Abstract
Effect of alternative splicing (AS) on diffuse large B-cell lymphoma (DLBCL) pathogenesis and survival has not been systematically addressed. Here, we compared differentially expressed genes and exons in association with survival after chemoimmunotherapy, and between germinal center B-cell like (GCB) and activated B-cell like (ABC) DLBCLs. Genome-wide exon array-based screen was performed from samples of 38 clinically high-risk patients who were treated in a Nordic phase II study with dose-dense chemoimmunotherapy and central nervous system prophylaxis. The exon expression profile separated the patients according to molecular subgroups and survival better than the gene expression profile. Pathway analyses revealed enrichment of AS genes in inflammation and adhesion-related processes, and in signal transduction, such as phosphatidylinositol signaling system and adenosine triphosphate binding cassette transporters. Altogether, 49% of AS-related exons were protein coding, and domain prediction showed 28% of such exons to include a functional domain, such as transmembrane helix domain or phosphorylation sites. Validation in an independent cohort of 92 DLBCL samples subjected to RNA-sequencing confirmed differential exon usage of selected genes and association of AS with molecular subtypes and survival. The results indicate that AS events are able to discriminate GCB and ABC DLBCLs and have prognostic impact in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid neoplasm in adults, comprising 30–40% of all malignant lymphomas. It is an aggressive disease, where ∼60% of patients can be cured with a combination of rituximab and anthracycline-based CHOP or CHOP-like chemoimmunotherapy.1, 2 However, responses to treatment are still largely unpredictable, and survival of the patients, who experience disease relapse or have primary refractory disease, is dismal. Thus, there is a growing need to further understand the molecular mechanisms underlying the disease that would not only provide information to predict the survival, but also enable the design of more targeted therapeutic strategies.
Based on gene expression profiling, three molecular subtypes showing germinal center B-cell (GCB), activated B-cell (ABC) and primary mediastinal B-cell lymphoma signatures have been identified. These subtypes differ in their genotypic, phenotypic and clinical features.3, 4, 5, 6 During the past few years, studies applying next-generation sequencing techniques have provided further insights into the heterogeneity and pathogenesis of DLBCL.7, 8, 9 Especially, a number of genetic alterations have been shown to be characteristic of GCB or ABC subtypes. However, despite the rapidly growing number of genetic aberrations reported in DLBCL, association of these findings with treatment outcome remains to be shown.
Alternative splicing (AS) is a common regulatory mechanism generating multiple RNA transcripts from a single gene and allowing enormous functional diversity in protein isoforms. The vast majority of the human genes, evaluates ranging from 70 to 95%, are alternatively spliced.10 Growing evidence suggests that AS is closely associated with cancer pathogenesis and progression.11, 12, 13 Alternatively spliced signatures derived from gene expression profiling have been shown to be more reliable for diagnostic purposes than signatures derived from expression profiling, for example in prostate cancer.14 Furthermore, isoform-level expression profiles can discriminate cancer cell lines from noncancer cells better than gene-level expression profiles.15
In DLBCL, the role of AS remains largely unexplored. Previous studies have mainly focused on AS isoforms of individual genes, such as FOXP1, where contradictory findings concerning its prognostic role can partly be explained by smaller and potentially oncogenic FOXP1 isoforms primarily expressed in ABC-type DLBCL.16 Other previously reported genes with AS in DLBCL include a developmentally regulated B-lymphoid phosphatase, PTPRO, regulating G0/G1 arrest17 and a leukocyte homing and hyaluronidase receptor CD44.18, 19, 20, 21
In this study, we have evaluated genome-wide gene and exon expression profiles in DLBCL. Differential exon usage was found to be a common event in DLBCL. Splicing events affected pivotal genes involved in DLBCL signaling, and were able to discriminate the patients according to cell of origin (COO) and survival.
Materials and methods
Patients
Prospectively collected discovery cohort consisted of 38 DLBCL patients <65 years old with clinically high-risk (age-adjusted International Prognostic Index Score 2–3) disease (Table 1). The patients were treated in a Nordic phase II NLG-LBC-04 protocol with six courses of dose-dense R-CHOEP-14 chemoimmunotherapy followed by systemic central nervous system prophylaxis with high-dose methotrexate and high-dose cytarabine.22 Further information on the patient cohort is provided in the Supplementary Materials and methods. The protocol and sampling were approved by Institutional Review Boards, National Medical Agencies and Ethics Committees in Finland, Denmark, Sweden and Norway, and the trial was registered at ClinicalTrials.gov, identifier number NCT01502982. All patients gave informed consent.
Table 1. Patient characteristics of the discovery and validation cohorts.
| Number of patients | Discovery cohort, n (%) | Validation cohort, n (%) |
|---|---|---|
| Total | 38 (100) | 92 (100) |
| Gender | ||
| Female | 14 (37) | 31 (34) |
| Male | 24 (63) | 61 (66) |
| Age | ||
| <60 | 28 (74) | 39 (42) |
| 60–65 | 10 (26) | 15 (16) |
| >65 | 0 (0) | 38 (41) |
| Subtype | ||
| GCB | 19 (50) | 51 (55) |
| ABC | 12 (32) | 32 (35) |
| Other/unclassified | 7 (18) | 9 (10) |
| Performance status | ||
| 0–1 | 24 (63) | 64 (70) |
| 2–3 | 14 (37) | 28 (30) |
| B-symptoms | 22 (58) | |
| Bulky disease | 22 (58) | |
| Elevated LDH | 37 (97) | |
| Stage | ||
| I–II | 1 (3) | 44 (48) |
| III–IV | 37 (97) | 48 (52) |
| IPI | ||
| 0 | 0 (0) | 16 (17) |
| 1 | 0 (0) | 24 (26) |
| 2 | 17 (45) | 22 (24) |
| 3 | 10 (26) | 18 (20) |
| 4 | 10 (26) | 10 (11) |
| 5 | 1 (3) | 2 (2) |
Abbreviations: ABC, activated B-cell; GCB, germinal center B-cell; IPI, International Prognostic Index; LDH, lactate dehydrogenase.
Validation cohort consisted of RNA sequencing (RNAseq) and clinical data from 92 R-CHOP-like chemotherapy-treated DLBCL patients generated by the National Cancer Institute (NCI) Cancer Genome Characterization Initiative (CGCI; dbGaP database applied study accession: phs000532.v3.p1)7, 8 (Table 1).
Samples and gene/exon expression analysis
Total RNA was extracted with Qiagen (Hilden, Germany) AllPrep DNA/RNA/Protein Mini kit and examined using Affymetrix Human Exon 1.0 ST arrays according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA). Hybridization protocols and raw expression microarray data are available at ArrayExpress archive (http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-3488). Gene- and exon-level expression data for the NLG-LBC-04 discovery cohort were quantified by MEAP (Multiple Exon Array Preprocessing) algorithm23 with MEAP probe annotation version 70.
In the CGCI validation cohort, adaptors and low-quality bases were trimmed from the samples before alignment. The reads were aligned with TopHat 2.0.8b using the hg19 human reference genome with Ensembl gene annotation version 70. HTSeq was then used to obtain the exon- and gene-level read counts. Differential expression at exon and gene levels was estimated using DEXSeq (version 1.7.14) and DESeq2 (version 1.4.5), respectively (https://bioconductor.org/packages/3.5/bioc/).
Molecular subgroup prediction
Samples with exon array data were classified into GCB, ABC and nonclassified subgroups using the gene predictor from Lymphochip data as previously described.24 More details are provided in the Supplementary Materials and methods.
Pathway and GO analysis
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway functional enrichment analyses were performed with the Pathway-Express (Intelligent Systems and Bioinformatics Laboratory, Detroit, MI, USA). Only enrichment results with false discovery rate <0.05 were considered significant. Gene annotation and gene ontology (GO) term enrichment for biological processes was performed with Gene Set Enrichment Analysis (GSEA)25 for the 289 genes with differentially expressed exons (DEEs) log2 fold change >1.5. This resulted into altogether 38 separate ontology terms with false discovery rate <0.05. To reduce the number of GO terms, we regrouped the related ontologies into 9 larger groups.
Statistical analyses
Data were analyzed using IBM SPSS Statistics 22.0 (IBM, Armonk, NY, USA). P-values of <0.05 were considered statistically significant and all P-values are two tailed. The χ2 test was used to assess the differences in the frequency of the prognostic factors. Cox univariate and multivariate analyses were performed to study the prognostic value of the factors. Kaplan–Meier method was used to estimate survival rates and their differences were compared with log-rank test. Overall survival (OS) was determined from the date of study entry or diagnosis until the last follow-up or death from any cause. Disease-specific survival was calculated as a period between the registration date and the date of death due to lymphoma. Progression-free survival (PFS) was measured as the period between the date of registration or diagnosis and progression or death from any cause. OS, disease-specific survival and PFS were reported in months. A web-based cutoff finder tool at http://molpath.charite.de/cutoff analysis26 was used to determine the most prognostic cutoff level for survival outcomes.
Exon-specific domain identification
DEEs were annotated by their genomic locations (5′ untranslated region (UTR), 3′ UTR, coding, noncoding and unknown). Domain analysis was done for all coding DEEs by translating coding exonic regions into peptide sequences using Ensembl API (version 70) and fetching domain information (Pfam, Smart, SignalP and TMHMM) for all peptide sequences with InterProScan (version 5).27 Phosphorylation sites within peptide sequence of each exon were searched against all known phosphorylation motifs downloaded from the PhosphoSitePlus.28
Results
Clinical characteristics of the patients
Baseline characteristics of the discovery cohort of 38 patients treated in the NLG-LBC-04 protocol22 are presented in Table 1. Median age of the patients was 55 years (range 21–65 years). During the median follow-up time of 66 months, 9 patients had relapsed and a total of 9 died. Four of the deaths were not lymphoma related. The estimated 5-year OS, disease-specific survival and PFS rates were 76%, 82% and 74%, respectively. In the CGCI validation cohort of 92 patients, the median age was 61 years (range 17–75 years). During the follow-up time of 59 months, 24 patients had relapsed and 21 died. Of the deaths, three were not lymphoma related. The estimated 5-year OS, disease-specific survival and PFS rates were 79%, 80% and 74%, respectively. The main differences between the discovery and validation cohort was that the patients in the discovery cohort were younger and had clinically higher risk disease (Table 1).
Identification of splicing events
Global mRNA and AS variations with prognostic impact were identified by comparing gene and exon expression profiles between the patients who experienced relapse (n=9, poor prognosis group) or remained in long-term remission (>24 months;29 n=29, good prognosis group) after chemoimmunotherapy (Figure 1). No significant differences were observed in baseline characteristics between the groups (Table 2). Using the gene-level expression data, 220 differentially expressed genes (DEGs) between the groups were identified (Supplementary Table S1). Of these, 59% were suppressed and 41% upregulated in patients with poor prognosis. The most upregulated genes included not only many nonprotein coding genes such as microRNAs and long noncoding RNAs but also MME, which is coding for CD10 and is included in the COO classification,24 and TRAF4, which has been shown to downregulate the innate immunity signaling.30 Innate immunity was also affected because of suppression of other genes in the pathway such as HLA-DQB1, and especially genes involved in T cell-mediated immune responses (FOS, IL2RB) or B-cell maturation (IL7 and KLF3). The pathways significantly enriched among the DEGs included immune response-related processes, such as antigen processing and presentation, JAK/STAT (Janus kinase/signal transducer and activator of transcription) signaling, circadian rhythm and hematopoietic cell lineage (Table 3 and Supplementary Table S2).
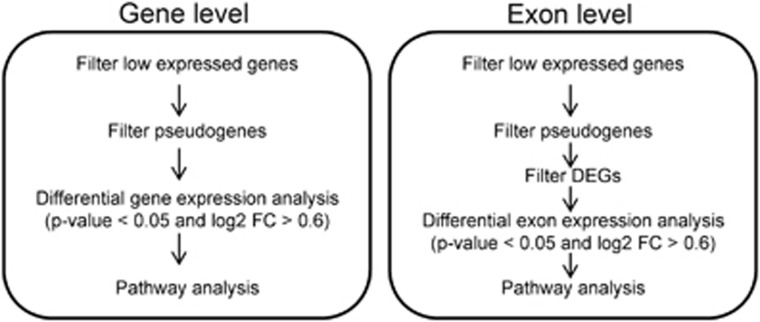
Figure 1.
Flowchart: identification of differentially expressed genes in DLBCL.
Table 2. Baseline characteristics of the discovery cohort according to good and poor prognosis.
| Patients | Good prognosis, n (%) | Poor prognosis, n (%) | P-value |
|---|---|---|---|
| Total | 29 (76) | 9 (24) | |
| Gender | |||
| Female | 19 (66) | 5 (56) | 0.699 |
| Male | 10 (34) | 4 (44) | |
| Age | |||
| <60 | 23 (79) | 6 (67) | 0.655 |
| >60 | 6 (21) | 3 (33) | |
| Subtype | |||
| GCB | 17 (59) | 7 (88) | 0.438 |
| Non-GCB | 12 (41) | 2 (22) | |
| Performance status | |||
| 0–1 | 20 (69) | 4 (44) | 0.245 |
| 2–3 | 9 (31) | 5 (56) | |
| B-symptoms | |||
| No | 10 (34) | 6 (67) | 0.128 |
| Yes | 19 (66) | 3 (33) | |
| Bulky disease | |||
| No | 13 (45) | 3 (33) | 0.706 |
| Yes | 16 (55) | 6 (67) | |
| Elevated LDH | |||
| No | 0 (0) | 1 (11) | 0.237 |
| Yes | 29 (100) | 8 (89) | |
| Stage | |||
| I–II | 1 (3) | 0 (0) | 1.000 |
| III–IV | 28 (97) | 9 (100) | |
| aaIPI | |||
| 3–4 | 21 (72) | 6 (67) | 1.000 |
| 8 (28) | 3 (33) | ||
| BCL2 | |||
| Low | 27 (93) | 7 (78) | 0.233 |
| Higha | 2 (7) | 2 (22) | |
| cMYC | |||
| Low | 28 (97) | 9 (100) | 1.000 |
| High | 1 (3) | 0 (0) |
Abbreviations: aaIPI, age-adjusted International Prognostic Index; GCB, germinal center B-cell; LDH, lactate dehydrogenase.
Z-score >2.0.
Table 3. Pathways significantly enriched among the DEGs and DEEs between good and poor prognosis groups.
| Pathway name | Input/total no. of genes | FDR |
|---|---|---|
| Gene level | ||
| Antigen processing and presentation | 2/89 | <0.001 |
| Circadian rhythm | 2/13 | <0.001 |
| JAK/STAT signaling pathway | 7/155 | 0.001 |
| Hematopoietic cell lineage | 5/87 | 0.003 |
| Biosynthesis of unsaturated fatty acids | 3/22 | 0.004 |
| Exon level (top 20 pathways) | ||
| Antigen processing and presentation | 17/89 | <0.001 |
| Leukocyte transendothelial migration | 22/119 | <0.001 |
| Cell adhesion molecules (CAMs) | 26/134 | <0.001 |
| Adherens junction | 19/78 | <0.001 |
| Phosphatidylinositol signaling system | 21/76 | <0.001 |
| ECM receptor interaction | 33/84 | <0.001 |
| Focal adhesion | 59/203 | <0.001 |
| Pathways in cancer | 84/330 | <0.001 |
| Calcium signaling pathway | 50/182 | <0.001 |
| ABC transporters | 19/44 | <0.001 |
| MAPK signaling pathway | 64/272 | <0.001 |
| Long-term depression | 25/75 | <0.001 |
| Tight junction | 36/135 | <0.001 |
| Amyotrophic lateral sclerosis (ALS) | 19/56 | 0.001 |
| Small cell lung cancer | 25/86 | 0.001 |
| VEGF signaling pathway | 22/74 | 0.002 |
| Axon guidance | 33/129 | 0.002 |
| Type I diabetes mellitus | 14/44 | 0.002 |
| Regulation of actin cytoskeleton | 47/217 | 0.003 |
| Thyroid cancer | 11/29 | 0.003 |
Abbreviations: ABC, adenosine triphosphate-binding cassette; ECM, extracellular matrix; DEE, differentially expressed exon; DEG, differentially expressed gene; FDR, false discovery rate; JAK/STAT, Janus kinase/signal transducer and activator of transcription; MAPK, mitogen-activated protein kinase; VEGF, vascular endothelial growth factor.
To identify AS events between the good and poor prognosis groups, we applied similar criteria to exon-level expression data with additional filtering of the DEGs from the gene-level analysis (Figure 1). As a result, 8785 unique exonic regions from 3888 genes with differential expression were found (Supplementary Table S3). Among the genes with DEEs were genes with known alternative isoforms associated with cancer, including RUNX1,31 TERT32, 33 and VEGFA,34 as well as CD44 and PTPRO that have previously been shown to be alternatively spliced in DLBCL.17, 19, 20, 35 In the case of CD44, differential exon inclusion was seen particularly in the variable region of the gene (exons v2–v10), involved in variant isoforms of CD44 that have been shown to substantiate to poor prognosis in DLBCL in the pre-rituximab era.18, 21 In the PTPRO gene, the DEE was localized in the catalytic domain region, present also in the lymphoid-predominant truncated isoform, shown to modulate SYK phosphorylation and B-cell receptor activity.36 Other genes with the most suppressed exon expression in the poor prognosis group included EPN1 and ARAP1, which are involved in endocytosis, and HLA-DQA1, CD4, EBI3, ICAM1 and UBA5, which are related to the immune system. Differential splicing was also observed in many splicing factors and regulators, such as NOVA1, TRA2B, SF3B1, ESRP1, RBFOX1 and CELF5, suggesting that the splicing mechanism might be affected through modulation of splicing factors themselves. In line with the pathways significantly enriched among the DEGs, the AS genes were significantly enriched for immune response-related pathways, such as antigen processing and presentation, and leukocyte transendothelial migration. In addition, the AS genes were enriched for the pathways related to adhesion, extracellular matrix interactions and signal transduction such as phosphatidylinositol signaling system and adenosine triphosphate (ATP)-binding cassette transporters (Table 3).
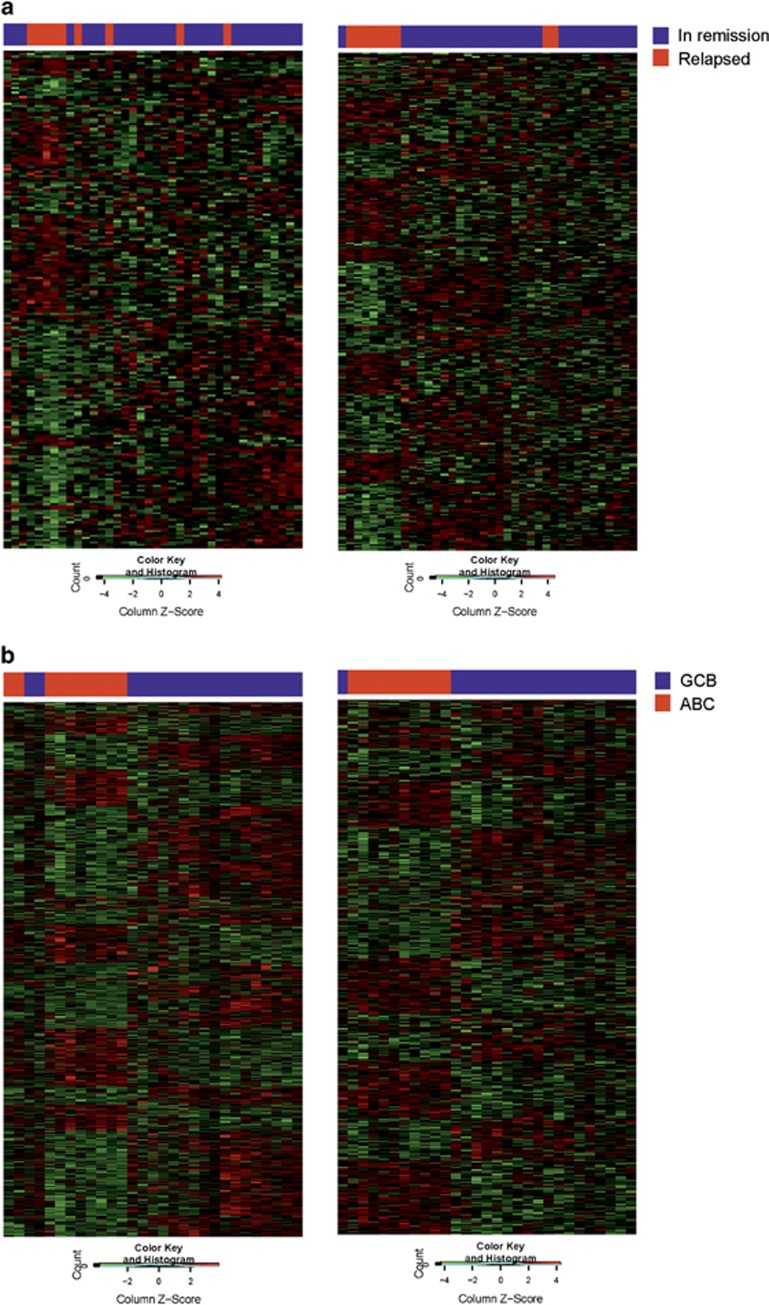
Interestingly, when the patient groups with poor and favorable outcomes were clustered according to the DEGs and DEEs, the expression profile of the most variable exons showed better separation of the groups than the profile of the most variable genes (Figure 2a). This indicates that AS plays an important role in DLBCL pathogenesis.
Figure 2.
Clustering of the patients according to differentially expressed genes or exons. (a) A total of 220 DEGs and 315 DEEs were used for clustering the DLBCL patients who relapsed or patients who remained in remission. (b) A total of 1045 DEGs and 590 DEEs were used for clustering the ABC and GCB subgroups.
Gene- and exon-level profiles according to COO subtypes
The observed differences in gene and exon levels between the patient groups with different outcomes raised the question of whether a similar phenomenon occurs when the patients are divided according to their molecular subtypes. When the gene expression-based COO classification was performed using previously described gene predictor,24 19 of the cases were predicted as GCB, 12 as ABC and 7 as other DLBCL subtypes (Supplementary Figure S1). In this cohort, no significant association between the molecular subtypes and survival was found (not shown). With similar criteria as previously used in the screen for the outcome-related DEGs and genes with DEEs, 1012 unique DEGs (Supplementary Table S4) and 20 386 exonic regions from 6726 unique genes (Supplementary Table S5) were discovered to be differentially expressed between the GCB and the ABC DLBCLs. As expected, the DEGs included many of the genes used in the ‘Wright’ subtype classification (SERPINA9, MYBL1, FUT8, IRF4), as well as other genes (FOXP1, MAPK10) shown to have subtype preferential expression profile.37, 38 On the contrary, the DEEs corresponded for only two genes (CCND2, DDB1) used in the subtype classification. In the pathway analysis, the genes from differential gene expression and exon usage between GCB and ABC subtypes were enriched in, for instance, phosphatidylinositol signaling system, regulation of actin cytoskeleton, adherens junction, focal adhesion and pathways in cancer (Supplementary Table S6). Altogether, the number of DEGs and DEEs was higher in the COO classification than in the survival-related profiles. Clustering of the patient groups with molecular subtypes according to the DEGs or DEEs was able to separate the ABC and GCB subtypes (Figure 2b).
Functional relevance of the alternative splicing
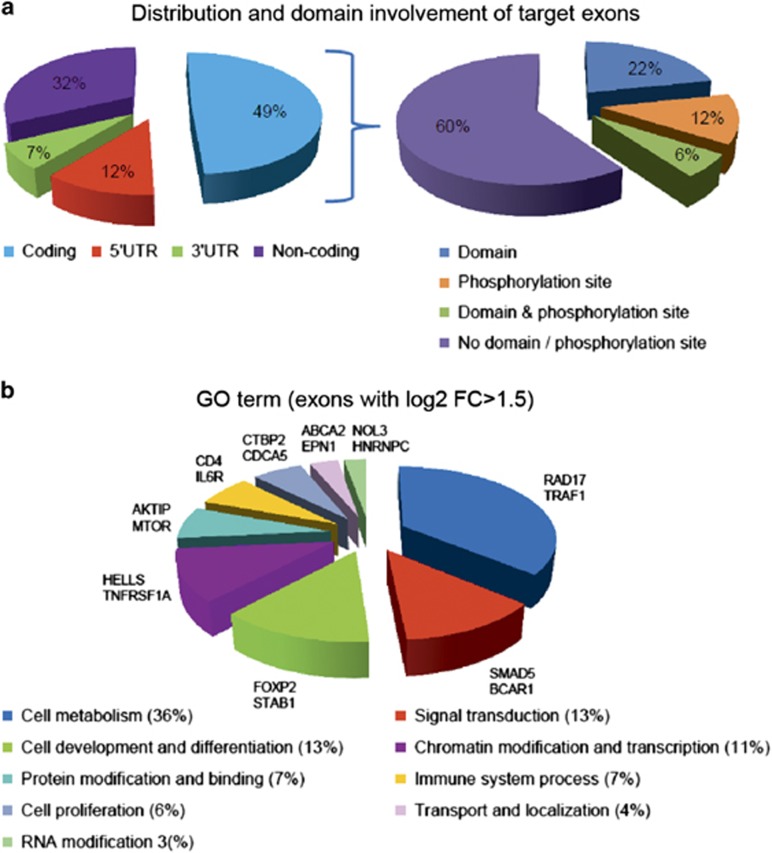
To analyze in more detail the functional effect of the DEEs identified from the comparison of the groups with poor and favorable outcomes, we studied the association of the exonic regions with their corresponding protein domains using our exon-specific functional analysis pipeline. Altogether, 49% (4351) of the AS-related exons were protein coding, whereas 12% were mapped to 5′ UTR, 7% to the 3′ UTR and 32% to other noncoding regions (Figure 3a and Supplementary Table S7). Domain prediction showed 28% of translated exons (1193) to include a functional protein domain. In addition, 18% of the coding exons were predicted to include phosphorylation sites, of which 53% were serines, 25% treonines and 22% tyrosines (not shown).
Figure 3.
Distribution of the splicing events and characteristics of the target exons. (a) Distribution and domain involvement of the target exons. (b) Relative distribution of the target exons by Gene Ontology (GO) categories. Representative genes from each group are presented next to the pie chart.
In order to define the biological functions potentially affected by AS, we performed GO annotations for biological processes with the genes with most differentially expressed exons (log2 fold change ⩾∣1.5∣, n=289). To reduce the redundancy among several categories, related ontologies were regrouped into larger groups (Figure 3b). From the separate ontologies, the highest number of genes was annotated to signal transduction (for example, SMAD5 and BCAR1). After regrouping, different cellular metabolic processes formed the most commonly implicated processes (for example, RAD17 and TRAF1). Transport and localization annotation included, for example, EPN1 that is involved in receptor-mediated endocytosis39 and ATP-binding cassette transporters ABCA2, ABCA3 and ABCA7 that are mostly known for their role in evolving drug resistance.40 Closer examination of ATP-binding cassette transporters revealed that many of the affected exons included a predicted functional protein domain or phosphorylation sites (not shown), suggesting that AS events might affect drug resistance by regulating the function and activity of the ATP-binding cassette transporters.
Validation of exon inclusion events
To validate the outcome-associated DEEs, we used data from a publicly available CGCI (RNAseq cohort of 92 patients).8 Of the 3888 AS genes, 547 were validated in the CGCI cohort. Thirty-seven DEEs (33 unique genes), corresponding to 17 protein coding transcripts, were exactly in the same location with the expression difference in the same direction (Supplementary Table S8). According to Cox univariate analysis, 29 out of the 37 DEEs were associated with PFS (P⩽0.05) and 20 with OS in the CGCI cohort (Table 4). In Cox multivariate analyses with International Prognostic Index score, 22 DEEs retained independence for prediction of PFS (Supplementary Table S9). These included several interesting genes, such as BCAR1 (coding for p130Cas, a docking protein involved in many intracellular signaling pathways), APH1A (anterior pharynx defective-1α, a component of the γ-secretase complex that cleaves integral membrane proteins such as Notch receptors and β-amyloid precursor protein), KCNH6 (member of a potassium voltage-gated channel family, shown to be associated with drug sensitivity) and CUL3 (cullin 3, a core component of an E3 ubiquitin ligase complex).
Table 4. Cox univariate analysis of the DEEs common in the discovery and validation cohorts (significant P<0.05 are in bold).
| Gene |
Discovery cohort |
Validation cohort |
||||||
|---|---|---|---|---|---|---|---|---|
|
PFS |
OS |
PFS |
OS |
|||||
| P-val exon | P-val gene | P-val exon | P-val gene | P-val exon | P-val gene | P-val exon | P-val gene | |
| CUL3 | 0.070 | 0.975 | 0.167 | 0.728 | 0.007 | 0.393 | 0.024 | 0.399 |
| DKK3 | 0.091 | 0.330 | 0.099 | 0.427 | 0.037 | 0.497 | 0.064 | 0.623 |
| BCAR1 | 0.022 | 0.179 | 0.108 | 0.499 | 0.005 | 0.439 | 0.002 | 0.449 |
| SLC9A3 | 0.112 | 0.154 | 0.084 | 0.016 | 0.002 | 0.164 | 0.002 | 0.139 |
| GAL | 0.051 | 0.938 | 0.468 | 0.475 | 0.003 | 0.687 | 0.033 | 0.659 |
| ABCB1 | 0.116 | 0.525 | 0.204 | 0.806 | 0.019 | 0.343 | 0.045 | 0.895 |
| NAMPT | 0.016 | 0.757 | 0.138 | 0.401 | 0.015 | 0.003 | 0.121 | 0.004 |
| CUBN | 0.035 | 0.285 | 0.038 | 0.246 | 0.047 | 0.024 | 0.079 | 0.018 |
| APH1A | 0.048 | 0.123 | 0.039 | 0.043 | <0.001 | 0.011 | <0.001 | 0.016 |
| SSUH2 | 0.010 | 0.201 | 0.015 | 0.113 | 0.022 | 0.396 | 0.400 | 0.094 |
| RHOT1 | 0.099 | 0.597 | 0.126 | 0.610 | 0.001 | 0.023 | 0.002 | 0.027 |
| NHSL1 | 0.012 | 0.732 | 0.037 | 0.700 | <0.001 | 0.153 | 0.007 | 0.354 |
| TACC2 | 0.091 | 0.973 | 0.177 | 0.728 | 0.002 | 0.183 | 0.087 | 0.245 |
| FRAS1 | 0.002 | 0.039 | 0.001 | 0.051 | 0.001 | 0.107 | 0.033 | 0.085 |
| PTPRQ | 0.013 | 0.862 | 0.170 | 0.861 | 0.130 | 0.404 | 0.018 | 0.032 |
| CYP4B1 | 0.051 | 0.327 | 0.054 | 0.150 | 0.013 | 0.269 | 0.136 | 0.268 |
| HMGCLL1 | 0.030 | 0.060 | 0.597 | 0.299 | 0.011 | 0.834 | 0.023 | 0.395 |
| FAM83A | 0.018 | 0.207 | 0.002 | 0.348 | 0.003 | 0.865 | 0.029 | 0.671 |
| CPB1 | 0.020 | 0.330 | 0.024 | 0.228 | 0.006 | 0.394 | 0.002 | 0.185 |
| SORBS2 | 0.104 | 0.418 | 0.285 | 0.962 | 0.101 | 0.151 | 0.417 | 0.149 |
| C2orf65 | 0.042 | 0.273 | 0.361 | 0.583 | <0.001 | 0.238 | 0.010 | 0.173 |
| CAMK2N1 | 0.011 | 0.433 | 0.009 | 0.794 | 0.489 | 0.028 | 0.986 | 0.094 |
| NOS3 | 0.022 | 0.246 | 0.031 | 0.687 | 0.034 | 0.753 | 0.064 | 0.760 |
| KCNH6 | 0.015 | 0.019 | 0.080 | 0.008 | 0.001 | 0.050 | 0.001 | 0.002 |
| TRIML2 | 0.005 | 0.005 | 0.008 | 0.003 | 0.015 | 0.338 | 0.019 | 0.859 |
| MAP3K15 | 0.011 | 0.024 | 0.142 | 0.025 | 0.001 | 0.578 | 0.003 | 0.217 |
| TMEM232 | 0.030 | 0.359 | 0.172 | 0.298 | 0.001 | 0.034 | 0.001 | 0.003 |
| CYHR1 | 0.041 | 0.167 | 0.086 | 0.019 | 0.001 | 0.724 | 0.001 | 0.586 |
| VEPH1 | 0.125 | 0.092 | 0.282 | 0.04 | <0.001 | 0.025 | 0.006 | 0.062 |
| AC018705.5 | 0.013 | 0.061 | 0.012 | 0.045 | 0.018 | 0.920 | 0.078 | 0.876 |
| SPANXA2_OT1 | 0.029 | 0.048 | 0.151 | 0.076 | 0.124 | 0.053 | 0.153 | 0.200 |
| RP11_69C17.1 | 0.007 | 0.400 | 0.029 | 0.636 | 0.012 | 0.246 | 0.131 | 0.561 |
| RP11_696N14.1 | 0.068 | 0.160 | 0.614 | 0.447 | 0.017 | 0.982 | 0.190 | 0.691 |
Abbreviations: DEE, differentially expressed exon; OS, overall survival; PFS, progression-free survival.
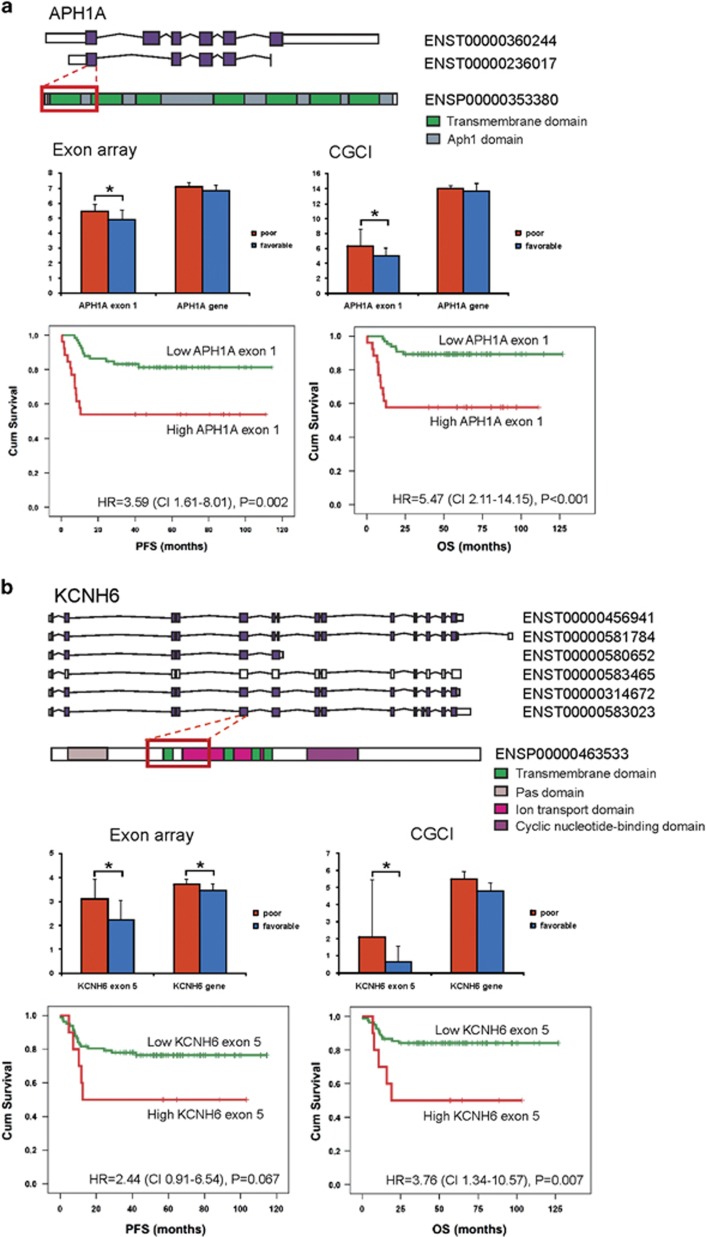
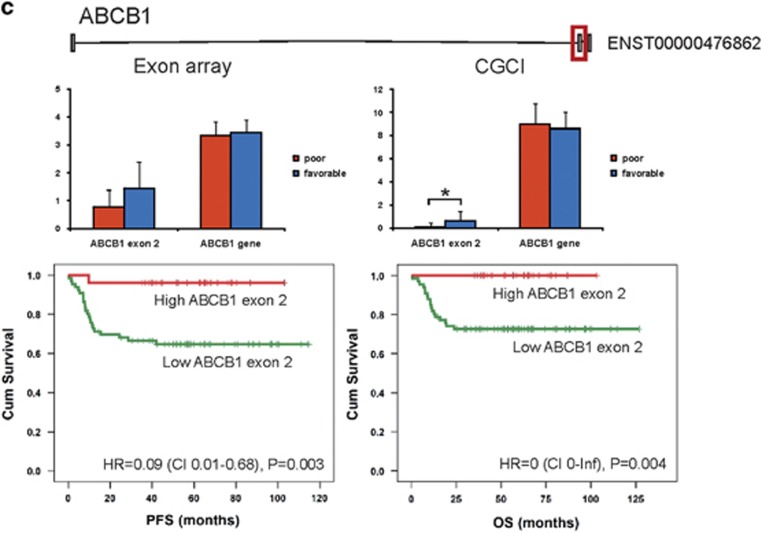
Some of the validated survival-associated DEEs that targeted protein coding regions could be linked to their protein-level functions. For example, the first coding exon of APH1A overexpressed in patients with poor prognosis corresponded to the entire first APH-1 transmembrane domain (Figure 4a). Similarly, the exon with differential expression in the KCNH6 gene mapped to the amino acids 226–367 including three transmembrane helix regions, and exon skipping was associated with better survival (Figure 4b). This suggests that differential coding exon usage could alter the protein functions and thus result in drug resistance and disease progression. On the contrary, ABCB1 represents a gene, where exon 2 skipping occurring in the promoter region is an unfavorable event and correlates with poor survival (Figure 4c).
Figure 4.
Differentially expressed exons may affect the functional properties of the protein and are associated with survival. (a–c) The upper panels show the domain information, the middle panels show the exon and gene expression in the discovery and validation cohorts and the lower panels show Kaplan–Meier survival plots of the exons in DLBCL patients (validation cohort).
Discussion
There has been an increasing interest in studying whole genome-level alterations in DLBCL during the recent years.9 The contribution of these studies in the identification of key elements in DLBCL pathogenesis has been crucial, yet the impact of AS on DLBCL pathogenesis and survival has to date remained largely unexplored. In this work, we have performed microarray-based exon and whole transcriptome profiling on freshly frozen lymphoma tissue collected prospectively from DLBCL patients treated homogenously in a Nordic phase II study.22 We show association of AS genes with molecular subtypes and survival. Interestingly, exon-level profile can separate clinically high-risk DLBCL patients into subgroups with poor and favorable survival more accurately than gene-level profile. We observe that many of the genes are differentially expressed only at the exon level but not at the gene level and would have been missed if only gene-level analysis or conventional 3′ oligonucleotide microarrays had been used. Finally, we validate molecular subtype and survival association of the key DEGs and DEEs in an independent DLBCL cohort. Our observations provide important basis for understanding the mechanisms of DLBCL pathogenesis and prognosis, and development of novel therapeutic strategies.
Approximately half of the AS events target noncoding regions. A significant proportion of the alterations affected 5′ and 3′ UTRs, suggesting modulation of epigenetic regulatory pathways in DLBCL. Although alternative exon usage in the coding regions can generate different protein isoforms, splicing in the noncoding 3′ UTRs can compromise microRNA-dependent gene regulation and change the composition of translational regulatory elements, or result in differential promoter usage in 5′ UTRs that in turn can further alter protein expression.41 Moreover, the upstream promoter site may be selectively affected and differential promoter usage produce N-terminal splice variants.42 Our results further suggest that the splicing mechanism might be affected through modulation of splicing factors themselves, providing yet another regulatory level for gene expression.
A novel finding from the pathway analysis of the DEGs was the enrichment of the genes in the circadian rhythm pathway. Disturbances of the mammalian clock genes have been linked to tumorigenesis. In DLBCL, circadian genes CEBPA and its downstream target PER2 are highly deregulated,43 and BMAL1 (ARNTL) is epigenetically inactivated.44 Furthermore, genetic variants of CRY2 have been associated with a risk of non-Hodgkin’s lymphoma. In our exon array results, the expression of the circadian clock genes PER1 and ARNTL was reduced in relapsed patients and thus associated with dismal prognosis.
ATP-binding cassette transporters, in turn, represent a novel group of genes we found to be regulated at the exon level and thereby potentially affected by AS. To date, ATP-binding cassette transporters have been mostly recognized from their contribution to drug resistance. The highly conserved ATP-binding cassette domains of these transporters provide the ATP-powered translocation of many substrates across the membranes, whereas the transmembrane domains creating the translocation pathway are more variable. According to our domain prediction analysis of splicing-associated ATP-binding cassette transporters, many of the affected domains included either a transmembrane domain or an ATP-binding cassette domain that might directly have an effect to the proper function of the transporter. Most of the AS-associated ATP-binding cassette transporters belonged to ABCA family. These included genes encoding ABCA2 and ABCA3 that have been shown to contribute to drug resistance in T-cell acute lymphoblastic leukemia.45 In addition, increased expression of ABCA3 has been linked to enhanced exosomal evasion of humoral immunotherapy.46
We validated the DEGs and DEEs in an independent patient cohort consisting of RNAseq data. APH1A was one of the interesting examples of the AS genes that could be validated by RNAseq and linked to protein-level functions. It encodes an essential component of the multi-transmembrane γ-secretase complex that is required for the cleavage and activation of integral membrane proteins, including Notch.47 Considering the increasing evidence of the deregulated Notch signaling in cancer progression, γ-secretases playing an important role in Notch activation and that APH1A is critically required for γ-secretase activity, our finding showing that alternative exon usage of APH1A gene has prognostic impact in DLBCL may have therapeutic implications. Thus, future studies should be directed to enhance our understanding of the function of APH1A isoforms and their relation to Notch signaling in DLBCL that may further improve opportunities for the design of selective lymphoma therapeutics.
KCNH6, also known as HERG2, encodes a pore-forming subunit of a voltage-gated potassium channel. Potassium channels comprise the largest family of ion channels encoded by genes with phenotypic diversity generated through alternative splicing, variable association of channel subunits and posttranslational modifications. Recent data indicate that blocking of the ion channel activity can impair cancer cell growth.48 Interestingly, HERG potassium channels are constitutively active in acute myeloid leukemia,49, 50 and have favorable prognostic impact on survival. In our study, no differences were observed in HERG2 gene expression levels between poor and good prognosis patients. However, low expression of exon 5, which corresponds to the first transmembrane and ion transport domains, was associated with favorable survival. The result suggests that these functional domains mediate prosurvival signals that can be overcome by exon 5 skipping. ABCB1 in turn represents a gene where exon 2 skipping occurring in the promoter region is an unfavorable event correlating with lymphoma progression.
Taken together, the results presented herein are promising and novel, and suggest a significant role for alternative exon usage in the molecular profiles of DLBCL. The importance of studying connections between splicing and DLBCL is emphasized by the possibility that some AS isoforms drive progression and may represent attractive therapeutic targets even if total gene expression is not affected. Moreover, unique or sets of isoforms may be used as biomarkers for disease progression.
Acknowledgments
Dr Ping Chen is thanked for help in analyzing exon array data. The diffuse large B cell lymphoma (DLBCL) data set is part of the Cancer Genomics Characterization Initiative (CGCI), supported by NCI contract N01-C0-12400 (http://cgap.nci.nih.gov/cgci.html).
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
The authors declare no conflict of interest.
Supplementary Material
References
- Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med 2002; 346: 235–242. [DOI] [PubMed] [Google Scholar]
- Pfreundschuh M, Trumper L, Osterborg A, Pettengell R, Trneny M, Imrie K et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. Lancet Oncol 2006; 7: 379–391. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008; 359: 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002; 346: 1937–1947. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003; 198: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011; 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 2013; 122: 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol 2015; 52: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene 2015; 34: 1–14. [DOI] [PubMed] [Google Scholar]
- Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol 2007; 8: 349–357. [DOI] [PubMed] [Google Scholar]
- Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays 2006; 28: 378–386. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li HR, Fan JB, Wang-Rodriguez J, Downs T, Fu XD et al. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics 2006; 7: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pal S, Bi Y, Tchou J, Davuluri RV. Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med 2013; 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood 2008; 111: 2816–2824. [DOI] [PubMed] [Google Scholar]
- Aguiar RC, Yakushijin Y, Kharbanda S, Tiwari S, Freeman GJ, Shipp MA. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood 1999; 94: 2403–2413. [PubMed] [Google Scholar]
- Nagel S, Hirschmann P, Dirnhofer S, Gunthert U, Tzankov A. Coexpression of CD44 variant isoforms and receptor for hyaluronic acid-mediated motility (RHAMM, CD168) is an International Prognostic Index and C-MYC gene status-independent predictor of poor outcome in diffuse large B-cell lymphomas. Exp Hematol 2010; 38: 38–45. [DOI] [PubMed] [Google Scholar]
- Stauder R, Eisterer W, Thaler J, Gunthert U. CD44 variant isoforms in non-Hodgkin's lymphoma: a new independent prognostic factor. Blood 1995; 85: 2885–2899. [PubMed] [Google Scholar]
- Terpe HJ, Koopmann R, Imhof BA, Gunthert U. Expression of integrins and CD44 isoforms in non-Hodgkin's lymphomas: CD44 variant isoforms are preferentially expressed in high-grade malignant lymphomas. J Pathol 1994; 174: 89–100. [DOI] [PubMed] [Google Scholar]
- Tzankov A, Pehrs AC, Zimpfer A, Ascani S, Lugli A, Pileri S et al. Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell-like types: a tissue microarray analysis of 90 cases. J Clin Pathol 2003; 56: 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holte H, Leppa S, Bjorkholm M, Fluge O, Jyrkkio S, Delabie J et al. Dose-densified chemoimmunotherapy followed by systemic central nervous system prophylaxis for younger high-risk diffuse large B-cell/follicular grade 3 lymphoma patients: results of a phase II Nordic Lymphoma Group study. Ann Oncol 2013; 24: 1385–1392. [DOI] [PubMed] [Google Scholar]
- Chen P, Lepikhova T, Hu Y, Monni O, Hautaniemi S. Comprehensive exon array data processing method for quantitative analysis of alternative spliced variants. Nucleic Acids Res 2011; 39: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA 2003; 100: 9991–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012; 7: e51862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014; 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40(Database issue): D261–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer MJ, Ghesquieres H, Jais JP, Witzig TE, Haioun C, Thompson CA et al. Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy. J Clin Oncol 2014; 32: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinis JM, Hutti JE, Homer CR, Cobb BA, Cantley LC, McDonald C et al. IkappaB kinase alpha phosphorylation of TRAF4 downregulates innate immune signaling. Mol Cell Biol 2012; 32: 2479–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattenloehner S, Chuvpilo S, Langebrake C, Reinhardt D, Muller-Hermelink HK, Serfling E et al. Novel RUNX1 isoforms determine the fate of acute myeloid leukemia cells by controlling CD56 expression. Blood 2007; 110: 2027–2033. [DOI] [PubMed] [Google Scholar]
- Amor S, Remy S, Dambrine G, Le Vern Y, Rasschaert D, Laurent S. Alternative splicing and nonsense-mediated decay regulate telomerase reverse transcriptase (TERT) expression during virus-induced lymphomagenesis in vivo. BMC Cancer 2010; 10: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meeker AK, Kowalski J, Tsai HL, Somervell H, Heaphy C et al. Telomere length is related to alternative splice patterns of telomerase in thyroid tumors. Am J Pathol 2011; 179: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biselli-Chicote PM, Oliveira AR, Pavarino EC, Goloni-Bertollo EM. VEGF gene alternative splicing: pro- and anti-angiogenic isoforms in cancer. J Cancer Res Clin Oncol 2012; 138: 363–370. [DOI] [PubMed] [Google Scholar]
- Akisik E, Bavbek S, Dalay N. CD44 variant exons in leukemia and lymphoma. Pathol Oncol Res 2002; 8: 36–40. [DOI] [PubMed] [Google Scholar]
- Chen L, Juszczynski P, Takeyama K, Aguiar RC, Shipp MA. Protein tyrosine phosphatase receptor-type O truncated (PTPROt) regulates SYK phosphorylation, proximal B-cell-receptor signaling, and cellular proliferation. Blood 2006; 108: 3428–3433. [DOI] [PubMed] [Google Scholar]
- Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood 2004; 104: 2933–2935. [DOI] [PubMed] [Google Scholar]
- Blenk S, Engelmann J, Weniger M, Schultz J, Dittrich M, Rosenwald A et al. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer Inform 2007; 3: 399–420. [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH et al. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 1998; 394: 793–797. [DOI] [PubMed] [Google Scholar]
- Tamaki A, Ierano C, Szakacs G, Robey RW, Bates SE. The controversial role of ABC transporters in clinical oncology. Essays Biochem 2011; 50: 209–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Bartsch L, Berkmann JC, Frose K, Heitmann J, Hoppe C et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med 2016; 8: 442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y et al. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res 2003; 13: 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoennissen NH, Thoennissen GB, Abbassi S, Nabavi-Nouis S, Sauer T, Doan NB et al. Transcription factor CCAAT/enhancer-binding protein alpha and critical circadian clock downstream target gene PER2 are highly deregulated in diffuse large B-cell lymphoma. Leuk Lymphoma 2012; 53: 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 2009; 69: 8447–8454. [DOI] [PubMed] [Google Scholar]
- Efferth T, Gillet JP, Sauerbrey A, Zintl F, Bertholet V, de Longueville F et al. Expression profiling of ATP-binding cassette transporters in childhood T-cell acute lymphoblastic leukemia. Mol Cancer Ther 2006; 5: 1986–1994. [DOI] [PubMed] [Google Scholar]
- Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M et al. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci USA 2011; 108: 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Shah S, Li H, Yu C, Han W, Yu G. Mammalian APH-1 interacts with presenilin and nicastrin and is required for intramembrane proteolysis of amyloid-beta precursor protein and Notch. J Biol Chem 2002; 277: 45013–45019. [DOI] [PubMed] [Google Scholar]
- Jehle J, Schweizer PA, Katus HA, Thomas D. Novel roles for hERG K(+) channels in cell proliferation and apoptosis. Cell Death Dis 2011; 2: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillozzi S, Masselli M, De Lorenzo E, Accordi B, Cilia E, Crociani O et al. Chemotherapy resistance in acute lymphoblastic leukemia requires hERG1 channels and is overcome by hERG1 blockers. Blood 2011; 117: 902–914. [DOI] [PubMed] [Google Scholar]
- Smith GA, Tsui HW, Newell EW, Jiang X, Zhu XP, Tsui FW et al. Functional up-regulation of HERG K+ channels in neoplastic hematopoietic cells. J Biol Chem 2002; 277: 18528–18534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.