Abstract
Recent studies have suggested increased plasticity of differentiated cells within the intestine to act both as intestinal stem cells (ISCs) and tumour-initiating cells. However, little is known of the processes that regulate this plasticity. Our previous work has shown that activating mutations of Kras or the NF-κB pathway can drive dedifferentiation of intestinal cells lacking Apc. To investigate this process further, we profiled both cells undergoing dedifferentiation in vitro and tumours generated from these cells in vivo by gene expression analysis. Remarkably, no clear differences were observed in the tumours; however, during dedifferentiation in vitro we found a marked upregulation of TGFβ signalling, a pathway commonly mutated in colorectal cancer (CRC). Genetic inactivation of TGFβ type 1 receptor (Tgfbr1/Alk5) enhanced the ability of KrasG12D/+ mutation to drive dedifferentiation and markedly accelerated tumourigenesis. Mechanistically this is associated with a marked activation of MAPK signalling. Tumourigenesis from differentiated compartments is potently inhibited by MEK inhibition. Taken together, we show that tumours arising in differentiated compartments will be exposed to different suppressive signals, for example, TGFβ and blockade of these makes tumourigenesis more efficient from this compartment.
Since the description of ‘top-down’ adenoma formation of colorectal tumours there has been discussion over the cell of origin of colorectal cancer (CRC). Here, human dysplastic crypts, carrying mutations of the APC tumour suppressor gene, were detectable exclusively at the top part of colonic crypts in patients with adenomatous polyps.1 Importantly, cells at the base of the crypts, on which the polyps were located, were morphologically normal and did not carry an APC mutation. However, histopathological analyses have also suggested a stem cell route through to the identification of monocryptal adenomas in patients who develop familial adenomatous polyposis due to a germline mutation in APC, supporting the ‘bottom-up’ model of intestinal tumourigenesis.2
To elucidate the cell of origin of human CRC, many studies have used ‘proof-of-concept’ approaches using genetically engineered mice to target different cell types and assess if they initiate tumourigenesis. This extensive work has also suggested two routes of intestinal cancer. In support of the ‘bottom-up’ model, one efficient route is where intestinal stem cells (ISCs) are transformed via Apc loss or stable expression of β-catenin and very rapidly form adenomas.3, 4, 5 At the same time, experimental evidence also supports the ‘top-down’ model of intestinal tumourigenesis, where non-ISCs are transformed and serve as cells of origin. However, non-ISCs alongside Apc loss require additional events to initiate tumourigenesis.6, 7, 8 Activation of oncogenic KrasG12D/+ or NF-κB activation concomitantly with Apc loss can drive a dedifferentiation programme within the villus epithelium.6 It has also been shown that the long-lived differentiated DCLK1+ve Tuft cells can act as a cell of origin for intestinal cancer8 if Apc loss was combined with inflammation. Moreover, elegant work by Davis et al.9 showed that the stromal microenvironment has a critical role in suppressing dedifferentiation. Disruption of the Gremlin-1 morphogen gradient within the intestinal epithelium led to the formation of ‘crypts in villus’, indicating that Apc-deficient enterocytes indeed dedifferentiated.
Recent sequencing studies have confirmed that in CRC alongside APC mutation there are common mutations in KRAS, TP53 and PI3KCA genes. Additionally, the TGFβ family members are also very commonly mutated or deregulated.10 Deletion, downregulation or inactivating mutations can occur in TGFBR1, TGFBR2 and the SMAD effectors (SMAD2–4).11, 12, 13 Frameshift mutations in TGFBR2 have been predominantly identified in CRC with mismatch repair (MMR) defects.14 In tumours with functional MMR, TGFβ signalling is commonly abolished by loss of SMAD4.11, 15
In addition to this clear tumour-suppressive role, TGFβ signalling also has tumour-promoting effects in the late stages of CRC. Enhanced levels of TGFβ ligand expression has been associated with advanced tumours,16 and the activation of an epithelial-to-mesenchymal transition (EMT) programme.17, 18 High levels of TGFβ target genes, in the stromal cells of CRC patients, are associated with poor prognosis,19 and in the recent publication of the consensus molecular subtypes of human CRC, high levels of TGFβ signalling define a mesenchymal subtype with the poorest prognosis.20
Since TGFβ signalling is commonly deregulated in CRC, we functionally explored the importance of this pathway and found that the likely driving force, behind these mutations, is the inhibition of its prodifferentiation tumour-suppressive effect. Mechanistically further deregulation of MAPK signalling is important for this phenotype.
Results
Transcriptome profiling of tumours derived from crypts versus villi failed to reveal major differences
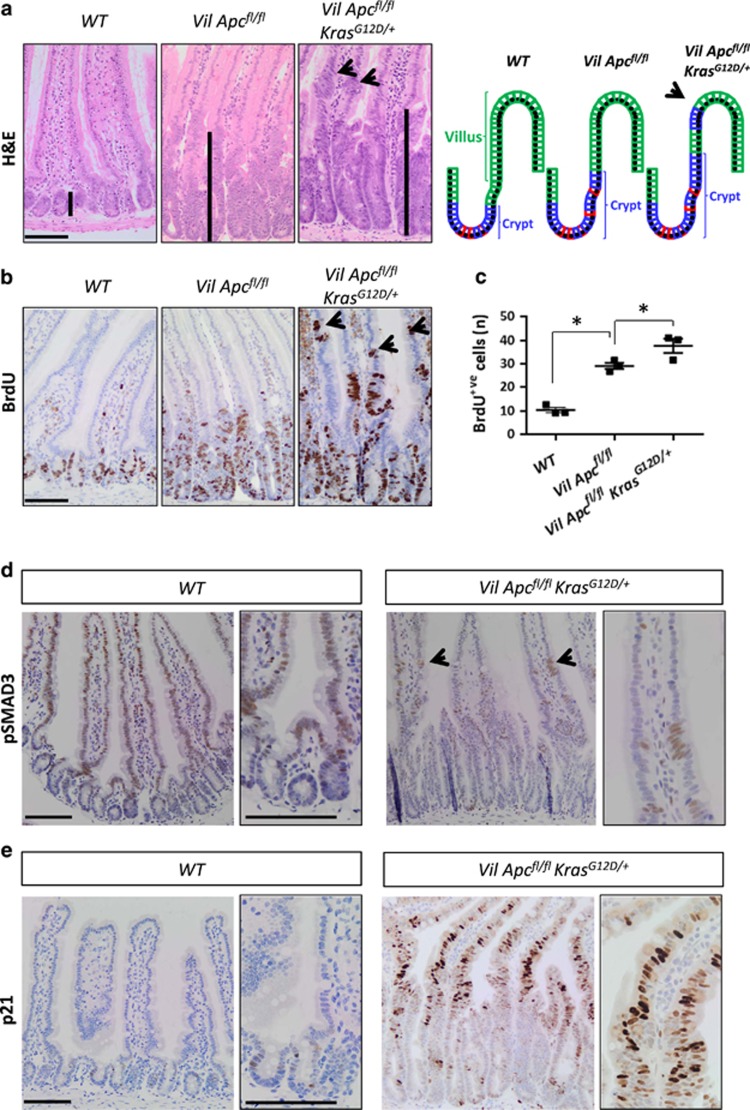
Acute deletion of Apc in the intestinal epithelium, using VillinCreER (referred to from now on as VilCreERApcfl/fl), causes a crypt-progenitor phenotype characterized by enlarged intestinal crypts when compared with the wild-type intestine (Figure 1a). An additional oncogenic mutation in Kras (referred to from now on as VilCreERApcfl/flKrasG12D/+) results in a marked increase in crypt size (Figure 1a) with the presence of proliferative cells in the villus, which are not detected in wild-type or VilCreERApcfl/fl mice (Figures 1b and c), showing that the additional mutation of Kras can drive proliferation in differentiated enterocytes.
Figure 1.
KrasG12D/+ enhances both the crypt-progenitor phenotype and transforming growth factor-β (TGFβ) pathway activation following Apc loss. (a) Haematoxylin and eosin (H&E) staining and a schematic model of the crypt-progenitor phenotype of wild-type (WT), VilCreERApcfl/fl (Vil Apcfl/fl) and VilCreERApcfl/flKrasG12D/+ (Vil Apcfl/flKrasG12D/+) small intestine. Note the presence of dedifferentiated cells (black arrow) in the villus of the Vil Apcfl/flKrasG12D/+genotype. Black bars highlight the differences in crypt length. (b) Bromodeoxyuridine (BrdU) staining of WT, Vil Apcfl/f and Vil Apcfl/flKrasG12D/+ small intestine. Note the presence of proliferating cells (black arrow) in the villi of the Vil Apcfl/flKrasG12D/+genotype. (c) Quantification of total BrdU+ cells demonstrating increased proliferation in the Vil Apcfl/flKrasG12D/+ genotype. Error bars represent mean± S.E.M., *P=0.04 Mann–Whitney test, one-tailed, n=3 biological replicates. (d) pSMAD3 and (e) p21 IHC analysis of intestine from WT and Vil Apcfl/flKrasG12D/+ mice. Black arrows indicate positive cells in the villi. High magnification insets highlight the presence of positive cells. Scale bars, 100 μm
In our previous studies, we have demonstrated that villi purified from VilCreERApcfl/flKrasG12D/+ mice grow in vitro as spheroids with tumourigenic capacity (unlike villi purified from either wild-type or VilCreERApcfl/fl mice).6 To investigate this process further, we analysed the transcriptomes of tumours derived through subcutaneous injection of either spheroids derived from crypts (CDS) or Villi (VDS) purified from VilCreERApcfl/flKrasG12D/+ mice. Surprisingly, our microarray analysis revealed no consistent differences between tumours derived from injection of CDS or VDS (Supplementary Data 1), indicating that in these models the cell of origin did not modify the transcriptional profile of the tumours. This also suggested that dedifferentiation, provoked by WNT activation and KrasG12D/+ mutation, was complete and that, after transplantation, the cell of origin does not affect subsequent gene expression. To investigate this in an autochthonous environment, we then compared gene expression when WNT pathway activation (using a β-catenin mutant that cannot be targeted for degradation – referred to as CtnnbloxEx3/+), coupled with an oncogenic KrasG12D/+ mutation, was targeted to just differentiated cells, using Xbp1sCreER, or to all cells, using VilCreER. Remarkably, expression profiles from epithelial extractions (performed when intestines showed a similar crypt-progenitor phenotype, 7 days for VilCreER and 21 days for Xbp1sCreER) did not reveal any significant differences, again suggesting that once cells had undergone dedifferentiation the gene expression pattern of the initiating cells is lost (Supplementary Data 2a and b).
TGFβ signalling pathway is upregulated following WNT and oncogenic KRAS activation
We reasoned that to have a better understanding of the pathways responsible for dedifferentiation, we would need to investigate transcriptional changes as the process was occurring. For this reason, we profiled VDS versus CDS, purified from VilCreERApcfl/flKrasG12D/+ mice, while they were still in culture. This approach proved more successful, and we were able to observe significant deregulation of a number of different growth signalling pathways (fold-change cutoffs⩾1.5, P⩽0.05; Supplementary Figure 1a and Supplementary Data 3). It was particularly notable that numerous members of the TGFβ family and superfamily were upregulated in the VDS compared with CDS (fold-change cutoffs⩾1.5; Supplementary Table 1 and Supplementary Data 3). To assess whether upregulation of TGFβ signalling was also occurring in vivo, we examined the activity of TGFβ signalling immediately after combined acute loss of Apc and KrasG12D/+ mutation. We detected phosphorylated SMAD3 (pSMAD3) positivity mainly in the villus aberrant foci when compared with the adjacent normal epithelium in VilCreERApcfl/flKrasG12D/+ mice (Figure 1d and Supplementary Figure 1b). Furthermore, p21 (a mediator of TGFβ signalling) was highly expressed in the VilCreERApcfl/flKrasG12D/+ villi when compared with wild-type villi (Figure 1e). We also observed increased levels of Smad7 (a canonical TGFβ target gene) in VilCreERApcfl/flKrasG12D/+ mice along the crypt–villus axis compared with wild-type intestine (Supplementary Figure 1c).
We then assessed the activation state of the TGFβ signalling pathway in intestinal tumours developed in VilCreERApcfl/+KrasG12D/+ mice. These mice develop tumours in both the small and large intestine upon loss of the remaining wild-type Apc allele (Supplementary Figure 2a). Importantly, tumours from these mice resemble both the top-down (Supplementary Figures 2b and d) and bottom-up morphologies (Supplementary Figures 2c and d). Importantly, we detected high levels of pSMAD3 and p21 in the top-down lesions, indicating an activation of TGFβ signalling (Supplementary Figures 2e and f). Tumours generated according to the bottom-up model show a morphogenic pSMAD3 gradient along the crypt–villus axis (Supplementary Figure 2g).
Thus, these results suggest that WNT and KRAS signalling can promote TGFβ signalling, particularly in cells that are dedifferentiating at crypt–villus junctions.
TGFβ signalling in wild-type intestine is higher as cells differentiate but is not essential for homeostasis
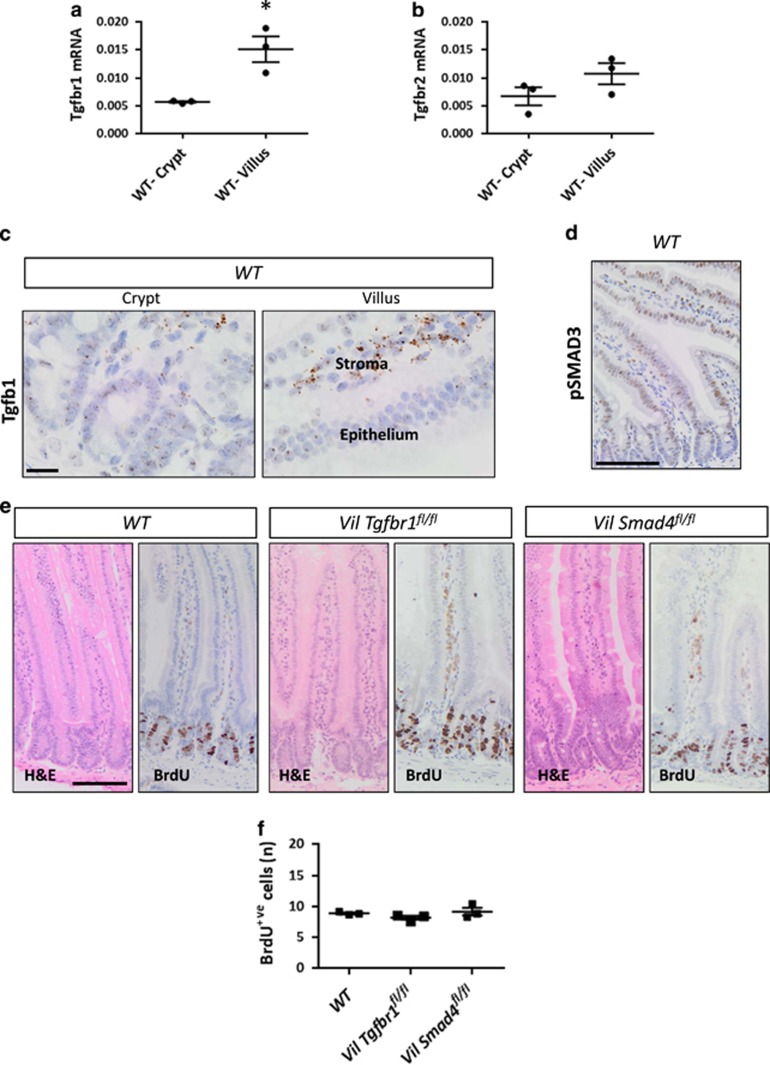
The TGFβ pathway has been previously reported to form a gradient along the vertical axis of the intestinal epithelium with the highest activity in differentiated cells and lowest activity in the crypt.21 However, studies that have genetically ablated TGFβ signalling either via receptor or SMAD4 deletion, have not reported marked effects on homeostasis.22, 23 For our studies if tumour cells arise further up the crypt–villus axis, they may be exposed to a high level of TGFβ signalling that might prevent dedifferentiation and limit proliferation. We first examined the expression of TGFβ signalling components in wild-type intestine. We compared Tgfbr1 and Tgfbr2 mRNA expression in freshly purified crypts with villi (which include intravillus stromal cells). mRNA of both receptors was detectable in both compartments with higher levels in the villus (Figures 2a and b). The Tgfb1 ligand expression was mainly detected in the intravillus region (Figure 2c), suggesting that the differentiated compartment might be exposed to more ligand and therefore activate TGFβ signalling to a higher extent. We then stained wild-type intestines for pSMAD3. Nuclear pSMAD3 was mainly detected in the late transit-amplifying and differentiated cells (Figure 2d).
Figure 2.
Loss of transforming growth factor-β (TGFβ) signalling does not affect the normal homeostatic proliferation. (a) Quantitative real-time-PCR (qRT-PCR) analysis of Tgfbr1 and (b) Tgfbr2 expression in wild-type freshly purified crypts (WT-Crypt) versus villi (WT-Villus). Data are shown as ratios to the internal Gapdh control with error bars representing mean±S.E.M., *P=0.04 by Mann–Whitney test, one-tailed, n=3 biological replicates. (c) RNAscope analysis of Tgfb1 in WT crypt and villus. Scale bars, 20 μm. (d) IHC analysis of pSMAD3 on WT small intestine. (e) Haematoxylin and eosin (H&E) and bromodeoxyuridine (BrdU) staining of WT, VilCreERTgfbr1fl/fl (Vil Tgfbr1fl/fl) and VilCreERSmad4fl/fl (Vil Smad4fl/fl) small intestine 4 days post induction with tamoxifen. Scale bars, 100 μm. (f) Quantification of total BrdU+ cells in WT, Vil Tgfbr1fl/fl and Vil Smad4fl/fl small intestine. Error bars represent mean±S.E.M., n=3 biological replicates
In line with previous studies, deletion of either Tgfbr1 (VilCreERTgfbr1fl/fl) or the key downstream transcription factor SMAD4 (VilCreERSmad4fl/fl) had no impact on intestinal proliferation. Normal levels of proliferation were observed 4 days after conditional deletion when compared with wild-type intestine (Figures 2e and f). Taken together, these data indicate that canonical TGFβ signalling is higher in the differentiated compartment of the intestine and could have a role in suppressing dedifferentiation even in the absence of a homeostatic role.
Loss of TGFβ signalling is not sufficient to drive dedifferentiation if only Apc is deleted
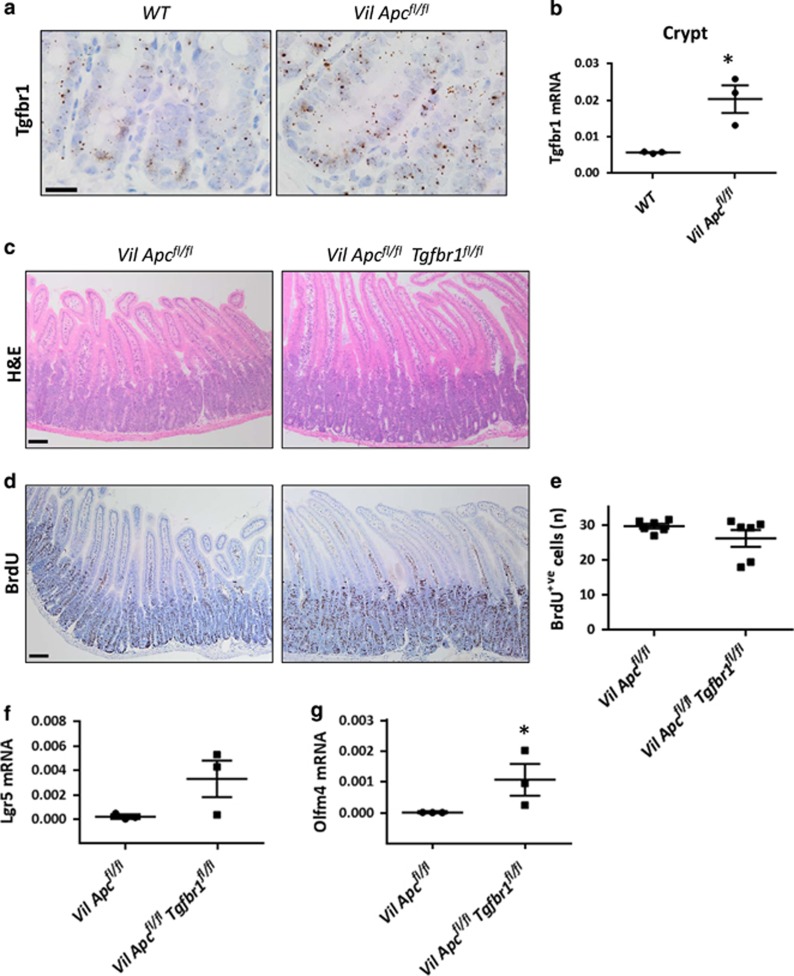
Analysis of our previous microarray,24 comparing the transcriptomes of wild-type intestine to intestine where Apc has been acutely deleted, showed that Tgfbr1 is upregulated following acute Apc loss. To confirm this, we analysed the expression of TGFβ signalling components in VilCreERApcfl/fl mice 4 days post induction and compared this with wild-type mice. We observed a marked increase in Tgfbr1 mRNA levels in the crypts but not the villi (Figures 3a and b and Supplementary Figure 3a).
Figure 3.
Tgfbr1 is upregulated following Apc loss. (a) RNAscope of Tgfbr1 on wild-type (WT) and VilCreERApcfl/fl (Vil Apcfl/fl) crypts. Note the increased positivity following Apc loss. Scale bars, 20 μm. (b) Quantitative real-time-PCR (qRT-PCR) analysis of Tgfbr1 on WT and Vil Apcfl/fl crypts. Data are shown as ratios to the internal Gapdh control with error bars representing mean±S.E.M., *P=0.04 by Mann–Whitney test, one-tailed, n=3 biological replicates. (c) Haematoxylin and eosin (H&E) and (d) bromodeoxyuridine (BrdU) staining on Vil Apcfl/fl and VilCreERApcfl/flTgfbr1fl/fl (Vil Apcfl/flTgfbr1fl/fl) intestine 4 days post induction with tamoxifen. Scale bars, 100 μm. (e) Quantification of total BrdU+ cells of Vil Apcfl/fl and Vil Apcfl/fl Tgfbr1fl/fl intestine. Error bars represent mean±S.E.M., n=6 biological replicates. (f) qRT-PCR analysis of the stem cell markers Lgr5 and (g) Olfm4 on CDS generated from Vil Apcfl/fl and Vil Apcfl/flTgfbr1fl/fl small intestine. Data are shown as ratios to the internal Gapdh control with error bars representing mean±S.E.M., *P=0.04 by Mann–Whitney test, one-tailed, n=3 biological replicates
Similar to the wild-type intestine, stromal cells were the main source of TGFβ1 (Supplementary Figure 3b). After Apc deletion, Smad7 mRNA was detectable in both crypts and villi (Supplementary Figure 3c) and p21 was strongly induced at the crypt–villus transition zone (Supplementary Figure 3d).
To test the functional importance of TGFβ signalling upregulation following Apc loss, we generated VilCreERApcfl/flTgfbr1fl/fl mice and analysed the crypt-progenitor phenotype 4 days post induction. Surprisingly, crypt size and the number of proliferative cells were not increased by Tgfbr1 loss (Figures 3c–e), suggesting that in the absence of Kras mutation, Tgfbr1 deficiency was insufficient to drive dedifferentiation. Accordingly, villi purified from VilCreERApcfl/flTgfbr1fl/fl mice were unable to form spheroids in vitro (Supplementary Figure 3e). Interestingly, Tgfbr1 loss increased the mRNA expression levels of the stem cells markers Lgr5 and Olfm4 in CDS (Figures 3f and g). To functionally test this, we then generated single-cell clones and observed that Tgfbr1 loss increased the number of cells with clonogenic potential. (Supplementary Figure 3f). These data are consistent with work by Batlle and co-workers25 who suggested that loss of TGFβ signalling increases stem cell properties and that TGFBR1/ALK5 inhibitors are required for human colonic organoid cultures.
We then aged VilCreERApcfl/+Tgfbr1fl/fl mice until they developed clinical signs. We observed a significant decrease in tumour latency compared with VilCreERApcfl/+ mice (Supplementary Figure 3g). However, in contrast to VilCreERApcfl/+KrasG12D/+ mice, there was no presence of top-down lesions. Consistent with previous studies,22, 26 the major consequence of Tgfbr1 loss on tumourigenesis was the development of invasive adenocarcinomas with E-cadherin+ve cells at the invasive front (Supplementary Figures 3h and i). Overall, these data show that while loss of TGFβ can drive progression of adenomas to adenocarcinoma, it is not sufficient to drive dedifferentiation in the absence of the KrasG12D mutation.
Loss of TGFβ signalling increases the crypt-progenitor phenotype and accelerates tumourigenesis when both Apc and Kras are mutated
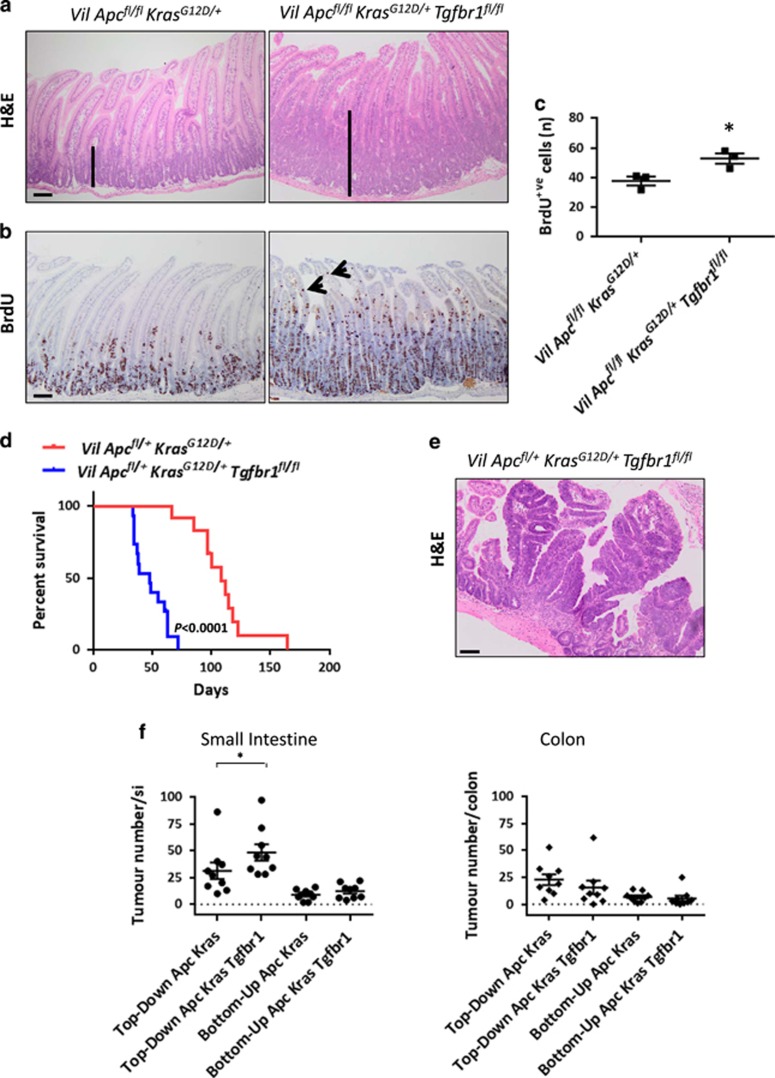
Given that Kras mutation appears to be critical for dedifferentiation and that we observed increased TGFβ signalling in VDS compared with CDS, we generated the VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl mouse model to assess if this modified the acute crypt-progenitor phenotype of VilCreERApcfl/flKrasG12D/+ mice. Tgfbr1 loss now significantly increased the crypt-progenitor phenotype of VilCreERApcfl/flKrasG12D/+ mice (Figure 4a). Importantly, proliferative cells were observed even further into the top part of the villus, suggesting enhanced proliferation (Figures 4b and c). Loss of TGFβ signalling was again associated with an increased expression of Lgr5 and Olfm4 in CDS (Supplementary Figures 4a and b) and with an increased clonogenic potential (Supplementary Figure 4c). We also observed increased Lgr5 and Olfm4 expression levels in the crypts of VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl mice when compared with VilCreERApcfl/flKrasG12D/+ mice. Whereas Lgr5 and Olfm4 levels in the villi were negligible at an early time point (Supplementary Figures 4d and e). To assess whether the increased crypt-progenitor phenotype observed in VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl mice translated into enhanced tumourigenesis, we generated cohorts of VilCreERApcfl/+KrasG12D/+ and VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice and aged them until they showed clinical signs. Tgfbr1 deletion accelerated intestinal tumourigenesis (Figure 4d). VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice developed tumours both in the small intestine and colon. Importantly, many lesions were now observed with histological characteristics reminiscent of top-down lesions, suggesting that they had arisen from differentiated compartments (Figures 4e and f), and 50% of mice developed in situ invasive tumours (Supplementary Figure 4f). Overall, these data suggest that TGFβ signalling acts as a potent tumour suppressor in the presence of WNT and RAS pathway activation, limiting proliferation, stem cell marker expression and potentially plasticity/dedifferentiation.
Figure 4.
Loss of Tgfbr1 cooperates with WNT and KRAS activation to accelerate intestinal tumourigenesis. (a) Haematoxylin and eosin (H&E) and (b) bromodeoxyuridine (BrdU) IHC on VilCreERApcfl/flKrasG12D/+ (Vil Apcfl/flKrasG12D/+) and VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl (Vil Apcfl/flKrasG12D/+Tgfbr1fl/fl) small intestine 3 days post induction with tamoxifen. Black bars highlight the difference in the crypt length. Black arrows indicate the presence of proliferating cells along the villus in the VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl genotype. Scale bars, 100 μm. (c) Quantification of the total BrdU+ cells per half crypt of the Vil Apcfl/flKrasG12D/+ and Vil Apcfl/flKrasG12D/+Tgfbr1fl/fl small intestine. Error bars represent mean±S.E.M., *P=0.04 by Mann–Whitney test, one-tailed, n=3 biological replicates. (d) Kaplan–Meier survival curve of VilCreERApcfl/+KrasG12D/+ (n=12) and VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl (n=15) mice, ****P<0.0001 by log-rank (Mantel–Cox) test. (e) H&E of top-down tumours in VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl (Vil Apcfl/+KrasG12D/+ Tgfbr1fl/fl) mice. Scale bars, 100 μm. (f) Quantification of total tumour number per small intestine and colon developed from crypt (bottom-up) or villus (top-down) in VilCreERApcfl/+KrasG12D/+ (Apc Kras) and VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl (Apc Kras Tgfbr1) mice. Error bars represent mean±S.E.M., *P=0.023 by Mann–Whitney test, one-tailed
Loss of TGFβ signalling promotes dedifferentiation
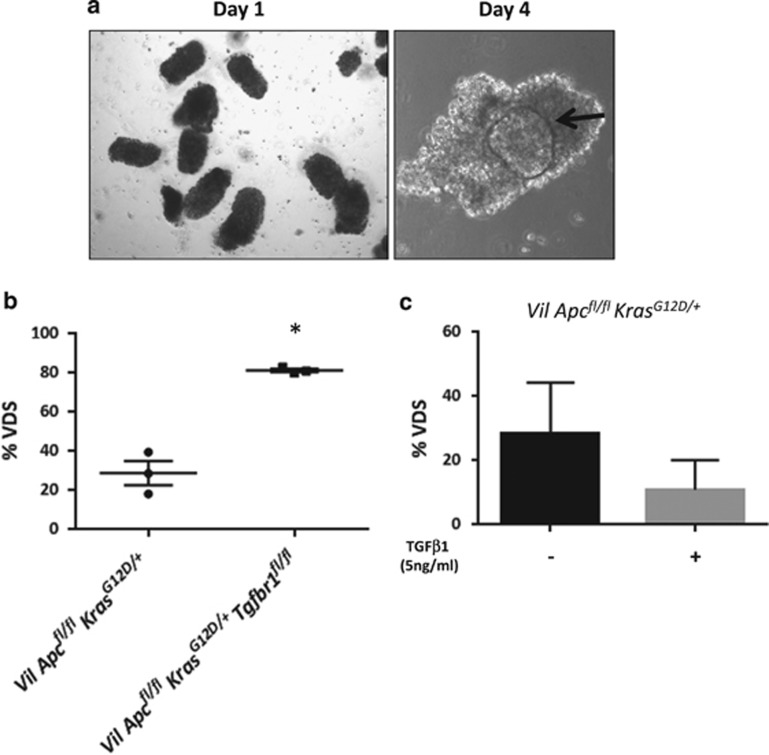
To study the role of TGFβ on dedifferentiation in more detail, we took advantage of the villus culture system. We purified villi from VilCreERApcfl/flKrasG12D/+ and VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl mice and monitored the formation of organoids in vitro. Tgfbr1 loss increased the efficacy of spheroid formation from villi (Figures 5a and b), confirming the role of TGFβ in restraining dedifferentiation. VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl VDS can be propagated long term in culture and have tumourigenic potential. Allograft tumours showed strong expression of the stem cell markers Lgr5 and CD44v6 (Supplementary Figures 5a–c).
Figure 5.
Transforming growth factor-β (TGFβ) signalling attenuates the capacity of villi to dedifferentiate in vitro. (a) Morphology of villi freshly purified from VilCreERApcfl/flKrasG12D/+ mice at day 1 (left panel) and day 4 (right panel) in culture. Black arrow indicates a spheroid inside the villus at day 4. (b) Quantification of VDS generated from Vil Apcfl/flKrasG12D/+ and VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl (Vil Apcfl/flKrasG12D/+Tgfbr1fl/fl) intestine. Error bars represent mean±S.E.M., *P=0.04 by Mann–Whitney test, one-tailed, n=3 biological replicates. (c) Quantification of VDS derived from VilCreERApcfl/flKrasG12D/+ villi treated with TGFβ1 (5 ng/ml) or Vehicle for 72 h. Plot represents mean with standard deviation (s.d.), n=3 biological replicates
To test whether increasing TGFβ would further suppress dedifferentiation, we treated VilCreERApcfl/flKrasG12D/+ villi with TGFβ1 and found that exogenous TGFβ1 reduced the capacity to form spheroids in vitro (Figure 5c). Taken together, these data confirmed that loss of TGFβ signalling increases dedifferentiation.
Loss of TGFβ pathway increases pERK levels and induces sensitivity to a MEK1/2 inhibition
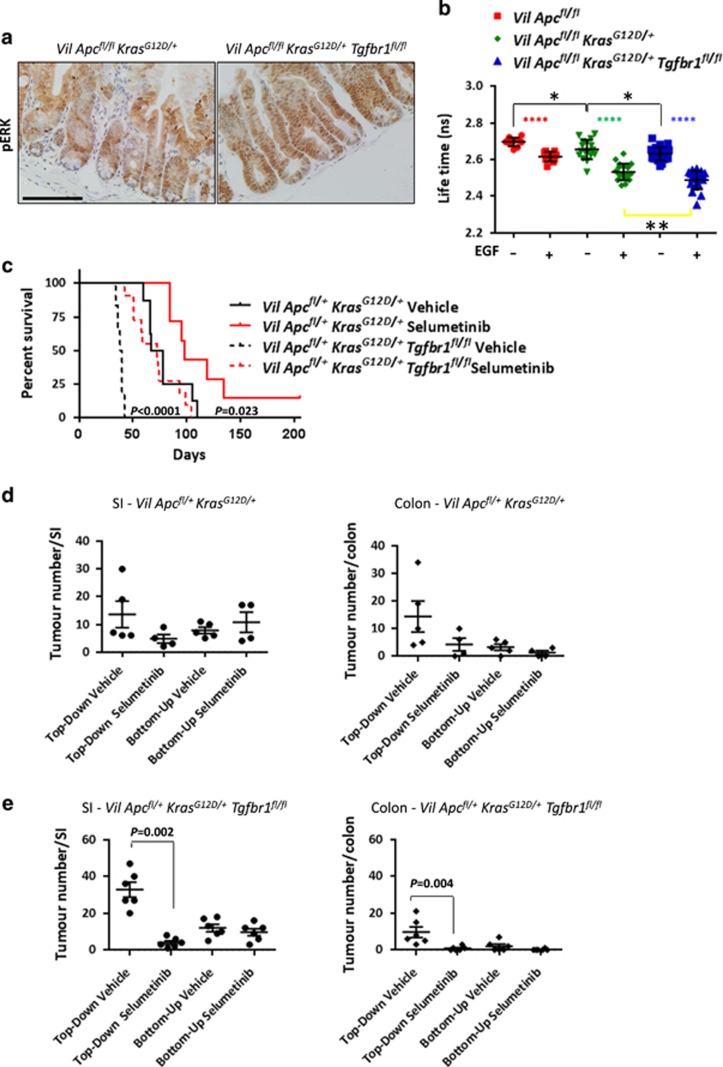
We next examined the mechanism by which Tgfbr1 loss caused increased dedifferentiation and accelerated tumourigenesis. Previous studies have shown crosstalk between the MAPK and TGFβ pathways.27, 28 pERK immunohistochemistry (IHC) showed a striking upregulation in the VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl mice compared with the VilCreERApcfl/flKrasG12D/+ mice (Figure 6a). To measure precisely ERK activation, VilCreERApcfl/fl, VilCreERApcfl/flKrasG12D/+ and VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl CDS were transfected with an ERK-FRET activity biosensor (we used the EKAREV biosensor consisting of an msECFP donor and YPet acceptor29) and treated with EGF for 45 min. A decreased lifetime indicates an activation of the nuclear biosensor. As expected, the fluorescence lifetime decreased following EGF administration in all three genotypes. Importantly, the activation of ERK was higher in the absence of Tgfbr1 (Figure 6b).
Figure 6.
Loss of transforming growth factor-β (TGFβ) signalling causes sensitivity to MEK1/2 inhibition. (a) pERK IHC on VilCreERApcfl/flKrasG12D/+ (Vil Apcfl/flKrasG12D/+) and VilCreERApcfl/flKrasG12D/+Tgfbr1fl/fl (Vil Apcfl/flKrasG12D/+Tgfbr1fl/fl) intestine. Scale bars, 100 μm. (b) Fluorescence lifetime changes for EKAREVnucl in VilCreERApcfl/fl (Vil Apcfl/fl), Vil Apcfl/flKrasG12D/+ and Vil Apcfl/flKrasG12D/+Tgfbr1fl/f CDS±EGF (50 ng/ml). (Vil Apcfl/fl Δlifetime: −0.08±0.023, ****P<0.0001 by paired t-test, one-tailed; Vil Apcfl/flKrasG12D/+: Δlifetime: −0.12±0.07, ****P<0.0001 by paired t-test, one-tailed; Vil Apcfl/flKrasG12D/+Tgfbr1fl/fl Δlifetime: −0.14±0.07, ****P<0.0001 by paired t-test, one-tailed). Data for each condition are mean of differences±S.D. for ⩾10 cells, n=1. VilCreERApcfl/flKrasG12D/++EGF versus VilCreERApcfl/flKrasG12D/+Tgfbr1fl/f +EGF, **P=0.003 by unpaired t-test, one-tailed. VilCreERApcfl/fl−EGF versus VilCreERApcfl/flKrasG12D/+−EGF, *P=0.014 by unpaired t-test, one-tailed. VilCreERApcfl/flKrasG12D/+−EGF versus VilCreERApcfl/flKrasG12D/+Tgfbr1fl/f –EGF, *P=0.049 by unpaired t-test, one-tailed. (c) Kaplan–Meier survival curve of VilCreERApcfl/+KrasG12D/+ treated with Selumetinib (Vil Apcfl/+KrasG12D/+, n=7) or Vehicle (Vil Apcfl/+KrasG12D/+, n=8), *P=0.023 by log-rank (Mantel–Cox) test and VilCreERApcfl+KrasG12D/+Tgfbr1fl/fl mice treated with Selumetinib (Vil Apcfl/+KrasG12D/+Tgfbr1fl/fl, n=11) or Vehicle (Vil Apcfl/+KrasG12D/+Tgfbr1fl/fl, n=6). ****P<0.0001 by log-rank (Mantel–Cox) test. Treatment started 1day post induction with tamoxifen. (d) Quantification of top-down or bottom-up tumours in the small intestine (SI, left panel) and colon (right panel) of VilCreERApcfl/+KrasG12D/+ (Vil Apcfl/+KrasG12D/+) mice treated with Selumetinib or Vehicle. Error bars represent mean±S.E.M. (e) Quantification of top-down or bottom-up tumours in the SI (left panel) and colon (right panel) of VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl (Vil Apcfl/+KrasG12D/+Tgfbr1fl/fl) mice treated with Selumetinib or Vehicle. Treatment started 1 day post induction with tamoxifen. Error bars represent mean±S.E.M., Mann–Whitney test, two-tailed
To test functionally whether MAPK pathway activation was required for faster tumourigenesis, we treated VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice with the potent allosteric MEK1/2 inhibitor Selumetinib (AZD-6244, ARRY-142886). Treatment started 1 day post induction and was able to reverse the acceleration of tumourigenesis conferred by Tgfbr1 loss so that VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice now had a median survival similar to vehicle-treated VilCreERApcfl/+KrasG12D/+ (Figure 6c). Interestingly, this effect was not restricted to the VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice as tumourigenesis was also delayed in VilCreERApcfl/+KrasG12D/+ mice treated with AZD-6244.
To treat the VilCreERApcfl/+KrasG12D/+ mice for a similar timeframe as the VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice, we started the treatment of the VilCreERApcfl/+KrasG12D/+ mice with Selumetinib 21 days post induction. Again, we were able to delay significantly tumourigenesis (Supplementary Figure 6a). Therefore, independent of the time of treatment (initiation or tumour outgrowth), Selumetinib could delay but not suppress tumourigenesis.
To assess if the reason for this is that Selumetinib affected the compartment from which tumours arose, we quantified top-down versus bottom-up lesions within the small intestine and colon of both genotypes (Figures 6d and e and Supplementary Figures 6b and c). MEK1/2 inhibition suppressed the formation of top-down lesions, suggesting that in both genotypes MAPK is important in the formation of this lesion type. However, mice still develop lesions from the crypt compartment, which led to the clinical signs. Thus, the MAPK pathway, following oncogenic Kras activation, is in part contributing to the increased plasticity of Apc-deficient cells.
Discussion
The TGFβ pathway is known to have both tumour-suppressing and -promoting effects in cancer. This makes it complex when trying to predict the outcome of therapies targeting the TGFβ pathway. Here, we show a tumour-suppressive role of TGFβ through its ability to limit tumour growth and dedifferentiation in the early stages of CRC. Recent studies have highlighted that plasticity of cells within the intestine act as stem cells and as potential tumour-initiating cells. Ours and other studies have highlighted that oncogenic Kras and inflammation can both drive plasticity in part via the NF-κB pathway.6, 8 We were able to show that reducing NF-κB directly or indirectly via Rac1 loss could further limit transformation and reduce stem cell capacity.6, 30 An excellent study using a carbonic anhydrase Cre recombinase to target colonic enterocytes showed that Apc loss and Kras activation, but not Apc loss alone, can drive tumour growth in differentiated cells and generate ‘top-down’ tumours.31 Apart from these studies, very little is known about the pathways that confer plasticity in adult intestinal epithelium. Here, we show that loss of the TGFβ signalling pathway induces the formation of more aggressive tumours, which are mainly responsible for the significant reduction of the overall survival. Interestingly, Tgfbr1 loss also increases the ability of cells to dedifferentiate facilitating an increased number of top-down lesions.
It is important to note that, at least in our systems, Kras mutation is still necessary for dedifferentiation. Loss of TGFβ signalling alone following Apc loss was not sufficient to drive dedifferentiation. Moreover, although Tgfbr1 loss increases both the number of top-down lesions and presumably their speed of formation, top-down lesions still arise in VilCreERApcfl/+KrasG12D/+ mice that have functional TGFβ signalling. We believe this might reflect a hierarchy of dedifferentiation capacity. Thus, dependent on the precise location where the lesions arise, they may be exposed to different levels of TGFβ signals and this may be sufficient to block tumourigenesis from some cells, while others (potentially exposed to lower levels of TGFβ signals) are able to form top-down tumours that once formed can grow regardless of these signals. It is interesting to note that despite high levels of p21 in many lesions that arise following Apc loss, deletion of p21 does not modify Apc loss-induced tumourigenesis.32
It is interesting to note that other studies have suggested that activation of TGFβ signalling might induce a stem cell phenotype18 and an EMT programme.33, 34, 35 However, our studies here indicate that exogenous TGFβ reduces organoid-forming capacity in vitro in agreement with Alitalo laboratory’s work, where exogenous TGFβ killed Apc-deficient crypts and reduced ISC markers in vitro.36
Previously, it has been shown that TGFβ can positively regulate MAPK signalling.37, 38 In contrast, other studies fit with our model where the TGFβ pathway can antagonize MAPK signalling. TGFβ can regulate the expression of MKP2, which in turn attenuates ERK.28 Moreover, oncogenic KrasG12D along with TGFβ pathway inactivation has been associated with increased expression of epiregulin and ERBB1.27 Recently, it has been reported that Tgfbr2 loss increases Ras/MAPK/ERK activation in primary keratinocytes leading to squamous cell carcinoma formation.39 Although our data support the model that TGFβ signalling restrains pERK levels, the mechanism by which this is achieved requires further investigation.
The functional requirement for upregulated MAPK pathway following Tgfbr1 loss was highlighted by MEK inhibition, which markedly reduced the number of tumours arising with top-down morphology. However, it is important to note that both VilCreERApcfl/+KrasG12D/+ and VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice still developed tumours (adenomas in the case of VilCreERApcfl/+KrasG12D/+ mice and both adenoma/adenocarcinoma in VilCreERApcfl/+KrasG12D/+Tgfbr1fl/fl mice) that have a bottom-up morphology presumably originating from the stem cell zone. This is an important finding showing that even in a simple mouse model system, where only Apc and Kras are mutated, Kras mutant tumours originating from the stem cell zone are resistant to MEK inhibition. These data are consistent with the lack of efficacy of MEK inhibitors as a single agent in advanced CRC carrying KRAS mutation (in both preclinical and clinical situations) as we would predict that inhibition of MEK would only work at a much earlier stage of tumourigenesis (e.g. during dedifferentiation).
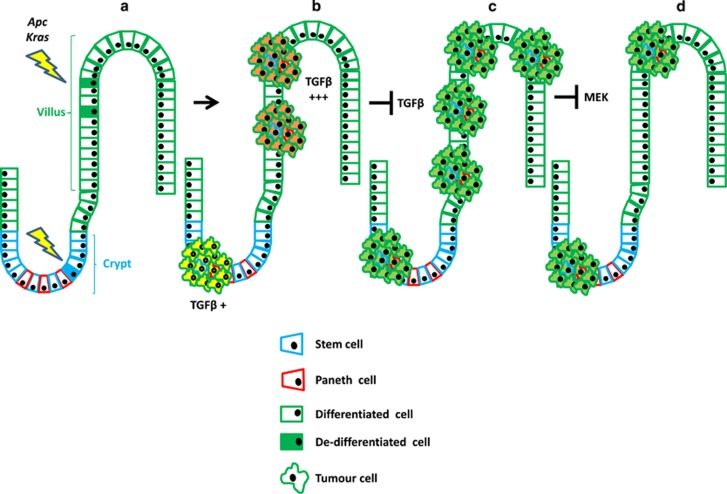
In summary, we have defined that TGFβ signalling limits dedifferentiation and tumourigenesis in cells carrying both Apc and Kras mutations (model in Figure 7). This is in part due to the gradients of TGFβ signalling in the intestine. Higher levels of TGFβ in the stroma further up the crypt–villus axis means that tumour-initiating cells in these regions are exposed to tumour-suppressive signals. Importantly, when tumours grow out from dedifferentiated cells, they seemed indistinguishable from tumours arising from the crypt. This suggests that once tumours have escaped the initial tumour-suppressive mechanism, the cell of origin does not affect the ongoing progression. Hence, mutational spectra might be a better way of predicting the initial cells of origin rather than transcriptomic data.
Figure 7.
Schematic model of intestinal tumourigenesis. (a) Aberrant activation of WNT and KRAS signalling drives intestinal tumour formation both from crypts (bottom-up model) and villi (top-down model). (b) High levels of transforming growth factor-β (TGFβ) signalling are present in the top-down model and this restrains tumourigenesis. (c) Loss of Tgfbr1 promotes intestinal tumourigenesis. (d) Top-down tumours lacking of Tgfbr1 are sensitive to MEK1/2 inhibition
Materials and methods
Mouse experiments
All experiments were performed in accordance with UK Home Office regulations (licence 70/8646), which undergoes local ethical review at the Glasgow University (Glasgow, UK). VillinCreER experiments were performed on C57BL/6J mice and on mixed background (C57BL/6J/S129). A mix of males and females were used. Mice were induced at >20 g and between 6 and 12 weeks of age. The alleles used for this study were as follows: VilCreER-T2,40 Apc580S,41 KrasG12D,42 Tgfbr1fl,43 Smad4fl,44 CtnnbloxEx3/+45 and Xbp1s-CreER-T2.6 Recombination in the tumour model was induced using a single intraperitoneal injection of 80 mg/kg tamoxifen. Mice were aged until they showed clinical signs (anaemia, hunching and/or weight loss).
Recombination in the short-term model was induced using a single intraperitoneal injection of 80 mg/kg tamoxifen for 2 consecutive days. If KrasG12D allele was present, recombination was induced using a single intraperitoneal injection of 80 mg/kg tamoxifen. VilCreERCtnnbloxEx3/+KrasG12D/+ and Xbp1sCreERCtnnbloxEx3/+KrasG12D/+ animals were induced with 1 mg tamoxifen for 5 consecutive days and sampled at day 7 (for VilCreER) and day 21 (for Xbp1sCreER). For the villi purification, mice were induced with intraperitoneal injection of 80 mg/kg tamoxifen for 2 consecutive days and sampled at day 1 post induction. The cultured villi treated with TGFβ1 were isolated from mice that had been induced with a single intraperitoneal dose of 80 mg/kg tamoxifen.
In accordance with the 3Rs, the smallest sample size was chosen that could give a significant difference. Given the robust phenotype of the Apcfl/fl, and our prediction of the role of TGFβ signalling, the minimum sample size assuming no overlap in control versus experimental is three animals. No randomization was used and the experimenter was blinded to drugs and genotypes.
In vivo treatment
Selumetinib (AZD6244, ARRY-142886; AstraZeneca, Cambridge, UK) was administered in a concentration of 25 mg/kg two times daily by gavage in a vehicle of 0.5% hydroxymethylpropylcellulose and 0.1% Tween-80.
For proliferation analysis, mice were injected intraperitoneally with 250 μl of BrdU (Amersham Biosciences/GE Heathcare, Buckinghamshire, UK) 2 h before being sampled.
Crypt/villus purification, propagation and treatment
Small intestines were washed with PBS and opened longitudinally. Villi were removed with a glass coverslip, washed in PBS and centrifuged at 60 × g for 3 min to separate villi from single cells. A total of 50–70 villi, mixed with 20 μl of Matrigel (BD Bioscience/Corning, Bedford, MA, USA), were plated in 24-well plates in Advanced DMEM/F12 supplemented with penicillin–streptomycin, 10 mM HEPES, 2 mM glutamine, N2, B27 (all from Gibco, Life Technologies, Paisley, UK), and EGF (50 ng/ml) and Noggin (100 ng/ml, both from Peprotech, London, UK). Crypts were isolated as described previously.46 Crypts mixed with 20 μl of Matrigel were plated in 24-well plates and cultured in the medium described above.
Recombinant TGFβ1 (Peprotech) was used at a final concentration of 5 ng/ml.
Clonogenic assay
Organoid cultures were dissociated into single cells using TrypLE Express (Gibco, Life Technologies) for 30–45 min at 37 °C and 5000 cells were cultured in a 24-well plate in 20 μl of Matrigel (BD Bioscience). The clonogenic potential was quantified after 72 h by microscope.
Transplantation
For transplantation experiments, 50 spheroids were suspended in 100 μl Matrigel (BD Bioscience) and injected subcutaneously into the flank of 6–8-week-old female athymic (CD-1) mice.
Immunohistochemistry
IHC was performed on formalin-fixed intestinal sections. Standard IHC techniques were used throughout this study. Primary antibodies used for immunohistochemistry were as follows: pSMAD3 (1:50; 52903; Abcam, Cambridge, UK), BrdU (1:200; 347580; BD Biosciences), p21 (1/4; CNIO Madrid, Spain), pERK (1:100; 9101; Cell SignalingTechnology, Leiden, The Netherland) and CD44v6 (1:50; BMS145; Invitrogen/Thermo Fisher Scientific, Vienna, Austria). For each antibody, staining was performed on at least three mice of each genotype. Representative images are shown for each staining. BrdU scoring was performed in a blinded manner. The number of positive cells per half crypt was scored. For each analysis, at least 30 crypts were scored from at least three mice for each genotype. Representative images were selected for each scoring.
RNAscope
In situ hybridization detection for Tgfb1, Tgfbr1, Smad7, Lgr5 and Olfm4 mRNA was performed using RNAscope 2.5 LS (Brown) Detection Kit (Advanced Cell Diagnostics, Hayward, CA, USA) on a Bond Rx autostainer (Leica, Milton Keynes, UK) strictly according to the manufacturer's instructions.
Image analysis
For RNAscope and IHC analysis, intestinal slides were scanned on an SCN400F slide scanner (Leica, Milton Keynes, UK). The files were exported and analysed in Halo v2.0 Image Analysis Software (Indica Labs, Corrales, NM, USA). For pSMAD3 quantification, areas were manually defined around the more distinctly altered villus areas and the adjacent normal epithelium at day 3 post induction. For Lgr5 and Olfm4 quantification, areas were manually defined around the entire crypt and the more distinctly altered villus areas at day 3 post induction.
RNA purification
RNA was isolated using a Qiagen RNeasy Mini Kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions. DNA-Free Kit (Ambion/Thermo Fisher Scientific, Warrington, UK) was used to remove genomic DNA contamination, according to the manufacturer’s instructions.
Microarray analysis
One microgram of total RNA isolated from intestinal tissue was reverse transcribed to cDNA and hybridized to Affymetrix Mouse Genome 430 2.0 Microarrays (Santa Clara, CA, USA). The data were preprocessed using affy package47 in R language, version 2.15.0. Background correction, quantile normalization and probe summarization were performed using the Robust Multiarray Analysis algorithm. Normalized data were analysed using the Limma package.48 Gene Ontology annotation was added using the GO.db package. Affymetrix Mose Gene ST 1.0 Chip (Santa Clara, CA, USA) has been used to analyse the VilCreERCtnnbloxEx3/+KrasG12D/+ and Xbp1sCreERCtnnbloxEx3/+KrasG12D/+ genotype. Data have been deposited in NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession numbers: GSE99077, GSE99100, GSE99408.
Quantitative PCR (qRT-PCR)
One microgram of RNA was reverse transcribed to cDNA using a DyNAmo SYBR Green 2-Step qPCR Kit (Finnzymes, Espoo, Finland) in a reaction volume of 20 μl. The cycling conditions were as follows: 95 °C for 15 min, followed by 40 cycles of three steps consisting of denaturation at 94 °C for 15 s, primer annealing at the optimal temperature for 30 s and primer extension at 72 °C for 30 s. A melting-curve analysis was performed to demonstrate the specificity of each amplicon and to identify the formation of primer dimers. β-Actin and Gapdh primers were used to normalize for differences in RNA input.
Transfection
For transfection experiments, intestinal spheroids were seeded in 96-well plates and transfected with EKAREV construct and Super PiggyBac Transposase expression vector (System Biosciences, Palo Alto, CA, USA) by using Magnetofectamine (OZ Biosciences, Marseille, France) according to the manufacturer's instructions. Subsequently, cells were incubated for 48 h at 37 °C and 5% CO2. After 48 h, organoids were selected using blasticidin.
FLIM analysis
Fluorescence lifetime microscopy (FLIM) measurements were conducted on a spinning disk system, as described previously.49 Briefly, a Nikon Eclipse TE 2000-U microscope (Surrey, UK) with a Lambert Instruments LIFA attachment was used with a Yokogawa CSU 22 confocal scanner unit and a × 60 1.49 NA oil objective. FLIM-FRET for the msECFP donor was measured by frequency domain, with a 445 nm laser modulated at 40 mHz (100 mW, Deepstar; Omicron, Rodgau-Dudenhofen, Germany) was used together with a CFP filterblock (470/40) to detect CFP emission. Fluorescein (10 μM in 0.1 M Tris-Cl, pH>10) was used as a reference standard with a known lifetime of 4.0 ns. The donor lifetime (msECFP29), τ, was analysed using the FLIM Software (version 1.2.12; Lambert Instruments, Groningen, Netherlands) drawing ROIs around the cell nuclei and recording the phase lifetime. Graph represents the lifetime variation (ns) before and after (45 min)±EGF (50 ng/ml) administration. In both cases, the mean of differences and standard deviation is calculated and represented P-values were calculated using Student’s t-test.
Statistical analysis
Statistical analysis was performed with Minitab 17 Statistical Software (State College, PA, USA) or GraphPad Prism V6 Software (La Jolla, CA, USA) using one- or two-tailed Mann–Whitney tests and paired or unpaired Student’s t-tests (where appropriate).
Data availability
The authors declare that all relevant data supporting the findings of this study are available within the article and its Supplementary Information files. Additional information can be obtained from the corresponding author (OJS).
Acknowledgments
OJS is supported by a Cancer Research UK core grant (A21139) and an ERC starting grant (311301). GJI was supported by Cancer Research UK (A4132, A10419) and by an ERC grant (250170). PC was supported by FP7 Health CP-IP – Large-scale integrating project grant (278568), and DFV is supported by ERC starting grant (311301). We thank Prof. M Matsuda for kindly providing the ERK biosensor. We thank the Head of the Transcriptome and Genome Analysis Laboratory (Microarray and Deep-Sequencing Facility at Göttingen University) Dr Gabriela Salinas-Reister for the analysis of the CtnnbloxEx3/+KrasG12D/+ microarray. We also thank the research and scientific services at the CRUK Beatson Institute in general.
Author contributions
OJS and PC contributed to study design; PC, DFV, RAR, CM, MN, AH, CS, MCH, JV, DJH, KLHP and CN contributed to the data acquisition; PC, DFV, RAR, CM, MN, MCH, JV, AH, GJI and OJS contributed to the data analysis and interpretation of the data; STB provided Selumetinb; PC, ADC, DFV, MCH, JV, STB, FRG, GJI and OJS to drafting the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
The authors declare no conflict of interest.
Supplementary Material
References
- Shih IM, Wang TL, Traverso G, Romans K, Hamilton SR, Ben-Sasson S et al. Top-down morphogenesis of colorectal tumors. Proc Natl Acad Sci USA 2001; 98: 2640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston SL, Wong WM, Chan AO, Poulsom R, Jeffery R, Goodlad RA et al. Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res 2003; 63: 3819–25. [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009; 457: 608–11. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 2008; 40: 915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012; 149: 146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152: 25–38. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet 2013; 45: 98–103. [DOI] [PubMed] [Google Scholar]
- Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 2014; 124: 1283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat Med 2015; 21: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med 2009; 361: 2449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku JL, Park SH, Yoon KA, Shin YK, Kim KH, Choi JS et al. Genetic alterations of the TGF-beta signaling pathway in colorectal cancer cell lines: a novel mutation in Smad3 associated with the inactivation of TGF-beta-induced transcriptional activation. Cancer Lett 2007; 247: 283–92. [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006; 314: 268–74. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 1995; 268: 1336–8. [DOI] [PubMed] [Google Scholar]
- Salovaara R, Roth S, Loukola A, Launonen V, Sistonen P, Avizienyte E et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut 2002; 51: 56–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima H, Ito N, Tamura S, Matsuda Y, Inada M, Yabuuchi I et al. Circulating transforming growth factor beta 1 as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res 2001; 7: 1258–62. [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–90. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial–mesenchymal transition in cancer progression. Curr Opin Oncol 2013; 25: 76–84. [DOI] [PubMed] [Google Scholar]
- Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell 2012; 22: 571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015; 21: 1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene 2005; 24: 5775–89. [DOI] [PubMed] [Google Scholar]
- Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res 2006; 66: 9837–44. [DOI] [PubMed] [Google Scholar]
- Kim BG, Li C, Qiao W, Mamura M, Kasprzak B, Anver M et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature 2006; 441: 1015–9. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 2004; 18: 1385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015; 47: 320–9. [DOI] [PubMed] [Google Scholar]
- Principe DR, DeCant B, Staudacher J, Vitello D, Mangan RJ, Wayne EA et al. Loss of TGFbeta signaling promotes colon cancer progression and tumor-associated inflammation. Oncotarget 2017; 8: 3826–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge P, Knoblaugh S, Washington MK, Munoz NM, Tsuchiya KD, Rojas A et al. TGF-beta receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a beta-catenin-independent pathway. Gastroenterology 2009; 136: 1680–8 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh S, Qi XJ, Wildey GM, Robinson J, Molkentin J, Letterio J et al. TGF beta-mediated BIM expression and apoptosis are regulated through SMAD3-dependent expression of the MAPK phosphatase MKP2. EMBO Rep 2008; 9: 990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Miyawaki A, Nagai T. Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nat Methods. 2008; 5: 339–45. [DOI] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 2013; 12: 761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Kretzschmar K, Begthel H, van den Born M, Korving J, Morsink F et al. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc Natl Acad Sci USA 2016; 113: 11859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AM, Ridgway RA, Derkits SE, Parry L, Barker N, Clevers H et al. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO Mol Med 2010; 2: 472–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial–mesenchymal transition. PLoS ONE 2008; 3: e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011; 145: 926–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener Z, Band AM, Kallio P, Hogstrom J, Hyvonen V, Kaijalainen S et al. Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc Natl Acad Sci USA 2014; 111: E2229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KM. Role of Ras and Mapks in TGFbeta signaling. Cytokine Growth Factor Rev 2000; 11: 23–35. [DOI] [PubMed] [Google Scholar]
- Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J 2007; 26: 3957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Schramek D, Adam RC, Keyes BE, Wang P, Zheng D et al. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. Elife 2015; 4: e10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004; 39: 186–93. [DOI] [PubMed] [Google Scholar]
- Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 1997; 278: 120–3. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001; 15: 3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Goumans MJ, Sjostrand LJ, van Rooijen MA, Ward D, Leveen P et al. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J 2001; 20: 1663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis 2002; 32: 80–1. [DOI] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 1999; 18: 5931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–5. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–15. [DOI] [PubMed] [Google Scholar]
- Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006; 7: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack NA, Porter AP, Whalley HJ, Schwarz JP, Jones RC, Khaja AS et al. Beta2-syntrophin and Par-3 promote an apicobasal Rac activity gradient at cell-cell junctions by differentially regulating Tiam1 activity. Nat Cell Biol 2012; 14: 1169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant data supporting the findings of this study are available within the article and its Supplementary Information files. Additional information can be obtained from the corresponding author (OJS).