Abstract
Transforming growth factor (TGF)-β is one of the major inducers of epithelial to mesenchymal transition (EMT), a crucial program that has a critical role in promoting carcinoma’s metastasis formation. MicroRNAs-143 and -145, which are both TGF-β direct transcriptional targets, are essential for the differentiation of vascular smooth muscle cells (VSMC) during embryogenesis, a TGF-β-dependent process reminiscent of EMT. Their role in adult tissues is however less well defined and even ambiguous, as their expression was correlated both positively and negatively with tumor progression. Here we show that high expression of both miRs-143 and -145 in mouse mammary tumor cells expressing constitutively active STAT3 (S3C) is involved in mediating their disrupted cell–cell junctions. Additionally, miR-143 appears to have a unique role in tumorigenesis by enhancing cell migration in vitro and extravasation in vivo while impairing anchorage-independent growth, which may explain the contradictory reports about its role in tumors. Accordingly, we demonstrate that overexpression of either miRNA in the non-transformed mammary epithelial NMuMG cells leads to upregulation of EMT markers and of several endogenous TGF-β targets, downmodulation of a number of junction proteins and increased motility, correlating with enhanced basal and TGF-β-induced SMAD-mediated transcription. Moreover, pervasive transcriptome perturbation consistent with the described phenotype was observed. In particular, the expression of several transcription factors involved in the mitogenic responses, of MAPK family members and, importantly, of several tight junction proteins and the SMAD co-repressor TGIF was significantly reduced. Our results provide important mechanistic insight into the non-redundant role of miRs-143 and -145 in EMT-related processes in both transformed and non-transformed cells, and suggest that their expression must be finely coordinated to warrant optimal migration/invasion while not interfering with cell growth.
Metastases at distant sites represent the main cause of death for cancer patients, underlining the importance of unraveling the molecular mechanisms that govern cancer progression to metastasis. Local invasion is an essential step in the metastasis process, and it is controlled by a coordinated series of cellular and molecular events known as EMT,1, 2 which endows cells with enhanced migration and invasion potential, resistance to anoikis and production of extracellular matrix components,2 facilitating the invasion at both local and distant tissues. Successful colonization of the target tissue occurs thanks to the inverse process, that is, mesenchymal to epithelial transition, leading to the formation of metastases with features of the tumor/tissue of origin.3 EMT main outcome is the acquisition of a spindle-like morphology due to the remodeling of the actin cytoskeleton to form stress fibers, a progressive loss of epithelial-specific adhesion molecules, like E-cadherin, Zonula Occludens-1 (ZO1) and Integrin β-1, and the expression of mesenchymal-specific adhesion and cytoskeletal proteins such as N-cadherin and Vimentin.2 This process is orchestrated by the upregulation of a number of EMT-inducing transcription factors and the modulation of specific miRNAs.4, 5, 6, 7
TGF-β, a main inducer of EMT, has a dual role in cancer progression. While its cytostatic activity initially inhibits proliferation of tumor cells, EMT induction favors progression to metastatic disease. Via its complex and highly modulated signaling network, TGF-β leads to the phosphorylation and activation of SMAD factors that in turn contribute to the activation or repression of hundreds of targets, both coding and non-coding.3, 7 Among others, TGF-β was shown to strongly induce the expression of miRs-143 and -145, which are clustered at an intergenic locus and subjected to coordinated transcriptional regulation.8, 9 Both were shown to have a role in the differentiation of vascular smooth muscle cells (VSMC) during development,8, 10 and their expression was sufficient to induce the differentiation of multipotent neural crest stem cells into VSMCs, a TGF-β-dependent process considered as a form of EMT.8, 10, 11 In adult tissues, inactivation of both miRNAs lead to impaired neointima formation upon vascular injury, due to disorganized actin stress fibers and reduced migratory ability of smooth muscle cells.11 Despite related biological functions, these two miRNAs do not share sequence similarity and mostly recognize different sets of target genes.8, –11 They are both considered as oncosuppressors, since they are frequently downregulated in epithelial tumors such as colon,12 prostate13 and ovary,14, 15 as well as in B-cell malignancies.16 Additionally, their locus is often deleted in tumors,17 and they both have been shown to exert anti-proliferative activities.18, 19, 20, 21, 22 Recently however, several reports have challenged this concept for both miR-143 and miR-145, whose expression was shown to correlate with invasion, disease grade and progression in specific types of tumors,23, 24, 25, 26 suggesting that their role in cancer may vary according to cell type and tumor stage, similar to what has been described for TGF-β itself.27
Here, we show that miRs-143 and -145 contribute to the invasive phenotype of cells derived from STAT3C/NeuT transgenic mice mammary tumors. These tumors, which develop thanks to the ectopic expression of the rat Neu oncogene in the mammary epithelium, are more aggressive and invasive when mice also carry a constitutively active form of the transcription factor STAT3 (S3C).28 Moreover, we show that miR-143 and -145 overexpression in the non-transformed NMuMG mammary epithelial cells elicits global gene expression changes including the downmodulation of several junction proteins.
Results
MiRs-143 and -145 contribute to the EMT phenotype of S3C mammary tumor cells
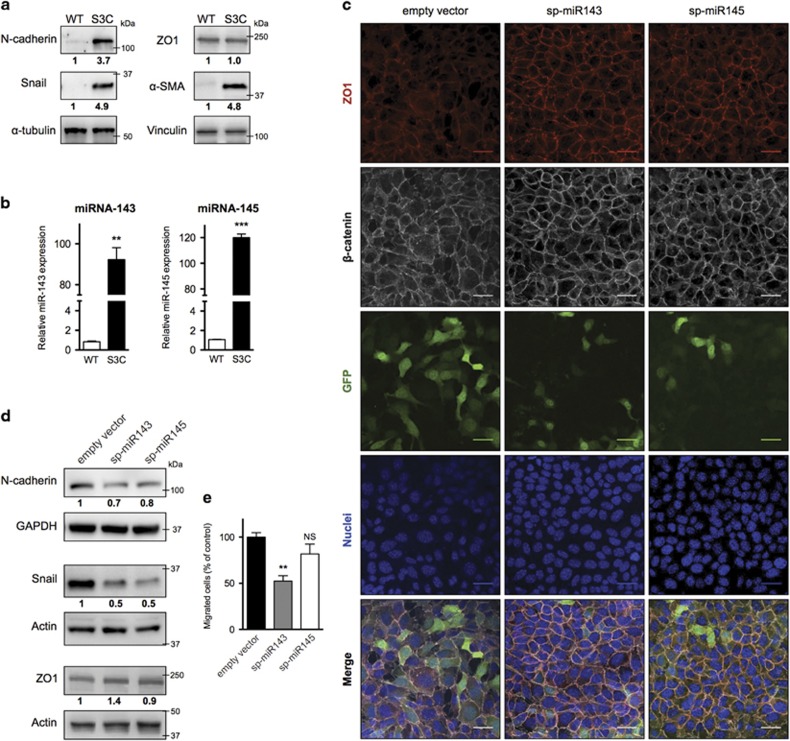
S3C cell lines from mammary tumors of NeuT-STAT3C transgenic mice, which express the MMTV-driven rat NeuT oncogene in the mammary epithelium and carry a knocked-in constitutively active STAT3 allele, display enhanced migration, invasion and in vivo tumorigenic potential correlating with disorganized cell-cell contacts, including delocalization of the tight junctions component ZO1.28 Accordingly, these cells also exhibit strongly increased expression of the EMT markers N-cadherin, Snail and α-SMA (Figure 1a), suggesting transition towards a mesenchymal status. In search for potential mediators of this phenotype, we observed that miRs-143 and -145 are dramatically upregulated in STAT3C cells (Figure 1b and Supplementary Figure 1). To assess the potential contribution of miR-143 and -145 to the EMT status of S3C cells, we decided to interfere with their activity by means of sponge constructs. Lentiviral vectors were engineered to carry a GFP expression cassette displaying multimerized miR-143 or miR-145 target sites in its 3′UTR region (Supplementary Figure 2). Upon stable transduction of S3C cells, both sponge vectors caused a reduction of GFP levels (as shown in Figure 1c and quantified in Supplementary Figure 3), demonstrating their efficacy. Inhibition of both miRNAs could revert the cell junction defects of S3C cells, triggering relocalization of β-catenin and ZO1 to the cell membrane and leading to a more organized, epithelial-like morphology (Figure 1c). Quantification of the sub-cellular distribution of the two markers performed as described in Methods section (Supplementary Figure 3), confirmed highly significant increase of membrane-localized β-catenin and ZO1 in the sponged cells (P<0,006). Additionally, both miR-143 and miR-145 sponges triggered a decrease in N-cadherin and Snail levels (Figure 1d). These changes correlated with impaired migration of miR-143-sponged cells as measured by Transwell assay (Figure 1e). Therefore, both miR-143 and -145 appear to be involved in regulating EMT features and cell-cell contacts in S3C cells, significantly affecting, in the case of miR-143, also in vitro cell migration. We thus decided to extend miR-143 analysis to in vivo assays.
Figure 1.
MiRs-143 and -145 modulate cell–cell contacts and migration of S3C cells. (a,b) S3C and WT cells were analyzed by western blot (a) or qRT-PCR (mean±S.E.M. of expression values relative to WT cells, n=3) (b). (c-e) sp-miR-143, sp-miR-145 or empty vector control S3C cells were analyzed by immunofluorescence or GFP detection (scale bar=30 μm) (c), western blot (d) and Transwell assay (mean±S.E.M. of the area covered by migrated cells, relative to the empty vector control, n=3) (e). Numbers below the lanes indicate signal intensity, normalized for loading and relative to the control sample. P-values: **P<0.01; ***P<0.001
Interference with miR-143 activity impairs in vivo cell extravasation
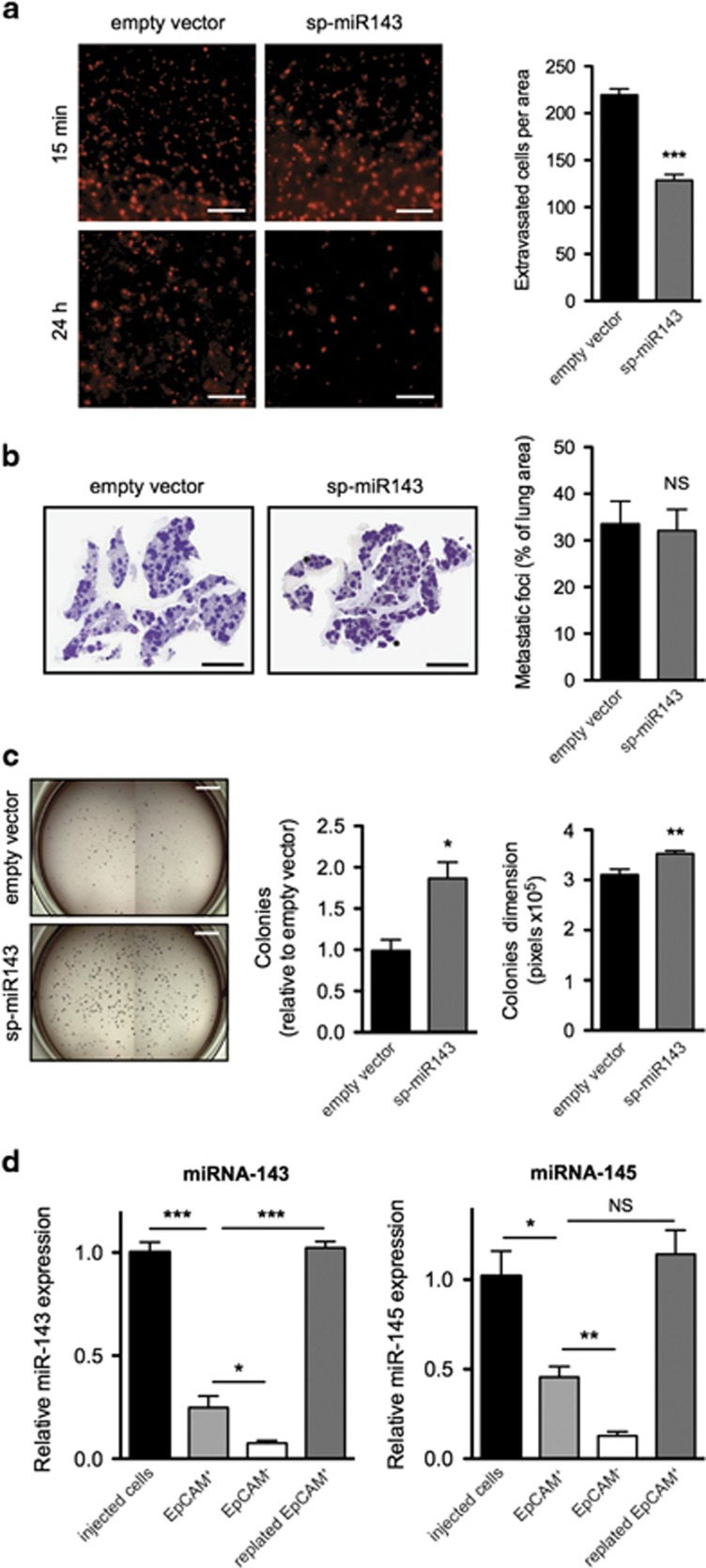
The effects of miR-143 on in vivo tumorigenic potential were assessed by comparing the ability of sponged or control S3C cells to extravasate into the lung parenchyma upon i.v. injection. Cells were labeled with a fluorescent dye (CMRA) to allow cell tracking, and injected in the tail vein of NSG immunocompromised mice, followed by evaluation of cell numbers in the tissue. Confirming equivalent loading, comparable numbers of both cell types were observed 15 min after injection, predominantly still associated to the blood vessels (Figure 2a, upper panel). Remarkably, sponged miR-143 S3C cells were impaired in their ability to extravasate into the lung parenchyma after 24 h (Figure 2a, lower panel and histograms). Despite this, they could give rise to an equivalent number of metastases as compared with the control cells (Figure 2b), suggesting enhanced ability to survive in the lung parenchyma. In the same vein, miR-143 inhibition significantly enhanced anchorage-independent growth, as shown by the increased number and size of sp-miR-143 S3C soft agar colonies (Figure 2c), despite normal proliferation rates of adherent cells and equivalent sensitivity to anoikis (Supplementary Figures 4a and b). That inhibiting miR-143 functions may improve the fitness of extravasated cells is also suggested by the observation that miR-143 and miR-145 are downregulated in the S3C empty vector cells upon metastasis formation, and become upregulated again in colture (Figure 2d). Accordingly, sp-miR-143 S3C cells regain GFP expression levels similar to control cells in the lung metastases (Supplementary Figure 4c).
Figure 2.
miR-143 inhibition reduces extravasation but not metastases. (a) Fluorescently labeled sp-miR-143 or empty vector control S3C cells were injected i.v. into NSG mice (scale bar, 1 mm), followed by analysis of the lungs at the indicated times. Histograms show the number of extravasated cells (mean±S.E.M, n=7) at 24 h. (b) Lung metastases were assessed three weeks after injection by H&E staining of lung sections (left panel; scale bar, 5 mm). Histograms show the metastatic area percentage (mean±S.E.M, n=13). (c) Sp-miR-143 or empty vector S3C cells were assessed for soft agar growth (left panel; scale bar, 5 mm), and the colony number and size scored (right panel, mean±S.E.M, n=3). (d) MiRs-143 and -145 are downregulated during metastatic growth in vivo. Empty vector S3C cells isolated from the lungs three weeks after injection were sorted in fractions positive or negative for the epithelial EpCAM marker. miRNAs expression levels were measured by qRT-PCR in both fractions, and compared with those detected in S3C cells at the time of injection or in cells from the EpCAM+ fraction plated and cultured for 5 days. Mean±S.E.M. of expression values relative to injected S3C empty vector cells, n=4. P-values: *P<0.05; **P<0.01; ***P<0.001
MiRs-143 and -145 expression in mammary epithelial cells is induced by TGF-β
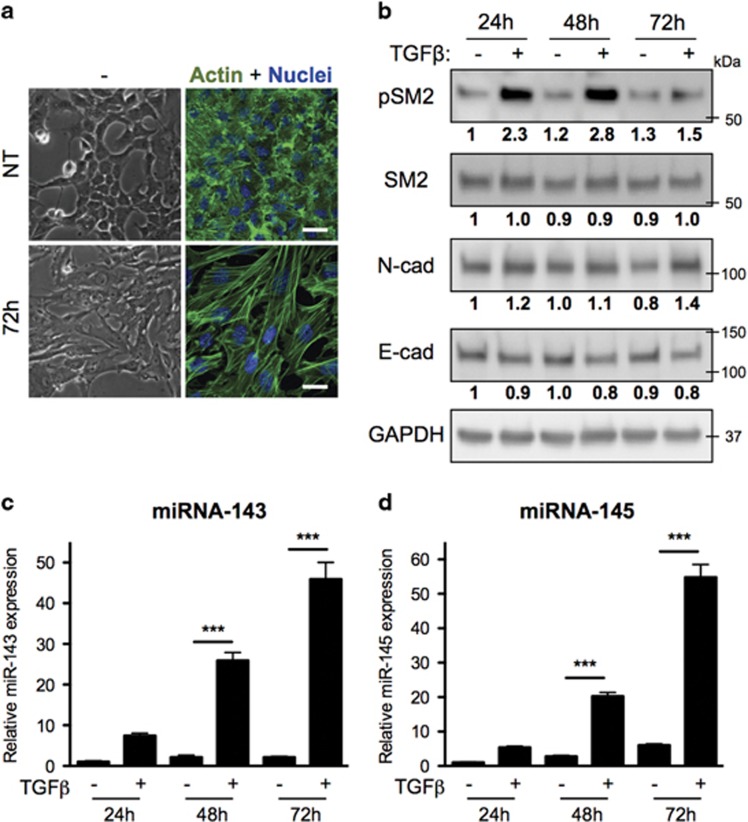
To confirm the involvement of miR-143 and -145 in the induction of EMT, we tested their activity in the normal Namru Murine Mammary Gland (NMuMG) cell line model of TGF-β-induced EMT.29 Indeed, treatment of NMuMG cells with TGF-β for 3 days triggered a transition to a mesenchymal-like morphology and the formation of abundant actin stress fibers (Figure 3a), concomitantly eliciting SMAD2 phosphorylation and reciprocal changes in the expression of N-cadherin and E-cadherin (Figure 3b), even more evident at five days (Supplementary Figure 5a). Strikingly, this correlates with a strong upregulation of the expression of both miR-143 and -145, already evident after 24 h (Figures 3c and d). Extending these findings, we observed TGF-β-dependent induction of miRNAs also in the human normal epithelial cells MCF10A and in the MDA-MB-231 mammary tumor cells, while these responses were completely abrogated in the SMAD4-null MDA-MB-468 tumor cells (Supplementary Figures 5b–e).
Figure 3.
MiRs-143 and -145 are induced by TGF-β treatment. NMuMG cells either untreated (NT) or treated with TGF-β for the indicated times were analyzed as follows. (a) Phase contrast and IF (scale bar, 30 μm). (b) Western blot for phospho-SMAD2, SMAD2, N-cadherin and E-cadherin (numbers indicate signal intensity, normalized for loading and relative to the control sample). (c,d) qRT-PCR assessing miR-143 and miR-145 expression (mean±S.E.M. of expression values relative to untreated cells, n=3). P-value: ***P<0.001
Overexpression of miRs-143 and -145 in NMuMG cells modulates the expression of junction proteins and induces EMT
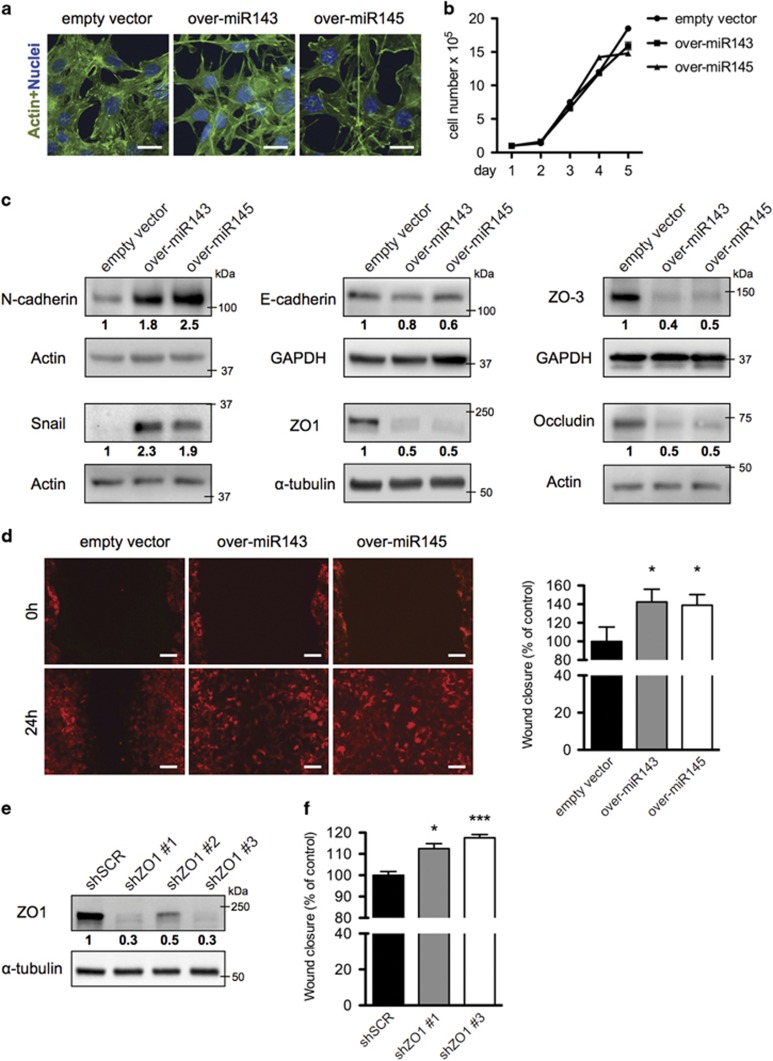
To assess their role in TGF-β-induced EMT, both miR-143 and -145 were stably overexpressed together with a RFP cassette in NMuMG cells by means of lentiviral vectors (Supplementary Figure 6). Overexpression of either miRNA triggered a transition in cell morphology and the formation of long, filamentous pseudopods, while cell proliferation was not altered as compared with control cells transduced with an empty vector (Figures 4a and b). Importantly, both miRNAs also triggered the upregulation of the N-cadherin and Snail EMT markers and a slight reduction of E-cadherin (Figure 4c). Strikingly, the expression of the tight junction proteins ZO1, ZO3 and Occludin was also dramatically downregulated at both the protein (Figure 4c) and the mRNA levels (Supplementary Figure 10c). These changes correlated with significantly increased motility in a wound healing assay (Figure 4d), which could be partly reproduced by ZO1 silencing (Figures 4e and f).
Figure 4.
MiRs-143 and -145 overexpression alters cell junctions enhancing cell motility. (a-d) miR-143, miR-145 overexpressing and empty vector control NMuMG cells were analyzed by: (a) fluorescence with phalloidin-FITC (scale bar, 20 μm); (b) cell proliferation assay (mean cell counts±S.E.M, n=3); (c) western blot for EMT and junction proteins; (d) scratch assay in a confluent cell monolayer. Images (left panel) take advantage of the endogenous RFP encoded by the miRNA expression constructs (scale bar, 100 μm); histograms (right panel) show the % of wound closure relative to the empty vector control (mean±S.E.M, n=3). (e,f) NMuMG cells were silenced for ZO1 with three independent shRNA constructs, or infected with a scrambled control (e), and analyzed by scratch assay assessed as above (f). P-values: *P<0.05; ***P<0.001
miRs-143 and -145 overexpression leads to increased SMAD-mediated transcription
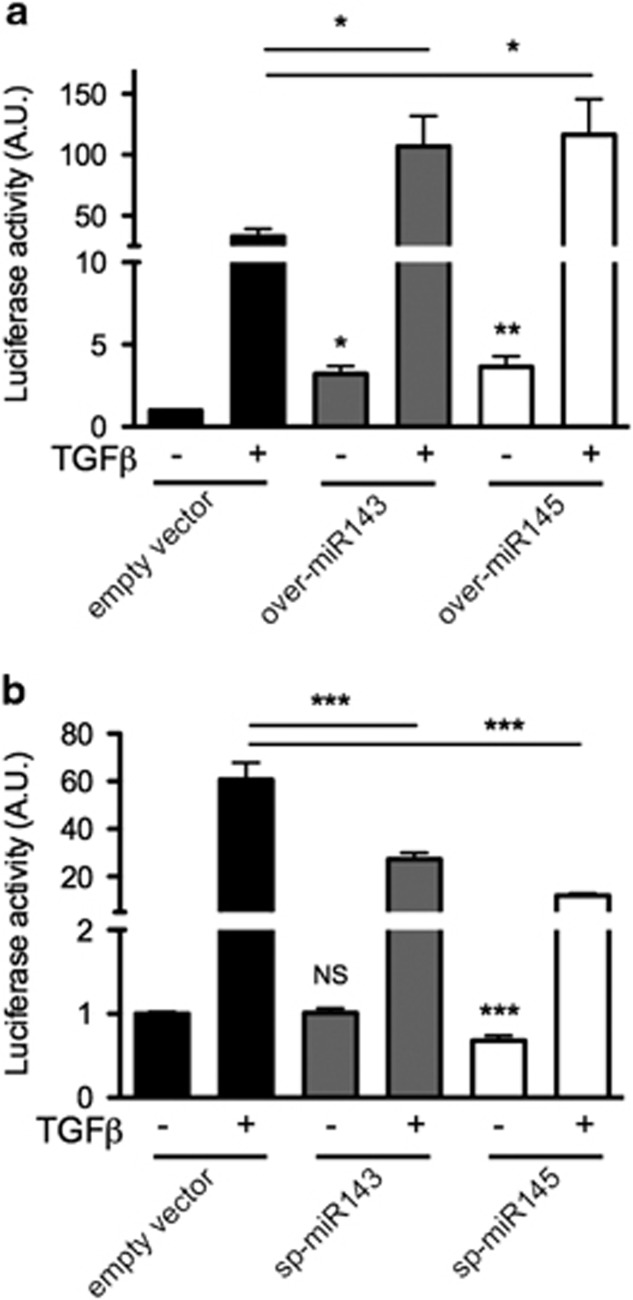
To assess the impact of miR-143 and miR-145 on TGF-β/SMAD signaling, we generated a SMAD-luciferase reporter construct, which was dose-dependently activated by TGF-β upon transient transfection in NMuMG cells (Supplementary Figures 7a,b). Interestingly, basal luciferase activity of the SMAD reporter was 3.2 and 3.6 fold higher, respectively, in NMuMG cell overexpressing miR-143 or miR-145 as compared with empty vector cells (Figure 5a). This was apparently independent from the expression or phosphorylation levels of SMAD family members, which were not significantly altered (Supplementary Figures 7c,d). Moreover, TGF-β treatment elicited about a three-fold higher luciferase induction in overexpressing cells with respect to controls (Figure 5a), suggesting enhanced SMAD-mediated transcription. Additionally, the expression of several endogenous genes involved in the TGF-β pathway, including the EMT drivers Snail and Slug, was increased upon miRNAs overexpression (Supplementary Figure 8a). The observation that the expression of Slug, which is not a direct SMAD target gene, was similarly enhanced under basal and TGF-β-induced conditions, suggests the involvement of TGF-β-independent mechanisms as well. In line with this also E-cadherin mRNA, which is negatively modulated by TGF-β, was downregulated in overexpressing cells and further reduced after TGF-β stimulation (Supplementary Figure 8a). Conversely, sponge-mediated interference with the activity of either miRNA significantly reduced TGF-β-dependent reporter activity in NMuMG cells (Figure 5b), correlating with lower basal and TGF-β-induced expression of N-cadherin, a direct SMAD target (Supplementary Figure 8b).
Figure 5.
MiRs-143 and -145 enhance SMAD-dependent transcription. NMuMG cells either overexpressing (a) or sponged (b) for miR-143 or miR-145 were transiently transfected with a SMAD-luciferase reporter sensor, followed by 24 h treatment with TGF-β and assessment of luciferase activity upon Renilla normalization (mean±S.E.M. relative to untreated empty vector cells, n=3). P-values: *P<0.05; **P<0.01; ***P<0.001
Overexpression of miRs-143 and -145 strongly perturbs the transcriptome of NMuMG cells
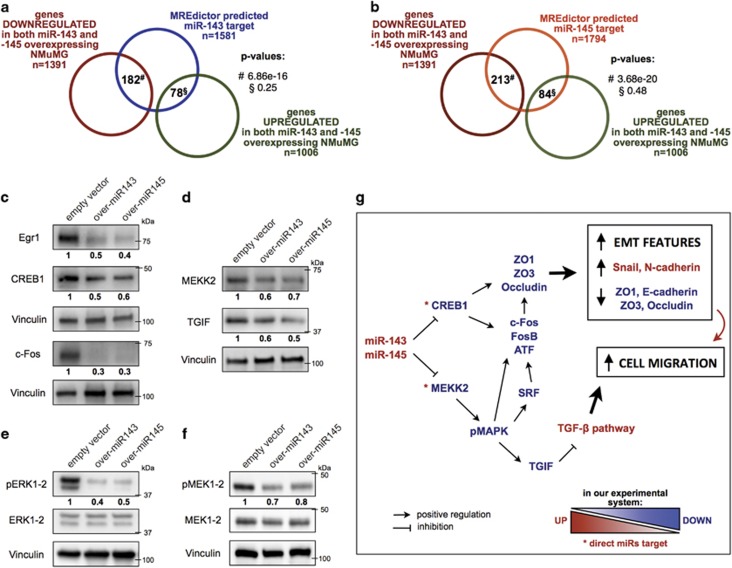
As shown above, overexpression of both miRNAs in NMuMG cells leads to remarkably similar alterations in the expression of relevant EMT-related genes, and in particular downregulation of E-cadherin and of the tight junction proteins ZO1, ZO3 and Occludin, which are not however predicted as common miRNA targets. We thus decided to assess the transcriptome alterations of the overexpressing cells by RNA sequence analysis. In keeping with their superimposable phenotypes, miR-143 and miR-145 overexpressing NMuMG cells displayed very similar mRNA expression profiles, with 1391 and 1006 genes, respectively, commonly down- and upregulated by at least 2 fold with respect to control cells (adjusted P-value <0.05, Supplementary Table 1). Importantly, predicted targets of both miRNAs were significantly enriched among the down-, but not the upregulated genes, (Figures 6a, b, Supplementary Figures 9a and b). Nevertheless, the number of downmodulated genes greatly exceeded the number of common miRNA target mRNAs, sixty according to the miRNA target prediction software MREdictor30 (Supplementary Table 2). Such a strong transcriptome perturbation may stem from the downregulation of key transcription factors such as Fos (c-Fos), Fosb, Creb, ATF, Elk4, Srf, and several Egr and C/EBP family members (Supplementary Figure 10a). Among these, Creb1 is a putative direct target of both miRNAs (Supplementary Table 2). Creb1, c-Fos and Egr1 downmodulation was also confirmed at the protein level (Figure 6c).
Figure 6.
Overexpression of miRs-143 or -145 profoundly perturbates the transcriptome of NMuMG cells. (a,b) Venn diagrams representing the overlap between MREdictor predicted targets of miR-143 (a) or miR-145 (b), and the genes commonly modulated by the overexpressed miRNAs in NMuMG cells. Note statistically significant enrichment only among downregulated genes. (c–f) Whole lysates from the indicated cells were analyzed by western blot. (g) Model explaining the EMT phenotype triggered by overexpression of either microRNA
KEGG analysis of downregulated genes revealed significant disruption of the MAP kinase pathway, and in particular strongly reduced levels of Map3k2 (MEKK2), Map4K5 and Mapk8ip3 kinases (Supplementary Figure 10b). Downregulation of MEKK2 kinase (Figure 6d), predicted as a common target of both miRNAs, correlated with significantly decreased phosphorylation of both ERK1/2 and MEK1/2 (Figures 6e and f).
RNA sequencing and RT-PCR data confirmed the downregulation of the RNAs encoding for the junction proteins ZO1 (Tjp1), ZO3 (Tjp3) and Occludin, whose downmodulation was already observed by western blot (Figure 4c and Supplementary Figure 10c). Finally, we also observed significant downregulation of the well know SMAD transcriptional co-repressor Tgif1 (TGIF) (Figure 6d and Supplementary Figure 10c), which may well explain the enhanced TGF-β-induced transcription observed in the miRNA overexpressing cells.
Discussion
MiRNAs are well-known players in tumor transformation and progression, being able to exert both tumor suppressive and promoting roles.31, 32, 33 However, the definition of their specific functions often escapes rigid classification, due to cell- and stage-specific effects such as differential expression and accessibility of their putative targets. MiRs-143 and -145 are no exception. Indeed, both were initially classified as tumor-suppressors based on both expression analysis and functional experiments.34, 35, 36 However, several recent publications suggested that their expression can also correlate with tumor progression, metastasis and recurrence in colorectal cancer, esophageal tumors and HBV-related hepatocellular carcinoma, where their functional role was confirmed by in vitro and in vivo experiments.23, 24, 25 Additionally, expression of both miRNAs was found to be much higher in mesenchymal cells, including the tumor stroma, than in epithelial cells, suggesting that their assessment in whole tumor biopsies may be misleading.35
Our data demonstrate that miRs-143 and -145 contribute to promote EMT and cell migration in both transformed and non-transformed mammary gland cells, consistent with a pro-metastatic, rather than oncosuppressive, role. This function is consistent with the phenotype observed in adult smooth muscle cells from miR-143 and miR-145 null mice. These cells display defective rearrangement of the actin cytoskeleton and impaired migration upon vascular injury, consequently failing to give origin to the scar tissue known as neointima.10, 11, 37 Thus, despite a general consensus that the expression of miRs-143 and -145 triggers the differentiation of embryonal stem and precursor cells,38 correlating with proliferation arrest as also observed in several tumor models,18, 19, 20, 22 their physiological function in adult tissues appears to be less well defined. Particularly intriguing is the observation that interference with miR-143 in the aggressive breast tumor S3C cells significantly reduced their ability to extravasate into the lung parenchyma, suggesting that miR-143 activity can enhance the invasion of cancer cells at distant tissues. On the other hand the observation that, despite less extravasates cells, metastasis number was not decreased in mice injected with sponged S3C cells suggests that high miR-143 activity may negatively affect the ability of tumor cells to survive in the hostile environment of the lung parenchyma. Thus, miR-143 activity likely requires a tight temporal regulation, since enhanced expression favors extravasation while impairing fitness in the colonized tissue. In agreement with this idea, our data show spontaneous downregulation of both miR-143 and -145 expression during the process of metastasis formation.
TGF-β is the major inducer of EMT, both during development and in adult cells including tumors.3 Our data suggest that both miRs-143 and -145 are involved in conferring at least some EMT features also independently of TGF-β in normal mammary epithelial cells. Indeed, both miRNAs are SMAD-dependent TGF-β transcriptional targets,9 and their ectopic expression is sufficient to up-regulate the EMT inducers Snail and Slug and to modulate several EMT markers and junction proteins, enhancing cell migration independently of cytokine treatment. This effect correlates with the ability of both miRNAs to enhance SMAD-dependent transcription.
RNA sequencing analysis has revealed that overexpression of either miRNA pervasively perturbs the transcriptome, in keeping with the knowledge that miRNAs can directly and indirectly modulate thousands of target genes, partly by regulating a high number of transcription factors.39, 40 Also in our cells transcription factors are highly represented among the downregulated genes, among them are Srf and Elk4, the AP-1 family members c-Fos, Fosb and ATF3 and several Egr family members. Although none of these is predicted as a direct miR-143 and/or miR-145 target, several are transcriptionally regulated by Creb1,41 which is instead a putative direct target of both miRNAs and is also downregulated in the overexpressing cells (see also scheme in Figure 6g). On the other hand, Creb1 is the main transcriptional activator of Occludin, and is predicted to have a role in the transcription of Tjp1/ZO1 as well, whose promoter additionally carries several functional AP-1 binding sites.42, 43 Thus, the reduced levels of Creb1 and of AP-1 family members may explain the observed downregulation of the tight junction proteins ZO1, Occludin and possibly also ZO3. In turn, altered expression of tight junction proteins can at least partly account for the enhanced migration elicited by the two miRNAs in NMuMG cells, as confirmed by the enhanced motility triggered by ZO1 silencing.
In agreement with previous work demonstrating that miRs-143 and -145 can modulate MAPK signaling by targeting several genes in the signaling cascade,44 our results revealed significant disruption of the MAPK pathway. Indeed, downregulation of the putative direct common target MEKK2 (and possibly of Map4k5 and Mapk8ip3) correlates with decreased phosphorylation of both ERK1/2 and MEK1/2, without affecting their protein levels. This can in turn contribute to the observed reduced expression of the AP-1- and Srf-family transcription factors. Although the activity of MAP kinases is generally considered as a marker of proliferation and tumor transformation, the many feedback interactions with other important signaling pathways forbid simplistic interpretations. For example, Kim et al. reported that miR26a-mediated downregulation of MEKK2 enhances the growth of glioblastoma cells both in vitro and in vivo.45 Additionally, ERK-mediated phosphorylation is known to stabilize TGIF, a well-known co-repressor of SMADs transcriptional activities, thus interfering with TGF-β-mediated functions.46, 47, 48 Although Tgif1 mRNA is not predicted to be a direct miRNA target and its transcriptional regulation is poorly characterized, TGIF downregulation may at least partly explain the enhanced SMAD-mediated transcription and consequent increased migration and EMT markers expression elicited by miR-143 and -145.
In conclusion, our results clearly demonstrate that miRs-143 and -145 enhance EMT features of both transformed and normal mammary epithelial cells, increasing motility but impairing anchorage-independent proliferation (miR-143). These effects occur via both direct and indirect regulation of the MAPK pathway and of transcription factors impacting on the expression of EMT markers and on SMADs transcriptional activity (Figure 6g). MicroRNAs expression is known to be tightly regulated in time and space under physiological conditions, in agreement with their ability to fine-tune gene expression. The evidences here reported support this idea, suggesting that the consequences of therapeutic approaches aimed at interfering with miRNAs activity have to be carefully evaluated.
Materials and methods
Ethics statement
The present investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and to national and international guidelines, and has been approved by the Faculty Ethical Committee and by the Italian Ministry of Health.
Animal treatments
Twelve weeks old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) female mice were maintained in the pathogen free transgenic unit of the Molecular Biotechnology Center, under a 12 h light-dark cycle and provided food and water ad libitum.
Cell lines
Murine mammary tumor cell lines (WT and S3C) were isolated from MMTV-NeuT mice and cultured as previously described.28 WT1 and S3C1 cells are used throughout the paper and referred to as WT, S3C. NMuMG cells were kindly provided by Prof. C. Watson (University of Cambridge, UK), and were maintained as previously described.29 hTGF-β1 (Peprotech, Rocky Hill, NJ, USA) was used at a final concentration of 10 ng/ml for the indicated time.
Plasmid construction and generation of stable cell lines
The genomic regions containing the murine pre-miRNA-143 or -145 were amplified by PCR and cloned under the control of a CMV promoter into the pLemiR-tRFP vector (GE Healthcare, Little Chalfon, UK) to obtain the over-miR-143 and over-miR-145 lentiviral constructs. For the preparation of the miR-143 and miR-145 sponges, 292 nts long sequences (Supplementary Table 3), containing eight bulged target sites specific for miR-143 or miR-145 interrupted by linkers, were designed as described in Krol et al.,49 synthesized by DNA2.0 (Newark, CA, USA) and cloned in the 3′UTR of a GFP cassette in a pLenti-CMV-GFP-Puro vector (Addgene, Cambridge, MA, USA), generating the sp-miR-143 and sp-miR-145 vectors. The pLemiR-tRFP and pLenti-CMV-GFP-Puro empty vectors were used as controls. All stable cell lines were generated via lentiviral infection. Lentiviral particles were produced according to Trono’s lab protocol (http://tronolab.epfl.ch). Supernatant was harvested 48 h post-transfection, filtered with 0.45 mm filters, diluted and used to transduce cells, in the presence of Polybrene (Sigma-Aldrich, Saint Louis, MI, USA). For the transduction of S3C cells, lentiviral particles were concentrated by supernatant ultracentrifugation (5 × 104 × g for 2 h at 4 °C), resuspended in PBS and then used to transduce cells in low adhesion conditions. To generate the SMAD reporter a DNA oligonucleotide sequence containing three SMAD3-4 binding sites50 (Supplementary Table 3) was cloned upstream of the TATA box in the pTA-Luc vector (Addgene). The SMAD reporter was co-transfected with a pRL-TK Renilla plasmid in NMuMG cells by means of Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. Firefly and Renilla Luciferase activities were measured 48 h upon transfection, with the Dual-Glo Luciferase Assay system (Promega, Madison, WI, USA) and a 96 microplates GloMax luminometer (Promega).
Western blots
Total protein extracts were obtained and Western blots performed as described in Schiavone et al.51 The following antibodies were used: N-cadherin (Abcam, Cambridge, UK); E-cadherin, β-catenin (BD, Franklin Lakes, NJ, USA); Snail, pSMAD2, SMAD2, pSMAD3-1, SMAD3, pERK1/2, ERK1/2, pMEK1/2, MEK1/2, Creb1 (Cell Signaling Technology, Danvers, MA, USA); α-SMA, α-tubulin (Sigma-Aldrich); ZO1, ZO3, Occludin (Thermo Fisher Scientific); SMAD4, Egr1, c-Fos, MEKK2, TGIF, Actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA); GAPDH (Merck-Millipore); Vinculin and GFP (home made). Western blot chemilumiscent signals were acquired with a ChemidocTouch and analyzed with the ImageLab software (BioRad). All blots were repeated at least three times. One representative image is shown.
RNA isolation and qRT-PCR for miRNA or mRNA detection
Total RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific). qRT-PCR reactions for miRNA detection were performed with the indicated TaqMan MicroRNA Assay (Thermo Fisher Scientific) on total RNA, according to manufacturer’s instructions, and normalized on U6 RNA levels. mRNAs expression levels were evaluated by means of qRT-PCR assays based on the Universal Probe Library (Roche, Basel, Switzerland) technology, normalized on 18 s RNA levels. Primers and probes used are listed in Supplementary Table 4. The relative expression levels between samples were calculated using the comparative delta CT (threshold cycle number) method (2−ΔΔCT) with a control sample as the reference point.
Immunofluorescence
Cells plated on glass coverslips were treated as reported in Barbieri et al.,28 incubated with primary antibodies at room temperature for 1 h, followed by anti-rabbit or anti-mouse conjugated AlexaFluor 568 antibody (Thermo Fisher Scientific) or phalloidin-FITC (Sigma-Aldrich) and then by Hoescht-dye. An Axiovert 200M Zeiss microscope or the Axio-Observer-Z1 Zeiss microscope with the ApoTome system for optical sectioning were used. Images were acquired with the MetaMorph software (Molecular Devices, Sunnivale, CA, USA) or the AxioVision release 4.6.3 software (Carl Zeiss, Oberkochen, Germany), respectively. Confocal microscopy imaging was performed using a Leica TCS SP5-AOBS 5-channel system (Leica Microsystems, Wetzlar, Germany) equipped with a 405-nm diode, an argon ion, a 561-nm DPSS, and a 633-nm HeNe laser. Images were acquired using a HCX PL APO 40 × /1.25 NA oil immersion objective (0.38 × 0.38 × 0.5 μm3 voxel size). 0.5 μm Z-stacks were acquired and showed as projections.
Quantification of cell contacs distribution
For the whole image approach, the score was designed to quantify the regularity of distribution of ZO1 or β-catenin on the cell surface, visible as a continuous line along the edges of cells, in the Z-projection of several Z-stack sections acquired by confocal imaging. For each image we measured the signal of DAPI and ZO1/β-catenin staining across 79 horizontal and equally spaced lines. We reasoned that a uniform distribution of ZO1/β-catenin along the cell contacts should lead to the detection of regularly alternating DAPI and ZO1/ β-catenin signal peaks across each line. Therefore, the score measures the regularity of this alternation. Peaks were identified using a signal intesity threshold (arbitrary units) of 40 for ZO1 or β-catenin and of 20 for the nuclei. Narrow DAPI peaks – corresponding to a single x position – were discarded, while peaks only one position distant were merged. ZO1/β-catenin peaks overlapping with DAPI staining were discarded. For each transition between ZO1/β-catenin and DAPI peaks detected along the line, the score was increased by one. Finally, the score obtained for each line was normalized on the number of total peaks detected on that line, and the final score was calculated as the mean±S.E.M. of 79 lines on eight individual images.
For the single cell approach, 10 cells were randomly selected from 3 independent images per condition, and the mean intesity of the signal (ZO1 or β-catenin) was measured both along the cell edge (membrane) and in the rest of the cell. The ratio between the two values was calculated, and represents the relative distribution of the signal between the membrane and the whole cell.
Scratch assay and transwell migration assays
In vitro scratch assays were performed as described by Liang et al.,52 with some modifications. In brief, cells were grown to confluence and starved overnight with 0% FCS. Two hours before the scratch, cells were treated with 10 μg/ml mitomycin C (Sigma-Aldrich). The cell monolayer was scraped in a straight line with a pipet tip in order to create a linear wound. Debris were removed by washing the cells with PBS and then replaced with complete medium plus mitomycin C. Photographs of the wounds (10 fields per experiment) were taken with a phase contrast Olympus IX70 microscope. For each image cells migrated into the original wound were measured, normalized to the original wound area, then the control condition is imposed as reference and other conditions expressed as relative. Transwell assays were performed as described in Barbieri et al.,28 after 24 h of migration pictures of the cells on the lower surface were taken and then quantified with the ImageJ software (NIH, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/).
In vivo extravasation and lung metastases
For extravasation experiments, 5 × 105 cells were labeled with CellTracker Orange CMRA (Thermo Fisher Scientific), resuspended in PBS and injected into the tail vein of NSG mice. After 15 min or 24 h, mice were sacrificed and 4% paraformaldehyde was injected into the trachea. Total lungs were dissected and photographed using a Leica MZ16F fluorescence stereomicroscope. Red-fluorescent (CMRA) cells were counted using the ImageJ software. For long term metastases, 1 × 105 cells were resuspended in PBS and injected into the tail vein of 7-week-old female NSG mice. After 3 weeks mice were sacrificed, lungs collected and fixed for paraffin embedding, semiserial sections at 100 μm intervals were stained with H&E and neoplastic lesions quantified with the ImageJ software.
Recovery of cells from metastatic lesions and cell sorting
Freshly isolated lungs were digested with collagenase A (Roche, Basel, Switzerland), filtered with cell strainer (70 μm), and stained with EpCAM-FITC coniugated antibody (Biolegend, San Diego, CA, USA), after red blood cell lysis. Positive and negative EpCAM cell populations (Supplementary Figure 3d) were sorted with FACS Aria III (BD). Only the lungs from injected mice showed EpCAM positivity, indicating that EpCAM+ cells derive from the S3C cells.
Anchorage-independent growth assay and anoikis
1 × 104 cells were resuspended in complete DMEM containing 0.3% low gelling agarose (Sigma-Aldrich) and seeded in plates containing a layer of solid 0.6% agar (Sigma-Aldrich). After 3 weeks colonies were stained with nitroblue tetrazolium (Sigma-Aldrich), photographed with a Nikon SMZ1000 stereomicroscope and colonies were counted using the ImageJ Software. For suspension-induced anoikis, confluent adherent cells were detached from regular culture plates with EDTA, seeded onto poly-HEMA coated plates (poly-HEMA, 10 mg/ml, Sigma-Aldrich), and cultured for 24 h in complete culture medium. Apoptosis was assessed by annexin V staining (BD) with aFACScalibur flow citometer (BD) and CellQuest software. Positive cells are expressed as percentage of the total.
RNA-seq library preparation
For RNA-seq library preparation, approximately 2 μg of total RNA from the different NMuMG cell lines was subjected to poly(A) selection, and libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Sequencing was performed on the Illumina NextSeq 500 platform.
RNA-seq data analysis
Illumina BCL files were demultiplexed to FastQ format using the bcl2fastq tool from Illumina. Before mapping, the reads quality was estimated using the FastQC tool v0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The nucleotide positions with a quality score below 30 (Phred33 scale) were trimmed using the fastx_trimmer tool from the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). After low-quality position trimming, the reads in which sequencing continued through the 3’ adapter sequence were subjected to adapter clipping using the fastx_clipper tool from the FASTX Toolkit, and the reads shorter than 35 nt were discarded. Reads were mapped to mouse mm9 assembly using TopHat v2 (http://tophat.cbcb.umd.edu/, parameters: —bowtie1 -a 5 —library-type fr-unstranded). Genes counts were computed using the htseq-count tool from the HTSeq package (http://www.huber.embl.de/HTSeq/doc/overview.html) and differential expression analysis was performed using R and DESeq2 package (http://www.bioconductor.org/packages/devel/bioc/html/DESeq2.html). Genes with adjusted P-value<0.05 and abs(log2(Fold Change)) ⩾1 were considered significantly modulated, and kept for downstream analyses. RNA-seq data has been deposited on Gene Expression Omnibus database, under the accession GSE98689.
MiRNA targets prediction
For microRNA targets prediction, we used either MREdictor30 or TargetScan algorithms. Annotated 3′-UTRs for up- or downregulated genes in either miR-143a or miR-145a overexpressing cells, were scanned for strict miRNA targets (8mer, 7mer-m8, and 7mer-A1). The significance of the measured overlaps were calculated using a hypergeometric test.
Statistical analyses
Unless otherwise noted, data were analyzed by one-way ANOVA or Student’s t-test, using the Prism (GraphPad software, Inc., San Diego, CA, USA). For qRT-PCR statistical analyses were performed on 2−ΔΔCT values.
Acknowledgments
We thank E Monteleone for bioinformatics support, F Orso for the generous gift of reagents and for technical advice, D Taverna for useful discussion, M Forni for help with histological analysis, A Camporeale and C Bologna for help with cell sorting, F Orso, D Taverna and M Santoro for critically reading the manuscript. This work was supported by grants from the Italian Cancer Research Association (AIRC IG 13009, 16930 to VP, and 15217 to SO), the Italian Ministry of University and Research (MIUR PRIN, prot. 20129JLHSY), the Ateneo/Compagnia di San Paolo (TO_Call2_2012_0062), and the Truus and Gerrit van Riemsdijk Foundation, Lichtestein to VP. MBS was supported by the MetastasiX project of the Swiss Initiative for Systems Biology (SystemsX.ch). LA was the recipient of a FIRC postdoctoral fellowship.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by BD Dynlacht
The authors declare no conflict of interest.
Supplementary Material
References
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7: 415–428. [DOI] [PubMed] [Google Scholar]
- Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006; 66: 8319–8326. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J et al. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 2002; 21: 3241–3246. [DOI] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 2008; 68: 537–544. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927–939. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 2008; 7: 3112–3118. [DOI] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009; 460: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Miano JM. Transforming growth factor-beta1 (TGF-beta1) utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem 2011; 286: 30119–30129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MVG et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ 2009; 16: 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 2009; 23: 2166–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 2010; 17: 398–408. [DOI] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008; 27: 1788–1793. [DOI] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res 2008; 14: 2690–2695. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007; 67: 8699–8707. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci 2007; 98: 1914–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 2010; 17: 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 2010; 24: 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res 2010; 2: 170–180. [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 2009; 106: 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 2013; 32: 61–69. [DOI] [PubMed] [Google Scholar]
- Derouet MF, Liu G, Darling GE. MiR-145 expression accelerates esophageal adenocarcinoma progression by enhancing cell invasion and anoikis resistance. PloS One 2014; 9: e115589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009; 50: 490–499. [DOI] [PubMed] [Google Scholar]
- Yuan W, Sui C, Liu Q, Tang W, An H, Ma J. Up-regulation of microRNA-145 associates with lymph node metastasis in colorectal cancer. PloS One 2014; 9: e102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Liang J, Yu F, Zhou Z, Tang J, Li K. miR-145 promotes osteosarcoma growth by reducing expression of the transcription factor friend leukemia virus integration 1. Oncotarget 2016; 7: 42241–42251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. J Pathol 2011; 223: 205–218. [DOI] [PubMed] [Google Scholar]
- Barbieri I, Pensa S, Pannellini T, Quaglino E, Maritano D, Demaria M et al. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res 2010; 70: 2558–2567. [DOI] [PubMed] [Google Scholar]
- Robson EJ, Khaled WT, Abell K, Watson CJ. Epithelial-to-mesenchymal transition confers resistance to apoptosis in three murine mammary epithelial cell lines. Differentiation 2006; 74: 254–264. [DOI] [PubMed] [Google Scholar]
- Incarnato D, Neri F, Diamanti D, Oliviero S. MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets. Nucleic Acids Res 2013; 41: 8421–8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 2009; 9: 293–302. [DOI] [PubMed] [Google Scholar]
- Cioce M, Strano S, Muti P, Blandino G. Mir 145/143: tumor suppressor, oncogenic microenvironmental factor or...both? Aging 2016; 8: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, McCall MN, Cornish TC, Halushka MK. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 2014; 42: 7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang L, Yang A-G. Is microRNA‐143 really a turncoat of tumor suppressor microRNA in hepatitis B virus–related hepatocellular carcinoma? Hepatology 2009; 50: 987. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res 2009; 105: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009; 137: 647–658. [DOI] [PubMed] [Google Scholar]
- Kaller M, Hermeking H. Interplay between transcription factors and microRNAs regulating epithelial-mesenchymal transitions in colorectal cancer. Adv Exp Med Biol 2016; 937: 71–92. [DOI] [PubMed] [Google Scholar]
- Lui P-Y, Jin D-Y, Stevenson NJ. MicroRNA: master controllers of intracellular signaling pathways. Cell Mol Life Sci 2015; 72: 3531–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2001; 2: 599–609. [DOI] [PubMed] [Google Scholar]
- Lui W-Y, Wong EWP, Guan Y, Lee WM. Dual transcriptional control of claudin-11 via an overlapping GATA/NF-Y motif: Positive regulation through the interaction of GATA, NF-YA, and CREB and negative regulation through the interaction of Smad, HDAC1, and mSin3A. J Cell Physiol 2007; 211: 638–648. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Zhang B, Eum SY, Toborek M. HIV-1 Tat triggers nuclear localization of ZO-1 via rho signaling and cAMP response element-binding protein activation. J Neurosci 2012; 32: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013; 32: 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci USA 2010; 107: 2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Ono K, Kinoshita M, Nishi H, Nagao K, Kawamura T et al. TG-interacting factor is required for the differentiation of preadipocytes. J Lipid Res 2008; 49: 1224–1234. [DOI] [PubMed] [Google Scholar]
- Lo RS, Wotton D, Massague J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J 2001; 20: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Liu Y. Hepatocyte growth factor antagonizes the profibrotic action of TGF-beta1 in mesangial cells by stabilizing Smad transcriptional corepressor TGIF. J Am Soc Nephrol 2004; 15: 1402–1412. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 2010; 141: 618–631. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1998; 1: 611–617. [DOI] [PubMed] [Google Scholar]
- Schiavone D, Dewilde S, Vallania F, Turkson J, Di Cunto F, Poli V. The RhoU/Wrch1 Rho GTPase gene is a common transcriptional target of both the gp130/STAT3 and Wnt-1 pathways. Biochem J 2009; 421: 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007; 2: 329–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.