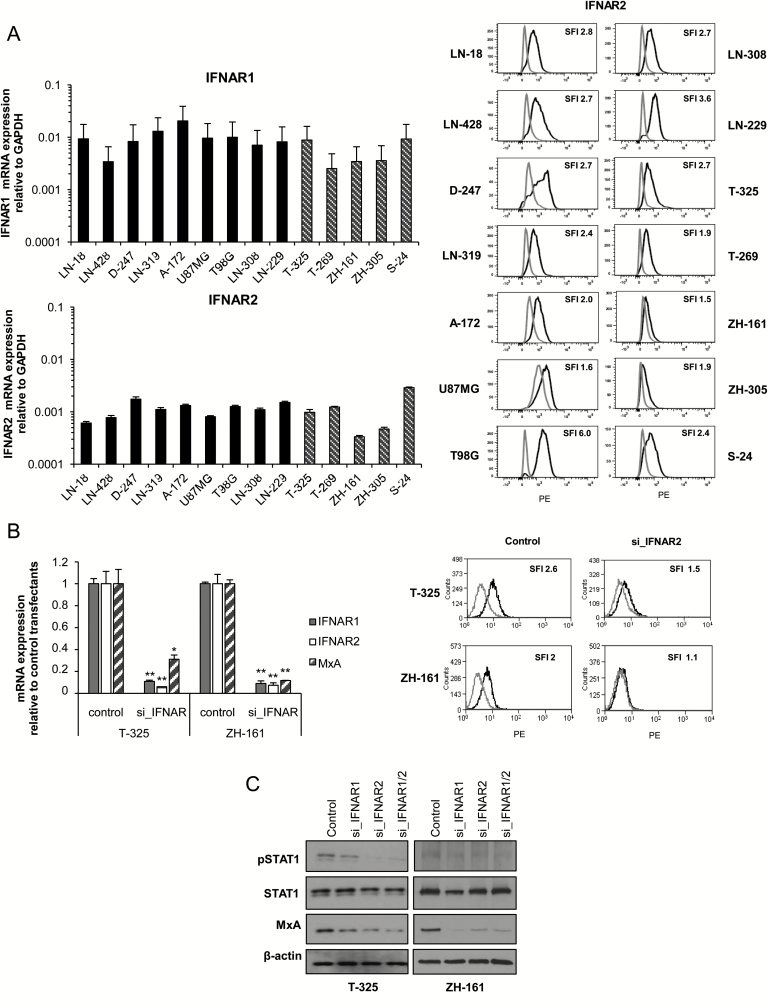
Fig. 3.
MxA expression in human GICs depends on signaling through IFNAR1 and IFNAR2. (A) Basal expression levels of IFNAR1 and IFNAR2 were assessed in LN-18, LN-428, D247MG, LN-319, A172, U87MG, T98G, LN-308 and LN-229, T-325, T-269, ZH-161, ZH-305, or S-24 cells by real-time PCR (left) (median expression levels ± SE are shown from 2 independent experiments). Cell surface IFNAR2 protein was analyzed by flow cytometry (1 out of 2 independent experiments is shown). Isotype control antibody (gray) and specific antibody (black) are shown in the histograms (right). (B) SiRNA-mediated gene silencing of IFNAR1 (siIFNAR1), IFNAR2 (siIFNAR2), or IFNAR1 and IFNAR2 in parallel (siIFNAR1/2) in T-325 or ZH-161 cells was performed by electroporation and confirmed for IFNAR1, IFNAR2, and MxA by real-time PCR 24 h posttransfection (median expression levels ± SE are shown from 3 independent experiments) and for IFNAR2 by flow cytometry at 48 h following transfection. Isotype control antibody (gray) and specific antibody (black) are shown in the histograms. (C) Phospho-STAT1, STAT1, and MxA levels of control, siIFNAR1, siIFNAR2, or double knockdown cells (siIFNAR1/2) were determined 48 h after transfection by immunoblot (1 out of 3 independent experiments is shown).
To confirm that constitutive MxA expression in GICs depends on signaling through IFNAR, we silenced both receptor subunits using RNA interference in T-325 and ZH-161 cells. The knockdown efficacy was confirmed at the mRNA level for both receptor subunits (Fig. 3B, left) and additionally at the protein level for IFNAR2 (Fig. 3B, right).
Silencing of IFNAR1 or IFNAR2 resulted in reduced MxA mRNA levels (Fig. 3B, left). In line with these findings, STAT1 phosphorylation and MxA protein levels were reduced upon silencing of IFNAR1 or IFNAR2 in both cell lines (Fig. 3C).
Identification of Ligands to the Type I IFN Receptor in Glioma Cells
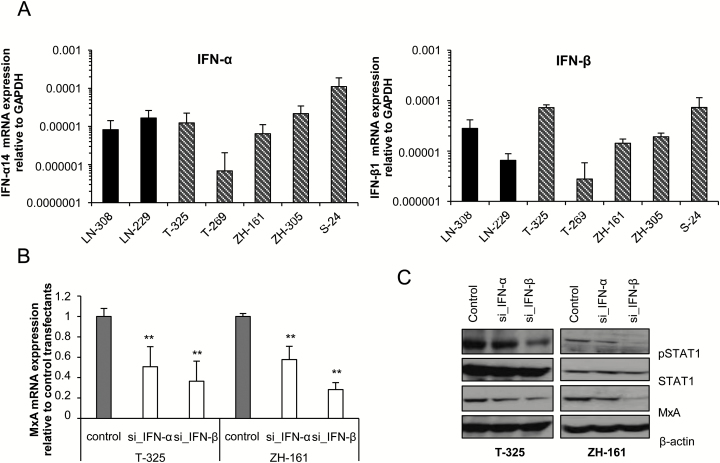
Based on the results obtained with IFN receptor silencing, it seemed most likely that endogenous IFN propagates the signaling through IFNAR. To identify the ligands, we determined the expression of IFN-α and IFN-β at mRNA level in glioma cell lines. Both ligands were expressed at variable levels in the panel of glioma cells tested (Fig. 4A). Silencing of IFN-α or IFN-β by RNA interference resulted in decreased MxA mRNA expression (Fig. 4B), whereas on the protein level, silencing of IFN-α or IFN-β led to reduced MxA and pSTAT1 protein levels, more prominently by silencing of IFN-β than IFN-α, confirming an autocrine IFN signaling loop in glioma cells (Fig. 4C).
Fig. 4.
Glioma-derived IFN-α or IFN-β induces autocrine signaling. (A) IFN-α or IFN-β mRNA expression levels were determined in LN-308, LN-229, T-325, T-269, ZH-161, ZH-305, or S-24 cells using real-time PCR (median expression levels ± SE are shown from 2 independent experiments). (B, C) SiRNA-mediated gene silencing of IFN-α (si_IFN-α) or IFN-β (si_IFN-β) was performed using electroporation in T-325 or ZH-161 cells. MxA mRNA expression was determined 24 h posttransfection by real-time PCR (median expression levels ± SE are shown from 2 independent experiments) (B), while pSTAT1, STAT1, and MxA protein levels were assessed at 48 h following transfection by immunoblot. Actin was used as a loading control (1 out of 3 independent experiments is shown) (C).
Autocrine IFN Signaling Impairs the Immunogenicity of Glioma Cells
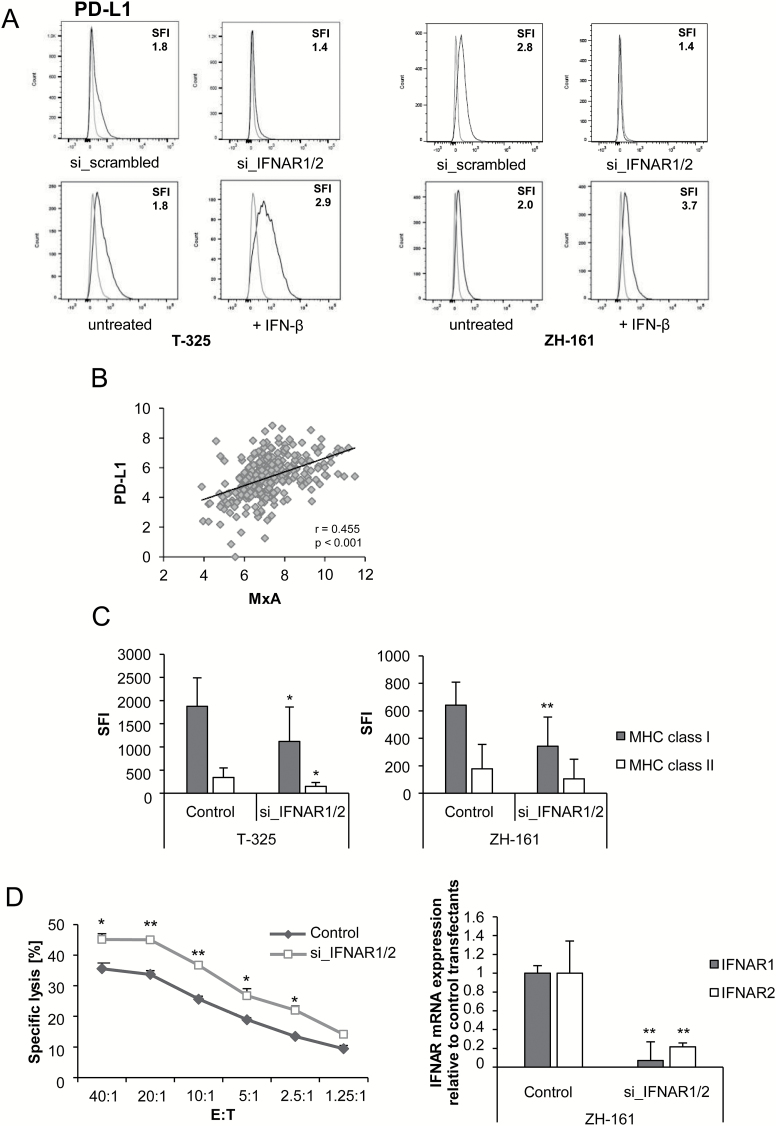
Finally, we defined the role of constitutive IFN signaling in human GICs on a functional level focusing on immunological aspects. Disruption of autocrine IFN signaling using siRNA-mediated silencing of IFNAR1 and IFNAR2 in T-325 or ZH-161 cells resulted in decreased PD-L1 cell surface expression, whereas addition of exogenous recombinant IFN-β increased PD-L1 cell surface levels (Fig. 5A). Accordingly, using a TCGA database interrogation, we observed a significant correlation between MxA and PD-L1 mRNA expression levels in vivo in glioma patients (Fig. 5B). Moreover, MxA levels correlated with the T-cell activation marker CD69, but not with CD25 or PD-1 (Supplementary Fig. 2A). In order to define whether MxA is a regulator of PD-L1 expression, we silenced MxA using RNA interference, which, however, did not affect PD-L1 cell surface levels (Supplementary Fig. 2B). Silencing of IFNAR1 and IFNAR2 by RNA interference did not influence NKG2DL levels but resulted in decreased major histocompatibility complex (MHC) class I and class II cell surface expression (Supplementary Fig. 3A, Fig. 5C). In line with these data, we found a significant correlation between MxA and MHC class I and II mRNA expression in vivo when data from glioblastoma patients deposited in the database of TCGA were analyzed (Supplementary Fig. 3B). MxA mRNA levels also correlated with the expression of the nonclassical MHC molecules HLA-E and HLA-G, which have been described as mediators of glioma immune escape (Supplementary Fig. 3C).25,26 Most importantly, ZH-161 glioma cells were more susceptible to NKL cell-mediated immune cell lysis upon IFNAR gene silencing, corroborating the assumption that constitutive IFN signaling acts as a negative regulator of antitumor immune responses (Fig. 5D). Inhibition of the PD-1 pathway using blocking PD-L1 antibodies did not alter NKL cell-mediated lysis of glioma cells (Supplementary Fig. 3D).
Fig. 5.
Constitutive IFN signaling impairs the immunogenicity of glioma cells. (A) SiRNA-mediated silencing of IFNAR1/2 or transfection with control oligonucleotides was performed in T-325 or ZH-161 using electroporation (top), or the cells were exposed to 150 IU/mL IFN-β or not (bottom). Cell surface PD-L1 protein levels were assessed after 72 h by flow cytometry (1 out of 3 independent experiments is shown). Isotype control antibody (gray) and specific antibody (black) are shown in the histograms. (B) Correlation analysis of MxA with PD-L1 mRNA expression was performed for glioma patients within the database of TCGA. Two-tailed Pearson test coefficients (r) and significances (P) are indicated. (C, D) SiRNA-mediated gene silencing of IFNAR1/2 in T-325 or ZH-161 cells was performed by electroporation. MHC class I or II cell surface protein levels were determined 48 h posttransfection by flow cytometry (median expression levels ± SD are shown from 8 independent experiments) (C). ZH-161 cells were used as target cells in a 4 h NKL cell lysis assay at various effector:target (E:T) ratios as indicated 72 h after transfection. The percentage of target cell lysis corrected for spontaneous background lysis is shown (left) (1 out of 4 independent experiments is shown). The knockdown efficiency for IFNAR1 and IFNAR2 gene silencing was confirmed by real-time PCR (right) (D).
Discussion
IFN-β has been used as a therapeutic agent against glioblastoma in small clinical trials which were largely interpreted as promising.27 However, the molecular mechanisms responsible for potential IFN-mediated effects on glioma cells in vivo have remained uncertain and the effect of IFN-β on the more resistant GIC population is the subject of current research. Importantly, it has been demonstrated that the efficacy of type I IFNs as antitumor drugs is affected by constitutive IFN signaling in that the expression and activation of the IFN-stimulated gene factor 3a correlated with cellular IFN responsiveness.15,16 Moreover, chronic IFN signaling induces the proliferation of dormant hematopoietic stem cells, which might also be relevant for cancer stem cells and their impact on sustained tumor growth.16 Based on these considerations, we investigated the role of autocrine IFN-β signaling in glioma cells, including GICs.
We observed constitutive STAT1 phosphorylation in most glioma cell lines and increased phosphorylation after exposure to exogenous IFN-β (Fig. 1B). Constitutively activated STAT proteins have been attributed a role in the malignant phenotype of many tumors, including glioblastoma,28 and constitutively activated STATs have been implicated in radioresistance.29,30 While STATs can be activated by IFN but also other cytokines or growth factors, MxA may be an ideal candidate to specifically assess the activity of the IFN signaling pathway.12,28 Although single studies suggested a potential role of α-defensin31 and interleukins32 in the induction of MxA, it is widely accepted that it is specifically induced by IFN.33 Thus, as a suitable readout for the presence of IFN activity, we examined the levels of MxA protein in glioma cells. Constitutive MxA expression was mainly observed in GIC lines (Fig. 1B). These results correlated with quantitative PCR results, which demonstrated higher MxA mRNA expression levels in T-325 and ZH-161 GIC lines than in other cell lines (Fig. 1A). Since LTC and GIC lines are cultured under different conditions, we asked whether the cell culture medium would influence constitutive MxA expression. Culturing LTCs in stem cell medium neither induced nor repressed MxA levels in any cell line, while FCS-containing complete medium increased (T-325) or decreased (ZH-161) MxA expression in GIC lines (Fig. 1D). Importantly, GICs change their phenotype in FCS-containing medium and become adherent, thus the effect of this medium on MxA expression might be a consequence of induced cell differentiation rather than a direct response to medium exchange. Based on these findings, and taking into account that not all GIC lines express MxA, we concluded that MxA expression is not an effect of cell culture medium conditions.
We then analyzed MxA levels in vivo and observed higher expression in gliomas than in normal tissue, with highest levels in glioblastoma, suggesting that IFN signaling is constitutively active also in gliomas in vivo (Fig. 2). No differential expression between the different subtypes classified by Verhaak22 was observed (Supplementary Fig. 1), suggesting that this is a rather general pathway. Survival analyses of glioblastoma patients from the database of TCGA revealed inferior survival for patients with tumors with increased MxA expression, suggesting an impact of constitutively active IFN signaling on the malignant phenotype of these tumors. Similarly, overexpression of IFN/STAT1-related genes may be predictive for poor survival in the proneural subtype of glioblastoma.28,34
We then confirmed that IFNAR1 and IFNAR2 are expressed in all cell lines and subsequently performed siRNA-mediated gene silencing of IFNAR1, IFNAR2, or both subunits in T-325 and ZH-161 cells. Receptor silencing led to reduced expression of MxA at mRNA and protein levels (Fig. 3B, C). Moreover, gene silencing of the ligands, IFN-α and IFN-β, also resulted in decreased MxA expression, confirming an autocrine signaling loop in which type I IFNs mediate the induction of MxA through IFNAR1/2 (Fig. 4B, C). There was no correlation of IFN receptor or IFN-β expression with MxA levels in glioblastomas in the database of TCGA in vivo. Hence, a variable activation status of the receptor subunits and/or their ligands and the involvement of other, yet undescribed, signaling pathways might explain this observation. Further limitations of TCGA analyses include the inability to determine whether mRNA species stem from tumor cells or infiltrating host cells, which is particularly relevant when exploring immunological aspects of the disease. On a TMA, we observed generally low but increasing numbers of tumor-infiltrating lymphocytes with the WHO grade, suggesting that IFN signaling in glioma cells might be induced in an autocrine and paracrine manner (Fig. 2D).
Although typically expressed only at low levels, IFN may exert strong effects on various tissues.16 Besides their role in the protection of cells from viral infections, constitutive IFN signaling seems to be involved in various cellular responses to other cytokines and growth factors as well as the proper regulation of immune responses.35–37 Therefore, we aimed at defining the functional impact of the observed constitutive IFN signaling on the immunogenicity of glioma cells. Here, we noticed reduced PD-L1 as well as MHC class I and II expression upon IFNAR1 or IFNAR2 silencing in GICs, suggesting that IFN signaling in these cells may be involved in the interaction between glioma cells and the microenvironment (Fig. 5). Additionally, MxA mRNA levels correlated with MHC class I and II molecules and with the nonclassical MHC molecules HLA-E and HLA-G in vivo (Supplementary Fig. 3B, C). PD-L1 is expressed by glioma cells in vitro and in vivo and confers immunosuppressive effects by promoting T-cell apoptosis and induction of regulatory T cells.38 Similarly, high MHC class I and II expression can inhibit innate immune responses against tumor cells.39 In line with our findings, a role for endogenous IFN-β in the regulation of PD-L1 expression has been described in neurons.40 The observation that RNA interference-mediated silencing of MxA did not alter PD-L1 expression indicates that PD-L1 expression is regulated by the IFN pathway independently of MxA (Supplementary Fig. 2B). Moreover, IFNs strongly activate monocytes/macrophages and upregulate MHC class I and II expression.41,42 Furthermore, the expression of the nonclassical MHC molecules HLA-E and HLA-G has been linked to immune-inhibitory effects in gliomas. Here, HLA-E mainly inhibits NK cell-mediated lysis, while HLA-G interferes with CD4 and CD8 T-cell activity.25,26 Accordingly, abrogation of IFN signaling in glioma cells by IFNAR1 and IFNAR2 gene silencing made these cells more susceptible to immune cell killing (Fig. 5). Since inhibition of the PD-1 pathway using blocking PD-L1 antibodies did not alter NKL cell-mediated lysis of glioma cells (Supplementary Fig. D), it is tempting to speculate that the increased susceptibility to NKL cell-mediated lysis upon IFNAR gene silencing results from decreased MHC class I and class II expression. Hence, our findings suggest that the upregulation of MHC molecules due to constitutive IFN signaling may impair antitumor immune responses by NK cells.
Interruption of endogenous IFN signaling did not result in increased NKG2DL expression, which is in line with previous findings indicating that exposure to exogenous IFN-β had no effect or decreased NKG2DL cell surface levels43 (Supplementary Fig. 3A). Thus, whether the immunological changes induced by treatment with recombinant exogenous IFN-β result in net immune stimulating or net immune inhibitory effects might be dependent on the cell line and context.
In summary, we describe the presence of constitutive IFN signaling in glioma cells, which may have an important role in the interaction of these cells with the microenvironment. A more detailed understanding of this intrinsic IFN activity, particularly in the stem cell compartment of gliomas, may lay a basis for the therapeutic targeting of this pathway in order to interfere with the continuous growth of these tumors.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was supported by a grant from the Swiss National Science Foundation (SNF 310030_146213) to M.W., a Swiss Government Research Scholarship for Foreign Scholars to S.N., and a grant by the Canton of Zurich (HSM-2) to M.W. and P.R.
Conflict of interest statement. P.R. has received honoraria for advisory boards and lectures from BMS, Roche, MSD, Novartis, and Molecular Partners. M.W. has received research grants from Actelion, Alpinia Institute, Bayer, Isarna, MSD, Merck Serono, Piqur, and Roche and honoraria for lectures or advisory board participation from Celldex, Immunocellular, Isarna, Magforce, MSD, Merck Serono, Pfizer, Roche, and Teva. C.H. has received honoraria for advisory boards from MSD. M.S., S.N., and H.S. have nothing to disclose.
Supplementary Material
Acknowledgments
We thank K. Frei (Department of Neurosurgery, Zurich, Switzerland), I. Burghardt (Department of Neurology, Zurich, Switzerland), and E. Rushing (Institute of Neuropathology, Zurich, Switzerland) for providing tissue samples and support with immunohistochemistry; J. Pavlovic for providing the MxA antibody; and A. Keller (Chair for Clinical Bioinformatics, Saarbrücken, Germany) for help with statistical analyses.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 2. Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. [DOI] [PubMed] [Google Scholar]
- 3. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Essers MA, Offner S, Blanco-Bose WE, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–908. [DOI] [PubMed] [Google Scholar]
- 5. Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15(6):696–700. [DOI] [PubMed] [Google Scholar]
- 6. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. [DOI] [PubMed] [Google Scholar]
- 7. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. [DOI] [PubMed] [Google Scholar]
- 8. Lim JK, Xiong J, Carrasco N, Langer JA. Intrinsic ligand binding properties of the human and bovine alpha-interferon receptors. FEBS Lett. 1994;350(2–3):281–286. [DOI] [PubMed] [Google Scholar]
- 9. Cutrone EC, Langer JA. Contributions of cloned type I interferon receptor subunits to differential ligand binding. FEBS Lett. 1997;404(2–3):197–202. [DOI] [PubMed] [Google Scholar]
- 10. Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250(1):317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cull VS, Tilbrook PA, Bartlett EJ, Brekalo NL, James CM. Type I interferon differential therapy for erythroleukemia: specificity of STAT activation. Blood. 2003;101(7):2727–2735. [DOI] [PubMed] [Google Scholar]
- 12. Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res. 2011;31(1):79–87. [DOI] [PubMed] [Google Scholar]
- 13. Bracarda S, Eggermont AM, Samuelsson J. Redefining the role of interferon in the treatment of malignant diseases. Eur J Cancer. 2010;46(2):284–297. [DOI] [PubMed] [Google Scholar]
- 14. Landolfo S, Guarini A, Riera L, et al. Chronic myeloid leukemia cells resistant to interferon-alpha lack STAT1 expression. Hematol J. 2000;1(1):7–14. [DOI] [PubMed] [Google Scholar]
- 15. Wong LH, Krauer KG, Hatzinisiriou I, et al. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J Biol Chem. 1997;272(45):28779–28785. [DOI] [PubMed] [Google Scholar]
- 16. Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Codo P, Weller M, Meister G, et al. MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget. 2014;5(17):7651–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowalewski DJ, Stevanovic S. Biochemical Large-Scale Identification of MHC Class I Ligands. In: van Endert P, ed. Antigen Processing: Methods and Protocols, Methods in Molecular Biology. Vol 960: Springer Science+Business Media; 2013:145–157. [DOI] [PubMed] [Google Scholar]
- 19. Wirsching HG, Krishnan S, Florea AM, et al. Thymosin β 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain. 2014;137(Pt 2):433–448. [DOI] [PubMed] [Google Scholar]
- 20. Kraus JA, Dabbs DJ, Beriwal S, Bhargava R. Semi-quantitative immunohistochemical assay versus oncotype DX(®) qRT-PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod Pathol. 2012;25(6):869–876. [DOI] [PubMed] [Google Scholar]
- 21. Silginer M, Burghardt I, Gramatzki D, et al. The aryl hydrocarbon receptor links integrin signaling to the TGF-β pathway. Oncogene. 2016;35(25):3260–3271. [DOI] [PubMed] [Google Scholar]
- 22. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Codó P, Weller M, Kaulich K, et al. Control of glioma cell migration and invasiveness by GDF-15. Oncotarget. 2016;7(7):7732–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Happold C, Roth P, Silginer M, et al. Interferon-β induces loss of spherogenicity and overcomes therapy resistance of glioblastoma stem cells. Mol Cancer Ther. 2014;13(4):948–961. [DOI] [PubMed] [Google Scholar]
- 25. Wolpert F, Roth P, Lamszus K, Tabatabai G, Weller M, Eisele G. HLA-E contributes to an immune-inhibitory phenotype of glioblastoma stem-like cells. J Neuroimmunol. 2012;250(1–2):27–34. [DOI] [PubMed] [Google Scholar]
- 26. Wiendl H, Mitsdoerffer M, Hofmeister V, et al. A functional role of HLA-G expression in human gliomas: an alternative strategy of immune escape. J Immunol. 2002;168(9):4772–4780. [DOI] [PubMed] [Google Scholar]
- 27. Motomura K, Natsume A, Kishida Y, et al. Benefits of interferon-β and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: a multicenter study. Cancer. 2011;117(8):1721–1730. [DOI] [PubMed] [Google Scholar]
- 28. Swiatek-Machado K, Kaminska B. STAT signaling in glioma cells. Adv Exp Med Biol. 2013;986:189–208. [DOI] [PubMed] [Google Scholar]
- 29. Khodarev NN, Minn AJ, Efimova EV, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67(19):9214–9220. [DOI] [PubMed] [Google Scholar]
- 30. Khodarev NN, Roach P, Pitroda SP, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4(6):e5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahanonda R, Sa-Ard-Iam N, Rerkyen P, Thitithanyanont A, Subbalekha K, Pichyangkul S. MxA expression induced by α-defensin in healthy human periodontal tissue. Eur J Immunol. 2012;42(4):946–956. [DOI] [PubMed] [Google Scholar]
- 32. Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine. 2005;31(2):109–118. [DOI] [PubMed] [Google Scholar]
- 33. Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J Biol Chem. 2010;285(37):28419–28424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuki K, Natsume A, Yokoyama H, et al. Induction of oligodendrogenesis in glioblastoma-initiating cells by IFN-mediated activation of STAT3 signaling. Cancer Lett. 2009;284(1):71–79. [DOI] [PubMed] [Google Scholar]
- 35. Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2(5):378–386. [DOI] [PubMed] [Google Scholar]
- 36. Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8(6):907–922. [DOI] [PubMed] [Google Scholar]
- 37. Chen HM, Tanaka N, Mitani Y, et al. Critical role for constitutive type I interferon signaling in the prevention of cellular transformation. Cancer Sci. 2009;100(3):449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nduom EK, Wei J, Yaghi NK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18(2):195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27(45):5932–5943. [DOI] [PubMed] [Google Scholar]
- 40. Liu Y, Carlsson R, Ambjørn M, et al. PD-L1 expression by neurons nearby tumors indicates better prognosis in glioblastoma patients. J Neurosci. 2013;33(35):14231–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bekisz J, Baron S, Balinsky C, Morrow A, Zoon KC. Antiproliferative properties of type I and type II interferon. Pharmaceuticals (Basel). 2010;3(4):994–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao W, Cha EN, Lee C, Park CY, Schindler C. Stat2-dependent regulation of MHC class II expression. J Immunol. 2007;179(1):463–471. [DOI] [PubMed] [Google Scholar]
- 43. Wolpert F, Happold C, Reifenberger G, et al. Interferon-β modulates the innate immune response against glioblastoma initiating cells. PLoS One. 2015;10(10):e0139603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.