Abstract
Our group recently employed genome-wide transcriptional profiling in tandem with machine-learning based analysis to identify a ten-gene pattern of differential expression in peripheral blood which may have utility for detection of stroke. The objective of this study was to assess the diagnostic capacity and temporal stability of this stroke-associated transcriptional signature in an independent patient population. Publicly available whole blood microarray data generated from 23 ischemic stroke patients at 3, 5, and 24 h post-symptom onset, as well from 23 cardiovascular disease controls, were obtained via the National Center for Biotechnology Information Gene Expression Omnibus. Expression levels of the ten candidate genes (ANTXR2, STK3, PDK4, CD163, MAL, GRAP, ID3, CTSZ, KIF1B, and PLXDC2) were extracted, compared between groups, and evaluated for their discriminatory ability at each time point. We observed a largely identical pattern of differential expression between stroke patients and controls across the ten candidate genes as reported in our prior work. Furthermore, the coordinate expression levels of the ten candidate genes were able to discriminate between stroke patients and controls with levels of sensitivity and specificity upwards of 90% across all three time points. These findings confirm the diagnostic robustness of the previously identified pattern of differential expression in an independent patient population, and further suggest that it is temporally stable over the first 24 h of stroke pathology.
Keywords: GA/kNN, Genetic algorithm, Triage, Biomarker, Immunology, Pattern recognition, Cerebrovascular disease, Brain injury
1. Introduction
The ability of clinicians to confidently recognize stroke during triage increases access to interventional treatments and affords patients improved odds for favorable outcome [1], [2]. However, the diagnostic tools currently available to emergency medical technicians, paramedics, and hospital staff for identification of stroke have significant limitations [3], [4]. Biomarker-based tests are clinically used to aid in the diagnosis of acute cardiovascular conditions such as myocardial infarction [5], however no such assay currently exists for the detection of stroke. This diagnostic limitation has resulted in a push for the identification of peripheral blood stroke biomarkers which could be rapidly measured in either the field or emergency department to guide early triage decisions [3], [6].
Our group recently employed high-throughput transcriptomics in combination with a machine learning technique known as genetic algorithm/k-nearest neighbors (GA/kNN) to identify a panel of ten candidate genes whose peripheral blood expression levels were able to differentiate between 78 ischemic stroke patients and 74 control subjects with a high degree of accuracy [7]. These candidate genes include seven whose expression levels were elevated in stroke patients relative to controls (CD163, ANTXR2, PDK4, PLXDC2, STK3, ID3, CTSZ, KIF1B), and three whose expression levels were down regulated (MAL, ID3, GRAP); their coordinate pattern of differential expression was able to discriminate between groups with levels of sensitivity and specificity approaching 100%. While the levels of diagnostic performance observed in this discovery investigation were unprecedented, limitations in study design necessitate further evaluation of the candidate genes in a validation analysis before definitive conclusions can be made regarding their true diagnostic efficacy.
Stroke patients and control subjects in this discovery investigation were not well matched in terms of cardiovascular disease (CVD) risk factors, leaving open the possibility that the pattern of differential expression which we observed across the ten candidate genes was driven by underlying CVD, and not by the acute event of stroke itself. Furthermore, subjects in this discovery study were almost exclusively Caucasian, and it is currently unknown whether ethnicity impacts the diagnostic efficacy the candidate genes, a possibility which deserves consideration due to the fact that there can be notable inter-ethnic differences in the pathophysiology of cardiovascular conditions [8], [9], [10], [11]. A further limitation in of this discovery study was the fact that blood samples were only collected at a single time point, making the temporal stability of candidate gene differential expression unclear with regards to the progression of stroke pathology. While post hoc statistical analyses were used to address these potential confounds as best possible, it would be reassuring to observe similar levels of diagnostic performance across multiple time points in a more ethnically diverse subject pool which is better matched in terms of CVD risk factors.
Stamova et al. recently used microarray to examine gender differences in the response of the peripheral immune system to stroke [12]. This investigation produced a publicly available data set which includes genome-wide whole blood expression data generated from 23 cardioembolic ischemic stroke patients at three replicate time points post-symptom onset (3, 5, and 24 h), as well as from 23 neurologically asymptomatic control subjects; this patient population was ethnically diverse and groups were well matched in terms of risk factors for CVD. In the study reported here, we assessed the diagnostic robustness of the ten previously identified candidate genes in the aforementioned publicly available data set.
2. Methods
2.1. Data procurement
Raw whole blood microarray data (Affymetrix Human Genome U133 Plus 2.0 Array) were downloaded as .CEL files from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) via accession number GSE58294 (Supplementary File 1). Patient clinical and demographic characteristics were aggregated from the gender-wise information reported by Stamova et al. [12].
2.2. Microarray analysis
Analysis of microarray data was performed using the ‘affy’ package for R (R project for statistical computing) [13], [14]. Raw perfect match probe intensities were background corrected, quantile normalized (Fig. 1), and summarized at the set level via robust multi-array averaging using the rma() function [15]. Probe set level data associated with the ten candidate genes were then extracted for differential expression analysis; in the case of candidate genes with more than one associated probe set, data were further summarized at the gene level via simple averaging. Gene level summarized expression levels were then compared between stroke patients and controls across all three post-onset time points.
Fig. 1.
Normalization of microarray data.
Distributions of pre and post-normalization perfect match probe intensities. Boxplots indicate standard five number summary values.
2.3. Diagnostic evaluation
The diagnostic robustness of candidate gene expression levels was tested in terms of their ability to discriminate between stroke patients and controls using k-nearest neighbors (kNN) at each time point post-symptom onset. Classification was performed using standardized expression values, five nearest neighbors, and majority rule via the knn.cv() function of the ‘class’ package for R [16]. Same-set leave one out cross-validation was performed, and the resultant prediction probabilities were used to generate receiver operator characteristic (ROC) curves using the roc() function of the ‘pROC’ package for R [17]. Areas under curves were then compared between time points via the roc.test() function according the non-parametric method described by DeLong et al. [18].
2.4. Statistics
All statistics were performed using R 3.3. Fisher's exact test was used for comparison of dichotomous variables. t-Test or one-way ANOVA was used for comparisons of continuous variables where appropriate. The null hypothesis was rejected when p < 0.05. In the case of multiple comparisons, p-values were false discovery rate adjusted using the Benjamini-Hochberg procedure [19].
3. Results
3.1. Clinical and demographic characteristics
Stroke patients were significantly older than control patients, but well matched in terms of gender and ethnicity. In terms of cardiovascular disease risk factors, groups were well matched with regards to rates of hypertension and diabetes, however control subjects displayed a significantly higher prevalence of dyslipidemia relative to stroke patients. All stroke patients received thrombolytic intervention via recombinant tissue plasminogen activator (rtPA) following 3 h blood collection, but prior to 5 h blood collection (Table 1).
Table 1.
Clinical and demographic characteristics.
| Cardiovascular disease (n = 23) | Ischemic stroke (n = 23) | p | |
|---|---|---|---|
| aAge mean ± SD | 57.9 ± 3.3 | 57.9 ± 7.9 | < 0.001⁎ |
| bFemale n(%) | 11 (47.8) | 11 (47.8) | 1.000 |
| bNon-caucasian n(%) | 4 (17.4) | 8 (34.8) | 0.314 |
| bDyslipidemia n(%) | 16 (69.6) | 6 (26.1) | 0.007⁎ |
| bHypertension n(%) | 16 (69.6) | 16 (69.6) | 1.000 |
| bDiabetes n(%) | 5 (21.7) | 4 (17.4) | 1.000 |
| aBaseline NIHSS mean ± SD | 0.0 ± 0.0 | 15.4 ± 7.4 | < 0.001⁎ |
| brtPA n(%) | 0 (0.0) | 23 (100.0) | < 0.001⁎ |
Compared via two sample two way t-test.
Compared via Fisher's exact test.
Statistically significant.
3.2. Microarray data processing
Distributions of perfect match probe intensities were visually similar following normalization, providing indication that normalized expression data were suitable for inter-sample comparison (Fig. 1). Probe sets extracted for differential expression analysis are listed in Table 2.
Table 2.
Probe sets extracted for differential expression analysis.
| Gene | Affy probe set ID | Target transcriptsa |
|---|---|---|
| CD163 | 203645_s_at | NM_004244 |
| 215049_x_at | NM_203416 | |
| 216233_at | XM_005253528 | |
| XM_005253529 | ||
| XR_429039 | ||
| ANTXR2 | 1555536_at | NM_001145794 |
| 225524_at | NM_001286780 | |
| 228573_at | NM_001286781 | |
| 238050_at | NM_058172 | |
| MAL | 204777_s_at | NM_002371 |
| NM_022438 | ||
| NM_022439 | ||
| NM_022440 | ||
| PDK4 | 205960_at | NM_002612 |
| PLXDC2 | 214807_at | NM_001282736 |
| 226865_at | NM_032812 | |
| 227276_at | ||
| 227995_at | ||
| 236297_at | ||
| 238455_at | ||
| STK3 | 204068_at | NM_001256312 |
| 211078_s_at | NM_001256313 | |
| NM_006281 | ||
| XM_005251034 | ||
| ID3 | 207826_s_at | NM_002167 |
| CTSZ | 210042_s_at | NM_001336 |
| 212562_s_at | ||
| GRAP | 206620_at | NM_006613 |
| 229726_at | XM_005256425 | |
| XM_005256426 | ||
| KIF1B | 209234_at | NM_015074 |
| 225878_at | NM_183416 | |
| 226968_at | ||
| 228657_at |
NCBI RefSeq ID.
3.3. Candidate gene differential expression
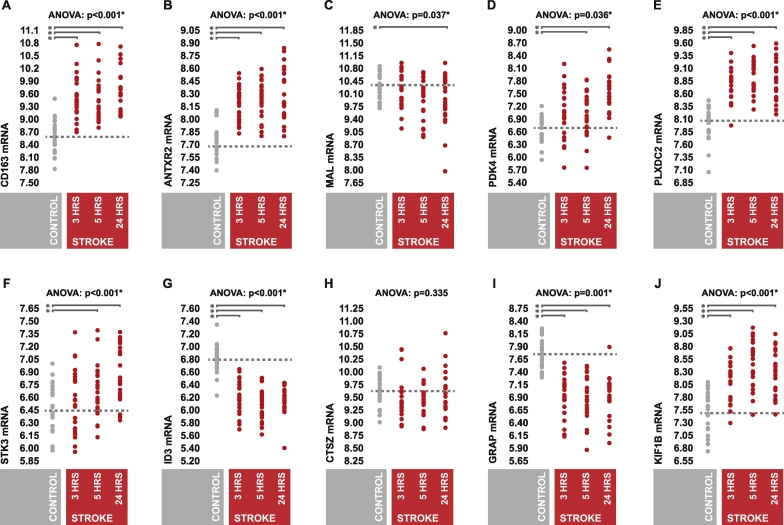
Six of the seven candidate genes which we had previously reported as being elevated in stroke in our prior investigation displayed similar up-regulation in stroke patients relative to controls (Fig. 2A, B, D, E, F, J), however one exhibited no significant differences in expression levels at any time point post-symptom onset (Fig. 2H). In terms of the candidate genes which we had previously reported as being down regulated in stroke, all three displayed significantly lower expression levels in stroke patients relative to controls (Fig. 2C, G, I). Collectively, these observations largely confirmed the pattern of candidate gene differential expression reported in our prior investigation.
Fig. 2.
Candidate gene differential expression.
(A–J) Peripheral whole blood expression levels of candidate genes in stroke patients and controls at 3, 5, and 24 h post-symptom onset. Expression values represent gene-level summarized Log2 perfect match probe intensities. Expression levels were statistically compared between stroke patients and controls across time points using one-way AVOVA; p-values were false discovery rate adjusted via the Benjamini-Hochberg procedure to account for multiple comparisons. In the case of a significant test, post hoc comparisons were made via two-sample two-tailed t-test.
3.4. Temporal profile of candidate differential expression
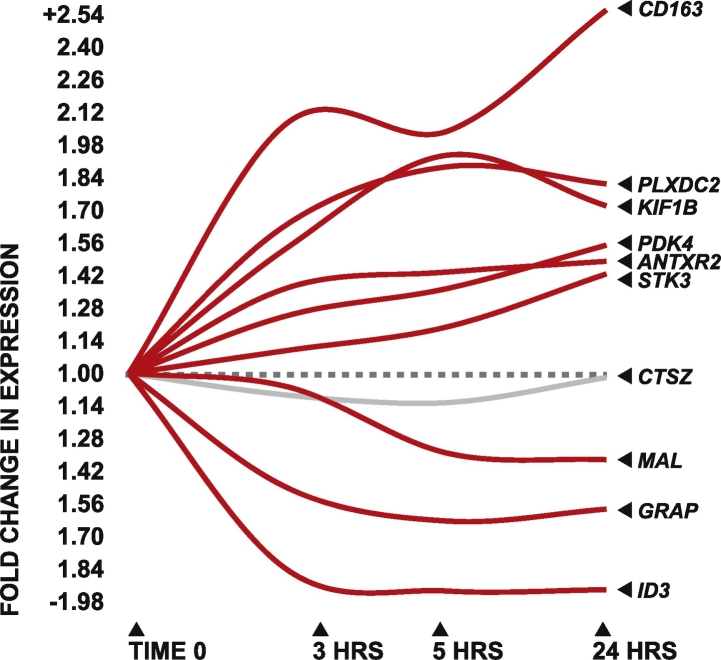
Most candidate genes displayed some degree of differential expression by 3 h post-symptom onset, and the magnitude of the overall response appeared to increase over time. Several candidate genes appeared to achieve maximal differential expression at 5 h post-onset and then plateau, while a few displayed steady increases in the degree of differential expression through 24 h (Fig. 3), providing evidence that the expression levels of the candidate genes are likely directly responsive to acute stroke pathology.
Fig. 3.
Temporal profile of candidate gene differential expression.
Magnitude of candidate gene differential expression between stroke patients and controls at 3, 5, and 24 h post-symptom onset, indicated as fold difference relative to control.
3.5. Candidate gene diagnostic performance
In terms of diagnostic ability, the coordinate expression levels of the ten candidate genes were able to discriminate between stroke patients and controls using kNN with levels of sensitivity and specificity upwards of 90% at all three time points post-symptom onset (Fig. 4A, B, C). While the overall diagnostic capacity of the ten candidate genes appeared slightly more robust at five and 24 h, no statistically significant differences in area under ROC curve were observed between time points (Fig. 4D). Taken together, these observations supported the high levels of diagnostic performance reported in our prior work, and suggest that the diagnostic capacity of the ten candidate genes is temporally stable over the first 24 h post-symptom onset.
Fig. 4.
Candidate gene diagnostic performance.
(A–C) Two-dimensional projections of the kNN feature spaces generated by the coordinate expression levels of the ten candidate genes at 3, 5, and 24 h post-symptom onset. Levels of sensitivity and specificity are associated with class predications generated via five nearest neighbors using a probability cutoff of 0.50. (D) ROC curves associated with the prediction probabilities generated in kNN. Shaded areas indicate 95% confidence intervals. Areas under curves were statistically compared between time points using the DeLong method.
4. Discussion
There has been a recent push for the identification of molecular biomarkers which could be used to aid clinicians in the recognition of stroke during patient triage. Our group recently employed high-throughput transcriptomics in combination with a machine-learning technique known at GA/kNN to identify a ten gene pattern of differential expression in peripheral blood which has potential utility for the detection of stroke [7]. However, patients in this discovery investigation were almost exclusively Caucasian, groups were not well matched in terms of CVD risk factors, and blood was only sampled at a single time point post-symptom onset. In the study reported here, we leveraged a publicly available microarray dataset to evaluate the previously identified candidate pattern of gene expression at multiple pathological time points in a more ethnically diverse subject pool which was better matched in terms of CVD risk factors.
The overall pattern of differential expression which we previously reported between stroke patients and controls was largely confirmed in the analysis described here, as nine of the ten candidate genes were identically differentially regulated. Furthermore, the candidate genes displayed similar levels of diagnostic robustness as described previously. This suggests that it is unlikely that our prior findings were substantially driven by intergroup differences in CVD risk factors; this notion is accentuated by the fact that the overall pattern of differential expression across the ten candidate genes was temporally dynamic with regards to time from symptom onset, providing evidence that the candidate genes are directly responsive to stroke pathology. The fact that our prior observations were largely recapitulated in the analysis reported here also suggests that ethnicity likely has little influence on the overall transcriptional response of the candidate genes to stroke.
One possible exception with this regard is CTSZ, which was the only candidate gene which failed to exhibit a similar response to stroke as previously reported. Thus, it is possible that the differential regulation of CTSZ which we observed in our discovery investigation was indeed driven primarily by underlying CVD, or that there are interethnic differences in the responsiveness of CTSZ to stroke. However, to our knowledge, there are no associations reported in the literature to support either conclusion, and is possible that the discrepancy in response between investigations has other explanation. The samples analyzed in this study were obtained exclusively from patients presenting with ischemic strokes of cardioembolic etiology, while the samples used in our prior discovery study were obtained from patients presenting with ischemic strokes of multiple etiologies, including a large number which were thrombotic in nature; thus it is possible that the disagreement in findings is due to an etiology-specific response. The disagreement in findings could also be driven by a technical confound, as the gene expression data used in this analysis were generated using a different gene chip then that which was used in our discovery investigation, and the chips do not have completely overlapping transcriptional coverage of CTSZ.
In addition to providing a general validation of the overall pattern of candidate gene differential expression, this study also afforded us an opportunity to evaluate its temporal stability with regards to stroke pathophysiology. The overall pattern of differential expression was modestly detectable at 3 h post-symptom onset and appeared to increase in magnitude though 24 h. Despite the modest magnitude, the levels of differential expression present at 3 h post-onset were still adequate to differentiate between groups with similarly high levels of diagnostic performance as those observed at the subsequent two time points. Overall, our findings suggested that the diagnostic ability of the candidate pattern of gene expression is relatively temporally stable over the first 24 h of stroke pathophysiology, which is encouraging from a translational standpoint in that the first clinical contact with stroke patients tends to vary across a wide time range with regards to time from onset, depending in the overtness of symptom presentation.
It is relevant to note that the 5 and 24 h blood samples which we analyzed in this study were collected from stroke patients following thrombolytic intervention, leaving open the possibility that the differential expression which we observed across the candidate genes at these time points was driven by the effect of rtPA, and not the ischemic event itself. However, we find this unlikely, as the differential expression pattern which we observed was highly similar to the one reported in our previous discovery investigation in which all blood samples were collected prior to the administration of thrombolytics. Furthermore, the fact we have now observed a similar pattern of differential expression both before and following thrombolytic intervention suggests that the response of the candidate markers is not largely influenced by rtPA; this property leaves open the possibility that the candidate markers could be clinically useful not only for triage, but also for non-acute post-treatment indications, such as to molecularly confirm pathology as means of determining clinical trial eligibility.
A potential limitation with regards to this study lies in that the stroke patients and controls associated with the samples used in this analysis were not well matched in terms of age. Ideally, multiple regression could be used to statistically control for such a potential confound, however non-aggregated demographic information was not available for the dataset, making such an analysis impossible. However, we explored the relationship between the expression levels of the ten candidate genes and age as part of our previously reported discovery investigation, and observed no significant associations. Thus, we feel that it is unlikely that the results reported here are confounded by intergroup age differences.
It is also important to note that a significant translational limitation in our analysis lies in that we built and tested a de novo classification model using only the candidate gene expression data contained in the dataset generated by stamova et al. Ideally, the classification model we generated in our previously-published discovery analysis could have been tested in the Stamova et al. dataset, however this was infeasible due to the fact that different microarray platforms were used between the two investigations (Illumina versus Affymetrix) and accurate cross-platform normalization is difficult. Nonetheless, this does not diminish the fact that we observed a highly identical pattern of differential expression across the candidate markers as reported in our prior discovery investigation, which provides compelling evidence that these makers are reliably altered in stroke pathology and have true potential for clinical biomarker use.
However, for such clinical use to be realized, there are several further developmental hurdles which need to be overcome. Most notable from this regard is the development of an assay which could measure these markers rapidly and accurately at the point of care with minimal specimen processing, which would be essential for triage use in the acute care setting. Unfortunately, a commercially available platform capable of rapid nucleic acid quantification with high enough fidelity to detect relatively modest levels of differential expression, such as those we have described across the candidate markers, does not currently exist. However, research into rapid detection of nucleic acids is ongoing, and promising new advances such as those regarding direct RNA nanodetection and thermoneutral amplification suggest that suitable technologies will be available in the near future [20], [21], [22].
Collectively, the findings of this analysis confirm the diagnostic robustness of the previously identified stroke-associated pattern of gene expression, and further suggest that it is temporally stable over the first 24 h of stroke pathology. Due to fact that this transcriptional signature has now demonstrated levels of diagnostic performance which well exceed those of the triage tools currently available to clinicians for identification of stroke in two independent investigations, we feel that it has legitimate translational potential and a path towards clinical implementation should be further explored.
Disclosures
GCO and TLB have a patent pending re: markers of stroke and stroke severity. TLB serves as chief scientific officer for Valtari Bio Incorporated. Work by GCO is part of a pending licensing agreement with Valtari Bio Incorporated. The remaining authors report no potential conflicts of interest.
Transparency document
Transparency document.
Acknowledgments
Acknowledgements
Work was funded via a Robert Wood Johnson Foundation nurse faculty scholar award to TLB (70319) and a National Institutes of Health CoBRE sub-award to TLB (P20 GM109098).
Footnotes
The Transparency document associated with this article can be found, in online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2017.08.006.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Lees K.R., Bluhmki E., von Kummer R., Brott T.G., Toni D., Grotta J.C., Albers G.W., Kaste M., Marler J.R., Hamilton S.A. Time to treatment with intravenous alteplase and outcome in stroke: an updated pooled analysis of ECASS, ATLANTIS, NINDS, and EPITHET trials. Lancet. 2010;375:1695–1703. doi: 10.1016/S0140-6736(10)60491-6. [DOI] [PubMed] [Google Scholar]
- 2.Marler J.R., Tilley B.C., Lu M., Brott T.G., Lyden P.C., Grotta J.C., Broderick J.P., Levine S.R., Frankel M.P., Horowitz S.H. Early stroke treatment associated with better outcome: the NINDS rt-PA stroke study. Neurology. 2000;55:1649–1655. doi: 10.1212/wnl.55.11.1649. [DOI] [PubMed] [Google Scholar]
- 3.Saenger A.K., Christenson R.H. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin. Chem. 2010;56:21–33. doi: 10.1373/clinchem.2009.133801. [DOI] [PubMed] [Google Scholar]
- 4.Purrucker J.C., Hametner C., Engelbrecht A., Bruckner T., Popp E., Poli S. Comparison of stroke recognition and stroke severity scores for stroke detection in a single cohort. J. Neurol. Neurosurg. Psychiatry. 2015;86:1021–1028. doi: 10.1136/jnnp-2014-309260. [DOI] [PubMed] [Google Scholar]
- 5.Keller T., Zeller T., Peetz D., Tzikas S., Roth A., Czyz E., Bickel C., Baldus S., Warnholtz A., Fröhlich M. Sensitive troponin I assay in early diagnosis of acute myocardial infarction. N. Engl. J. Med. 2009;361:868–877. doi: 10.1056/NEJMoa0903515. [DOI] [PubMed] [Google Scholar]
- 6.Hill M.D. Diagnostic biomarkers for stroke: a stroke neurologist's perspective. Clin. Chem. 2005;51:2001–2002. doi: 10.1373/clinchem.2005.056382. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell G.C., Petrone A.B., Treadway M.B., Tennant C.S., Lucke-Wold N., Chantler P.D., Barr T.L. Machine-learning approach identifies a pattern of gene expression in peripheral blood that can accurately detect ischaemic stroke. npj. Genome Med. 2016;1:16038. doi: 10.1038/npjgenmed.2016.38. The Author(s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi N. Ethnic differences in cardiovascular disease. Heart. 2003;89:681–686. doi: 10.1136/heart.89.6.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajat C., Tilling K., Stewart J.A., Lemic-Stojcevic N., Wolfe C.D.A. Ethnic differences in risk factors for ischemic stroke: a European case-control study. Stroke. 2004;35:1562–1567. doi: 10.1161/01.STR.0000131903.04708.b8. [DOI] [PubMed] [Google Scholar]
- 10.Markus H.S., Khan U., Birns J., Evans A., Kalra L., Rudd A.G., Wolfe C.D.A., Jerrard-Dunne P. Differences in stroke subtypes between black and white patients with stroke: the South London Ethnicity and Stroke Study. Circulation. 2007;116:2157–2164. doi: 10.1161/CIRCULATIONAHA.107.699785. [DOI] [PubMed] [Google Scholar]
- 11.Johnson J.A. Ethnic differences in cardiovascular drug response: potential contribution of pharmacogenetics. Circulation. 2008;118:1383–1393. doi: 10.1161/CIRCULATIONAHA.107.704023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamova B., Jickling G.C., Ander B.P., Zhan X., Liu D.Z., Turner R., Ho C., Khoury J.C., Bushnell C., Pancioli A. Gene expression in peripheral immune cells following cardioembolic stroke is sexually dimorphic. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0102550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross I., Robert G., Ihaka R., Gentleman R.R. A language for data analysis and graphics. Vol. 5. J. Comput. Graph. Stat. 1996:299–314. [Google Scholar]
- 14.Parmigiani G., Garrett E.S., Irizarry R.A., Zeger S.L. The Analysis of Gene Expression Data: Methods and Software New York. Springer New York; NY: 2003. An R package for analyses of affymetrix oligonucleotide arrays. (Statistics for Biology and Health) [Google Scholar]
- 15.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Venables W.N., Ripley B.D. Springer New York; New York, NY: 2002. Modern Applied Statistics with S. (Statistics and Computing) [Google Scholar]
- 17.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinf. 2011;12:1–8. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 19.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995:289–300. [Google Scholar]
- 20.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 21.Azzazy H.M.E. Nanodiagnostics: a new frontier for clinical laboratory medicine. Clin. Chem. 2006;52:1238–1246. doi: 10.1373/clinchem.2006.066654. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Lang H.P., Huber F., Bietsch A., Grange W., Certa U., Mckendry R., Güntherodt H.-J., Hegner M., Gerber C. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nat. Nanotechnol. 2006;1:214–220. doi: 10.1038/nnano.2006.134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary material.