Abstract
The introduction of next-generation sequencing (NGS) had a significant effect on the availability of genomic information, leading to an increase in the number of sequenced genomes from a large spectrum of organisms. Unfortunately, due to the limitations implied by the short-read sequencing platforms, most of these newly sequenced genomes remained as “drafts”, incomplete representations of the whole genetic content. The previous genome sequencing studies indicated that finishing a genome sequenced by NGS, even bacteria, may require additional sequencing to fill the gaps, making the entire process very expensive. As such, several in silico approaches have been developed to optimize the genome assemblies and facilitate the finishing process. The present review aims to explore some free (open source, in many cases) tools that are available to facilitate genome finishing.
Keywords: microbial genetics, molecular microbiology, genomics, microbiology, draft genomes
Introduction
The advent of second generation of sequencing platforms, usually referred to as Next Generation Sequencing (NGS) technologies, promoted an expressive growth in the availability of genomic data in public databases, mainly due to the drastic reduction in the cost-per-base (Chain et al., 2009). Compared to the Sanger sequencing technique (Sanger et al., 1977), NGS platforms, like Illumina HiSeq, IonTorrent PGM, Roche 454 FLX, and ABI SOLiD, are able to generate a significantly higher throughput, resulting in a high sequencing coverage. However, they typically have a lower accuracy (in terms of average Phred-score in the raw data) and, in most cases, can only generate short-length reads (short-reads) (Liu et al., 2012). The term “short-read” is commonly used to refer to the data generated by platforms such as Illumina, IonTorrent and SOLiD, as the length of their reads usually range from 30 bp (eg: SOLiD) to ~120 bp (e.g., Illumina HiSeq), which is smaller than the length usually obtained by Sanger sequencing (~1 kb), and by the PacBio and Oxford Nanopore platforms (commonly referred to as “long-reads”).
The higher throughtput obtained by NGS has stimulated the development of new algorithms and tools that are capable of dealing with a larger volume of data and generating genomic assemblies in a reasonable time. Traditional sequence assemblers, like Phrap (http://www.phrap.org/phredphrap/) and CAP3 (Huang and Madan, 1999), were replaced with new ones, such as Velvet (Zerbino and Birney, 2008), ABySS (Simpson et al., 2009), Ray (Boisvert et al., 2010), SPAdes (Bankevich et al., 2012) and SOAPdenovo (Luo et al., 2012), many of which were based on the De Bruijn graph algorithm (Pevzner et al., 2001; Compeau et al., 2011). A large number of these software offerings are capable of assembling data from different sequencing platforms, called “hybrid assembly”; however, all of them exhibit some limitations and, even after several corrections and optimization steps, the generation of a finished genome continues to represent a complex task (Alkan et al., 2011).
Genome finishing is achieved by converting a set of contigs or scaffolds into complete sequences that represent the full genomic content of the organism without unknown regions (gaps) (Mardis et al., 2002), and aims a “closed” and full representation of the chromosome organization. On the contrary, a draft genome may be composed of a set of a few or several contigs or scaffolds (Land et al., 2015). Although useful for many studies, the unfinished and fragmented nature of draft genomes may difficult analysis on comparative genomics and structural genomics (Ricker et al., 2012). In addition, some genes may be missed if located in a region without coverage (e.g., edges of contigs/scaffolds), or due to assembly errors (e.g., repetitive regions that are “collapsed” into a single one) (Klassen and Currie, 2012).
Genome finishing represents a relevant step to reduce data loss and lead to a more accurate representation of the genomics features of the organism of interest. The traditional way to close gapped genomes includes: (1) design primers based on the edge of adjacent contigs, (2) PCR amplification, (3) Sanger sequencing, and (4) local assembly, usually with (5) manual curation. As this process is very time consuming, new in vitro approaches were developed to speed it up, including multiplex PCR (Tettelin et al., 1999), optical mapping (Latreille et al., 2007), and hybrid sequencing and assembly (Ribeiro et al., 2012). This, however, usually results in a drastic increase in the cost of the sequencing project. Therefore it is not surprising that from the 87,956 prokaryote genomes available in GenBank until December 2016 (GenBank release 217), only a small fraction (~6,586) is finished (http://www.ncbi.nlm.nih.gov/genbank/).
In the last years, the availability of third-generation sequencing technologies, such as PacBio SMRT and Oxford Nanopore, has also provided another way to achieve finished genomes (Ribeiro et al., 2012). As these platforms usually generate reads with length > 10 kb, the assembly algorithms have to deal with less ambiguities and problematic regions (Koren and Phillippy, 2015). This makes the reconstruction of the chromosome sequences easier, but just like second generation technologies, both platforms have their own limitations. In the earlier versions of the PacBio SMRT platform, for example, it used to present a high error-rate, so it was recommended to correct part of these errors using short-reads data (e.g., Illumina) (Koren et al., 2012; Au et al., 2012; Salmela and Rivals, 2014) before using its result for any analysis. Although this problem has been minimized with the improvements in the sequencing chemistry and base-calling process over the last years, the time required for the sequencing, the cost of each run, and the price of the equipment itself are still drawbacks, and these are some of reasons as to why it is not more frequently applied. In the case of Oxford Nanopore, there is a limited number of tools that can be used to pre-process and analyze its data, and some types of errors (e.g., under-representation of specific k-mers) are still recurrent (Deschamps et al., 2016). Therefore, second-generation platforms are still the most popular ones for a wide variety of applications, and remain as the cheaper alternative to obtain genomic data.
Previous reviews that focused on genome assembly and whole-genome sequencing analysis have already described some of the in silico tools that have the potential to improve a genome assembly without the need for experimental data. Edwards and Holt (2013), Dark (2013) and Vincent et al. (2016) have provided very comprehensive descriptions of some of the main steps of data analysis in microbial genomes sequenced by next-generation technologies, including de novo assembly, reference-based contig ordering, genome annotation and comparative genomics, and these are useful starting points for those that are new to microbial genomics with NGS. Nagarajan et al. (2010) have also made some useful considerations regarding genome assembly and presented some methodologies for genome finishing, but limited to a small set of approaches that they have developed to improve assemblies. Ramos et al. (2013) exemplified some assembly and post-assembly methods for genomes sequenced with IonTorrent platforms. Finally, Hunt et al. (2014) have also made a complete and accurate benchmark analysis of different tools for assembly scaffolding based on paired-end/mate-pair data. However, although very instructive, these reviews provide descriptions of some specific procedures, but do not give an in-depth view of the different finishing strategies, nor of the algorithms that are used in background by different tools from each category. Therefore, a more complete description of these tools may be useful, especially for those researchers that are starting to work with microbial genome analysis and want to achieve better assemblies without relying on re-sequencing or manual gap-closing.
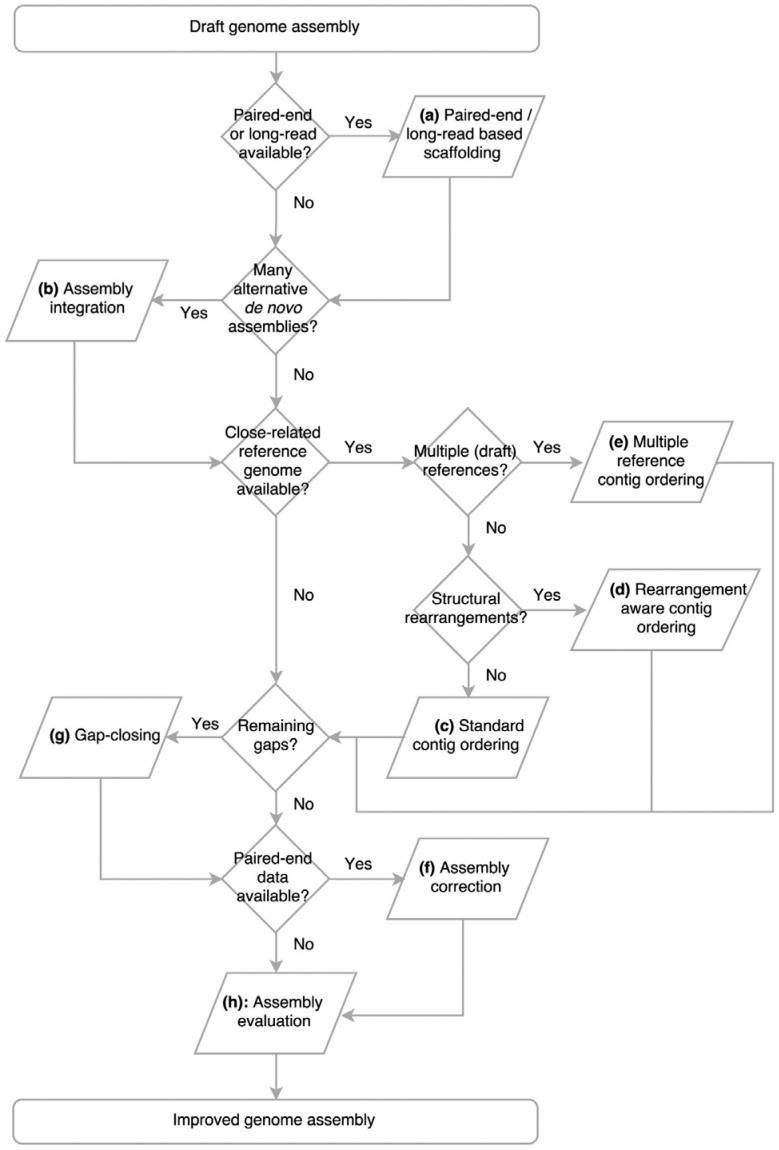
The present review aims at discussing some of the categories of software that may be applied in the process of assembly finishing, especially in the context of microbial genome sequencing projects. A particular focus is placed on those that do not require additional experimental and/or new sequencing data. The review intentionally focuses on tools that are freely available, at least for academic use, and omits proprietary software, like the CLC Genomics Workbench (http://www.clcbio.com/), for example. This was not due to anticipated differences in the performance or quality of the results, but to adhere to the original intention: an overview of tools that could be used by most researchers. As different approaches have been developed to improve genomic assemblies, the description of these programs was divided into four main categories: scaffolding (placement of contigs into larger sequences by using experimental data or information for closely-related genomes, and joining them by gap regions), assembly integration (generation of a consensus assembly using multiple assemblies for the same genome), gap closing (solving gaps by identifying their correct sequence), error correction (removal of misidentified bases or misassembled regions) and assembly evaluation (quantification of the reliability of a genome assembly and identification of its erroneous regions) (Table 1). These different categories of programs can be combined, according to the type of data that is available (e.g., sequencing platform and library that was used), and may help to reduce the fragmentation and improve the reliability of a genome assembly. Based on the categories of software that are discussed in the present review we have created the flowchart showed in Figure 1, which may of help in choosing the most appropriate approach for genome finishing, depending on the type of the data that is available. Examples of genome projects that used some of the tools discussed in the present review are shown in Supplementary material (Table S1 (179KB, pdf) ).
Table 1. Overview of the tools described in the present review.
| Category | Tool | Main features | Dependences* | Reference | Download link / webserver |
|---|---|---|---|---|---|
| Scaffolding | ABySS |
|
boost libraries: www.boost.org/ Open MPI: http://www.open-mpi.org sparse-hash library: http://goog-sparsehash.sourceforge.net/ |
(Simpson et al., 2009) | http://www.bcgsc.ca/platform/bioinfo/software/abyss |
| Scaffolding | Bambus 2 |
|
AMOS package (Treangen et al., 2011): http://amos.sourceforge.net/ |
(Koren et al., 2011) | https://sourceforge.net/projects/amos/ |
| Scaffolding | MIP |
|
lpsolve library: http://sourceforge.net/projects/lpsolve/ lemon library: http://lemon.cs.elte.hu/ |
(Salmela et al., 2011) | https://www.cs.helsinki.fi/u/lmsalmel/mip-scaffolder/ |
| Scaffolding | OPERA |
|
BWA (Li and Durbin 2009): http://bio-bwa.sourceforge.net/ Bowtie (Langmead et al., 2009): http://bowtie-bio.sourceforge.net/ Samtools (Li et al., 2009): http://samtools.sourceforge.net/ |
(Gao et al., 2011) | https://sourceforge.net/projects/operasf |
| Scaffolding | SCARPA |
|
None | (Donmez and Brudno, 2013) | http://compbio.cs.toronto.edu/hapsembler/scarpa.html |
| Scaffolding | SGA |
|
Bamtools (Barnett et al., 2011): https://github.com/pezmaster31/bamtools BWA (Li and Durbin, 2009): http://bio-bwa.sourceforge.net/ Samtools (Li et al., 2009): http://samtools.sourceforge.net/ Sparse-hash library: http://goog-sparsehash.sourceforge.net/ |
(Simpson and Durbin, 2012) | https://github.com/jts/sga |
| Scaffolding | SOPRA |
|
None | (Dayarian et al., 2010) | http://www.physics.rutgers.edu/~anirvans/SOPRA/ |
| Scaffolding | SSPACE |
|
None | (Boetzer et al., 2011) | http://www.baseclear.com/genomics/bioinformatics/basetools/ |
| Scaffolding | SSPACE-LongRead |
|
None | (Boetzer and Pirovano, 2014) | http://www.baseclear.com/genomics/bioinformatics/basetools/ |
| Scaffolding | MUMmer |
|
(Kurtz et al., 2004) | http://mummer.sourceforge.net/ | |
| Scaffolding | ABACAS |
|
MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012): http://primer3.ut.ee/ |
(Assefa et al., 2009) | http://abacas.sourceforge.net/ |
| Scaffolding | CONTIGuator |
|
ABACAS (Assefa et al., 2009): http://abacas.sourceforge.net/ BioPython (Python package): http://biopython.org/ BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012): http://primer3.ut.ee/ |
(Galardini et al., 2011) | http://contiguator.sourceforge.net/ |
| Scaffolding | Mauve |
|
Java: https://www.java.com/ |
(Darling et al., 2004; Rissman et al., 2009) | http://darlinglab.org/mauve/mauve.html |
| Scaffolding | FillScaffolds |
|
Java: https://www.java.com/ |
(Muñoz et al., 2010) | Supplementary data of Muñoz et al. (2010). http://dx.doi.org/10.1186/1471-2105-11-304 |
| Scaffolding | SIS |
|
MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Dias et al., 2012) | http://marte.ic.unicamp.br:8747. |
| Scaffolding | CAR |
|
MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ PHP: https://php.net/ |
(Lu et al., 2014) | http://genome.cs.nthu.edu.tw/CAR/ |
| Scaffolding | RACA |
|
None | (Kim et al., 2013): | http://bioen-compbio.bioen.illinois.edu/RACA/ |
| Scaffolding | Ragout |
|
Networkx (Python package): http://networkx.github.io/ Newick (Python package): http://www.daimi.au.dk/~mailund/newick.html Sibelia: http://github.com/bioinf/Sibelia |
(Kolmogorov et al., 2014) | https://github.com/fenderglass/Ragout |
| Scaffolding | MeDuSa |
|
BioPython (Python package): http://biopython.org/ Java: https://www.java.com/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Bosi et al., 2015) | https://github.com/combogenomics/medusa |
| Assembly integration | Minimus |
|
AMOS package (Treangen et al., 2011): http://amos.sourceforge.net/ |
(Sommer et al., 2007) | https://sourceforge.net/projects/amos/ |
| Assembly integration | Reconciliator |
|
MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Zimin et al., 2008) | http://www.genome.umd.edu/ |
| Assembly integration | MAIA |
|
Matlab: https://www.mathworks.com/ MUMmer: http://mummer.sourceforge.net/ GAIMC (Matlab toolbox): http://github.com/dgleich/gaimc |
(Nijkamp et al., 2010) | http://bioinformatics.tudelft.nl |
| Assembly integration | CISA |
|
BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Lin and Liao, 2013) | http://sb.nhri.org.tw/CISA/ |
| Assembly integration | GAA |
|
BLAT (Kent, 2002): https://genome.ucsc.edu/ GSMapper: http://454.com/ |
(Yao et al., 2012) | http://sourceforge.net/projects/gaa-wugi/ |
| Assembly integration | Mix |
|
Networkx (Python package): http://networkx.lanl.gov/ BioPython (Python package): http://biopython.org/ MUMmer(Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Soueidan et al., 2013) | https://github.com/cbib/MIX |
| Assembly integration | GAM / GAM-NGS |
|
cmake: https://cmake.org/ zlib library: http://www.zlib.net/ boost libraries: www.boost.org/ sparse-hash library: http://goog-sparsehash.sourceforge.net/ |
(Casagrande et al., 2009; Vicedomini et al., 2013) | https://github.com/vice87/gam-ngs |
| Assembly integration | Zorro |
|
AMOS (Treangen et al., 2011): http://amos.sourceforge.net/ BioPerl (Perl module): http://bioperl.org Bowtie (Langmead et al., 2009): http://bowtie-bio.sourceforge.net/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Argueso et al., 2009) | http://lge.ibi.unicamp.br/zorro/ |
| Gap closing | GapCloser |
|
None | (Li et al., 2010) | http://soap.genomics.org.cn/ |
| Gap closing | IMAGE |
|
None | (Tsai et al., 2010) | https://sourceforge.net/projects/image2 |
| Gap closing | GapFiller |
|
None | (Boetzer and Pirovano, 2012) | http://www.baseclear.com/genomics/bioinformatics/basetools |
| Gap closing | Enly |
|
BioPython (Python package): http://biopython.org/ BLAST and BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ Cdbfasta/cdbyank: http://compbio.dfci.harvard.edu/tgi/software/ EMBOSS: http://emboss.sourceforge.net/ Minimo assembler (Treangen et al., 2011): http://amos.sourceforge.net/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ Phrap: http://www.phrap.org/phredphrapconsed.html |
(Fondi et al., 2014) | http://enly.sourceforge.net/ |
| Gap closing | FGAP |
|
Matlab: https://www.mathworks.com/ |
(Piro et al., 2014) | http://www.bioinfo.ufpr.br/fgap/ |
| Gap closing | Sealer |
|
boost libraries: www.boost.org/ sparse-hash library: http://goog-sparsehash.sourceforge.net/ Open MPI: http://www.open-mpi.org |
(Paulino et al., 2015) | https://github.com/bcgsc/abyss/tree/sealer-release |
| Gap closing | GMCLoser |
|
MUMmer (Kurtz et al. 2004): http://mummer.sourceforge.net/ BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ Bowtie (Langmead et al., 2009): http://bowtie-bio.sourceforge.net/ YASS (Noé and Kucherov, 2005): http://bioinfo.lifl.fr/yass |
(Kosugi et al., 2015) | https://sourceforge.net/projects/gmcloser/ |
| Gap closing | MapRepeat |
|
BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ BioPython (Python package): http://biopython.org/ MIRA: http://mira-assembler.sourceforge.net MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(Mariano et al., 2015) | http://github.com/dcbmariano/maprepeat |
| Gap closing | GapBlaster |
|
BLAST and BLAST+ (Altschul et al., 1990;
Camacho et al.,
2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ |
(de Sá et al., 2016) | https://sourceforge.net/projects/gapblaster2015/ |
| Assembly evaluation | REAPR |
|
File::Basename, File::Copy,
File::Spec, File::Spec::Link, Getopt::Long and List::Util (Perl modules): http://www.cpan.org/ R: https://www.r-project.org/ |
(Hunt et al., 2013) | http://www.sanger.ac.uk/science/tools/reapr |
| Assembly evaluation | QUAST |
|
boost libraries: www.boost.org/ Java: https://www.java.com/ Matplotlib (Python package): http://matplotlib.org Time::HiRes (Perl module): http://www.cpan.org/ |
(Gurevich et al., 2013) | http://bioinf.spbau.ru/quast |
| Assembly evaluation | ALE |
|
Matplotlib (Python package): http://matplotlib.org Mpmath (Python package): http://mpmath.org Numpy (Python package): http://www.numpy.org Pymix (Python package): http://www.pymix.org/pymix Setuptools (Python package): https://github.com/pypa/setuptools |
(Clark et al., 2013) | http://www.alescore.org |
| Assembly evaluation | CGAL |
|
None | (Rahman and Pachter, 2013) | http://bio.math.berkeley.edu/cgal/ |
| Assembly evaluation | GMvalue |
|
MUMmer (Kurtz et al., 2004): http://mummer.sourceforge.net/ BLAST+ (Altschul et al., 1990; Camacho et al., 2009): ftp://ftp.ncbi.nlm.nih.gov/blast/ Bowtie (Langmead et al., 2009): http://bowtie-bio.sourceforge.net/ YASS (Noé and Kucherov, 2005): http://bioinfo.lifl.fr/yass |
(Kosugi et al., 2015) | https://sourceforge.net/projects/gmcloser/ |
| Assembly correction | iCORN |
|
SNP-o-matic (Manske and Kwiatkowski, 2009): https://snpomatic.svn.sourceforge.net/svnroot/snpomatic SSAHA Pileup (Ning et al., 2001): ftp://ftp.sanger.ac.uk/pub/zn1/ssaha_pileup/ |
(Otto et al., 2010) | http://icorn.sourceforge.net/ |
| Assembly correction | SEQuel |
|
Java: https://www.java.com/ JGraphT (Java library): http://jgrapht.org/ |
(Ronen et al., 2012) | http://bix.ucsd.edu/SEQuel/ |
| Assembly correction | GFinisher |
|
Java: https://www.java.com/ |
(Guizelini et al., 2016) | http://gfinisher.sourceforge.net/ |
= Considering a computer running UNIX, Linux or Mac OS operating systems (OSs). As Make, sed, awk, GCC, Perl, Bash, Python and the GNU/Unix standard utility set are already included in most of the distributions / versions of these OSs, these programs were not listed as dependences.
Figure 1. A flowchart demonstrating how and when the different genome finishing approaches can be combined according to the data that is available for the user. (a) Scaffolding using paired-end reads or long-reads, which is directly dependent on the way the genome was sequencing (platform, library), and sometimes performed as part of the de novo assembly process. (b) Assembly integration, which consists in the combination of different de novo assemblies and generation of a consensus/extended assembly. Some programs use only the assemblies as input, while others use also the sequencing reads. (c) The standard contig-ordering approach based on a single reference genome, which consists in the identification of synteny blocks that guide the orientation of the contigs in the draft genome, without taking into count the occurrence of genome inversions other rearrangements. (d) The rearrangement-aware contig-ordering, that identifies potential sites of inversion and translocations based on signatures on the alignment against the reference genome. (e) The multiple-reference contig ordering, that may be more appropriate in those cases where there is no finished reference genome, but there is a relatively high number of close-related drafts, or when there are no apparent closest reference to be used. (f) Assembly correction, which consists in the removing of short misassemblies, including base-substitutions and short insertions and deletions. (g) Gap-closing, which consists in the joining of adjacent contigs that used to be spaced by a gap. (h) Assembly evaluation, which may provide help to access the reliability of the assembly.
Scaffolding
By definition, a contig consists of a contiguous sequence has no unknown regions or assembly gaps (but may contain “N” that represent base-calling errors) (Staden, 1979). On the other hand, a scaffold consists of two or more contigs that have been joined according to some linkage information (e.g., paired-end reads, genome maps) (Huson et al., 2002). Paired-end or mate-pair libraries can be very useful in de novo genome assembly, and several tools use the relative position information to connect contigs into scaffolds (Hunt et al., 2014). In a similar way, with the increase in the availability of genomic sequences from a wide variety of species, other scaffolding alternatives were developed to use one or multiple genomes as reference to order the contigs.
Paired-end scaffolding
Most of the de novo genome assemblers usually integrate scaffolding steps after the contig constructions, although it is also possible to use third-party tools aiming a more reliable result. The A5 assembly pipeline (Tritt et al., 2012), for example, uses de novo assembler IDBA (Peng et al., 2010) to construct the contigs and SSPACE (Boetzer et al., 2011) to generate scaffolds. The scaffolding with paired-end reads usually consist of the alignment of reads to the contigs, followed by the identification of connections between different contigs using the relative-orientation information and the estimated insert-size. ABySS (Simpson et al., 2009), SOPRA (Dayarian et al., 2010), SOAPdenovo (Li et al., 2010), Bambus 2 (Koren et al., 2011), MIP (Salmela et al., 2011), Opera (Gao et al., 2011), SSPACE (Boetzer et al., 2011), SLIQ (Roy et al., 2012), SGA (Simpson and Durbin 2012), SCARPA (Donmez and Brudno 2013), WiseScaffolder (Farrant et al., 2015) and ScaffoldScaffolder (Bodily et al., 2016) are examples of scaffolding tools based on paired-end information. More recently, the use of long-reads was also incorporated into scaffolding tools such as AHA (Bashir et al., 2012) and SSPACE-LongRead (Boetzer and Pirovano, 2014).
ABySS (Simpson et al., 2009): The program abyss-scaffold, which comes with the ABySS assembly package (Simpson et al., 2009), uses the estimated mate-distance distribution in paired-end reads to connect contigs and generate scaffolds. The distance distribution can be calculated by DistanceEst, that is also part of the package, and is also used by other assembly pipelines, such as SGA (Simpson and Durbin, 2012). ABySS was developed to be used both with small and large genomes, and can be executed in a computer clustering by using the Message Passing Interface (MPI), this being useful also in case of high-coverage data and when dealing with multiple libraries. Like most scaffolding programs, abyss-scaffold can be used also in the scaffolding of contigs generated by third-party programs. Finally, ABySS also supports scaffolding with long-reads by using BWA-MEM for read alignment (Li and Durbin, 2009). The source code of the ABySS package is available at the address http://www.bcgsc.ca/platform/bioinfo/software/abyss, and is developed to work on the Linux operating system.
SOPRA (Dayarian et al., 2010): This scaffolding tool was designed to improve assemblies generated by Velvet (Zerbino and Birney, 2008) and SSAKE (Warren et al., 2007), and targets the earlier sequencing platforms from Illumina and ABI SOLiD. The program parses the read-placing file generated by these assemblers and extracts information of paired-end/mate-pair reads, that is used to calculate the mean distance between pairs and the correct orientation. Based on this file, SOPRA also infers the connections between contigs by searches of those pairs of reads where mates are in different contigs. The program is not fully automated, so each step of data processing must be executed by a different script before the main scaffolding process. Another drawback is the limited support for different de novo assemblers, as it requires read-placing files in AFG format, and this is only produced by a few assemblers nowadays (e.g., Velvet, Ray and AMOS). SOPRA can be obtained from the website http://www.physics.rutgers.edu/~anirvans/SOPRA/.
Bambus 2 (Koren et al., 2011): It is part of the AMOS package (Treangen et al., 2011) and is both a genome and metagenome scaffolding tool and an updated version of the Sanger-based program Bambus targeting NGS data (Pop et al., 2004). The program requires read-placing information to construct a contig-graph, and explore the graph to find consistent connections between the contigs. As Bambus2 can also be used to scaffold metagenomic assemblies, different from other programs, it considers the effect of DNA samples containing mixes of closely related organisms in the assembly processes and reduces the chance of fragmentation and miss-joining by analyzing the molecular variants. However, the use of Bambus 2 is not as simple as for other scaffolding tools, as it requires some experience with the AMOS tools to generate its input file and processes the outputs (Treangen et al., 2011). The AMOS package can be obtained from the SourceForge repository: https://sourceforge.net/projects/amos/.
MIP (Salmela et al., 2011): uses the concept of mixed integer programming to generate a set of scaffolds from a genome assembly and a set paired-ends/mate-pair reads. First, readaligner (Mäkinen et al., 2010) is used to map the read-pairs back to the contigs. Then, the pairs are filtered to remove inconsistent connections, and the distances between the contigs are estimated based on the mean distance between the mates, which is calculated for each library used in the assembly. The connections in the generated scaffold graph have a minimum and maximum estimated length, derived from the library information. The MIP source code, along with usage instructions, is available at https://www.cs.helsinki.fi/u/lmsalmel/mip-scaffolder/.
Opera (Gao et al., 2011): takes as inputs a collection of contigs and mapped reads and generates a scaffold graph based on the paired-end information. Frist, the program filters the connections between contigs to remove possible miss-joining errors caused by chimeric pairs. The graph is contracted, and the optimum orientation of the contigs inside the scaffolds is inferred by a dynamic programming algorithm that explores the search space. The algorithm can also infer the occurrence of repeated genomic regions, usually assembled into a single contig in case of short-reads. In this case, repeated regions are identified by comparing the coverage of the contigs to the mean coverage of the whole genome and selection those with value greater than 1.5 times the genomic mean. The identification of these regions allows a contig to be used in more than one scaffolds, which can provide a better assembly of repeated regions, but can also result in misassemblies. Opera can be obtained from its SourceForge repository https://sourceforge.net/projects/operasf/.
SGA (String Graph Assembler) (Simpson and Durbin 2012): is a de novo genome assembler developed for the memory-efficient assembly of small and large genomes by applying the method proposed by Myers (2005). As part of its assembly pipeline, SGA also provide a scaffolding tool that uses information from read alignment (in .BAM format), that can be generated by a wide variety of mapping tools (Li et al., 2008; Langmead et al., 2009; Lunter and Goodson, 2011), and estimated distance between mates, generated by DistanceEst, from the ABySS package, to connect contigs into scaffolds. SGA also supports scaffolding from multiple libraries, with different insert sizes, and was optimized to work with Illumina data. SGA is available from the GitHub repository https://github.com/jts/sga.
SCARPA (Donmez and Brudno, 2013): uses paired-end information to generate scaffolds, but takes into account that not only chimeric reads may be to responsible for inconsistent linkages between contigs but also misassembled sequences. It estimates the mean and standard deviation of the distance between the mates, but only uses information from those contigs with length greater than the assembly N50. The connections between the contigs are estimated based on the mate information and the calculated metrics, and if more than one library is provided, SCARPA process the scaffolding iteratively starting from the library with smaller insertion size. The program can be obtained from the URL http://compbio.cs.toronto.edu/hapsembler/scarpa.html.
SSPACE (Boetzer et al., 2011) and SSPACE-LongRead (Boetzer and Pirovano, 2014): these scaffolding programs are currently distributed by BaseClear (http://www.baseclear.com), which also distributes the gap-closing program GapFiller (Boetzer and Pirovano, 2012). SSPACE requires information about paired-end library, including mean and standard deviation of distance between the mates and the expected orientation, whose values can be predicted with the script “estimate_insert_size.pl”, distributed along with the program. The user may choose between BWA (Li and Durbin, 2009) and Bowtie (Langmead et al., 2009) for read mapping, the minimum number of connections to link two contigs, the number of bases that will be removed from the border of the contigs (as they usually contain errors), and the number of iterations. For SSPACE-LongRead, the target assembly is aligned to a collection of long-reads using BLASR (Chaisson and Tesler, 2012) and the alignments are filtered and refined to find the best orientation. Both SSPACE and SSPACE-LongRead can be requested from the BaseClear website http://www.baseclear.com/genomics/bioinformatics/basetools/.
Hunt el al. (2014) have performed an extensive comparison of the scaffolding tools and demonstrated that the quality of the resulting scaffolds is directly affected by the read-mapping program and the complexity of the genome. For the tested datasets, the best results were obtained by SGA, SOPRA and SSPACE, although all tested tools presented a certain percentage of miss-joined scaffolds in their outputs. Some scaffolding tools (e.g., SGA) use pre-aligned reads as input, so the user is able to test and choose the read mapper. In this case, it is important to try different read mappers, taking into account that platform-specific bias and read quality may have a drastic effect on the quality of the alignment (Hatem et al., 2013; Caboche et al., 2014). When using mate-pair libraries (long-insert paired-end reads) it is also important to check if the scaffolder was designed to support it, or if it was just designed for standard, short-insert, paired-end reads. As mate-pairs may present a relatively high rate of “false mates”, special care may be taken when working with this type of data.
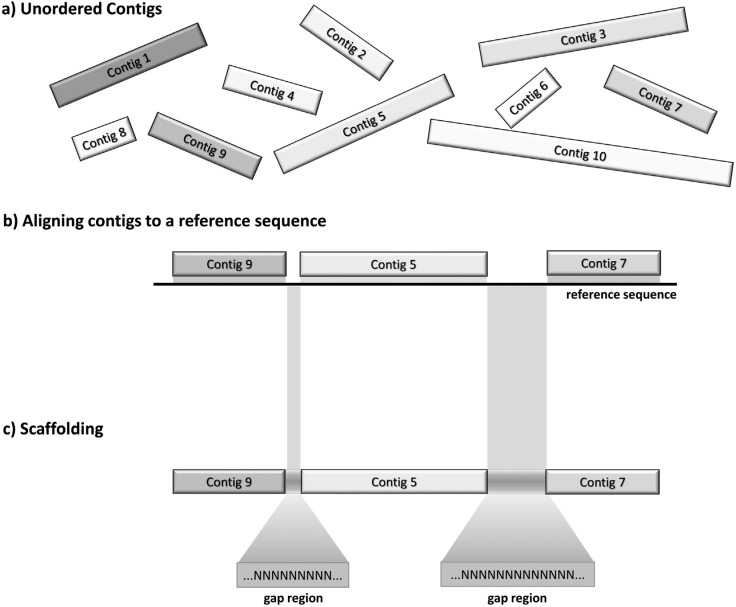
Single reference-based scaffolding
In many cases the pairing information is not enough to generate a reliable reconstruction of the genome's structure, or simply, the genome was not sequenced using paired-reads, but with single-end sequencing. In order to overcome this, some tools were developed to use a reference genome as a template to perform the contig ordering and relative positioning (Figure 2). Software like MUMmer (Kurtz et al., 2004), ABACAS (Assefa et al., 2009), CONTIGuator (Galardini et al., 2011) and Mauve (Darling et al., 2004; Rissman et al., 2009) are able to identify the most probable orientation of the contigs, but may generate incorrect results in the case of genome inversions of translocations. On the other hand, SIS (Dias et al., 2012), CAR (Lu et al., 2014), and FillScaffolds (Muñoz et al., 2010) consider the occurrence of changes in the genomic structure and take these phenomena into account during the analysis, generating a more accurate reconstruction. All these tools use information from a single genome as reference, however more recently, some tools, such as Ragout (Kolmogorov et al., 2014) and MeDuSa (Bosi et al., 2015), were developed to use information from multiple reference genomes, allowing an evolutionary-based inference of structural re-arrangements. These multiple reference-based contig ordering tools will be discussed in the next section.
Figure 2. Reference-based contig ordering. (a) The program takes a set of contigs (or scaffolds) and (b) aligns these to a reference genome to identify the most probable relative orientation of the sequences in the draft genome. (c) Regions not covered by the contigs represent gaps and may be sequencing/assembling artifacts or natural deletions. Based on the relative position of each contig, a scaffold is created.
MUMmer (Kurtz et al., 2004): is a genome-scale sequence alignment tool which can be applied to perform the alignment of a set of contigs/scaffolds to a reference genome, allowing a wide variety of applications in genomic analysis and NGS data processing, including reference-guided scaffolding. The two main algorithms of the MUMmer package are NUCmer, which performs a standard DNA-DNA alignment, and PROmer, which performs an alignment of the six reading frames of both sequences (leading to a more sensitive result, especially in the case of highly divergent organisms). The package also includes other tools, such as delta-filter, that can be used to remove the ambiguities in the alignments and select those that are more relevant for the analysis. Many scaffolding tools, like ABACAS (Assefa et al., 2009), CONTIGuator (Galardini et al., 2011) and MeDuSa (Bosi et al., 2015), are built on top of MUMmer and take advatange of its performance, but also add new features to improve the output. MUMmer itself does not provide the sequence of the scaffold, just the positions of the alignments. Therefore, it is necessary to perform a post-processing of the results to obtain the sequence of the scaffolds. MUMmer can be obtained from its SourceForge repository http://mummer.sourceforge.net/.
ABACAS (Algorithm-based Automatic Contiguation of Assembled Sequences) (Assefa et al., 2009): can use NUCmer or PROmer from the MUMmer (Kurtz et al., 2004) package to align the contigs against a reference genome. The regions that do not have an equivalent sequence in the contig set are filled with Ns, indicating gaps. ABACAS can also be used to design PCR primers to amplify the unknown regions by integrating Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012). ABACAS can be obtained from its SourceForge repository http://abacas.sourceforge.net/, and as part of the PAGIT package (Swain et al., 2012), available at http://www.sanger.ac.uk/science/tools/pagit.
CONTIGuator (Galardini et al., 2011): uses ABACAS (Assefa et al., 2009) to perform the contig ordering, but adds support to multiple references, which may be useful in the case of organisms that have more than one chromosome. BLAST (Altschul et al., 1990; Camacho et al., 2009) is used to align the contigs used as input with the reference sequences to identify the correct reference for each sequence. Then, ABACAS (Assefa et al., 2009) is used, and its results are integrated with the BLAST alignment to generate a final assembly. CONTIGuator can be obtained from its SourceForge repository http://contiguator.sourceforge.net/, and is also available as a webserver http://combo.dbe.unifi.it/contiguator.
Mauve (Darling et al., 2004; Rissman et al., 2009): is an alignment tool that can handle and align multiple genomes and identify regions of high similarity called Locally Collinear Blocks (LCBs). One of the program's features, Mauve Contig Mover, performs contig ordering using the same algorithm (Rissman et al., 2009). The program runs in an iterative mode, generating and optimizing the contig orientations based on the reference until no change is possible that can improve the model. A directory is generated for each iteration that contains inputs to visualize the genome in Mauve and a FASTA file with the sorted contigs. The Mauve aligner can be obtained from the URL http://darlinglab.org/mauve/mauve.html.
FillScaffolds (Muñoz et al., 2010): analyzes the genomic distance between the contig set and a reference genome and generates an ordered sequence through identifying orthologous genes. It considers the effects of the evolutionary distance in the case of missing genes, and then uses the position of the orthologos present in the reference to order the contigs. The source code of FillScaffolds is available as a supplementary data of the Muñoz et al. (2010) paper at: http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-304.
SIS (Scaffolds from Inversion Signatures) (Dias et al., 2012): takes as input a set of contigs in FASTA format and a coordinate file generated by NUCmer or PROmer (Kurtz et al., 2004) after these contigs have been aligned with the reference sequence. Using the coordinates, the program searches for inversion signatures and generates a collection of orientations of the sequences that can be used to construct the scaffolds. The source code of SIS can be obtained from the URL http://marte.ic.unicamp.br:8747.
CAR (Contig Assembly using Rearrangements) (Lu et al., 2014): uses NUCmer and PROmer in combination, unlike ABACAS (Assefa et al., 2009) and SIS (Dias et al., 2012), that use the result of only one. Based on the coordinates, CAR uses a block permutation model to generate the contig order by considering not only the effect of the genomic inversions, but also the occurrence of transpositions (Li et al., 2013). CAR can be used from the webserver http://genome.cs.nthu.edu.tw/CAR/, where the source code is also available for download.
Considering the main algorithm of each program, it is important to keep in mind that the most appropriate tool for a given task will depend on the organism and the availability of the reference genomes. ABACAS (Assefa et al., 2009) is very useful if the reference genome is larger than the target genome (considering the sum of the length of all contigs, and that all contigs have a homologous region in the reference), and the primer designing tools might be helpful in some cases; however, its sensibility decreases in cases of structural divergence. In such cases, other tools, like CONTIGuator (Galardini et al., 2011) and Mauve (Darling et al., 2004; Rissman et al., 2009), may be more effective. Finally, SIS (Dias et al., 2012) and CAR (Lu et al., 2014) are indicated if the draft genome may present genomic inversions or transpositions. For most of the applications, these tools usually provide reliable results, especially for organisms that do not show a very variable genomic organization, and/or when there are enough finished genomes to properly choose the best reference. However, in some situations it may be necessary to evaluate different tools and references to check which one provides the best results. Finally, a single-reference may also lead to an “overfitted” ordering, especially when the reference is smaller, or in case of genomic inversions and translocations.
Multiple reference scaffolding
Sometimes it is very difficult to identify the most appropriate reference genome to use for contig ordering, especially when structural rearrangements are common events in the genus/species of interests. Additionally, when using BLAST to identify the most “close-related” strain from a database of already finished genomes, it is not usual to find different strains as best hit for each contig. Finally, there are also those cases where no finished genome is available, but there are draft genomes of related strains. In these cases it would not be appropriate to use programs that take into account alignments against only one reference, so data from multiple organisms should be considered. The use of multi-references is relatively recent and another consequence of the advent of NGS, as there is more draft genomes than finished ones available in public databases. Examples of algorithms and programs that use this approach are RACA (Kim et al., 2013), Ragout (Kolmogorov et al., 2014) and MeDuSa (Bosi et al., 2015).
RACA (Reference-Assisted Chromosome Assembly) (Kim et al., 2013): uses local sequence alignment to identify co-linear synteny blocks. The synteny blocks are filtered using a length threshold, and based on the reference genomes, the probability of each synteny block adjacent to the others is calculated. This probability can also be combined with paired-end information to identify the most probable set of scaffold. The source code of RACA can be obtained from the URL http://bioen-compbio.bioen.illinois.edu/RACA/.
Ragout (Kolmogorov et al., 2014): uses phylogenetic information and synteny blocks to order a set of contigs from a target genome using multiple genome references. First, Sibelia (Minkin et al., 2013) is used to identify synteny blocks shared by the target and the reference sequences. Based on the synteny, the nucleotides of the genomes are represented as sequences of blocks, and the best “block orientation” is identified by a maximum parsimony, taking into account the block order in the reference genomes. The source code of Ragout can be obtained from its repository at GitHub https://github.com/fenderglass/Ragout.
MeDuSa (Multi-Draft based Scaffolder) (Bosi et al., 2015): is a graph-based scaffolder that uses information from multiples references, which can be finished or draft genomes. The program uses NUCmer to alignment the target genomes to the references and construct a weighted graph based on the alignments were the nodes of the graph are connected by identifying those contigs that aligned to the same sequence in the references. In the next step, the orientation of each contig is assigned based on the alignment information and the most-probable ordered identified in the graph. The source code of MeDuSa can be obtained from the repository at GitHub https://github.com/combogenomics/medusa.
Assembly integration
Different assemblers, or even the same assembler executed with different configurations, may produce different results. Minimum coverage, coverage cut-offs, minimum contig length, and k-mer size are examples of just some parameters that can affect the decisions of the assembler during the construction of the contigs (Baker, 2012). The way low-quality reads are treated, or how the correct paths in the assembly graph are constructed, is also different for each program. As different assemblies may present different representations of a given region in the genome, the construction of a “consensus assembly” can be an effective method of reducing assembly errors and generating an optimized set of contigs. This process, which is sometimes called “assembly reconciliation,” “assembly merging,” or “assembly integration,” can receive as input only a set of assemblies, as implemented in Minimus (Sommer et al., 2007), Reconciliator (Zimin et al., 2008), MAIA (Nijkamp et al., 2010), CISA (Lin and Liao 2013), GAA (Yao et al., 2012) and Mix (Soueidan et al., 2013), or both a set of assemblies and the reads used for the assembly, as is the case with GAM-NGS (Vicedomini et al., 2013) and Zorro (Argueso et al., 2009).
Minimus (Sommer et al., 2007): is an assembly tool from the AMOS package (Treangen et al., 2011). Initially conceived to perform assembly of small genomes, it was posteriorly adapted for assembly integration. The main algorithm is based on the overlap-layout-consensus paradigm (Peltola et al., 1984), which involves taking a set of sequences and performing several alignments to identify overlaps. The information provided by the alignments is used to construct a graph that is minimized by a combination of algorithms (Myers, 1995, 2005) to generate a final assembly. Minimus is available as part of the AMOS package, which can be obtained from the SourceForge repository https://sourceforge.net/projects/amos/.
Reconciliator (Zimin et al., 2008): uses NUCmer, from the MUMmer package (Kurtz et al., 2004), to identify assembly errors by comparing a template with a secondary assembly. With the alignments, the tool is able to identify the regions that have possibly suffered compression or expansion due to assembly errors in repetitive DNA sequences. The source code of Reconciliator is available from the URL http://www.genome.umd.edu/.
MAIA (Multiple Assembly Integrator) (Nijkamp et al., 2010): uses the overlap-layout-consensus paradigm in a similar way to Minimus to construct a graph based on the overlaps identified by MUMmer (Kurtz et al., 2004). The connections in the graph are used to construct a new assembly, and contigs that have no connection can be integrated with the assembly using a reference genome as a template. MAIA was implemented on top of the Matlab programming language and is available as a package for it that can be obtained from the URL http://bioinformatics.tudelft.nl.
GAA (Graph Accordance Assembly) (Yao et al., 2012): is an assembly integration software that is based on a homonymous data structure. Taking a set of contigs as input, the tool uses BLAT (Kent, 2002) to generate alignments, identify overlaps and then generate a graph that represents the connections between the contigs. GAA is available from the SourceForge repository http://sourceforge.net/projects/gaa-wugi/.
CISA (Contig Integrator for Sequence Assembly) (Lin and Liao, 2013): uses a four-step algorithm to generate the merged assembly. First, a set of representative contigs is chosen from the individual assemblies. Assembly errors are identified by aligning all sets to one another, and any regions that are present in only one sequence are considered to be erroneous. In the event of errors, the contigs are broken in the incorrect portion into smaller sequences. The third step consists of generating several alignments using BLAST (Altschul et al., 1990; Camacho et al., 2009), and NUCmer (Kurtz et al., 2004) to identify the optimal length of repetitive sequences. The information generated in the third step is used to construct the merged assembly in the final stage of the program. CISA can be obtained from the URL http://sb.nhri.org.tw/CISA/.
Mix (Soueidan et al., 2013): uses alignments generated by NUCmer (Kurtz et al., 2004) to generate an extension graph where the contigs are connected by their borders. The alignments are filtered to remove repetitive sequences, and this information is used to generate a graph. Finally, the algorithm parses the graph to identify the Maximal Independent Longest Path Set (MILPS) that represents the final assembly. The Mix source code is available at the GitHub repository https://github.com/cbib/MIX.
GAM (Genomic Assemblies Merger) (Casagrande et al., 2009) and GAM-NGS (Genomic Assemblies Merger for Next Generation Sequencing) (Vicedomini et al., 2013): GAM takes an assembly as a template, which is referred to as the “master”, and extends it using one or more sets of auxiliary assemblies (called “slaves”) by identifying blocks inside the sequences that were generated by the same reads. GAM was designed to run using contigs that are generated by Sanger sequencing, and it needs an AFG layout file that contains the position information of the reads used to construct the contigs. AFG files are only produced by a few NGS assemblers, like Velvet (Zerbino and Birney, 2008) and Ray (Boisvert et al., 2010). A newer version, called GAM-NGS (Vicedomini et al., 2013), was developed to work in the modern context of genome assembly and to avoid the requirement for a template file by using a read alignment file (BAM) (Li et al., 2009) instead. Additionally, GAM-NGS can also identify misassemblies and perform corrections before generating the final assembly. The source code of GAM-NGS is available from the GitHub repository https://github.com/vice87/gam-ngs.
Zorro (Argueso et al., 2009): Combines a preprocessing step based on the masking of repetitive DNA in the sequences, followed by the split of possible misassembled regions, and the assembly integration performed by Minimus (Sommer et al., 2007). The misassembled regions are identified by using Bowtie (Langmead et al., 2009) to remap the reads used for the assembly back to the contigs and by analyzing the coverage along the sequence. Zorro can be obtained from the URL http://lge.ibi.unicamp.br/zorro/.
The use of read alignment information, usually taken from BAM/SAM files, may help Assembly integrating tools to properly choose and identify bocks that are derived from the same genomic region, but, just like scaffolding, the quality of the process be directly affected by the sequencing platform and settings of the read mapper. Additionally, assembly integration with paired-end reads is usually much more time and memory consuming than scaffolding, so it is important to its important to check the system's availability before choose use this approach.
Gap closing
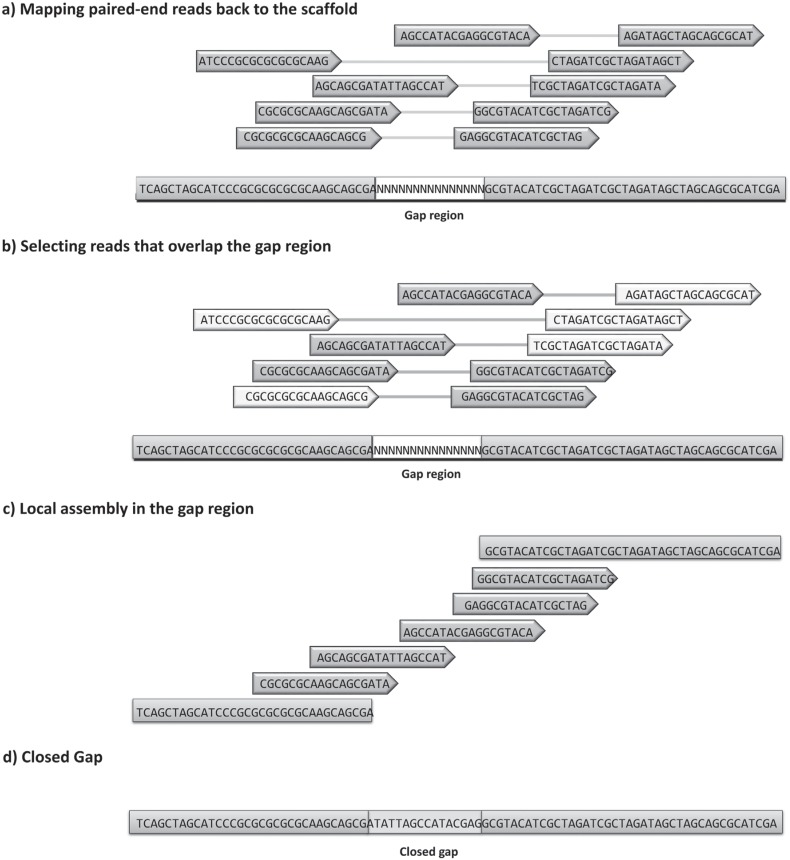
As described in the first section, different kinds of information may be used to generate scaffolds, like paired sequence, optical and/or genomics maps, and a reference genome. To improve the assembly, some algorithms were designed to close the gaps inside the scaffolds. One approach (Figure 3), which is used by tools like GapCloser (Li et al., 2010; Luo et al., 2012), IMAGE (Tsai et al., 2010), GapFiller (Boetzer and Pirovano, 2012), Enly (Fondi et al., 2014), MapRepeat (Mariano et al., 2015) and Sealer (Paulino et al., 2015), utilizes the information of single-end/paired-end reads to extend, and sometimes locally-reassemble, the contigs and close the gaps. Another approach, which is employed by FGAP (Piro et al., 2014) and GapBlaster (de Sá et al., 2016), uses an auxiliary set of contigs to find sequences that may fill the region based on local alignment. Finally, hybrid approaches, such as GMcloser (Kosugi et al., 2015), may use a combination of paired-end data, alternative assemblies and long reads in the gap-closing process.
Figure 3. Example of a gap-closing approach using paired-end reads. (a) Taking as example a scaffold constituted by two contigs joined by an assembly gap (a run of `N's) by remapping the reads back to the contigs (b) it is possible to identify reads that have at least one of the mates in the gap region. Finally, (c) the reads identified inside the gap can be de novo assembled to fill the region, resulting in a (d) closed gap.
GapCloser (Li et al., 2010): is part of the SOAPdenovo software package (Li et al., 2010). This module uses the alignment algorithms from the package to realign the reads to the scaffolds. Based on the reads located near the gap, a local de novo assembly is performed constructed using a De Bruijn algorithm. In SOAPdenovo2 (Luo et al., 2012), the GapCloser module was updated to deal with the possible errors caused by high divergent reads that might be used during the construction of the consensus sequence. GapCloser is distributed both as a stand-alone application or as part of the SOAPdenovo package, and can be obtained from the URL http://soap.genomics.org.cn/.
IMAGE (Iterative Mapping and Assembly for Gap Elimination) (Tsai et al., 2010): takes as inputs a library of Illumina paired-end reads and a set of scaffolds and performs a remapping of the reads back to the sequences using SSAH2 (Ning et al., 2001). Alternatively, the user can provide only the reads, and a de novo assembly is performed by Velvet (Zerbino and Birney, 2008) before the gap-closing step. The program maps the reads to the scaffolds and identifies those that are located at the border. Then, the reads pairs that have at least one of the mates on the border are selected, and a de novo assembly is performed by Velvet using the selected reads and contigs where the reads were mapped as inputs. After the assembly, the contigs are extended, and this increases the probability that the mates will align with the adjacent contigs or the gap will be filled after the assembly increases as more iterations are performed. At the end of the process, the contigs that were linked during the assembly can be ordered according to their previously known relative position. IMAGE https://sourceforge.net/projects/image2/ can be obtained from its SourceForge repository or as part of the PAGIT package www.sanger.ac.uk/science/tools/pagit.
GapFiller (Boetzer and Pirovano 2012): In a similar approach to that used by the previous tools, GapFiller performs a remapping of the paired-end reads back to the scaffolds. However, to run the program, it is necessary to know (or at less have an estimation of) the insert size and the orientation of the library used. If this information is not available, it is possible to calculate it using the script “estimate_insert_size.pl” that is distributed along with SSPACE. To perform the gap filling exercise, the program trims the contigs on both sides to reduce the possible assembly errors caused by low coverage and then applies an iterative process of read remapping and extension of the contigs. The alignment of the reads can be performed using BWA (Li and Durbin, 2009) or Bowtie (Langmead et al., 2009) and the pairs that have one of the mates on the border of the contig and the other inside the gap region are identified. The contig extension uses a k-mer based assembly according to the reads found inside the gap. GapFiller, in the same way than SSPACE, is available for download at the BaseClear website http://www.baseclear.com/genomics/bioinformatics/basetools/.
Enly (Fondi et al., 2014): is a simple gap-closing program that works by re-mapping reads, in FASTA format, back to a target assembly using BLAST (Altschul et al., 1990; Camacho et al., 2009), followed by a local assembly using Phrap (www.phrap.org/) or Minimo, from the AMOS package (Treangen et al., 2011). If a reference genome is provided, a modified version of CONTIGuator (Galardini et al., 2011) is used to order de target assembly and verify the accuracy of the gap-closing. The program can be obtained from the SourceForge repository http://enly.sourceforge.net/.
FGAP (Piro et al., 2014): Unlike the other gap-filling tools, which utilize paired-end reads, FGAP's algorithm uses a supplementary set of long sequences, which can be a library of long-reads or an alternative assembly of the genome. After trimming the contigs, BLASTn, from the NCBI-BLAST+ package (Altschul et al., 1990; Camacho et al., 2009), is used to identify sequences in the supplementary set that overlaps the gap. FGAP can be download and used as a webserver from the URL http://www.bioinfo.ufpr.br/fgap/.
Sealer (Paulino et al., 2015): The program was developed to be applied in large genomes, although can be applied in small prokaryote genomes as well. The main algorithm consists in the selection of nucleotides in the assembly that flank the gap-regions, followed by a local assembly by Konnector (Vandervalk et al., 2014) using data from paired-end reads. Konnector, which takes the paired-end reads and generates pseudo-long reads by applying a combination of Bloom filter and De Bruijn graph, is distributed alongside with the recent versions of ABySS (Simpson et al., 2009). Sealer evokes Konnector with different k-mers, what allows a more efficient closing of gaps caused by low coverage (usually closed by shorter k-mers) and repetitive elements (usually closed by longer k-mers). As it was developed aiming large genomes, Sealer tends to be more memory-efficient, but some steps of the program are executed serially, not in parallel, increasing the computation time. Sealer can be obtained from its GitHub repository https://github.com/bcgsc/abyss/tree/sealer-release.
GMCLoser (Kosugi et al., 2015): combines information from paired-end reads and an alternative assembly, or long-reads, to fill gaps inside scaffolds. Paired-end reads are mapped to the target, and alternative assembly with Bowtie (Langmead et al., 2009) and MUMmer (Kurtz et al., 2004) is used to align the alternative assembly to the target assembly. To reduce the effect of misassembled regions that may be present in alternative assemblies, the program uses a likelihood approach to evaluate the contig joining by verifying the consistence based on the mate information. The likelihood algorithm available to evaluate assemblies can be accessed with the GMValue program, distributed alongside with GMCloser. The program can be obtained from its SourceForge repository https://sourceforge.net/projects/gmcloser/.
MapRepeat (Mariano et al., 2015): is both a gap-closing and reference-guided scaffolding program. This software receives a genome assembly (in FASTA format), a reference genome (in Genbank format) and the sequencing reads (FASTA or FASTQ format). First, the assembly is ordered using the reference genome using CONTIGuator (Galardini et al., 2011), and for each gap the adjacent contigs are aligned back to the reference using BLAST (Altschul et al., 1990; Camacho et al., 2009), aiming the identification of the region in the reference that is analogous to the missing region inside the gap in the target assembly. MIRA (www.chevreux.org) is used to align the reads to the reference and generate a consensus sequence, and the regions analogous to the gap are selected and used to close it. The source code of MapRepeat can be obtained from its GitHub repository http://github.com/dcbmariano/maprepeat.
GapBlaster (de Sá et al., 2016): is a gap-closing programs that, in contrast with most of the other tools, allows a manual curation of the gaps in a draft assembly instead of providing an automated gap-closing algorithm. The user may choose between the legacy NCBI-BLAST package, NCBI-BLAST+ (Altschul et al., 1990; Camacho et al., 2009) or Nucmer (Kurtz et al., 2004) to perform alignments between a set of contigs and the draft genome of interest, and then verify which alignments identified as flanking the gap regions may be considered for gap-closing.
The algorithms implemented in GapCloser (Li et al., 2010; Luo et al., 2012), IMAGE (Tsai et al., 2010) and GapFiller (Boetzer and Pirovano, 2012) are based on paired-end information and are very useful for most of the genomes sequenced using Illumina or Roche 454 platforms. GapFiller requires not only the paired-end reads, but also an estimation of the size, of the insert (and its standard deviation) and the orientation of the read pairs (FR: forward-reverse, RF: reverse-forward, FF: forward-forward or RR: reverse-reverse), however, if such information is not available, it is possible to estimate it using the same script distributed along with SSPACE (Boetzer et al., 2011).
In comparison to Gapfiller, Gapcloser, which also uses paired-end information for gap-closing, requires less information and may provide a more straightforward way to fill gaps. However, GapCloser also has its own limitations, including the lack of support for reads longer than 155 bp (nowadays, 2×250 bp paired-ends reads are very common for whole-genome sequencing, especially for microbial genome sequenced using Illumina MiSeq).
IMAGE is also very easy to be executed. It does not require prior knowledge about the library construction, and only needs the scaffolds, the reads and the number of iterations to be run. As it uses SMALT (based on hashes) instead of BWA or Bowtie (based on Burrows-Wheeler compression), the read-mapping also tends to be more sensitive and able to identify alignments, even if only part of the reads matches the sequence (e.g., edges of contigs and gaps). However, as IMAGE does not handle insert size, it is unable to estimate the size of the gap, so the user must choose an arbitrary length after closing for those gaps that remained in the scaffolds. Although it is not problem when working with genomes assembled using only paired-end reads (with short-insert sizes), it may result in drawbacks if mate-pair reads where used for scaffolding, as long gaps would be confused with short ones.
More recently, FGAP (Piro et al., 2014), GMcloser (Kosugi et al., 2015) and GapBlaster (de Sá et al., 2016) have provided another way to close gaps by using information for alternative assemblies, long reads or merged paired-end reads. In fact, GMcloser may also use information from paired-end reads, but by combining data from alternative assemblies and longs reads, it is possible to achieve a more reliable result. Sealer (Paulino et al., 2015) also uses some of the advantages of long-reads, but by merging paired-end reads in artificial long one. Although it is not even close to PacBio or Oxford Nanopore reads, or not even to Sanger reads (~1 kb), this merging process may be helpful to solve some short unknown regions.
FGAP and GapBlaster are similar in the way they identify potential targets for gap-closing, but different in the sense that FGAP performs the gap-closing automatically whereas GapBlaster requires manual inspection. Although most of the tools are automated, the availability of tools for manual revision is also important, as some regions are very difficult to be assembled or corrected, and it is difficult to define generic rules to solve these situations.
Error-correction and assembly evaluation
The combination of different approaches employed to assemble and finish a genome can result in some artifacts, caused by the limitations of each software and/or the platform used for sequencing. Tools like QUAST (Gurevich et al., 2013), REAPR (Hunt et al., 2013), ALE (Clark et al., 2013) and GMvalue (Kosugi et al., 2015) were developed to evaluate the accuracy of an assembly. Additionally, other softwares, like iCORN (Otto et al., 2010) and SEQuel (Ronen et al., 2012), are able to correct assembly errors, including insertions, deletions and base substitutions.
Assembly evaluation
REAPR (Recognition of Errors in Assemblies using Paired Reads) (Hunt et al., 2013): takes as input a FASTA file containing the scaffolds and a BAM file generated by the remapping of the reads used in the assembly. First, a coverage analysis is performed using SNP-o-matic (Manske and Kwiatkowski 2009) or SMALT (https://www.sanger.ac.uk/resources/software/smalt), for genomes that are smaller or bigger than 100 Mb, respectively. The software analyzes the assembly base-per-base and uses the information per position in a metric called Fragment Coverage Distribution (FCD), where the expected coverage distribution is compared to the observed values. The discrepant regions are treated as possible misassemblies and REAPR can generate a new set of scaffolds splitting the erroneous regions into separate sequences. REAPR can be obtained from the URL http://www.sanger.ac.uk/science/tools/reapr.
QUAST (QUality ASsessment Tool for genome assemblies) (Gurevich et al., 2013): can be used to compare different assemblies of the same genome or to simply analyze an assembly. If more than one assembly is loaded, the program uses the MUMmer package to align the sequences and identify possible erroneous regions, count the number of aligned/unaligned bases, and also calculate, for each assembly, metrics like N50, L50, C+G% content, and other useful statistics. QUAST can be obtained from the URL http://bioinf.spbau.ru/quast, where it is also available through a webserver.
ALE (Assembly Likelihood Evaluation) (Clark et al., 2013): uses a combination of statistical analysis that are mainly based on probability distribution and Bayesian inference to determine the accuracy of an assembly without requiring a reference genome. To evaluate the assembly, the program analyzes the k-mer distribution, C+G% and the relative orientation of the mates (paired-end reads) in the BAM file. ALE can be obtained from its website http://www.alescore.org.
CGAL (Computing Genome Assembly Likelihood) (Rahman and Pachter 2013): evaluates the accuracy of the assembly using a probability distribution analysis that takes into consideration the expected coverage with that obtained after the reads are remapped to the scaffolds. CGAL can be obtained from the URL http://bio.math.berkeley.edu/cgal/.
GMvalue (Kosugi et al., 2015): is a program distributed along with the gap-closing program GMCloser, but can be used as a stand-alone application. The program uses NUCmer to align the assembly to a reference genome and identify misassemblies, such as insertions and deletions (INDELs). GMvalue can also generate “error-free” assemblies by splitting contigs in their erroneous regions. The program is distributed along with GMCloser and can be obtained from its SourceForge repository https://sourceforge.net/projects/gmcloser/.
When assembling a novel genome, the first metrics that are taken into account are the number of contigs, the length of the assembly and the N50. Although they can provide a good idea of how “contiguous” is the assembly, they do not measure its reliability, and may be easily distorted by inappropriate assembly procedures. Use of information from just one pair of reads to join two contigs into a scaffold, for example, may lead to an assembly error, even if the apparent fragmentation of the assembly is being reduced. If two regions are wrongly joined during the assembly, it may be misinterpreted as a natural biological event, and makes difficult further steps of assembly finishing.
Assembly correction
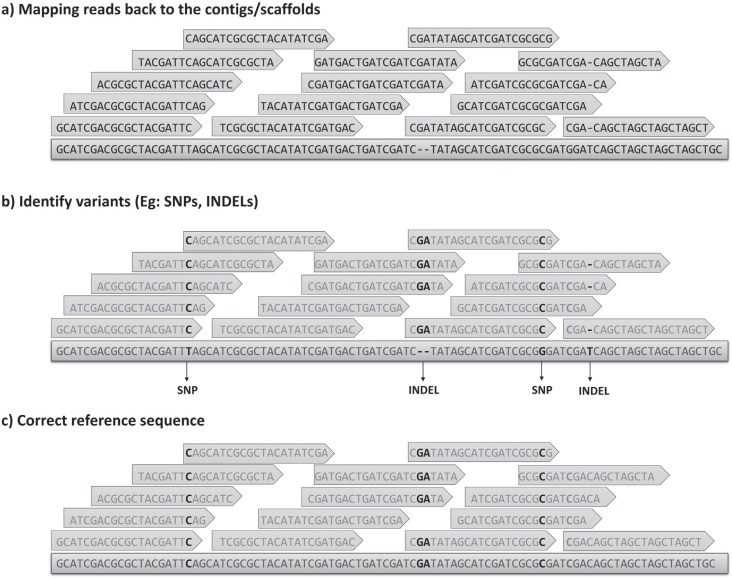
Assembly evaluation tools are very useful to identify structural inconsistences in a draft of apparently “finished” genome, but sometimes a manual revision and correction is inapplicable. Platform-specific errors, such as base-substitutions in Illumina data, or homopolymeric-sequence errors in IonTorrent data, may affect the annotation process, as some genes may be wrongly identified as frameshifted or mutated. As these types of errors may occur along the genome assembly, it is important to have automated tools to correct them automatically and reduce the chance of potential effects on the downstream analysis. ICORN and SEQuel are examples of programs that can be used to reduce these base-scale errors, and are based in the same principle that is used for variant calling analysis, consisting in the mapping of the reads against the sequence followed by the identification of those regions where there is a discrepancy, such as single-nucleotide polymorphism (SNPs) and Insertions and Deletions (INDELs) (Figure 4).
Figure 4. Example of a simplified assembly correction approach for base substitutions and insertion/deletion misassemblies. The process steps are (a) map the reads to the assembly, (b) identify variants (eg: SNPs and INDELs) in a similar way to the common variant calling analysis pipelines, and finally, (c) correct the regions in the assembly that show discrepancies. These steps may be reiterated several times until no further change be able to improve the assembly.
iCORN (Iterative Correction of Reference Nucleotides) (Otto et al., 2010): is an automated pipeline for assembly correction. Using a paired-end library, the program performs the read remapping using SSAH (Ning et al., 2001) and the variant calling and coverage analysis using SNP-o-matic (Manske and Kwiatkowski, 2009). The correction of the reference is followed by a new coverage analysis. If the correction promoted an improvement in the coverage, a new iteration commences, and the corrected sequence is used as the new reference; otherwise, the program stops and the last corrected sequence is returned as output. ICORN can be obtained from its SourceForge repository http://icorn.sourceforge.net/ and as part of the PAGIT package.
SEQuel (Ronen et al., 2012): uses a modification of the widely used De Bruijn graphs called positional De Bruijn graph. Using the BWA's output (Li and Durbin, 2009), the main algorithms combine the k-mer information with the relative position and orientation information of the mates in the paired-end library to construct a graph that is used to generate a new set of corrected contigs. SEQuel can be obtained from the URL http://bix.ucsd.edu/SEQuel/.
Both iCORN and SEQuel can be used to reduce base-scale errors in genome sequences, such as single nucleotide substitutions errors and small artificial INDELs, but are not able to identify and correct genome-scale misassemblies, such as those identified by REAPR or GMValue. In fact, both REAPR and GMValue can be used to break misassembled genomes and generate a correct set of contigs, but they are not able to generate an improved assembly, in terms of contiguity, and new rounds of scaffolding and gap-closing would be necessary for this purpose. To address the problem of genome-scale assembly correction, Guizelini et al. (2016) have developed the tool GFinisher, which integrates the detection and correction of assembly errors with the reference-guided scaffolding and the gap-closing processes.
GFinisher (Guizelini et al., 2016): is an assembly correction tool that also incorporates elements of assembly integration, reference-based scaffolding, and gap-closing in its internal pipeline. First, the program performs a reference-guided scaffolding using the module jContigSort (https://sourceforge.net/projects/jcontigsort/), which applies a HashMap-based algorithm using the k-mers from the draft genome and from a reference. Gaps between the contigs are then closed using jFGAP, a Java-implementation of the FGAP algorithm (Piro et al., 2014), by taking information form alternative assemblies of the target genome. To identify misassembled regions, GFinisher uses an adaptation of the GC-skew metric (Lobry, 1996), which allows the analysis of fluctuations in the distribution of C and G along the sequence by using a sliding window. Points in the sequence where the GC-skew seems to be discrepant when compared to its context are considered as assembly errors, and are used to split the contigs. Then, the processes of reference-guided scaffolding, gap-closing and analysis of the GC-skew are repeated, and a final assembly is generated. At the end, all intermediary assemblies and the final one are analyzed by QUAST (Gurevich et al., 2013). GFinisher can be obtained from its SourceForge repository http://gfinisher.sourceforge.net/.
While base-scale errors may affect the genome annotation by the presence of artificial frameshifts or non-synonymous mutations, the genome-scale misassemblies may lead to the erroneous identification of genome rearrangements or even the loss of relevant genes in the annotation, if they are located in the misassembled region. Therefore, the correction of base-scale errors and genome-scale misassemblies are crucial steps in the generation of high-quality finished assemblies.
Conclusion
NGS platforms have provided new ways to obtain large amounts of genomics data and reduced expressively the cost of the sequencing process itself, but also brought new challenges, especially to the data management and analysis. In the case of genome sequencing projects, for example, depending on the organism and the sequencing platform, the size of the FASTQ files containing the raw reads may vary from less than 1 gigabyte (Gb) to thousands of Gbs. Although nowadays it is perfectly possible to perform the de novo assembly of a microbial genome using a desktop computer, or even a notebook, when the volume of data is relatively low, many NGS analysis still require high-performance computing infrastructures (e.g., clusters, cloud computing, distributed systems) to be executed. In fact, even de novo genome assembly, which is in a certain way a well-established field, has been constantly renewed as more approaches are being developed to optimize memory usage (e.g., string-graphs, compressed data structures) and the assembly process itself, especially due to the limitations implied by the short reads sequencers and, more recently, the relatively higher error-rate observed in third-generation sequencing platforms.
The development of new bioinformatics tools in the last decade was highly influenced by the new generation of sequencing platforms, and many software offerings were specifically designed to overcome the limitations of the NGS platforms and the challenges they have brought. The use of whole genome shotgun (WGS) sequencing became a common practice, and the amount of draft genomes available rose exponentially. However, the process by which a finished genome is generated, even in the case of bacteria, is still somewhat flawed and challenging.
The present review aimed to describe some tools that might be useful for improving genome assembly and facilitating the finishing process. For didactical purposes, the available tools were divided into four main groups. However, there are many other forms of improvements that can be used to optimize an assembly. Additionally, in silico tools can be very helpful and may reduce the need of re-sequencing. However, they also have some limitations, and it is important to know that their efficiency is directly dependent on the quality of the assembly and the reads.
As there are many aspects that may affect the assembly, from the sequence itself (e.g., repetitive DNA, GC-rich or GC-poor) to technique artifacts (e.g., platform bias, assembler errors), it is very difficult to create a straightforward method to turn millions of short-reads to close and error-free chromosome sequences. Therefore, when working with a newly sequenced organism, it may be useful to try different strategies for de novo assembly, assembly integration, scaffolding and gap-closing, and check how they affect the reliability of the genome assembly to choose the best parameters. Knowing the characteristics of the genomic structure of an organism, the sequencing platform and the library construction may also be very useful when choosing the tools, as some of them are designed and optimized to deal with certain specific error patterns.
Although not always applicable, there are many genome finishing tools that are also available as webservers, what may be very convenient for those researchers that are not fully confortable with Linux and commandline interfaces. Additionally, as most of these tools do not directly require the sequencing data or mapping files, the uploading and processing time is usually very fast.
Genome announcement papers that describe sequencing projects using the same technology may facilitate the choice, especially when the organism is closely related to the one you are interested to analyze. As new tools are constantly being developed and released, it may also be useful to check not only bibliography databases, such as PubMed, but also bioinformatics software repositories, such as OMICtools (https://omicstools.com), and web-based discussion forums, such as Biostars (https://www.biostarts.org) and SEQanswers (https://seqanswers.com), to keep updated about new programs and techniques.
The new generation of DNA sequencing platforms, such as PacBio RS II and Oxford Nanopore, will certainly guide the development of new bioinformatics tools and analysis protocols for the next years, and may provide an easier way to generate high-quality finished genomes. These sequencing platforms intended to produce reads that are much longer than the older second-generation platforms, like Illumina, IonTorrent, and SOLiD, and may overcome most of the limitations associated with short-reads. One of the most common issues associated with genome assembly that uses short-reads is that repeated DNA regions, like simple sequence repetitions (SSRs) that are longer than the read length, are considered computationally impossible to assemble “exactly”. However, today, the throughput of these new long-read sequencers is still low and has a relatively high error rate compared to alternative platforms. This indicates that many aspects of genome sequencing still require improvement. Additionally, sequencing by PacBio SMRT still very time consuming and expensive when compared to other platforms, such as IonTorrent PGM and Illumina MiSeq. Finally, the size of the raw files generated by PacBio SMRT and Oxford Nanopore are substantially larger than the ones for second-generation platforms, even for microbial genomes, what reflects the growing need for computational and storage resources in the context of NGS.
Fifteen years since the Human Genome Sequencing Consortium released the first draft, and 10 years after Roche released its 454 platform, DNA sequencing has undergone many changes, and many technologies have been released. Furthermore, it is possible that, in the near future, only a de novo assembly, without any read preprocessing or post-assembly improvement, will be enough to generate high-quality finished genomes. However, such procedures are nowadays still necessary to achieve a reliable result.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Guilherme Corrêa de Oliveira
References
- Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OVC, Missawa SK, Galzerani F, Costa GGL, Vidal RO, Noronha MF, et al. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res. 2009;19:2258–2270. doi: 10.1101/gr.091777.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: Algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, Underwood JG, Lee L, Wong WH. Improving PacBio long read accuracy by short read alignment. PLoS One. 2012;7:e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. De novo genome assembly: What every biologist should know. Nat Methods. 2012;9:333–337. [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir A, Klammer AA, Robins WP, Chin C-S, Webster D, Paxinos E, Hsu D, Ashby M, Wang S, Peluso P, et al. A hybrid approach for the automated finishing of bacterial genomes. Nat Biotechnol. 2012;30:701–707. doi: 10.1038/nbt.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily PM, Fujimoto MS, Snell Q, Ventura D, Clement MJ. ScaffoldScaffolder: Solving contig orientation via bidirected to directed graph reduction. Bioinformatics. 2016;32:17–24. doi: 10.1093/bioinformatics/btv548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56–R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. SSPACE-LongRead: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211–211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi E, Donati B, Galardini M, Brunetti S, Sagot M-F, Lió P, Crescenzi P, Fani R, Fondi M. MeDuSa: A multi-draft based scaffolder. Bioinformatics. 2015;31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- Caboche S, Audebert C, Lemoine Y, Hot D, Soon W, Hariharan M, Snyder M, Fonseca N, Rung J, Brazma A, et al. Comparison of mapping algorithms used in high-throughput sequencing: Application to Ion Torrent data. BMC Genomics. 2014;15:264–264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421–421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande A, Del Fabbro C, Scalabrin S, Policriti A. GAM: Genomic Assemblies Merger: A graph based method to integrate different assemblies. IEEE Int Conf Bioinform Biomed. 2009;2009:321–326. [Google Scholar]
- Chain PSG, Grafham D V, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics. 2012;13:238–238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SC, Egan R, Frazier PI, Wang Z. ALE: A generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics. 2013;29:435–43. doi: 10.1093/bioinformatics/bts723. [DOI] [PubMed] [Google Scholar]
- Compeau PEC, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dark M. Whole-genome sequencing in bacteriology: State of the art. Infect Drug Resist. 2013;6:115–123. doi: 10.2147/IDR.S35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarian A, Michael TP, Sengupta AM. SOPRA: Scaffolding algorithm for paired reads via statistical optimization. BMC Bioinformatics. 2010;11:345–345. doi: 10.1186/1471-2105-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sá PHCG, Miranda F, Veras A, de Melo DM, Soares S, Pinheiro K, Guimarães L, Azevedo V, Silva A, Ramos RTJ. GapBlaster-A graphical gap filler for prokaryote genomes. PLoS One. 2016;11:e0155327. doi: 10.1371/journal.pone.0155327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, Mudge J, Cameron C, Ramaraj T, Anand A, Fengler K, Hayes K, Llaca V, Jones TJ, May G, et al. Characterization, correction and de novo assembly of an Oxford Nanopore genomic dataset from Agrobacterium tumefaciens . Sci Rep. 2016;6:28625–28625. doi: 10.1038/srep28625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Z, Dias U, Setubal JC. SIS: A program to generate draft genome sequence scaffolds for prokaryotes. BMC Bioinformatics. 2012;13:96–96. doi: 10.1186/1471-2105-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez N, Brudno M. SCARPA: Scaffolding reads with practical algorithms. Bioinformatics. 2013;29:428–434. doi: 10.1093/bioinformatics/bts716. [DOI] [PubMed] [Google Scholar]
- Edwards DJ, Holt KE. Beginner's guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp. 2013;3:2–2. doi: 10.1186/2042-5783-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant GK, Hoebeke M, Partensky F, Andres G, Corre E, Garczarek L. WiseScaffolder: An algorithm for the semi-automatic scaffolding of Next Generation Sequencing data. BMC Bioinformatics. 2015;16:281–281. doi: 10.1186/s12859-015-0705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondi M, Orlandini V, Corti G, Severgnini M, Galardini M, Pietrelli A, Fuligni F, Iacono M, Rizzi E, De Bellis G, et al. Enly: Improving draft genomes through reads recycling. J Genomics. 2014;2:89–93. doi: 10.7150/jgen.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. CONTIGuator: A bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 2011;6:11–11. doi: 10.1186/1751-0473-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Sung W-K, Nagarajan N. Opera: Reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol. 2011;18:1681–1691. doi: 10.1089/cmb.2011.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizelini D, Raittz RT, Cruz LM, Souza EM, Steffens MBR, Pedrosa FO. Gfinisher: A new strategy to refine and finish bacterial genome assemblies. Sci Rep. 2016;6:34963–34963. doi: 10.1038/srep34963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatem A, Bozdag D, Toland AE, Çatalyürek ÜV, Flicek P, Birney E, Cokus S, Feng S, Sultan M, Schulz M, et al. Benchmarking short sequence mapping tools. BMC Bioinformatics. 2013;14:184–184. doi: 10.1186/1471-2105-14-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. REAPR: A universal tool for genome assembly evaluation. Genome Biol. 2013;14:R47–R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Newbold C, Berriman M, Otto TD. A comprehensive evaluation of assembly scaffolding tools. Genome Biol. 2014;15:R42–R42. doi: 10.1186/gb-2014-15-3-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Reinert K, Myers EW. The greedy path-merging algorithm for contig scaffolding. J ACM. 2002;49:603–615. [Google Scholar]
- Kent WJ. BLAT: The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Larkin DM, Cai Q, Asan, Zhang Y, Ge R-L, Auvil L, Capitanu B, Zhang G, Lewin HA, et al. Reference-assisted chromosome assembly. Proc Natl Acad Sci U S A. 2013;110:1785–1790. doi: 10.1073/pnas.1220349110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen JL, Currie CR. Gene fragmentation in bacterial draft genomes: Extent, consequences and mitigation. BMC Genomics. 2012;13:14–14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Raney B, Paten B, Pham S. Ragout - a reference-assisted assembly tool for bacterial genomes. Bioinformatics. 2014;30:i302–i309. doi: 10.1093/bioinformatics/btu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Phillippy AM. One chromosome, one contig: Complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol. 2015;23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotech. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Treangen TJ, Pop M. Bambus 2: Scaffolding metagenomes. Bioinformatics. 2011;27:2964–2971. doi: 10.1093/bioinformatics/btr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hirakawa H, Tabata S. GMcloser: Closing gaps in assemblies accurately with a likelihood-based selection of contig or long-read alignments. Bioinformatics. 2015;31:3733–3741. doi: 10.1093/bioinformatics/btv465. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12–R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, Ahn T-H, Karpinets T, Lund O, Kora G, Wassenaar T, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25–R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille P, Norton S, Goldman BS, Henkhaus J, Miller N, Barbazuk B, Bode HB, Darby C, Du Z, Forst S, et al. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics. 2007;8:321–321. doi: 10.1186/1471-2164-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-L, Chen K-T, Lu CL. Assembling contigs in draft genomes using reversals and block-interchanges. BMC Bioinformatics. 2013;14(Suppl 5):S9–S9. doi: 10.1186/1471-2105-14-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-H, Liao Y-C. CISA: Contig integrator for sequence assembly of bacterial genomes. PLoS One. 2013;8:e60843. doi: 10.1371/journal.pone.0060843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364–251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substituion patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lu C, Chen K-T, Huang S-Y, Chiu H-T. CAR: Contig assembly of prokaryotic draft genomes using rearrangements. BMC Bioinformatics. 2014;15:381–381. doi: 10.1186/s12859-014-0381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18–18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen V, Välimäki N, Laaksonen A, Katainen R. Unified view of backward backtracking in short read mapping. In: Elomaa T, Mannila H, Orponen P, editors. Algorithms and Applications. Springer-Verlag; Berlin: 2010. pp. 182–195. [Google Scholar]
- Manske HM, Kwiatkowski DP. SNP-o-matic. Bioinformatics. 2009;25:2434–2435. doi: 10.1093/bioinformatics/btp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E, McPherson J, Martienssen R, Wilson RK, McCombie WR. What is finished, and why does it matter. Genome Res. 2002;12:669–671. doi: 10.1101/gr.032102. [DOI] [PubMed] [Google Scholar]
- Mariano DC, Pereira FL, Ghosh P, Barh D, Figueiredo HC, Silva A, Ramos RT, Azevedo VA. MapRepeat: An approach for effective assembly of repetitive regions in prokaryotic genomes. Bioinformation. 2015;11:276–9. doi: 10.6026/97320630011276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkin I, Patel A, Kolmogorov M, Vyahhi N, Pham S. Algorithms in Bioinformatics. In: Darling A, Stoye J, editors. Proc. 13th International Workshop. WABI. Springer; Berlin: 2013. pp. 215–229. [Google Scholar]
- Muñoz A, Zheng C, Zhu Q, Albert VA, Rounsley S, Sankoff D. Scaffold filling, contig fusion and comparative gene order inference. BMC Bioinformatics. 2010;11:304–304. doi: 10.1186/1471-2105-11-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW. Toward simplifying and accurately formulating fragment assembly. J Comput Biol. 1995;2:275–290. doi: 10.1089/cmb.1995.2.275. [DOI] [PubMed] [Google Scholar]
- Myers EW. The fragment assembly string graph. Bioinformatics. 2005;21(Suppl 2):i79–i85. doi: 10.1093/bioinformatics/bti1114. [DOI] [PubMed] [Google Scholar]
- Nagarajan N, Cook C, Di Bonaventura M, Ge H, Richards A, Bishop-Lilly KA, DeSalle R, Read TD, Pop M. Finishing genomes with limited resources: Lessons from an ensemble of microbial genomes. BMC Genomics. 2010;11:242–242. doi: 10.1186/1471-2164-11-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp J, Winterbach W, van den Broek M, Daran J-M, Reinders M, de Ridder D. Integrating genome assemblies with MAIA. Bioinformatics. 2010;26:433–439. doi: 10.1093/bioinformatics/btq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noé L, Kucherov G. YASS: Enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Sanders M, Berriman M, Newbold C. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics. 2010;26:1704–1707. doi: 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino D, Warren RL, Vandervalk BP, Raymond A, Jackman SD, Birol I. Sealer: A scalable gap-closing application for finishing draft genomes. BMC Bioinformatics. 2015;16:230–230. doi: 10.1186/s12859-015-0663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola H, Söderlund H, Ukkonen E. SEQAID: A DNA sequence assembling program based on a mathematical model. Nucleic Acids Res. 1984;12:307–321. doi: 10.1093/nar/12.1part1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. Research in Computational Molecular Biology. In: Berger B, editor. Proc. 14th Annual International Conference, RECOMB 2010. Springer Berlin; Heidelberg: 2010. pp. 426–440. [Google Scholar]
- Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro VC, Faoro H, Weiss VA, Steffens MBR, Pedrosa FO, Souza EM, Raittz RT. FGAP: An automated gap closing tool. BMC Res Notes. 2014;7:371–371. doi: 10.1186/1756-0500-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M, Kosack DS, Salzberg SL. Hierarchical scaffolding with Bambus. Genome Res. 2004;14:149–159. doi: 10.1101/gr.1536204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Pachter L. CGAL: Computing genome assembly likelihoods. Genome Biol. 2013;14:R8–R8. doi: 10.1186/gb-2013-14-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RTJ, Carneiro AR, Soares S de C, Santos AR dos, Almeida S, Guimarães L, Figueira F, Barbosa E, Tauch A, Azevedo V, et al. Tips and tricks for the assembly of a Corynebacterium pseudotuberculosis genome using a semiconductor sequencer. Microb Biotechnol. 2013;6:150–156. doi: 10.1111/1751-7915.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro FJ, Przybylski D, Yin S, Sharpe T, Gnerre S, Abouelleil A, Berlin AM, Montmayeur A, Shea TP, Walker BJ, et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 2012;22:2270–2277. doi: 10.1101/gr.141515.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker N, Qian H, Fulthorpe RR. The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics. 2012;100:167–175. doi: 10.1016/j.ygeno.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen R, Boucher C, Chitsaz H, Pevzner P. SEQuel: Improving the accuracy of genome assemblies. Bioinformatics. 2012;28:188–196. doi: 10.1093/bioinformatics/bts219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy RS, Chen KC, Sengupta AM, Schliep A. SLIQ: Simple linear inequalities for efficient contig scaffolding. J Comput Biol. 2012;19:1162–1175. doi: 10.1089/cmb.2011.0263. [DOI] [PubMed] [Google Scholar]
- Salmela L, Mäkinen V, Välimäki N, Ylinen J, Ukkonen E. Fast scaffolding with small independent mixed integer programs. Bioinformatics. 2011;27:3259–3265. doi: 10.1093/bioinformatics/btr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L, Rivals E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL, Pop M. Minimus: A fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64–64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soueidan H, Maurier F, Groppi A, Sirand-Pugnet P, Tardy F, Citti C, Dupuy V, Nikolski M. Finishing bacterial genome assemblies with Mix. BMC Bioinformatics. 2013;14(Suppl 1):S16–S16. doi: 10.1186/1471-2105-14-S15-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979;6:2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain MT, Tsai IJ, Assefa SA, Newbold C, Berriman M, Otto TD. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc. 2012;7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized multiplex PCR: Efficiently closing a whole-genome shotgun sequencing project. Genomics. 1999;62:500–507. doi: 10.1006/geno.1999.6048. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M. Next generation sequence assembly with AMOS. Curr Protoc Bioinformatics. 2011;33:11.8.1–11.8.18. doi: 10.1002/0471250953.bi1108s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritt A, Eisen JA, Facciotti MT, Darling AE. An integrated pipeline for de novo assembly of microbial genomes. PLoS One. 2012;7:e42304. doi: 10.1371/journal.pone.0042304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Otto TD, Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41–R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandervalk BP, Jackman SD, Raymond A, Mohamadi H, Yang C, Attali DA, Chu J, Warren RL, Birol I. Konnector: Connecting paired-end reads using a bloom filter de Bruijn graph; 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2014. pp. 51–58. [Google Scholar]
- Vicedomini R, Vezzi F, Scalabrin S, Arvestad L, Policriti A. GAM-NGS: Genomic assemblies merger for next generation sequencing. BMC Bioinformatics. 2013;14(Suppl 7):S6–S6. doi: 10.1186/1471-2105-14-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AT, Derome N, Boyle B, Culley AI, Charette SJ. Next-generation sequencing (NGS) in the microbiological world: How to make the most of your money. J Microbiol Methods. 2016;138:60–71. doi: 10.1016/j.mimet.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Warren RL, Sutton GG, Jones SJM, Holt RA. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Ye L, Gao H, Minx P, Warren WC, Weinstock GM. Graph accordance of next-generation sequence assemblies. Bioinformatics. 2012;28:13–16. doi: 10.1093/bioinformatics/btr588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Smith DR, Sutton G, Yorke JA. Assembly reconciliation. Bioinformatics. 2008;24:42–45. doi: 10.1093/bioinformatics/btm542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.