Abstract
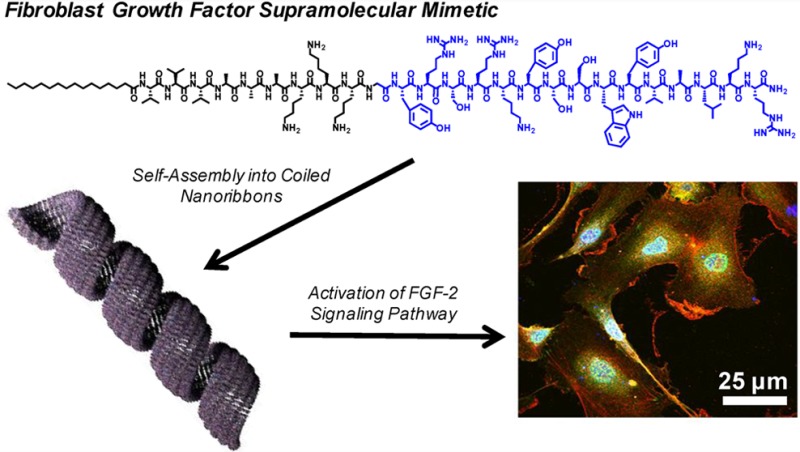
Fibroblast growth factor (FGF-2) is a multifunctional growth factor that has pleiotropic effects in different tissues and organs. In particular, FGF-2 has a special role in angiogenesis, an important process in development, wound healing, cell survival, and differentiation. Therefore, incorporating biological agents like FGF-2 within therapeutic biomaterials is a potential strategy to create angiogenic bioactivity for the repair of damaged tissue caused by trauma or complications that arise from age and/or disease. However, the use of growth factors as therapeutic agents can be costly and does not always bring about efficient tissue repair due to rapid clearance from the targeted site. An alternative would be a stable supramolecular nanostructure with the capacity to activate the FGF-2 receptor that can also assemble into a scaffold deliverable to tissue. We report here on peptide amphiphiles that incorporate a peptide known to activate the FGF-2 receptor and peptide domains that drive its self-assembly into supramolecular nanoribbons. These FGF2-PA nanoribbons displayed the ability to increase the proliferation and migration of the human umbilical vein endothelial cells (HUVECs) in vitro to the same extent as the native FGF-2 protein at certain concentrations. We confirmed that this activity was specific to the FGFR1 signaling pathway by tracking the phosphorylation of downstream signaling effectors such ERK1/2 and pH3. These results indicated the specificity of FGF2-PA nanoribbons in activating the FGF-2 signaling pathway and its potential application as a supramolecular scaffold that can be used in vivo as an alternative to the encapsulation and delivery of the native FGF-2 protein.
Keywords: supramolecular biomaterials, peptide amphiphiles, nanoribbons, fibroblast growth factor-2 mimetic peptide
Introduction
Growth factors (GFs) play a key role in regulating important cellular behaviors such as survival, proliferation, migration, and differentiation.1,2 These proteins are secreted to regulate a diverse number of cellular processes, such as tissue development or repair, and are usually bound and tightly regulated by their interactions with extracellular matrix (ECM) components. Multiple strategies to encapsulate and deliver GFs using biomaterials have been developed to replace or repair damaged tissues.3,1,4 GFs can be incorporated into biomaterials using various chemical conjugation and/or physicochemical methods to improve retention and delivery.5−8 For example, a mutant variant of VEGF (VEGF121-cys) was covalently attached through Michael addition chemistry to a PEG scaffold for the induction of in vivo vascularization.9 Another alternative is to chemically attach heparin, a glycosaminoglycan capable of binding multiple GFs, into the polymer scaffold, as a way to deliver growth factors without relying on recombinant GF variants.10,11 However, both of these methods can be synthetically challenging and require the use of significant amounts of protein. Other major drawbacks of GF delivery in polymeric scaffolds include short half-life and rapid tissue clearance of proteins, which prevents an effective GF signaling response in tissues of interest.8,12,13 In general, the delivery of therapeutic amounts of GFs can be costly and have serious side effects such as the emergence of angiogenic malignancies (tumors) and nonspecific responses in other organs such as the kidney or atheroma.14,15
An alternative would be to utilize short bioactive peptide sequences that signal GF receptors as native GFs do but contained within a stable supramolecular scaffold in order to avoid their rapid degradation by enzymes. In previous work, these protein-mimetic sequences have been identified by analyzing fragments of the native GF protein, using computational studies, or by using phage-display technology.16−19 The short peptide sequences can be readily incorporated into peptide-based matrices, which have proven to be effective biomaterials for a diverse number of in vitro and in vivo applications by being cost-effective, increasing the regenerative activity on the wounded area, and being nontoxic.20−24 One of the most important GFs for regenerative biomaterials is basic fibroblast growth factor (bFGF, also known as FGF-2).25,26 FGF-2 is a member of the receptor tyrosine kinase (RTK) family, which are GFs that control many essential cell activities important for tissue development, cell survival, cell differentiation, and homeostasis. Other important members of the RTK family include VEGF (vascular endothelial growth factor), EGF (epidermal growth factor), and PDGF (platelet-derived growth factor), which are also commonly encapsulated within biomaterials for in vivo applications.5,27 In particular, FGF-2 has been shown have a pleiotropic effect in different tissues and organs, including an important role in angiogenesis and wound healing as well as in embryonic development and neural survival and differentiation.28−30,26,31 Thus, signaling of the FGF-2 receptor (FGFR) by a bioactive biomaterial could have many important applications ranging from therapies for myocardial infarction32,29 to muscle regeneration and spinal cord injury,33 among many others. A short peptide sequence (YRSRKYSSWYVALKR) was identified by Baird et al.34 which activates the FGFR and thus mimics the bioactivity of FGF-2. The approach utilized to identify the bioactive peptide was to screen various sequences derived from FGF-2. They found that the peptide domain 106–120 in FGF-2 is a partial agonist of FGFR determined in a proliferation assay with 3T3 fibroblast cells.34 Later, Zamora et al. incorporated the sequence in a branched peptide construct termed F2A4-K-NS, which contains two copies on the FGF-2 mimetic peptide alongside a heparin binding sequence (NH2-YRSRKYSSWYVALKRK(NH2-YRSRKYSSWYVALKR)-(Ahx-Ahx-Ahx)-RKRLDRIAR-CONH2, where Ahx = aminohexanoic acid). This peptide mimetic was able to bind FGFR1 and activate the corresponding proliferation-signaling cascade via the phosphorylation of ERK1/2 and also enhanced angiogenesis in vivo when encapsulated within Matrigel implants.35
We report here on the incorporation of the FGF-2 mimetic sequence in a peptide amphiphile (PA) capable of self-assembling in aqueous media into one-dimensional nanostructures. The objective has been to create a bioactive supramolecular scaffold of filamentous structures that could be used to encapsulate cells. PAs that form filamentous networks were developed in the Stupp laboratory,36−40 and their in vitro and in vivo bioactivity has been demonstrated in several cell cultures and preclinical models for regenerative medicine. Some of these applications include the differentiation of neural progenitor cells into neurons,41 axon regeneration after spinal cord injury,42,43 hard tissue formation such as enamel44 and bone,45,46,24 vascularization on demand,47,48,21 and cartilage regeneration.49 PA molecules that form the filamentous nanostructures are composed of three main segments: a single lipid tail for hydrophobic collapse, a β-sheet domain that drives one-dimensional self-assembly, charged residues for solubility, and a bioactive signal at one terminus of the peptide. In this work, we have investigated by physical and biological experiments a filament-forming PA molecule containing the FGF-2 mimetic peptide.
Materials and Methods
Peptide Synthesis
FGF2-PA (C16V3A3K3GYRSRKYSSWYVALKR), mutant FGF2-PA (C16V3A3K3GYARSEKYSSVYVALSR), scrambled FGF2-PA (C16V3A3K3GWRSKKYSLYYVASRR), and the FGF-2 peptide mimetic (Ac-YRSRKYSSWYVALKR) were synthesized using standard 9-fluorenyl methoxycarbonyl (Fmoc) solid-phase peptide synthesis (SPPS) on Rink amide 4-methylbenzhydrylamine resin (Millipore, Billerica, MA) with the Liberty 12-Channel Automated Microwave Peptide Synthesizer (CEM, Matthews, NC). The standard conditions for synthesis involve loading the resin (0.50 mmol) and coupling all the desired Fmoc-amino acids, starting with Fmoc-Arg(Pbf)-OH (1 mmol) using 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, 0.95 mmol), and N,N-diisopropylethylamine (DIEA, 3 mmol) in N,N-dimethylformamide (DMF). Palmitic acid (4 mmol) using HBTU (0.95 mmol) and DIEA (3 mmol) in DMF was used to cap N-terminus of the PAs. In the case of the FGF-2 mimetic peptide, the N-terminus was capped using a 10:2.5:100 (v/v/v) mixture of acetic anhydride, DIEA, and DMF for 30 min. The PA was subsequently cleaved from the resin using a 95:5 TFA/TIPS (trifluoroacetic acid/triisopropyl silane) cocktail for 4 h. PAs and crude peptide were purified by reverse-phase high-performance liquid chromatography (HPLC) with a 2–95% ACN/H20 (0.1% TFA) gradient for 60 min using a Varian Modular HPLC system (Agilent, Santa Clara, CA). The desired PA fractions were collected, evaporated under vacuum, and lyophilized into a solid powder. Purified PAs were further characterized by analytical LC/MS using a 6520 Quadrupole Time-of-Flight (Q-TOF) LCMS (Agilent, Santa Clara, CA) (see Figure S1).
Peptide Amphiphile Preparation
The desired amount of PA powder was weighed out in an Eppendorf tube in order to make 500 μL of a 1 mM PA stock solution in 25 mM HEPES at pH 7.4 buffer. The PA solution was subsequently annealed in 80 °C water bath for 30 min and slowly cooled down overnight to room temperature as previously described.38
Transmission Electron Microscopy
Images for conventional and cryo-TEM were obtained using a Hitachi HT-7700 Biological TEM (Hitachi High Technologies America, Schaumburg, IL) equipped with a LaB6 filament working at an accelerating voltage of 100 kV. For Cryo-TEM, PA samples were plunged frozen using a Vitrobot Mark IV (FEI, Hillsboro, OR) operating at 25 °C with 100% humidity. The PA sample (8 μL) was deposited on 300 square mesh copper grids with a lacey carbon film (Ted Pella, Redding, CA), blotted, and plunged into a liquid ethane reservoir cooled by liquid nitrogen. Following vitrification, the sample was transferred to a Gatan 626 cryo-holder (Gatan, Pleasanton, CA) under liquid nitrogen with the aid of a transfer stage. Images were acquired using an Orius SC 1000A CCD camera. All PA formulations were imaged at concentrations of 500 μM in 25 mM HEPES at pH 7.4 buffer. For conventional TEM, PA samples were dried on carbon film 300 square mesh copper grids (Ted Pella, Redding, CA) and stained with 0.5% uranyl acetate (UA) solution. Banding pattern was analyzed using ImageJ by measuring the pixel value of the gray scaled microscopic images, where a value of 0 represents white, and a value of 200 or above represents black.
Atomic Force Microscopy
PA samples were diluted to 200 μM in DI H2O and spin coated on freshly cleaved mica substrates at a spin rate of 6000 rpm for 1 min. AFM characterization was performed using a Bruker Dimension ICON atomic force microscope (Bruker, Billerica, MA) at ambient conditions. Tapping mode was utilized with single-beam silicon cantilevers with a nominal oscillation frequency of 300 kHz.
Circular Dichroism
CD measurements were performed at a concentration of 1 mM PA after annealing in 25 mM HEPES at pH.74 buffer using a Jasco J-815 CD spectrophotometer (Jasco Analytic Instruments, Easton, MD) at 25 °C using a 0.01 mm path length demountable quartz cuvette over a wavelength range of 190–260 nm with a step size of 1 nm and a data accumulation of n = 3.
Cell Culture
HUVECs (pooled donor, LONZA, Allendale, New Jersey) were grown to 70–80% confluence for each experiment (P2–P4) in a T-75 cell culture flask using complete media (EndoGRO-VEGF Complete Culture Media Kit, Millipore, Billerica, MA) supplemented with 1% penicillin–streptomycin. Media were changed every 3 days.
Proliferation Assay
Confluent cells were washed with PBS and detached with 2 mL of 0.05% Trypsin for 1 min at 37 °C. Cells were then diluted with 8 mL of complete media and counted using a hemocytometer. Cells were pelleted by centrifuge for 5 min at 2,000 rpm and resuspended with 0.5% FBS starvation media (EBM-2 media, LONZA, Allendale, New Jersey) supplemented with 1% penicillin–streptomycin and to make a 1 × 105 cell/mL stock solution. 100 μL of this stock solution was then pipetted into each well of a 96-well plate, and cells were allowed to attach and spread for 6 h on a cell incubator at 37 °C with 5% CO2. Afterward, media were aspirated, and all PA samples diluted in 0.5% FBS starvation media were added to the cells. Cell number was quantified after 16 h of sample incubation using the Quant-iT PicoGreen dsDNA kit proliferation assay (ThermoFisher Scientific, Waltham, MA) following the manufacturer’s protocol. Fluorescence measurements were obtained with the Cytation 3 cell imaging multimode reader (BioTek Instruments, Winooski, VT). Fold increase calculations were made by normalizing the data of each sample to the t = 0 starvation condition. Experiments were performed in 3 sets of quadruplicates.
LIVE/DEAD Assay
After 16 h of PA sample incubation, cells were washed and incubated with the LIVE/DEAD (ThermoFisher Scientific, Waltham, MA) assay dyes following the manufacturer’s protocol (1 μM of calcein AM and ethidium homodimer-1). Cells were then washed and imaged with the Cytation 3 cell imaging multimode reader. Quantification of cell viability was performed as a percent ratio of calcein AM positive cells over total number of cells from the images of two wells per sample.
Metabolic Activity Assay
1 ×105 cells were plated in a 96-well plate and allowed to remain overnight in complete media. Cells were washed with PBS and 0.5% FBS starvation media before PA samples diluted in 0.5% FBS starvation media were incubated for 8 h. A solution of CellTiter MTS solution (Promega, Madison, WI) was added as recommended by the manufacturer’s protocol. After 2 h of incubation with the MTS reagent, absorbance measurements at 490 nm were taken using the Cytation 3 cell imaging multimode reader. Metabolic activity calculations for each sample were normalized to cells grown in starvation conditions. Experiments were performed in 2 sets of quadruplets.
Migration Assay
The 48-well NeuroProbe reusable multiwell chemotaxis chambers (NeuroProbe, Gaithersburg, MD) were used for this experiment following the manufacturer’s protocol. Briefly, HUVECs growing to 70–80% confluence on a T-75 flask were starved overnight, trypsinized, and diluted into 1 × 106 cell stock in 0.1% BSA RPMI media. Lower chambers were filled with 29 μL of the PA sample solution diluted in 0.1% BSA RPMI media, while 50 μL of the cell stock solution was added to the upper chambers. The chamber was incubated overnight at 37 °C with 5% CO2. Later, cells on the nonmigrated side of the membrane were removed with a wiper blade. Cells on the migrated side were stained with DAPI (1:10,000, Molecular Probes Thermo Scientific, Grand Island, NY) for 10 min, and fluorescent images were taken (5 images for each well) at 10× magnification using a Nikon Eclipse TE2000 inverted microscope (Nikon, Melville, NY) and analyzed with ImageJ. Chambers were separated using a polycarbonate membrane with 8 μm pores coated with 0.1 mg/mL collagen IV (Sigma-Aldrich, St. Louis, MO). Experiments were performed in 3 sets of triplicates.
Western Blot
1 ×105 cells were plated on 6-well plates and allowed to grow to 80–90% confluence over 3 days using complete media. Confluent cells were then washed with PBS and starved for 24 h using 0.5% FBS starvation media. Afterward, 1 mL of PA samples diluted in 0.5% FBS starvation media were incubated for 5 min, and cells were subsequently washed and lysed. Protein extracts obtained from cell cultures were separated by a SDS– polyacrylamide gel and electro-transferred to a nitrocellulose membrane (Bio-Rad). Membranes were blocked and incubated first with primary antibodies overnight at 4 °C and then with their corresponding secondary HRP-conjugated antibodies (1:3000; Santa Cruz Biotechnology, Dallas, TX). Protein signal was detected using the ECL chemiluminescent system (Amersham, GE Healthcare, Malborough, MA). Densitometry analysis, standardized to GADPH as a control for protein loading, was performed using ImageJ software. For quantification, two different sets of experiments in duplicate were analyzed. Finally, the preparations were placed in a coverslip with mounting solution for imaging. The following primary antibodies were used: mouse anti-FGFR1 (1:1000, Abcam, Cambridge, MA), mouse anti-phospho-FGFR1 (1:1000, Abcam, Cambridge, MA), mouse anti-GAPDH (1:1000, Sigma-Aldrich, St. Louis, MO), rabbit anti-ERK1/2 (1:2000, Abcam, Cambridge, MA), mouse anti-phospho-ERK1/2 (1:10000, Abcam, Cambridge, MA), and rabbit anti-PH3 (proliferation marker, 1:1000, Cell Signaling, Danvers, MA), 1:1000, Abcam).
Immunocytochemistry
1 ×105 cells were incubated in a 48-well cell culture plate containing 12 mm round coverslips and grown overnight with complete media. On the second day, media were exchanged for 0.5% FBS starvation media and incubated for 24 h. After this period, 200 μL PA samples diluted in 0.5% FBS starvation media were incubated in starvation media for 5 min. Cells were washed and fixed (4% PFA for 15 min at RT) and subsequently incubated with primary antibodies and appropriate Alexa 488 or Alexa 555 secondary antibodies (1:500, Molecular Probes Thermo Scientific, Grand Island, NY). Phospho FGFR1 (1:1000) was used to stain the activated FGF receptor 1, vinculin was used to stain the cytoskeleton (1:2000, Sigma-Aldrich, St. Louis, MO), and DAPI (1:500) was used to stain nuclei.
Fluorescent Imaging Analysis
Fluorescent preparations were viewed, and micrographs were captured with a Nikon A1R confocal laser-scanning microscope with GaAsP detectors. Images were assembled in Adobe Photoshop (v. 7.0), with adjustments for contrast, brightness, and color balance to obtain optimum visual reproduction of data.
Statistical Analysis
Statistical analysis was performed using Graphpad Prism v.6 software. Analysis of variance (ANOVA) with the posthoc test was used for all multiple group experiments. P values <0.05 were deemed significant. Values in graphs are the mean ± SEM.
Results and Discussion
Self-Assembly of FGF2-PA into Nanoribbons
We designed the FGF-2 mimetic PA (FGF2-PA) molecule using C16V3A3K3 as the nonbioactive backbone (K3-PA), known to form nanofibers with bioactive sequences displayed at the C-terminus.21,38,51 The FGF2-PA was synthesized with the sequence C16V3A3K3GYRSRKYSSWYVALKR, containing the FGF-2 peptide sequence separated by a single glycine from the K3-PA backbone. The FGF-2 PA was subsequently purified and characterized with HPLC and MALDI-TOF MS (see Materials and Methods and Figure S1). In order to promote self-assembly, the PA sample was prepared by annealing a 1 mM solution of FGF2-PA in 25 mM HEPES at pH 7.4 buffer in an 80 °C water bath for 30 min and cooled overnight to room temperature. This annealing procedure has proven to produce thermodynamically stable PA nanostructures.38 We found by TEM and AFM that the FGF2-PA forms after the annealing procedure coiled nanoribbons, micrometers in length with an average width of ∼32 nm (Figures 1A and S2). The extent of coiling varied widely in the nanostructures with a helical pitch that ranged from 30 to 215 nm. Also, the ribbons seemed to be dynamic in nature with no preference for left- or right-handed coiled structures (cryo-TEM on Figure 1A and Figure S2E). This type of coiling has been observed before in PA molecules possessing a high proportion of aromatic residues, and it is believed to arise from π–π interactions.53,54 Interestingly, however, the FGF2-PA nanoribbons contained an internal striation or banding pattern parallel to the length of the nanoribbons as revealed when stained with 0.5% uranyl acetate (UA) (conventional TEM on Figure 1A and Figure S2A and B). If the FGF2-PA solution was not annealed, bundles of aggregated ribbons would form otherwise but still maintaining the banding pattern (Figure S2D). Since the K3-PA backbone forms fibers in the absence of a bioactive sequence (Figure S8A), it is evident that intermolecular interactions between the FGF2 mimetic peptides influence the observed nanoribbon morphology.
Figure 1.
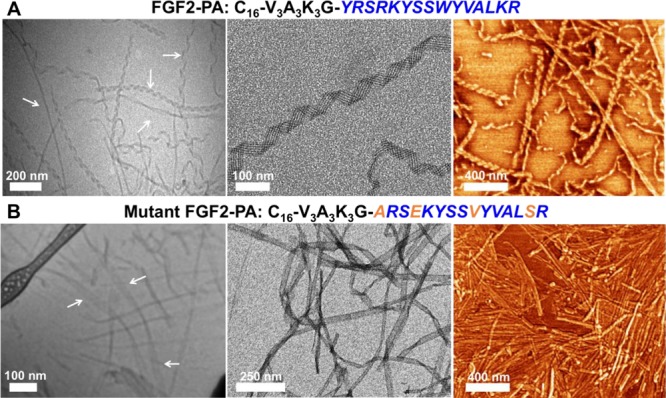
Nanostructure characterization of the FGF2-PAs. (A) Cryo-TEM (left), conventional TEM (middle), and AFM (right) images of FGF2-PA nanoribbons (in the cryo-TEM images, areas revealing helical turns along the nanoribbon are highlighted by white arrows). (B) Cryo-TEM (left), conventional TEM (middle), and AFM (right) images of mutant FGF2-PA nanoribbons (in the cryo-TEM images, areas where ribbons do not fold into helical structures are highlighted by white arrows).
To understand the biological specificity of FGF2-PA nanostructures, we also synthesized a PA molecule we refer to as the “mutant FGF2-PA”, containing the sequence C16V3A3K3GYARSEKYSSVYVALSR, where four amino acids from the FGF-2 peptide mimetic sequence were mutated (highlighted in orange on Figure 1B and Figure S3). The first two mutations, Y106A and W114V, replaced the bulky aromatic residues tyrosine and tryptophan by the smaller hydrophobic residues alanine and valine, respectively. The other two mutations, R109E and K119S, switched the positive charged residues, arginine and lysine, for a negatively charged one, glutamate, and a serine residue. The rationale behind these mutations was partly based on a previous study that reported decreased mitogenic activity when positively charged residues were exchanged with negatively charged ones, and bulky aromatic residues were exchanged with small hydrophobic ones in the receptor binding domain of FGF-2.55 TEM and AFM showed that the mutant FGF2-PA was able to still form micrometer length nanoribbons with widths similar to that of FGF2-PA (Figures 1 and S4). The mutant FGF2-PA also formed coiled nanoribbons but to a lesser extent than FGF-2 PA nanostructures (cryo-TEM in Figure 1 and Figure S4). The changes in coiling are likely due to a decrease of aromatic interactions due to the removal of one tyrosine residue and tryptophan. The banding pattern was also evident in this case when the nanoribbons were stained with 0.5% UA (conventional TEM on Figure 1B and Figure S4A). Overall, the FGF2-PA and mutant FGF2-PA formed very similar nanostructures and can therefore be compared in our biological experiments based on their differences in sequence. A third PA was synthesized to determine the effect of amino acid sequence on the FGF-2 mimetic peptide, and we refer to it here as the scrambled FGF2-PA; its sequence is C16V3A3K3GWRSKKYSLYYVASRR (Figure S10). Instead of replacing amino acid residues, we exchanged the position of six amino acids, Y106 was switched for W114, R109 switched for K119, and S113 switched for L118. We found that the scrambled FGF2-PA was able to form nanoscale ribbons but with a much thinner width of 16 nm on average. The nanostructures formed by this PA molecule did not exhibit a propensity to coil and also did not reveal the banding pattern observed in supramolecular assemblies of the other two molecules (Figure S11A and B). We again see that the supramolecular structure changes when the bioactive sequence is altered, and therefore in subsequent experiments, we focused on FGF2-PA and the mutant FGF2-PA given their similarities in nanoscale morphology.
Circular dichroism (CD) provided a way to investigate the intermolecular interactions within the PA nanostructures. From the CD data, we can determine the presence of a peptide secondary structure forming within the nanoribbons. Both the FGF2-PA and mutant FGF2-PA possessed CD minima around 218 nm, but only the mutant FGF2-PA had a significant CD maximum around 203 nm (Figure 2A). These CD signals typically correspond to β-sheet hydrogen bonding.56 However, the K3-PA without any bioactive sequence, displayed a stronger β-sheet signal. The FGF-2 mimetic peptide, which does not assemble into a supramolecular structure, has a CD spectrum characteristic of random coil peptides. This is expected since residues 106–120 in the actual native FGF-2 protein do not form any type of secondary structure.57,58 The data show how the random coil conformation of the FGF-2 mimetic peptide in FGF-2 PA nanoribbons prevents the formation of extensive H-bonding through its K3-PA backbone. Therefore, we hypothesize that the weak β-sheet signal of the FGF2-PA and the mutant FGF2-PA nanoribbons are mostly associated with the V3A3K3 PA backbone, not the bioactive or mutant peptide sequence. The scrambled FGF2-PA also revealed an even weaker β-sheet signal with a CD compared to that of the other PAs (Figure S12). With this information, we propose that both the bioactive sequence portion of FGF2-PA and mutant FGF2-PA have random coil conformation, as it would be in the native FGF-2 protein (Figure 2B).
Figure 2.
CD and TEM analysis of FGF2-PA and mutant FGF2-PA nanostructures. (A) CD spectra of FGF2-PA (blue line), mutant FGF2-PA (orange line), FGF-2 mimetic peptide (green line), and K3-PA backbone at 1 mM in 25 mM HEPES at pH 7.4 buffer after annealing. Inset: close-up of the β-sheet region for FGF2-PA, mutant FGF2-PA, and FGF-2 mimetic peptide, showing the weakness in β-sheet signal compared to that of the K3-PA backbone PA. (B) Proposed conformations of the FGF-2 peptide, FGF2-PA, and mutant FGF2-PA (length scale on the left side). (C and D) Banding pattern analysis of the section of a UA stained FGF2-PA nanoribbon and a mutant FGF2-PA nanoribbon (white double-headed arrows). The graphs on the right show that the low gray values (gray bars) correspond to the darker areas (UA deposition) in the TEM images and that the higher gray values correspond to the lighter areas.
By understanding the intermolecular interactions found in the PA nanostructures using CD, we envision that once the FGF2-PA molecules are in solution, hydrophobic collapse immediately occurs through the aliphatic carbon tails, causing the molecules to self-assemble into nanofiber-like structures. Then, growth of the supramolecular structure occurs parallel and perpendicular to the long axis of the ribbons via interactions among highly aromatic/hydrophobic residues of the FGF-2 mimetic peptide sequence. This would result in “fused” nanofiber-like structures within the nanoribbons, as indicated by the banding patterns in TEM images of FGF2-PA and mutant FGF2-PA (Figures 1 and 2C and D). This pattern is highly periodic in both nanoribbons, where the dark regions correspond to the UA deposition around the edges of each cylindrical fiber structure (see TEM images on Figure 2C and D). Given that approximately six dark regions are observed across the 32 nm-wide nanoribbons, we hypothesize that there are around five cylindrical fiber-like structures within each nanoribbon. This proposed supramolecular structure is consistent with a length of ∼4 nm of the PA molecules as shown in Figure 2B since the nanofibers within the nanoribbon have a diameter of approximately 8 nm. The graphs on Figure 2C and D quantify the periodicity of the nanoribbons by measuring the distance between the dark and lighter regions as depicted in the UA stained TEM images, where the lighter areas correspond to the cores of fibers, while the dark regions (gray bars) correspond to their edges. Other amphiphilic molecules that self-assemble into lamellar ribbons composed of aligned nanofibers have also shown similar banding patterns by negative staining in TEM.59 In our case, the hydrophobic and aromatic nature of the FGF-2 mimetic peptide seems to be favorable for intermolecular interactions that promote highly ordered nanoribbon structures, even though these types of residues are not always necessary to form ribbon-like nanostructures.60 Alternatively, the sequence of the mutant FGF2-PA has fewer aromatic residues but sufficient hydrophobic residues to also form banded nanoribbon structures.
Evaluating the Proliferation and Migration of HUVECs Using FGF2-PA Nanoribbons
One of the basic functions of the native FGF-2 protein is the ability to promote proliferation, survival, and migration of endothelial cells.26 Therefore, we introduced FGF2-PA nanoribbons in cell culture media of human umbilical vein endothelial cells (HUVECs) to evaluate their bioactivity relative to the native protein. We used these cells given their sensitive response to growth factors, especially those pertaining to the RTK family such as VEGF and PDGF. To assess the ability of FGF2-PA and mutant FGF2-PA nanostructures to promote proliferation of HUVECs, we first plated the cells in starvation media and allowed them to attach for 6 h (t = 0) before incubating the cells with the supramolecular nanostructures at various concentrations ranging from 1 μM to 250 nM for 16 h (proliferation was quantified using the PicoGreen dsDNA assay). The native FGF-2 protein, FGF-2 mimetic peptide, and starvation only conditions were used as controls. After this period, the dsDNA of each cell sample was quantified and normalized to cells grown at t = 0. We found that the FGF2-PA was able to enhance cell proliferation at 750, 500, and 250 nM concentrations (Figures 3A and S5A). Moreover, the proliferative activity of FGF2-PA was comparable to that of 1.50 nM of native FGF-2 protein. Neither the mutant FGF2-PA nor the FGF-2 mimetic peptide revealed any significant effect in proliferation at the concentrations screened (Figure S5B and C). The FGF-2 mimetic peptide was even incubated at higher concentrations (500 μM–0.0700 μM), but no proliferative effect was observed (Figure S9). When analyzing the activity of the scrambled FGF2-PA ribbons, only a modest activity was observed (Figure S11B). Given the observed dissimilarity in supramolecular structure between the scrambled FGF2-PA and the FGF2-PA, all subsequent experiments focus on comparisons between FGF2-PA and the mutant FGF2-PA. We also tested K3-PA nanofibers and did not observe any effect on proliferation (see Figure S8B). This confirmed that the backbone (C16V3A3K3) did not play a significant role in that activity and therefore that the bioactivity observed for FGF2-PA nanostructures has its origin in the presence of the FGF-2 mimetic sequence.
Figure 3.
Cell proliferation, migration, and viability assays. (A) HUVEC proliferation after 16 h of incubation with FGF2-PA (blue bars), mutant FGF2-PA (orange bars), and FGF-2 mimetic peptide at 750 and 500 nM with native FGF-2 protein at 1.50 and 0.150 nM as positive controls. (B) Fluorescent images of cells after being incubated with FGF2-PA and mutant FGF2-PA for 16 h stained with calcein AM and ethidium homodimer-1. (C) Graph showing >90% metabolic activity for HUVECS incubated with 750 and 500 nM of FGF2-PA, mutant FGF2-PA, and FGF-2 mimetic peptide. (D) Graph showing significant migration of HUVECs using the chemotaxis chamber setup with starved cells incubated with FGF2-PA, mutant FGF2-PA, and FGF-2 mimetic peptide at 750 and 500 nM and with native FGF-2 protein at 3.00, 1.50, and 0.150 nM (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, with starvation media used as a negative control).
After the FGF2-PAs were incubated for 16 h, cell survival was monitored using calcein AM and ethidium homodimer-1 dyes. As expected from the proliferation experiment results, an increase in viable cells was observed when cells were incubated with FGF2-PA, as indicated by green fluorescence (Figures 3B and S6A). Conversely, fewer viable cells were observed when cells were incubated with mutant FGF2-PA, FGF-2 mimetic peptide, and starvation media conditions. Overall, quantification of calcein AM positive cells incubated with all the samples confirms that cells were alive but only proliferated in the presence of FGF2-PA nanostructures (Figure S6B). The various PAs were also tested for metabolic activity using the Cell Titer MTS assay (Figures 3C and S7). Cells were shown to be metabolically active when incubated with FGF2-PA, mutant FGF2-PA, and the FGF-2 mimetic peptide at concentrations ranging from 2 μM to 250 nM (Figure S7). This indicates that at the concentrations used in our experiments, the PAs are nontoxic but that only the FGF2-PA seems to have a specific proliferative activity. Lastly, a fluorescent caspase activity assay was performed to confirm cell survival at the bioactive concentrations. In the assay, the FGF2-PAs did not promote the activation of the caspase-3 dependent apoptosis pathway, a key indicator of cell survival (Figure S7D).
The native FGF-2 protein is also an important growth factor in promoting the migration of endothelial cells, a key process in tissue development and repair. Therefore, a migration assay using a chemotaxis chamber setup was used to evaluate the ability of FGF2-PA nanostructures to have this type of bioactivity expected in the protein. FGF2-PAs, FGF-2 mimetic peptide, and native FGF-2 protein were incubated in the lower chambers and starved cells on the top chamber, separated by a collagen-coated porous membrane. After incubation overnight, migration bioactivity similar to that observed with 3.00 nM native FGF-2 protein was found using 750 and 500 nM of the FGF2-PA, mirroring our observations with the proliferation assay. As expected, the mutant FGF2-PA and the FGF-2 mimetic peptide were found to be inactive (Figure 3D). Thus, the FGF2-PA nanoribbons are able to effectively promote HUVEC survival, proliferation, and migration with the same degree as the native FGF-2 protein, while all of these elements of bioactivity are not observed in the mutant FGF2-PA and the FGF-2 mimetic peptide.
FGFR1 Signaling Pathway Activation with FGF2-PA Nanoribbons
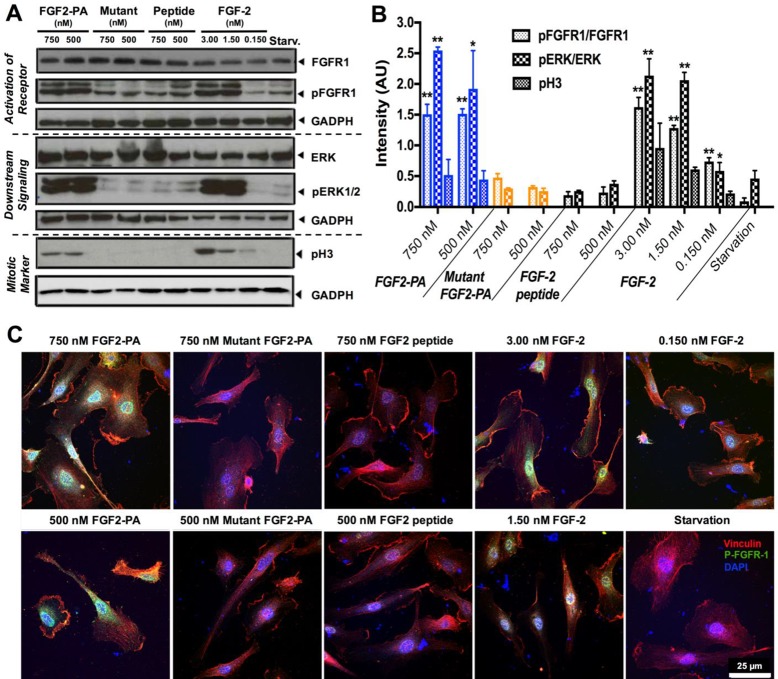
In order verify that the observed bioactivity of FGF2-PA nanostructures in vitro is indeed linked to the FGF-2 signaling pathway, we carried out Western blot protein analysis and immunocytochemistry experiments. For these experiments, HUVECs were starved for 24 h and then incubated for 5 min with FGF2-PA, mutant FGF2-PA, FGF-2 mimetic peptide, or native FGF-2 protein. First, we investigated the phosphorylation of FGFR1 (FGF receptor 1), one of the FGF-2 receptors which upon binding ligand activates proliferation and migration signaling pathways in endothelial cells.61 While the total FGFR1 expression remained constant under all conditions, phosphorylated FGFR1 (pFGFR1), indicating active signaling, was only observed when cells were incubated with 750 and 500 nM of FGF2-PA nanoribbons or at various concentrations of native FGF-2 protein (upper panel, Figure 4A). Expression of pFGFR1 under these conditions was significantly higher compared with starvation conditions, exposure to mutant FGF2-PA nanoribbons, or exposure to FGF-2 mimetic peptide (Figure 4B). Next, we examined the expression of phosphorylated ERK1/2 (pERK1/2), one of the most important downstream effectors of the FGFR1 signaling cascade and a clear indicator of a proliferative response. In similar fashion, pERK1/2 was significantly overexpressed when cells where incubated with FGF2-PA and the native FGF-2 protein (middle panel in Figure 4A and B). The presence of pERK1/2 was of course expected given the observed phosphorylation of the upstream effector FGFR1. The last signaling effector analyzed was the phospho-histone 3 (pH3) mitotic marker, a definite indicator that cells undergo mitosis and proliferate through the FGFR1 pathway.62 As expected, there was a clear expression of pH3 when the media contained FGF2-PA and native FGF-2 protein, whereas there was none detected when we used the mutant FGF2-PA or the FGF-2 mimetic peptide to modify the media and also in the starvation control (lower panel of Figure 4A and B). We conclude that the Western blot analysis of the FGFR1 signaling pathway confirms that proliferation observed during in vitro experiments induced by the FGF2-PA nanoribbons or the native FGF-2 protein is associated with the FGFR1 signaling pathway.
Figure 4.
Effect of FGF2-PA on activation of cell signaling. (A) Western blot analysis shows upregulation of the pFGFR1 receptor, pERK1/2 proliferation signaling pathway, and pH3 proliferation marker promoted by FGF2-PA at 750 and 500 nM, native FGF-2 protein at 3.00, 1.50, and 0.150 nM and starvation conditions. Mutant FGF2-PA and FGF-2 mimetic peptide at 750 and 500 nM failed to induce any significant FGF-2 signaling. (B) Bar graph presenting quantitative analysis of the Western blot data using densitometry (intensity values normalized to GADPH). (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, with starvation media used as a negative control). (C) Confocal images of vinculin staining (red), nuclei (DAPI, blue), and phosphorylated FGFR1 (green).
Additional immunocytochemistry experiments were performed to corroborate the presence of pFGFR1. FGF2-PA, mutant FGF2-PA, FGF-2 mimetic peptide, and native FGF-2 protein were incubated for 5 min in starvation media before cells were fixed and stained with the corresponding antibody. Vinculin staining revealed that HUVEC cells were attached and spread after the different treatments. Cells incubated with 750 and 500 nM of FGF2-PA and native FGF-2 protein revealed punctuated fluorescence from pFGFR1 (green) around the nucleus, whereas the other conditions did not reveal the expression of the activated receptor (Figure 4C). This type of nuclear localization of p-FGFR1 has been observed before when the receptor is activated.63 These images further confirm the effective activation of FGFR1 by FGF2-PA nanoribbons with complementary SEM images suggesting interactions between the FGF2-PA nanoribbons and cell surfaces (Figure S13).
Both the in vitro cell experiments and the protein Western blot analysis have confirmed that the bioactivity induced by FGF2-PA nanoribbons is specifically linked to FGFR1 signaling and is as potent as that associated with the native FGF-2 protein. Moreover, mutant FGF2-PA nanoribbons were not specific enough to induce significant bioactivity, ruling out any activity induced by the nanoribbon structure. Zamora et al. previously found that the FGF-2 mimetic peptide in its monomeric state cannot induce bioactivity in endothelial cells. Therefore, they synthesized a new molecule termed F2A4-K-NS, which contains two copies on the FGF-2 mimetic peptide in order to promote receptor binding and bioactivity.35 Similarly, we found that the FGF-2 mimetic peptide was only bioactive when displayed in a supramolecular nanostructure, most likely due to the multivalency of bioactive signals on the surfaces of the nanoribbons capable of dimerizing and activating the FGFR1 signaling cascade.
Conclusions
We have developed a PA molecule that self-assembles into coiled supramolecular nanoribbons that mimic the bioactivity of one of the most important growth factors in biological development and regeneration, FGF-2. The bioactivity of the coiled supramolecular structures was observed in human umbilical vein endothelial cells, promoting both their proliferation and migration. Moreover, we confirmed that the bioactivity is directly linked to signaling of the FGF receptor by the nanostructures and is comparable in intensity to the native protein. Bioactive supramolecular nanostructures such as the ones reported here offer an alternative to protein therapies which often have unacceptably short half-lives. The nanostructures could also be easily integrated into scaffolds for regenerative medicine therapies.
Acknowledgments
The studies reported here were supported by the National Institute of Health PPG (5P01HL108795-05) and by the National Institute of Health Bioengineering Research Partnership (BRP) (5R01EB003806-09) research grants. C.M.R.P. gratefully acknowledges support from the National Institute of Health NIBIB Supplement Award (3R01EB003806-09S1). Additional support for T.A. to perform atomic force microscopy experiments was provided by the U.S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences, under award no. DE- FG02-00ER45810. Z.A. received postdoctoral support from the Beatriu de Pinós Fellowship 2014 BP-A 00007 (Agència de Gestió d’Ajust Universitaris i de Recerca, AGAUR). PA synthesis was performed in the Peptide Synthesis Core Facility of the Simpson Querrey Institute at Northwestern University. The U.S. Army Research Office, the U.S. Army Medical Research and Materiel Command, and Northwestern University provided funding to develop this facility, and ongoing support is being received from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205). Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. CD experiments were performed using the Keck Biophysics Facility at Northwestern University. TEM and AFM experiments were performed at the EPIC facility of the NUANCE Center at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205); the MRSEC program (NSF DMR-1121262) at the Materials Research Center; the International Institute for Nanotechnology (IIN); the Keck Foundation; and the State of Illinois, through the IIN. We thank Mark McClendon for SEM experiments and Mark Seniw for schematic illustrations and helpful discussions.
Glossary
Abbreviations
- GF
growth factor
- ECM
extracellular matrix
- FGF-2
fibroblast growth factor
- RTK
receptor tyrosine kinase
- PA
peptide amphiphile
- HUVECs
human umbilical vein endothelial cells
- cryo-TEM
cryogenic transmission electron microscopy
- EBM
endothelial basal media
- Cryo-TEM
cryogenic transmission electron microscopy
- AFM
atomic force microscopy
- CD
circular dichroism
- UA
uranyl acetate
- pFGFR1
phosphorylated fibroblast growth factor 1
- pERK
phosphorylated extracellular-regulated kinase
- pH3
phospho-histone H3
- DAPI
(4′,6-diamidino-2-phenylindole)
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials.7b00347.
PA chemical structures and characterization as well as additional TEM and SEM micrographs, CD spectra, proliferation, and cell viability studies (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Barrientos S.; Stojadinovic O.; Golinko M. S.; Brem H.; Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen 2008, 16 (5), 585–601. 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- Behm B.; Babilas P.; Landthaler M.; Schreml S. Cytokines, chemokines and growth factors in wound healing. J. Eur. Acad. Dermatol Venereol 2012, 26 (7), 812–20. 10.1111/j.1468-3083.2011.04415.x. [DOI] [PubMed] [Google Scholar]
- Pashuck E. T.; Stevens M. M. Designing Regenerative Biomaterial Therapies for the Clinic. Sci. Transl. Med. 2012, 4 (160), 160sr4. 10.1126/scitranslmed.3002717. [DOI] [PubMed] [Google Scholar]
- Pashuck E. T.; Stevens M. M. Designing regenerative biomaterial therapies for the clinic. Sci. Transl. Med. 2012, 4 (160), 160sr4. 10.1126/scitranslmed.3002717. [DOI] [PubMed] [Google Scholar]
- Belair D. G.; Le N. N.; Murphy W. L. Design of growth factor sequestering biomaterials. Chem. Commun. 2014, 50 (99), 15651–15668. 10.1039/C4CC04317K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briquez P. S.; Clegg L. E.; Martino M. M.; Gabhann F. M.; Hubbell J. A. Design principles for therapeutic angiogenic materials. Nature Reviews Materials 2016, 1, 15006. 10.1038/natrevmats.2015.6. [DOI] [Google Scholar]
- Zhu J.; Marchant R. E. Design properties of hydrogel tissue-engineering scaffolds. Expert Rev. Med. Devices 2011, 8 (5), 607–26. 10.1586/erd.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Silva E. A.; Mooney D. J. Growth factor delivery-based tissue engineering: general approaches and a review of recent developments. J. R. Soc., Interface 2011, 8 (55), 153–70. 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisch A. H.; Lutolf M. P.; Ehrbar M.; Raeber G. P.; Rizzi S. C.; Davies N.; Schmokel H.; Bezuidenhout D.; Djonov V.; Zilla P.; Hubbell J. A. Cell-demanded release of VEGF from synthetic, biointeractive cell ingrowth matrices for vascularized tissue growth. FASEB J. 2003, 17 (15), 2260–2. 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- Zieris A.; Chwalek K.; Prokoph S.; Levental K. R.; Welzel P. B.; Freudenberg U.; Werner C. Dual independent delivery of pro-angiogenic growth factors from starPEG-heparin hydrogels. J. Controlled Release 2011, 156 (1), 28–36. 10.1016/j.jconrel.2011.06.042. [DOI] [PubMed] [Google Scholar]
- Mammadov R.; Mammadov B.; Toksoz S.; Aydin B.; Yagci R.; Tekinay A. B.; Guler M. O. Heparin Mimetic Peptide Nanofibers Promote Angiogenesis. Biomacromolecules 2011, 12 (10), 3508–3519. 10.1021/bm200957s. [DOI] [PubMed] [Google Scholar]
- Laham R. J.; Rezaee M.; Post M.; Sellke F. W.; Braeckman R. A.; Hung D.; Simons M. Intracoronary and Intravenous Administration of Basic Fibroblast Growth Factor: Myocardial and Tissue Distribution. Drug Metab. Dispos. 1999, 27 (7), 821–826. [PubMed] [Google Scholar]
- Post M. J.; Laham R.; Sellke F. W.; Simons M. Therapeutic angiogenesis in cardiology using protein formulations. Cardiovasc. Res. 2001, 49 (3), 522–531. 10.1016/S0008-6363(00)00216-9. [DOI] [PubMed] [Google Scholar]
- Jain R. K.; Au P.; Tam J.; Duda D. G.; Fukumura D. Engineering vascularized tissue. Nat. Biotechnol. 2005, 23 (7), 821–823. 10.1038/nbt0705-821. [DOI] [PubMed] [Google Scholar]
- Simons M.; Bonow R. O.; Chronos N. A.; Cohen D. J.; Giordano F. J.; Hammond H. K.; Laham R. J.; Li W.; Pike M.; Sellke F. W.; Stegmann T. J.; Udelson J. E.; Rosengart T. K. Clinical Trials in Coronary Angiogenesis: Issues, Problems, Consensus: An Expert Panel Summary. Circulation 2000, 102 (11), e73–e86. 10.1161/01.CIR.102.11.e73. [DOI] [PubMed] [Google Scholar]
- D’Andrea L. D.; Del Gatto A.; De Rosa L.; Romanelli A.; Pedone C. Peptides Targeting Angiogenesis Related Growth Factor Receptors. Curr. Pharm. Des. 2009, 15 (21), 2414–2429. 10.2174/138161209788682235. [DOI] [PubMed] [Google Scholar]
- Place E. S.; Evans N. D.; Stevens M. M. Complexity in biomaterials for tissue engineering. Nat. Mater. 2009, 8 (6), 457–470. 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- Shah R. N.; Shah N. A.; Del Rosario Lim M. M.; Hsieh C.; Nuber G.; Stupp S. I. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (8), 3293–3298. 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea L. D.; Iaccarino G.; Fattorusso R.; Sorriento D.; Carannante C.; Capasso D.; Trimarco B.; Pedone C. Targeting angiogenesis: Structural characterization and biological properties of a de novo engineered VEGF mimicking peptide. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (40), 14215–14220. 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubert Perez C. M.; Stephanopoulos N.; Sur S.; Lee S. S.; Newcomb C.; Stupp S. I. The powerful functions of peptide-based bioactive matrices for regenerative medicine. Ann. Biomed. Eng. 2015, 43 (3), 501–14. 10.1007/s10439-014-1166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber M. J.; Tongers J.; Newcomb C. J.; Marquardt K.-T.; Bauersachs J.; Losordo D. W.; Stupp S. I. Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (33), 13438–13443. 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C. E.; Dombrowski A. W.; Rubert Pérez C. M.; Bahnson E. S. M.; Tsihlis N. D.; Jiang W.; Jiang Q.; Vercammen J. M.; Prakash V. S.; Pritts T. A.; Stupp S. I.; Kibbe M. R. Tissue-Factor Targeted Peptide Amphiphile Nanofibers as an Injectable Therapy To Control Hemorrhage. ACS Nano 2016, 10 (1), 899–909. 10.1021/acsnano.5b06025. [DOI] [PubMed] [Google Scholar]
- Lin Y.-D.; Luo C.-Y.; Hu Y.-N.; Yeh M.-L.; Hsueh Y.-C.; Chang M.-Y.; Tsai D.-C.; Wang J.-N.; Tang M.-J.; Wei E. I. H.; Springer M. L.; Hsieh P. C. H. Instructive Nanofiber Scaffolds with VEGF Create a Microenvironment for Arteriogenesis and Cardiac Repair. Sci. Transl. Med. 2012, 4 (146), 146ra109–146ra109. 10.1126/scitranslmed.3003841. [DOI] [PubMed] [Google Scholar]
- Lee S. S.; Hsu E. L.; Mendoza M.; Ghodasra J.; Nickoli M. S.; Ashtekar A.; Polavarapu M.; Babu J.; Riaz R. M.; Nicolas J. D.; Nelson D.; Hashmi S. Z.; Kaltz S. R.; Earhart J. S.; Merk B. R.; McKee J. S.; Bairstow S. F.; Shah R. N.; Hsu W. K.; Stupp S. I. Gel Scaffolds of BMP-2-Binding Peptide Amphiphile Nanofibers for Spinal Arthrodesis. Adv. Healthcare Mater. 2015, 4 (1), 131–141. 10.1002/adhm.201400129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto J. B.; McClatchey A. I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 2012, 12 (6), 387–400. 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y. R.; Won J. E.; Jeon E.; Lee S.; Kang W.; Jo H.; Jang J. H.; Shin U. S.; Kim H. W. Fibroblast growth factors: biology, function, and application for tissue regeneration. J. Tissue Eng. 2010, 1, 218142. 10.4061/2010/218142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayalia P.; Mooney D. J. Controlled Growth Factor Delivery for Tissue Engineering. Adv. Mater. 2009, 21 (32–33), 3269–3285. 10.1002/adma.200900241. [DOI] [PubMed] [Google Scholar]
- Turner N.; Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 2010, 10 (2), 116–129. 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Murakami M.; Sakurai T. Role of fibroblast growth factor signaling in vascular formation and maintenance: orchestrating signaling networks as an integrated system. Wires Syst. Biol. Med. 2012, 4 (6), 615–629. 10.1002/wsbm.1190. [DOI] [PubMed] [Google Scholar]
- Raballo R.; Rhee J.; Lyn-Cook R.; Leckman J. F.; Schwartz M. L.; Vaccarino F. M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 2000, 20 (13), 5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury M. E.; Ikezu T. Fibroblast growth factor-2 signaling in neurogenesis and neurodegeneration. J. Neuroimmune Pharmacol 2014, 9 (2), 92–101. 10.1007/s11481-013-9501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H.; Jung Y.; Kim S.-H.; Sun K.; Choi J.; Kim H. C.; Park Y.; Kim S. H. The enhancement of mature vessel formation and cardiac function in infarcted hearts using dual growth factor delivery with self-assembling peptides. Biomaterials 2011, 32 (26), 6080–6088. 10.1016/j.biomaterials.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Tam R. Y.; Fuehrmann T.; Mitrousis N.; Shoichet M. S. Regenerative Therapies for Central Nervous System Diseases: a Biomaterials Approach. Neuropsychopharmacology 2014, 39 (1), 169–188. 10.1038/npp.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird A.; Schubert D.; Ling N.; Guillemin R. Receptor- and heparin-binding domains of basic fibroblast growth factor. Proc. Natl. Acad. Sci. U. S. A. 1988, 85 (7), 2324–8. 10.1073/pnas.85.7.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.; Takahashi K.; Campion S. L.; Liu Y.; Gustavsen G. G.; Pena L. A.; Zamora P. O. Synthetic peptide F2A4-K-NS mimics fibroblast growth factor-2 in vitro and is angiogenic in vivo. Int. J. Mol. Med. 2006, 17 (5), 833–9. 10.3892/ijmm.17.5.833. [DOI] [PubMed] [Google Scholar]
- Hartgerink J. D.; Beniash E.; Stupp S. I. Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (8), 5133–5138. 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Greenfield M. A.; Mata A.; Palmer L. C.; Bitton R.; Mantei J. R.; Aparicio C.; de la Cruz M. O.; Stupp S. I. A self-assembly pathway to aligned monodomain gels. Nat. Mater. 2010, 9 (7), 594–601. 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantakitti F.; Boekhoven J.; Wang X.; Kazantsev R. V.; Yu T.; Li J.; Zhuang E.; Zandi R.; Ortony J. H.; Newcomb C. J.; Palmer L. C.; Shekhawat G. S.; de la Cruz M. O.; Schatz G. C.; Stupp S. I. Energy landscapes and functions of supramolecular systems. Nat. Mater. 2016, 15 (4), 469–476. 10.1038/nmat4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhoven J.; Stupp S. I. 25th Anniversary Article: Supramolecular Materials for Regenerative Medicine. Adv. Mater. 2014, 26 (11), 1642–1659. 10.1002/adma.201304606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos N.; Ortony J. H.; Stupp S. I. Self-assembly for the synthesis of functional biomaterials. Acta Mater. 2013, 61 (3), 912–930. 10.1016/j.actamat.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G. A.; Czeisler C.; Niece K. L.; Beniash E.; Harrington D. A.; Kessler J. A.; Stupp S. I. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science 2004, 303 (5662), 1352–1355. 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- Tysseling-Mattiace V. M.; Sahni V.; Niece K. L.; Birch D.; Czeisler C.; Fehlings M. G.; Stupp S. I.; Kessler J. A. Self-Assembling Nanofibers Inhibit Glial Scar Formation and Promote Axon Elongation after Spinal Cord Injury. J. Neurosci. 2008, 28 (14), 3814–3823. 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L.; North H. A.; Sahni V.; Jeong S. J.; McGuire T. L.; Berns E. J.; Stupp S. I.; Kessler J. A. β1-Integrin and Integrin Linked Kinase Regulate Astrocytic Differentiation of Neural Stem Cells. PLoS One 2014, 9 (8), e104335. 10.1371/journal.pone.0104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.; Sargeant T. D.; Hulvat J. F.; Mata A.; Bringas P.; Koh C. Y.; Stupp S. I.; Snead M. L. Bioactive Nanofibers Instruct Cells to Proliferate and Differentiate During Enamel Regeneration. J. Bone Miner. Res. 2008, 23 (12), 1995–2006. 10.1359/jbmr.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata A.; Geng Y. B.; Henrikson K. J.; Aparicio C.; Stock S. R.; Satcher R. L.; Stupp S. I. Bone regeneration mediated by biomimetic mineralization of a nanofiber matrix. Biomaterials 2010, 31 (23), 6004–6012. 10.1016/j.biomaterials.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S.; Huang B. J.; Kaltz S. R.; Sur S.; Newcomb C. J.; Stock S. R.; Shah R. N.; Stupp S. I. Bone regeneration with low dose BMP-2 amplified by biomimetic supramolecular nanofibers within collagen scaffolds. Biomaterials 2013, 34 (2), 452–459. 10.1016/j.biomaterials.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. W.; Bitton R.; Webber M. J.; Carvajal D.; Shull K. R.; Sharma A. K.; Stupp S. I. A bioactive self-assembled membrane to promote angiogenesis. Biomaterials 2011, 32 (6), 1574–1582. 10.1016/j.biomaterials.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajangam K.; Arnold M. S.; Rocco M. A.; Stupp S. I. Peptide amphiphile nanostructure-heparin interactions and their relationship to bioactivity. Biomaterials 2008, 29 (23), 3298–3305. 10.1016/j.biomaterials.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R. N.; Shah N. A.; Lim M. M. D.; Hsieh C.; Nuber G.; Stupp S. I. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (8), 3293–3298. 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb C. J.; Sur S.; Ortony J. H.; Lee O. S.; Matson J. B.; Boekhoven J.; Yu J. M.; Schatz G. C.; Stupp S. I. Cell death versus cell survival instructed by supramolecular cohesion of nanostructures. Nat. Commun. 2014, 5, 3321. 10.1038/ncomms4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashuck E. T.; Stupp S. I. Direct Observation of Morphological Tranformation from Twisted Ribbons into Helical Ribbons. J. Am. Chem. Soc. 2010, 132 (26), 8819–8821. 10.1021/ja100613w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamley I. W.; Dehsorkhi A.; Castelletto V.; Furzeland S.; Atkins D.; Seitsonen J.; Ruokolainen J. Reversible helical unwinding transition of a self-assembling peptide amphiphile. Soft Matter 2013, 9 (39), 9290–9293. 10.1039/c3sm51725j. [DOI] [Google Scholar]
- Springer B. A.; Pantoliano M. W.; Barbera F. A.; Gunyuzlu P. L.; Thompson L. D.; Herblin W. F.; Rosenfeld S. A.; Book G. W. Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitogenesis. J. Biol. Chem. 1994, 269 (43), 26879–26884. [PubMed] [Google Scholar]
- Greenfield N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc 2007, 1 (6), 2876–2890. 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M.; Olsen S. K.; Ibrahimi O. A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005, 16 (2), 107–37. 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Schlessinger J.; Plotnikov A. N.; Ibrahimi O. A.; Eliseenkova A. V.; Yeh B. K.; Yayon A.; Linhardt R. J.; Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 2000, 6 (3), 743–50. 10.1016/S1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Lin Y. Y.; Qiao Y.; Tang P. F.; Li Z. B.; Huang J. B. Controllable self-assembled laminated nanoribbons from dipeptide-amphiphile bearing azobenzene moiety. Soft Matter 2011, 7 (6), 2762–2769. 10.1039/c0sm01050b. [DOI] [Google Scholar]
- Miravet J. F.; Escuder B.; Segarra-Maset M. D.; Tena-Solsona M.; Hamley I. W.; Dehsorkhi A.; Castelletto V. Self-assembly of a peptide amphiphile: transition from nanotape fibrils to micelles. Soft Matter 2013, 9 (13), 3558–3564. 10.1039/c3sm27899a. [DOI] [Google Scholar]
- Goetz R.; Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14 (3), 166–180. 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S. J.; Corces V. G. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004, 20 (4), 214–220. 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Esser J. S.; Rahner S.; Deckler M.; Bode C.; Patterson C.; Moser M. Fibroblast Growth Factor Signaling Pathway in Endothelial Cells Is Activated by BMPER to Promote Angiogenesis. Arterioscler., Thromb., Vasc. Biol. 2015, 35 (2), 358–367. 10.1161/ATVBAHA.114.304345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.