Abstract
The SOX4 transcription factor is a key regulator of embryonic development, cell-fate decision, cellular differentiation and oncogenesis. Abnormal expression of SOX4 is related to malignant tumor transformation and cancer metastasis. However, no reports are available regarding the clinical significance of SOX4 in acute myeloid leukemia (AML) and the role of SOX4 in leukemogenesis. In the current study, we found that AML patients with low bone marrow (BM) SOX4 expression had higher remission rates and longer overall survival than those with high SOX4 expression, regardless of age, white blood cell count at diagnosis, karyotype profile and NPM1/FLT3-ITD status. To elucidate the role of SOX4 in leukemogenesis, we generated a transgenic zebrafish model that overexpressed human SOX4 in the myeloid lineage Tg(spi1-SOX4-EGFP). These transgenic zebrafish showed, at 5 months of age, increased myelopoiesis with dedifferentiation in kidney marrow. At 9 months of age, their kidney structure was significantly effaced and distorted by increased infiltration of myeloid progenitor cells. These results suggest that SOX4 is not only an independent prognostic factor of AML, but also an important molecular factor in leukemogenesis.
Introduction
The SOX4 belongs to the SOX (Sry-related high-mobility group box) family and has been characterized as a transcription factor.1, 2 Over the past decade, multiple functions of SOX4 have been unveiled, and the protein is now known to play important roles in embryonic development, cell-fate decision and cellular differentiation.3, 4, 5, 6 Overexpression and amplification of SOX4 have been implicated in various cancers and are correlated with poor prognosis.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 In mouse models, previous studies demonstrated that the upregulation of Sox4 can be induced by and then cooperate with the aberrant expression of AML1-ETO,17 NUP98-DDX10(ref. 18) and PML-RARa;19 the overexpression of HOXA9(ref. 20) CREB21 and Evi1;22 and the haplosufficiency of PU.1(ref. 23) to trigger leukemogenesis. Conversely, mice lacking Sox4 have shown pro-B-lymphocyte expansion and defects in cardiac outflow tract formation.24 Furthermore, a previous study that employed retroviral transduction of Sox4 and bone marrow transplantation techniques revealed that increased Sox4 expression may cooperate with the deregulation of Mef2c expression to induce myeloid leukemia in recipient mice.25
Recently, Sox4 gene was reported to be a direct target of C/EBPα (CCAAT/enhancer-binding protein-α). C/EBPα is known to inhibit the self-renewal of leukemic cells and to restore cellular differentiation. The overexpression of Sox4 that results from C/EBPα inactivation contributes to the development of a type of leukemia that is characterized by a distinct leukemia-initiating cell phenotype. This recent work further indicated that Sox4 is a key oncogenic target and critical mediator of C/EBPα mutants in acute myeloid leukemia (AML), suggesting a potential novel therapeutic approach to the treatment of this disease.26, 27 However, the clinical implications of SOX4 expression and its role of AML leukemogenesis are not well understood.
The current study investigated the relationship between bone marrow (BM) SOX4 expression and clinicopathological parameters of de novo AML and also evaluated the prognostic value of SOX4 expression for AML patients. This is the first study to report on the prognostic implications of SOX4 expression for AML patients. Our immunohistochemical staining results illustrated that high BM SOX4 protein content is an independent unfavorable prognostic factor for overall survival. A meta-analysis that we conducted using an online data cohort retrieved from PrognoScan (a new database for meta-analysis of the prognostic value of genes; http://www.abren.net/PrognoScan/) revealed similar findings. Finally, as several zebrafish models have been proposed for study of hematopoiesis and myeloid malignancies in recent years,28, 29, 30, 31, 32 we used a transgenic SOX4 zebrafish line to demonstrate, for the first time, that SOX4 overexpression driven by spi-1 (a myeloid-specific promoter) can lead to expanded myelopoiesis with leukemic phenotype.
Materials and methods
Study patients
From March 2009 to December 2011, a total number of 112 adult patients were enrolled in this study. This study was approved by the institutional review board of the National Taiwan University Hospital and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. See Supplementary Materials and Methods for details.
Immunocytochemical staining of SOX4 protein
To assess SOX4 expression in leukemic cells, immunocytochemical staining was performed.33 See Supplementary Materials and Methods for details.
Generation and husbandry of transgenic zebrafish
Zebrafish (Danio rerio) embryos, larvae and adult fish were maintained at 28 °C under continuous flow and a 14 h light/10 h dark cycle.31 All experiments involving zebrafish were approved by the institutional animal care and use committee of the National Taiwan University. The myeloid-specific spi-1-driven SOX4 transgenic fish was generated as previously described.29, 34 The primer sequences are listed in Supplementary Table 1. See Supplementary Materials and Methods for details.
Isolation of RNA as well as reverse transcription-PCR
Total RNA from various tissues or a total number of 30 embryos was isolated using NucleoSpin and TRIzol (Invitrogen, Carlsbad, CA, USA), then reverse transcribed into complementary DNA (cDNA) with the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Carlsbad, CA, USA) and finally amplified by PCR with KOD-FX Taq polymerase (Toyobo, Osaka, Japan), in accordance with the manufacturer’s instructions. See Supplementary Materials and Methods for details.
Quantitative reverse transcription-PCR
Quantitative reverse transcription-PCR was carried out in an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR green as the detection dye (Power SYBR Green PCR Master Mix, Applied Biosystems). PCR conditions and subsequently cycle thresholds (Ct) calculations were according to standard protocols. The primer sequences of various target genes are listed in Supplementary Table 1. See Supplementary Materials and Methods for details.
Whole-mount in situ hybridization and whole mount immunohistochemical staining
Whole-mount in situ hybridization analysis was performed essentially as previously described32 using the antisense digoxigenin-labeled myeloperoxidase (mpo) RNA probe that was generated from the partial cDNA (901 bp) sequence of mpo was amplified by PCR with T3-mpo-F and T7-mpo-R primers (Supplementary Table 1). See Supplementary Materials and Methods for details.
For whole-mount immunohistochemical staining analysis, embryos were fixed, dehydrated and rehydrated as procedures for whole-mount in situ hybridization. Embryos were then incubated in the blocking solution, reacted with rabbit anti-GFP antibodies (1:100 dilutions; GTX113617, GeneTex, San Antonio, TX, USA), and then finally reacted with Alexa Fluor 488-labeled goat-anti rabbit secondary antibodies (1:100 dilutions; A-11008, Invitrogen). See Supplementary Materials and Methods for details.
Tissue collection and histochemical analysis
Tissue collection and histochemical analysis were performed as previously described.29 See Supplementary Materials and Methods for details.
Cytological analysis of kidney marrow and peripheral blood
Collection and cytological analysis of blood cells from kidney marrow (KM) and peripheral blood were performed as previously decribed.29 See Supplementary Materials and Methods for details.
Flow cytometric analysis
Collection and flow cytometric analysis of blood cells from KM and peripheral blood were performed as previously decribed.29 See Supplementary Materials and Methods for details.
Sudan black staining
Sudan black staining was performed as previously described.35 See Supplementary Materials and Methods for details.
Myeloperoxidase staining
KM smears were fixed with pH 6.6 formalin-acetone (Muto Pure Chemicals Co., Ltd, Tokyo, Japan) for 30 s, washed with double-distilled water and then dried. Slides were subsequently treated with 3,3’-diaminobenzidine-tetrahydrochloride (Sigma Chem. Co., St Louis, MO, USA) for 15 min at room temperature. After being washed and dried, slides were counterstained with hematoxylin for 3 min.
Statistical analysis
All statistical analyses performed for this study involved comparison between experimental and control groups using two-tailed Student’s t-tests, Mann–Whitney U-tests, one-way analysis of variance, χ2 test or Fisher’s exact test and multivariate analysis with Cox proportional hazards regression models as previously described.36 We used Kaplan–Meier estimation techniques to plot survival curves and log-rank tests to examine difference between groups as previously described.37 P-values of <0.05 were considered statistically significant. See Supplementary Materials and Methods for details.
Results
BM SOX4 expression as an independent prognostic factor of AML
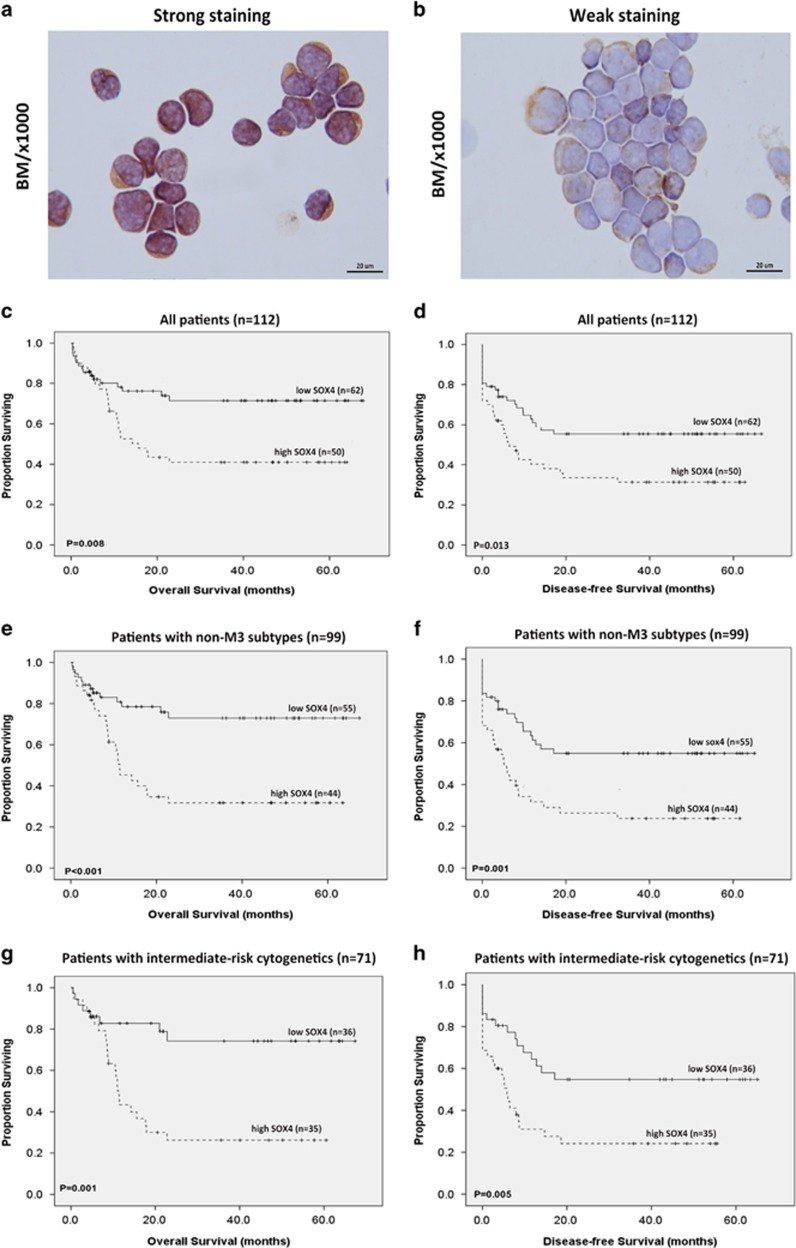
Results of immunocytochemical staining revealed that BM SOX4 expression varied greatly in AML patients. We divided patients into two groups according to the intensity and extent of SOX4 expression as follows: low expression group (score 0–2, n=62, Figure 1a) and high expression group (score 3–4, n=50, Figure 1b), respectively. The various clinical manifestations of AML did not show significant differences in terms of SOX4 expression (Supplementary Table 2); except that high SOX4 expression was somehow correlated with CD11b expression in leukemia cells (Supplementary Table 3), and AML patients with low SOX4 expression tended to have favorable-risk karyotyping (P=0.0866, Supplementary Table 2). We did not observe significant differences between the high and low expression groups in terms of age, gender, hemograms, nucleophosmin-1 (NPM1) mutation and FLT3-ITD (internal tandem duplication of the fms-like tyrosine kinase-3). In addition, of the 112 AML patients who underwent conventional intensive induction chemotherapy, 85 (75.9%) achieved complete remission, and the high and low expression groups showed similar probabilities of achieving first complete remission (36/50, 72 vs 49/62, 79%, P=0.3219). However, high SOX4 expression was associated with increased relapse rates compared with low SOX4 expression (19/36, 52.8 vs 13/49, 26.5%, P=0.028).
Figure 1.
Kaplan–Meier survival analysis according to SOX4 expression in AML patients. Representative immunocytochemical stain of SOX4 protein in bone marrow specimens from patients with low SOX4 protein (a) and with high SOX4 protein (b). Overall survival and disease-free survival were analyzed in all patients with de novo AML (c, d), in patients with non-M3 subtypes (e, f), patients with intermediate-risk cytogenetics (g, h) and patients with normal cytogenetics (i, j). Number of patients and median survival time are indicated in each subgroup.
Furthermore, with a median follow-up period of 46.7 months (range: 0.3–70.9 months), SOX4 expression was associated with overall survival and disease-free survival in all patients with de novo AML (P=0.008 and P=0.013, respectively, Figure 1c), patients with non-M3 subtypes (P<0.001 and P=0.001, respectively, Figure 1d), patients with intermediate-risk cytogenetics (P=0.001 and P=0.005 respectively, Figure 1e) or even in those with normal karyotype profile (P=0.022 and P=0.111, Figure 1f). In multivariate analysis, high SOX4 expression was found to be an independent poor prognostic factor of overall survival (relative risk 1.924, 95% confidence interval 1.020–3.628, P=0.043) irrespective of age, white blood cell count at diagnosis, karyotype profile and NPM1/FLT3-ITD status (Table 1).
Table 1. Multivariate analysis (Cox regression) of SOX4 expression on the overall survival and disease-free survival.
| Variables |
Overall survival |
Disease-free survival |
||||||
|---|---|---|---|---|---|---|---|---|
|
95% CI |
95% CI |
|||||||
| RR | Lower | Upper | P value | RR | Lower | Upper | P value | |
| Agea | 1.023 | 1.002 | 1.044 | 0.034 | 1.012 | 0.994 | 1.029 | 0.203 |
| WBCb | 1.004 | 1.000 | 1.007 | 0.029 | 1.004 | 1.001 | 1.006 | 0.012 |
| Karyotypec | 3.038 | 1.188 | 7.771 | 0.020 | 2.127 | 1.105 | 4.096 | 0.024 |
| NPM1/FLT3-ITDd | 1.197 | 0.510 | 2.811 | 0.680 | 1.345 | 0.651 | 2.778 | 0.423 |
| High SOX4e | 1.924 | 1.020 | 3.628 | 0.043 | 1.663 | 0.983 | 2.813 | 0.058 |
Abbreviations: CI, confidence interval; RR, relative risk; WBC, white blood cell.
The risk by 1 year older in age.
The risk by 1000/μl increase in WBC.
Unfavorable cytogenetics vs others.
NPM1mut/FLT3-ITDneg vs other subtypes.
High SOX4 expression vs low SOX4 expression.
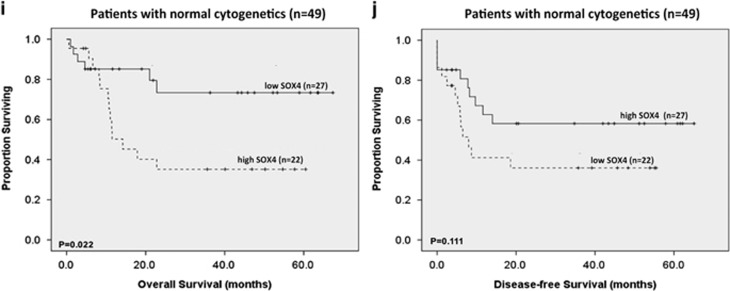
Generation of Tg(spi1:SOX4-EGFP) zebrafish lines by tol2 transposon system
To further investigate the role of SOX4 in leukemogenesis, we use Multisite Gateway (Invitrogen, Grand Island, NY, USA) and Tol2 transposon methods to generate pTolCG-spi1:SOX4-EGFP constructs. In these constructs, SOX4 and EGFP expression is controlled by the myeloid-specific spi1 (also known as pu.1) promoter. The pTolCG-spi1:SOX4-EGFP constructs were then coinjected with Tol2 transposase mRNA into one-cell embryos of wild-type AB strain zebrafish to generate transgenic zebrafish founder, namely Tg(spi1:SOX4-EGFP) (Figure 2a). Two transgenic SOX4 lines (TG1 and TG2) were generated in parallel. F1 fish were generated by crossing SOX4 founders and wild-type zebrafish, and SOX4 transgene expression was demonstrated by semi-quantitative PCR (Figure 2b). The expression of SOX4-EGFP fusion proteins was verified using whole-mount immunohistochemical staining with anti-EGFP antibodies. In contrast to wild-type fish, which showed no enhanced green fluorescent protein (EGFP), the Tg(spi1:SOX4-EGFP) fish exhibited EGFP in the yolk sac and in caudal hematopoietic tissue after 48 h post fertilization (Figure 2c). We further investigated SOX4 expression by applying immunohistochemistry staining anti-SOX4 on tissue sections of 5-month-old Tg(spi1:SOX4-EGFP), and found that SOX4 expression was restricted to a small fraction of cells in the KM (Figure 2d).
Figure 2.
Generation and characterization of the Tg(spi1:SOX4-EGFP) transgenic zebrafish. (a) Schematic diagram showing the LR recombination reaction used to generate the expression constructs, including three entry clones (p5E-spi1, pME-SOX4 and p3E-EGFPpA) and a destination vector (pDestTol2CG2) that contains the cmlc2:EGFP-pA expression cassette. The final construct (pTolCG-spi1:SOX4-EGFP) is shown at the bottom of the figure. (b) Results of semiquantitative PCR showing the expression of SOX4 in TGs and wild-type embryos at 20, 48 and 72 h post fertilization (HPF) and at 5 days. Negative control: non-template; Positive control: pTolCG-spi1:SOX4-EGFP plasmid. (c) Whole-mount immunohistochemical staining (IHC) showed EGFP-positive cells in the heart (yellow arrow) as well as around the yolk sac and caudal hematopoietic tissue (CHT) at 48 HPF. The panel shows GFP+ cells at a higher magnification. (d) Tissue sections showing SOX4-positive cells in the KM of Tg(spi1:SOX4-EGFP) fish but not in those of wild-type fish. The panel shows SOX4-positive cells at a higher magnification. (e) Results from quantitative reverse transcription-PCR (RT-PCR) analysis of hematopoietic marker genes in SOX4 transgenic zebrafish embryos and in wild-type fish embryos. Data are presented as the mean±s.e.m. from three independent experiments. (f, g) Results from whole-mount in situ hybridization (WISH) of mpo and Sudan black (SB) staining showing mpo-positive cells in CHT at 48 HPF and SB-positive cells in CHT at 72 HPF, respectively (magnification: × 40). The panel shows positive cells at a higher magnification (× 100), respectively. Quantification of mpo-positive cells (h) and SB-positive cells (i). Differences among variables were assessed using Student’s t-test.
Tg(spi1:SOX4-EGFP) zebrafish embryos underwent normal hematopoiesis
To assess whether SOX4 overexpression driven by spi1 has an effect on early hematopoietic processes in transgenic zebrafish, several primitive and definitive markers including cebpa, csf1r, gata1, l-plastin, mpo, mpeg1, spi1, c-myb and runx1 were examined by quantitative reverse transcription-PCR analysis. We found that there were no significant differences of these hematopoiesis-related transcription factors in transgenic zebrafish at both primitive and definitive stages (Figure 2e). Further whole-mount in situ hybridization experiments revealed that comparable numbers of mpo-positive cells were located in the aorta-gonad-mesonephros and caudal hematopoietic tissue regions in both SOX4 and wild-type fish at 48 h post fertilization (Figures 2f and h). Sudan black staining reveled similar results for granulocyte colonized in caudal hematopoietic tissue at 72 h post fertilization (Figures 2g and i). These results indicate that SOX4 transgenic zebrafish were undergoing normal hematopoietic processes during the larval stage.
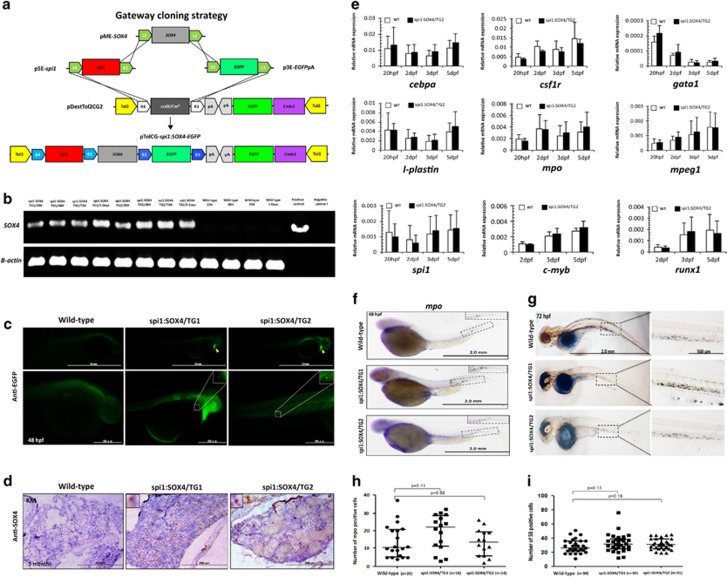
Myeloid-specific expression of SOX4 results in expanded myelopoiesis with poor differentiation in adult zebrafish
Myeloid malignancies are known to result from multiple long-term processes. Therefore, long-term examination of adult zebrafish was reasonable even though the overexpression of SOX4 did not influence early hematopoiesis. In this study, biopsy sections of kidney tissues showed mild vacuoles in renal tubule structure in 5-month-old SOX4 fish; moreover, kidneys showed mild or moderate effacement, a distorted structure and increased infiltration of myeloid cells in 9- and 12-month-old SOX4 fish (Supplementary Figure 1).
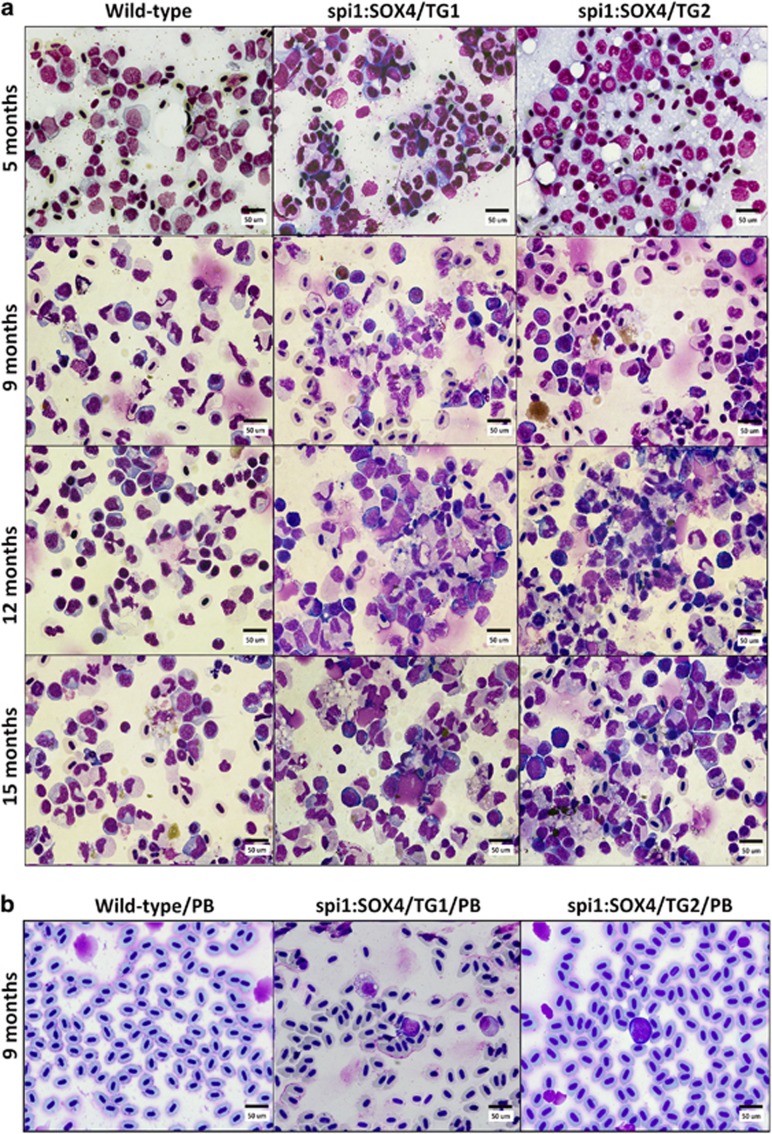
Microscopic observation of KM smears from Tg(spi1:SOX4-EGFP) fish were analyzed at the ages of 5, 9, 12 and 15 months. The KM of 5-month-old SOX4 zebrafish was comparable with the KM of age-matched 5-month-old AB wild-type fish; however, KM of SOX4 transgenic fish demonstrated a greater number of myeloid progenitors and an excess of blast cells with focal aggregation at 9, 12 and 15 months, implying that myeloid transformation is age dependent (Figure 3a). These blast cells in KM were generally of medium to large size and were characterized by a mildly basophilic cytoplasm with scanty or fewer granule as well as a high nucleus-to-cytoplasm ratio (Figure 3a). In addition, a few typical blast cells in peripheral blood were observed in 9-month-old SOX4 fish (Figure 3b).
Figure 3.
Morphological analysis of various blood cell types harvested from the KM and peripheral blood of Tg(spi1:SOX4-EGFP) zebrafish at indicated age. Tg(spi1:SOX4-EGFP) KM smears showed myeloid hyperplasia with increased production of myeloblasts in an age-dependent manner (a) and their peripheral blood smears showed immature myeloid cells (b). Conversely, wild-type fish showed normal hematopoiesis with adequate maturation (magnification: × 1000).
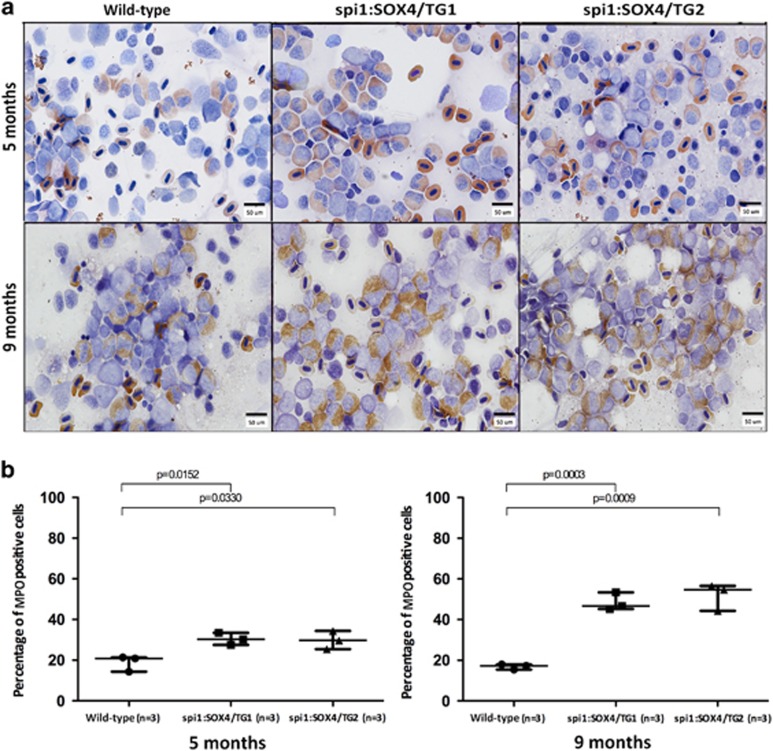
On microscopic examination of blood cells in KM smears, we found that compared with age-matched 9-month-old AB wild-type fish, the KM of 9-month-old SOX4 zebrafish showed a greater myeloid-to-erythroid ratio, a higher number of myeloid progenitors and decreased number of both immature and mature erythroid cells. Additional significant differences in myeloid-to-erythroid ratios and in all blood components of KM were found in 12- and 15-month-old transgenic SOX4 zebrafish (Table 2). Similar results were demonstrated using an additional flow cytometric analysis (Supplementary Figure 2). In addition, MPO staining further confirmed that there were significantly more myeloid cells in SOX4 fish compared with wild-type control fish of the same age (Figure 4). Taken together, our findings reveal that continuous overexpression of SOX4 led to an expansion of myeloid cells in the KM of adult fish, and that the majority of accumulated myeloid cells were immature.
Table 2. Morphological analysis of blood cell types in the kidney marrow from SOX4 transgenic fish compared with wild type.
| Total number of zebrafisha (n=65) | Myeloid progenitors (blasts/blast-like; %) | Myelomonocyte/neutrophil (%) | Lymphocyte (%) | Immature erythroid (%) | Mature erythroid (%) | M/E ratio |
|---|---|---|---|---|---|---|
| 5M-wild-type (n=5) | 5.27±0.74 | 45.00±5.36 | 15.33±1.36 | 10.27±1.82 | 24.13±3.19 | 5.08±1.57 |
| 9M-wild-type (n=5) | 5.73±0.64 | 39.67±5.01 | 14.39±2.97 | 12.27±0.83 | 27.93±6.03 | 3.72:1±0.53 |
| 12M-wild-type (n=5) | 7.00±1.10 | 42.07±3.18 | 16.20±1.12 | 10.06±2.55 | 24.67±4.16 | 4.61±0.64 |
| 15M-wild-type (n=6) | 7.11±1.88 | 40.95±2.33 | 15.95±2.04 | 11.28±1.04 | 25.44±4.06 | 4.30±0.47 |
| 5M-SOX4 (n=12)# | 7.64±2.56 | 37.53±5.22 | 13.33±1.65* | 11.95±3.02 | 29.08±6.40 | 4.10±1.59 |
| 9M-SOX4 (n=12)# | 14.86±7.07** | 50.53±10.33* | 9.69±1.65** | 7.22±3.46* | 17.78±5.19* | 11.62±7.25** |
| 12M-SOX4 (n=11)# | 16.24±7.25** | 58.33±7.14** | 6.09±3.61** | 6.21±2.16* | 13.12±4.38** | 13.55±5.45** |
| 15M-SOX4 (n=9)# | 14.19±5.16** | 50.74±9.15* | 9.59±1.38*** | 7.15±3.29* | 16.74±4.42** | 10.48±4.08** |
Abbreviations: M, month; M/E ratio, myeloid-to-immature erythroid ratio.
Significant differences between the SOX4 transgenic fish and wild-type are indicated (*P<0.05; **P<0.01; ***P<0.001) by Mann–Whitney U-test.
Significant difference of groups between the SOX4 transgenic fish and wild-type are indicated (#P<0.001) by one-way analysis of variance (ANOVA).
A total number of 300 cells per zebrafish was counted to classify the distribution and subtypes of these hematopoietic cells.
Figure 4.
Myeloperoxidase (MPO) staining analysis of spi1:SOX4-EGFP zebrafish. (a) Increased myeloperoxidase was detected in SOX4 transgenic fish at 5 and 9 months (magnification: × 1000). (b) Quantification of MPO-positive cells. Differences among variables were assessed using Student’s t-test.
Discussion
To the best of our knowledge, this is the first study to report that increased BM SOX4 expression is an independent poor prognostic factor of overall survival in AML patients, irrespective of age, white blood cell count, karyotype profile and other genetic markers. The poor prognosis associated with high BM SOX4 expression was also demonstrated in CN-AML patients. These results are consistent with an online transcriptome data (GSE12417-GPL96 cohort) from AML patients (Supplementary Figure 3). Their data were analyzed by PrognoScan, a new database in which meta-analyses to elucidate the prognostic value of genes can be performed. It demonstrated that higher SOX4 mRNA expression led to a worse overall survival in AML patients (hazard ratio 1.21, 95% confidence interval 1.01–1.47, P=0.043). Zhang et al.26 previously reported that SOX4 is a direct target and crucial mediator of C/EBPα mutants in AML. It is therefore possible that poor prognosis is associated with CEBPA mutants or CEBPA methylation. However, in our previous report, we found that AML patients with CEBPA double mutations38 or higher CEBPA methylation39 had a superior response to chemotherapy treatment, and similar findings were reported in studies by other researchers as well.40, 41, 42 The different prognoses associated with SOX4 expression and CEBPA gene status may be explained by the fact that SOX4 expression is also associated with other genetic abnormities including MOZ-TIF2, AML1-ETO, NUP-HOXA9 and FLT3-ITD.26, 43 Of these abnormalities, AML1-ETO was found to be a better prognostic factor for AML,44, 45 and FLT3-ITD was found to be a poor prognostic factor.46 Therefore, the conflicting results could result from the complexity, heterogeneity and multifactorial nature of AML. In addition, results from a cancer microarray database and integrated data (Oncomine; https://www.oncomine.org/resource/login.html) showed that leukemia cells of AML patients (n=542) expressed significantly more SOX4 than did normal cells from controls (n=74) (GSE13164 cohort; Supplementary Figure 4a). Significant differences between M1 and other FAB (French–American–British) categories (GSE14468 cohort) were also found (Supplementary Figure 4b). The mechanisms underlying the increase in SOX4 expression independently associated with poor prognosis and various genetic aberrations still require further study. Nonetheless, complementing chemotherapy with agents that target SOX4 could be beneficial treatment strategy for AML patients.
Zebrafish sox4 proteins (sox4a: accession number. NP_957195.1 and sox4b: accession number NP_998287.1) share a high level of amino acid similarity with human SOX4 (accession number: NP_003098.1) and mouse Sox4 (accession number: NP_033264.2), particularly within the HMG box (sox4a: 100%, sox4b: 97.4%) and the TAD/DD (sox4a: 91.2%, sox4b: 70.6%) functional domains (Supplementary Figure 4a). Phylogenetic analysis revealed that the zebrafish sox4a/b is homologous with SOX4 proteins from other species (Supplementary Figure 5b). The high conservation of SOX4 among humans, mice and zebrafish suggests that SOX4 proteins could share similar functions. We also characterized the mRNA expression of sox4a and sox4b in early developmental stages of transgenic zebrafish. In so doing, we found that the expression levels of sox4b and sox4a in transgenic zebrafish were comparable to those of wild-type fish (data not shown).
The kidneys of Tg(spi1:SOX4-EGFP) zebrafish began showing mild vacuoles in renal tubule structure at the age of the 5 months. At 9 and 12 months, the kidneys further showed mild or moderate effacement, a distorted structure and increased infiltration of myeloid cells. Microscopic examination revealed that the KM blood cells of 5-month-old Tg(spi1:SOX4-EGFP) zebrafish were comparable with those of age-matched AB wild-type fish. However, at 9, 12 and 15 months, KM cells from Tg(spi1:SOX4-EGFP) fish had a greater number of myeloid progenitors and an excess of blast cells with focal aggregation than did KM cells of age-matched AB wild-type fish, implying that myeloid transformation was age dependent. This is the first study to report on transgenic SOX4 zebrafish with myeloid transformation phenotype.
Results from previous research in mouse model studies support our findings. For example, in one study, 73% of mice developed myeloid leukemia after receiving a transplant of MSCV-Sox4-infected marrow, whereas none of the control mice developed leukemia.25 Other reports have also found that Sox4 leads to myeloid malignancies in mouse model as follows. Sox4 may upregulate CREB expression and cooperate with CREB to activate downstream cyclin D1, bcl-2 and c-fos that in turn increases cell proliferation.21 Sox4 may also downregulate Pu.1, leading to Pu.1 haploinsufficiency, which accelerates and increases the penetrance of Sox4-induced leukemia.23 A later study revealed that a novel PAD4/SOX4/PU.1 signaling pathway is involved in the differentiation of leukemic cells into granulocytes.47 Furthermore, in an AKXD23 mouse model of myeloid leukemia, proviral insertions to Sox4 and Evi1 were found to cause increases in Sox4 and Evi1 expressions, respectively; and Sox4 acted alone or cooperated with Evi1 to induce transcriptional activation and subsequent cell proliferation.22 Findings from that research also demonstrated that overexpression of Sox4 in 32Dcl3 cells markedly inhibited cytokine-induced granulocyte maturation and associated cell proliferation.22 Finally, Fung et al.27 suggested that activation of the Wnt pathway, activation of the TGF-FOXO pathway and epigenetic aberration of Sox4 could be responsible for myeloid leukemogenesis.
In conclusion, we reveal that BM SOX4 expression could serve as an informative new biomarker for the clinical prognosis of AML patients. Moreover, we demonstrate that in zebrafish, the myeloid-specific expression of SOX4 can induce leukemic phenotype. Therefore, we suggest that treating AML patients with agents that target SOX4 or its downstream molecules in addition to chemotherapy could be an effective therapeutic strategy. Furthermore, this zebrafish disease model could be useful for studying gene functions involved in myeloid malignancies and to be a valuable in vivo platform for screening antileukemic drugs.
Acknowledgments
This study was supported by the research grants from Taiwan Health Foundation (to L-IL), the National Taiwan University Hospital (NTUH105S-3065 to C-YC), the Ministry of Science and Technology (MOST-102-2628-B-002-029-MY3 and MOST-105-2320-B-002-051-MY3 to L-IL; MOST-103-2628-B-002-008-MY3 and MOST-104-2314-B-002-128-MY4 to H-AH) and the Ministry of Health and Welfare (MOHW105-TDU-B-211-134005 to H-FT), Taiwan. We acknowledge the services provided by Flow Cytometric Analysing and Sorting Core Facility at NTUH; and we also Taiwan Zebrafish Core Facility (TZeCF) at NHRI for providing fish lines and resources; TZeCF is supported by a grant (MOST-104-2321-B-001-045) from the Ministry of Science and Technology, Taiwan.
Author contributions
J-WL and L-IL designed and coordinated the study; J-WL and M-SH were responsible for establishing the transgenic fish and designing individual zebrafish experiments; J-WL, M-SH and H-AH were responsible for flow cytometric and cytomorphologic analysis of zebrafish KM and PB; H-AH performed immunocytochemical staining of human bone marrow; H-AH, C-YC and H-FT contributed patient samples and clinical data; and J-WL, M-SH, H-AH and L-IL wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
The authors declare no conflict of interest.
Supplementary Material
References
- Harley V, Lefebvre V. Twenty Sox, twenty years. Int J Biochem Cell Biol 2010; 42: 376–377. [DOI] [PubMed] [Google Scholar]
- Jafarnejad SM, Ardekani GS, Ghaffari M, Li G. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci 2013; 70: 2677–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, Gan L. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem 2013; 288: 18429–18438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene 2013; 32: 3397–3409. [DOI] [PubMed] [Google Scholar]
- Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol 1997; 27: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol 1999; 22: 295–304. [DOI] [PubMed] [Google Scholar]
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 2006; 66: 3434–3442. [DOI] [PubMed] [Google Scholar]
- Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet 2009; 18: 1343–1352. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhao H, Lu J, Yin J, Zang L, Song N et al. Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn Pathol 2012; 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Fang CL, Hseu YC, Chen CL, Wang JW, Hsu SL et al. Clinical and prognostic implications of transcription factor SOX4 in patients with colon cancer. PLoS One 2013; 8: e67128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Yang X, Chang YW, Qi M, Zhou Z et al. SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis 2013; 16: 301–307. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang J, Zhan X, Lin T, Yang M, Hu J et al. SOX4 is associated with poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun 2014; 452: 614–621. [DOI] [PubMed] [Google Scholar]
- Li L, Li Q, Chen X, Xu M, Li X, Nie L et al. SOX4 is overexpressed in diffusely infiltrating astrocytoma and confers poor prognosis. Neuropathology 2015; 35: 510–517. [DOI] [PubMed] [Google Scholar]
- Lu N, Lin T, Wang L, Qi M, Liu Z, Dong H et al. Association of SOX4 regulated by tumor suppressor miR-30a with poor prognosis in low-grade chondrosarcoma. Tumour Biol 2015; 36: 3843–3852. [DOI] [PubMed] [Google Scholar]
- Song GD, Sun Y, Shen H, Li W. SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour Biol 2015; 36: 4167–4173. [DOI] [PubMed] [Google Scholar]
- Wang D, Hao T, Pan Y, Qian X, Zhou D. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol Cell Biochem 2015; 402: 75–82. [DOI] [PubMed] [Google Scholar]
- Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia 2007; 21: 2495–2505. [DOI] [PubMed] [Google Scholar]
- Yassin ER, Abdul-Nabi AM, Takeda A, Yaseen NR. Effects of the NUP98-DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia 2010; 24: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidvar N, Maunakea ML, Jones L, Sevcikova S, Yin B, Himmel KL et al. PML-RARalpha co-operates with Sox4 in acute myeloid leukemia development in mice. Haematologica 2013; 98: 424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 2012; 119: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval S, Kraus C, Cho EC, Cho M, Bies J, Manara E et al. Sox4 cooperates with CREB in myeloid transformation. Blood 2012; 120: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood 2006; 107: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aue G, Du Y, Cleveland SM, Smith SB, Dave UP, Liu D et al. Sox4 cooperates with PU.1 haploinsufficiency in murine myeloid leukemia. Blood 2011; 118: 4674–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 1996; 380: 711–714. [DOI] [PubMed] [Google Scholar]
- Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood 2005; 106: 2498–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell 2013; 24: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TK, Leung AY, So CW. Sox4you: a new player in C/EBPalpha leukemia. Cancer Cell 2013; 24: 557–559. [DOI] [PubMed] [Google Scholar]
- Tregnago C, Manara E, Zampini M, Bisio V, Borga C, Bresolin S et al. CREB engages C/EBPdelta to initiate leukemogenesis. Leukemia 2016; 30: 1887–1896. [DOI] [PubMed] [Google Scholar]
- Lu JW, Hou HA, Hsieh MS, Tien HF, Lin LI. Overexpression of FLT3-ITD driven by spi-1 results in expanded myelopoiesis with leukemic phenotype in zebrafish. Leukemia 2016; 30: 2098–2101. [DOI] [PubMed] [Google Scholar]
- Liu W, Wu M, Huang Z, Lian J, Chen J, Wang T et al. c-myb hyperactivity leads to myeloid and lymphoid malignancies in zebrafish. Leukemia 2017; 31: 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JW, Hsieh MS, Liao HA, Yang YJ, Ho YJ, Lin LI. Zebrafish as a model for the study of human myeloid malignancies. Biomed Res Int 2015; 2015: 641475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BL, Shi X, Man CH, Ma AC, Ekker SC, Chow HC et al. Functions of flt3 in zebrafish hematopoiesis and its relevance to human acute myeloid leukemia. Blood 2014; 123: 2518–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HA, Chou WC, Lin LI, Tang JL, Tseng MH, Huang CF et al. Expression of angiopoietins and vascular endothelial growth factors and their clinical significance in acute myeloid leukemia. Leuk Res 2008; 32: 904–912. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007; 236: 3088–3099. [DOI] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 2008; 111: 132–141. [DOI] [PubMed] [Google Scholar]
- Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia 2014; 28: 50–58. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Hou HA, Tang JL, Liu CY, Lin CC, Chou WC et al. Genetic alterations and their clinical implications in older patients with acute myeloid leukemia. Leukemia 2016; 30: 1485–1492. [DOI] [PubMed] [Google Scholar]
- Hou HA, Lin LI, Chen CY, Tien HF. Reply to 'Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favorable prognosis'. Br J Cancer 2009; 101: 738–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TC, Hou HA, Chou WC, Ou DL, Yu SL, Tien HF et al. CEBPA methylation as a prognostic biomarker in patients with de novo acute myeloid leukemia. Leukemia 2011; 25: 32–40. [DOI] [PubMed] [Google Scholar]
- Poiana C, Neamtu MC, Avramescu ET, Carsote M, Trifanescu R, Terzea D et al. The poor prognosis factors in G2 neuroendocrine tumor. Rom J Morphol Embryol 2013; 54(3 Suppl): 717–720. [PubMed] [Google Scholar]
- Preudhomme C, Sagot C, Boissel N, Cayuela JM, Tigaud I, de Botton S et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 2002; 100: 2717–2723. [DOI] [PubMed] [Google Scholar]
- Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009; 113: 3088–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvinlaug BT, Chan WI, Bullinger L, Ramaswami M, Sears C, Foster D et al. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res 2011; 71: 4117–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield CD, Lawrence D, Byrd JC, Carroll A, Pettenati MJ, Tantravahi R et al. Frequency of prolonged remission duration after high-dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype. Cancer Res 1998; 58: 4173–4179. [PubMed] [Google Scholar]
- Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood 1998; 92: 2322–2333. [PubMed] [Google Scholar]
- Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 1999; 93: 3074–3080. [PubMed] [Google Scholar]
- Song G, Shi L, Guo Y, Yu L, Wang L, Zhang X et al. A novel PAD4/SOX4/PU.1 signaling pathway is involved in the committed differentiation of acute promyelocytic leukemia cells into granulocytic cells. Oncotarget 2016; 7: 3144–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.