SUMMARY
The regulatory effect auxin has on its own transport is critical in numerous self-organizing plant patterning processes. However, our understanding of the molecular mechanisms linking auxin signal transduction and auxin transport is still fragmentary, and important regulatory genes remain to be identified.
To track a key link between auxin signaling and auxin transport in development, we established an Arabidopsis thaliana genetic background in which fundamental patterning processes in both shoot and root were essentially abolished and the expression of PIN FORMED (PIN) auxin efflux facilitators was dramatically reduced.
In this background, we demonstrate that activating a steroid-inducible variant of the Auxin Response Factor (ARF) MONOPTEROS (MP) is sufficient to restore patterning and PIN gene expression. Further, we show that MP binds to distinct promoter elements of multiple genetically defined PIN genes.
Our work identifies a direct regulatory link between central, well-characterized genes involved in auxin signal transduction and auxin transport. The steroid-inducible MP system directly demonstrates the importance of this molecular link in multiple patterning events in embryos, shoots and roots, and provides novel options for interrogating the properties of self-regulated auxin-based patterning in planta.
Keywords: Arabidopsis development, AUXIN RESPONSE FACTOR (ARF), embryogenesis, organogenesis, PIN FORMED genes, root apical meristem, self-organized patterning, shoot apical meristem
INTRODUCTION
All stages of plant growth and development critically depend on the action of the phytohormone auxin. Auxin is required to establish the bodyplan during embryogenesis and later plays a key role in the initiation and outgrowth of new organs from stem cell regions called apical meristems (Vanneste & Friml, 2009). Many processes in both the shoot apical meristem (SAM) and root apical meristem (RAM) involve auxin distribution patterns. For example, in the shoot, the positioning and growth of new organs is dictated by auxin concentration maxima established by the PIN FORMED (PIN) family of membrane efflux facilitators, which mediate polar auxin transport between cells (Adamowski & Friml, 2015). The canonical auxin signaling pathway, which involves the Auxin Response Factor (ARF) family of transcriptional regulators, is required to elicit the appropriate developmental output in response to these local concentration maxima (Chapman & Estelle, 2009).
Many auxin-dependent patterning events have a self-organizing property consistent with a proposed ability of auxin to regulate and reinforce its own flow (Vanneste & Friml, 2009). In particular, an influence of auxin on the expression and subcellular localization of PIN efflux carriers is a central prerequisite of self-organization in plant patterning according to many mathematical models (Kuhlemeier, 2007). Consistent with this, some PIN genes appear to be primary auxin response genes (Vieten et al., 2005; Dello Ioio et al., 2008; Chen et al., 2015). Additionally, auxin can influence PIN subcellular localization to control auxin flow and, in turn, the positioning and growth of new tissues and organs (Benkova et al., 2003; Sauer et al., 2006).
Whereas modulation of PIN protein localization and stability has been thoroughly analyzed for years, molecular details regarding the direct transcriptional control of PINs are clearly incomplete. Most transcription factors that have been implicated in PIN regulation have not been linked to the ARF-mediated canonical auxin signal transduction pathway (Cui et al., 2013; Garay-Arroyo et al., 2013; Wang et al., 2015), but there are a few recent exceptions. For instance, among a group of CYTOKININ RESPONSE FACTORS (CRFs) that target PIN genes (Simaskova et al., 2015), one (CRF2) is itself regulated by ARF5/MONOPTEROS (MP) (Schlereth et al., 2010). Further, ARF7/NONPHOTOTROPIC HYPOCOTYL4 (NPH4) and FOUR LIPS/MYB124 have been shown to jointly and directly target PIN3 in the root (Chen et al., 2015). However, mutations in each of these direct regulators of PINs result in rather subtle defects in specific aspects of root development, suggesting that important molecular links between auxin signaling and auxin transport remain to be identified.
The functions of ARF5/MP and ARF7/NPH4 have been shown to be asymmetrically redundant (Hardtke et al., 2004). ARF7/NPH4, a regulator of phototropic auxin responses, has a gratuitous and dispensable function in patterning processes, visible in the nph4 mutant only through its enhancement of mp patterning defects. In the mp nph4 double mutant, structure and function of both apical meristems is abolished, suggesting a complete collapse in auxin-mediated patterning (Hardtke et al., 2004). Importantly, however, such defects are undetectable in nph4 single mutants, indicating that ARF5/MP is sufficient for all auxin-mediated patterning in both root and shoot. In this study, we have introduced an inducible variant of MP, MP-GR, into the mp nph4 double mutant (Krogan et al., 2014). This background demonstrated that MP is sufficient to restart auxin-mediated patterning processes from completely disorganized tissue in both shoot and root. As such, this establishes a genetic system that can provide insight into auxin-mediated self-organization by revealing the consequences of flexibly restarting patterning processes in diverse developmental stages. Further, we demonstrate that the expression of at least three PIN genes is strongly dependent on MP, which activates their transcription by binding to discrete elements in the promoters of each gene. Based on the dramatic phenotypes reported for multiple arf as well as multiple pin mutant combinations, our results indicate that ARF5/MP functions as a central connector between auxin signal transduction and auxin transport.
MATERIALS AND METHODS
Plant material and growth conditions
Unless stated otherwise, Arabidopsis thaliana (L.) Heynh seeds were plated and plants grown as described (Hardtke et al., 2004). Mutant alleles used were mpG12, mpBS1354 (Hardtke & Berleth, 1998) and nph4-1 (Harper et al., 2000). Transgenic lines mp nph4 MP-GR (Krogan et al., 2014), PIN1::PIN1-GFP (Benkova et al., 2003), MP::MP-GUS (Vidaurre et al., 2007) and MP::MP-GFP (Cole et al., 2009) have been described previously.
Transgene construction
To make PIN transcriptional reporter genes, 2011bp, 2020bp and 2108bp upstream of the translational start codons of PIN1, PIN3, and PIN7, respectively, were fused to the β-glucuronidase (GUS) reporter gene and transformed into the Columbia-0 ecotype.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
cDNA template preparation was performed as previously reported (Krogan et al., 2014). For shoot samples, PCR reactions of cDNA template included 1.2μCi Redivue [α-32P] dCTP (Amersham Biosciences, Mississauga, ON, Canada) to facilitate product quantification. Low cycle numbers (24 cycles) were used to prevent saturation of product amplification. Further, at least two concentrations of template were amplified in parallel to ensure that a doubling of the amount of starting template resulted in a proportional doubling of the final PCR product. Electrophoresed RT-PCR products were scanned by a Personal Molecular Imager FX Scanner and quantified by accompanying Quantity One Quantitation Software (Bio-Rad). Root samples were analyzed similarly, except that radioactive labeling was omitted and product intensity was instead quantified by ImageJ software (National Institutes of Health, MD, USA). Primer sequences are given in Supporting Information Table S1.
Electrophoretic mobility shift assays (EMSAs)
The purification of His-MP(432) protein and EMSA experimental conditions have been described (Krogan et al., 2014). Labeled probes were created by PCR reactions containing 20μCi of Redivue [α-32P] dCTP (Amersham Biosciences). Primer sequences are provided in Table S2. The nonspecific competitor used in EMSAs corresponded to −1870bp to −1729bp (relative to translational start) of PIN3.
Chromatin immunoprecipitation (ChIP)
ChIP experiments on floral tissue of MP::MP-GFP was performed as previously described (Krogan et al., 2012). Real-time PCR on ChIP samples was performed with a Mx3005P QPCR system (Agilent Technologies, Santa Clara, CA, USA) using PerfeCTa SYBR Green SuperMix (Quanta Biosciences, Inc., Beverly, MA, USA). Data analysis was carried out using MxPro QPCR Software (Agilent Technologies). Enrichment was calculated as a ratio of the signal from ChIP samples to that from input samples. Fold enrichment was calculated as the ratio of MP::MP-GFP sample enrichment to non-transgenic control sample enrichment, and was normalized using ACT7 data. Primer sequences are given in Table S3.
Microtechniques and microscopy
For low to medium magnification, samples were viewed under bright field and fluorescence illumination (GFP) with a Leica MZ FLIII (Leica Microsystems, Wetzlar, Germany) dissecting stereomicroscope. For high magnification, samples were viewed under differential interference contrast (DIC) optics with an Olympus AX70 microscope (Olympus Canada Inc., Richmond Hill, ON, Canada). For confocal laser scanning microscopy of roots, samples were mounted in water or 10μg/mL propidium iodide (PI) and observed with a Zeiss Axiovert 100M microscope equipped with a Zeiss LCM510 laser module confocal unit. Analysis of GUS activity was as described (Krogan et al., 2014) with modifications (Table S4a,b). ImageJ software was used to create rainbow spectrum look-up-table (LUT) images of GFP signal intensity and to quantify angles of root tropic growth.
RESULTS AND DISCUSSION
To investigate the influence of MP on postembryonic development, we sought to remove redundant contributions of NPH4 to MP-mediated processes by analyzing mp nph4 expressing an inducible MP-GR transgene driven by native MP regulatory sequence (Krogan et al., 2014). Further, since mp nph4 embryos fail to produce cotyledons and functional apical meristems (Hardtke et al., 2004) in some experiments we bypassed embryonic abnormalities by continually providing mp nph4 MP-GR parental plants with the synthetic glucocorticoid dexamethasone (DEX), which activates MP-GR function. This restored embryo patterning in seeds and led to the germination of seedlings with rescued apical-basal axis formation (Fig. 1a), but was followed by rapid deterioration of postembryonic development in the absence of DEX. In the following, we use this background as a genetic switch to restart auxin-driven patterning in individuals after either impaired or normalized embryogenesis and refer to these as mp nph4 MP-GR or mp nph4 MP-GRer (embryonically rescued), respectively.
Figure 1.
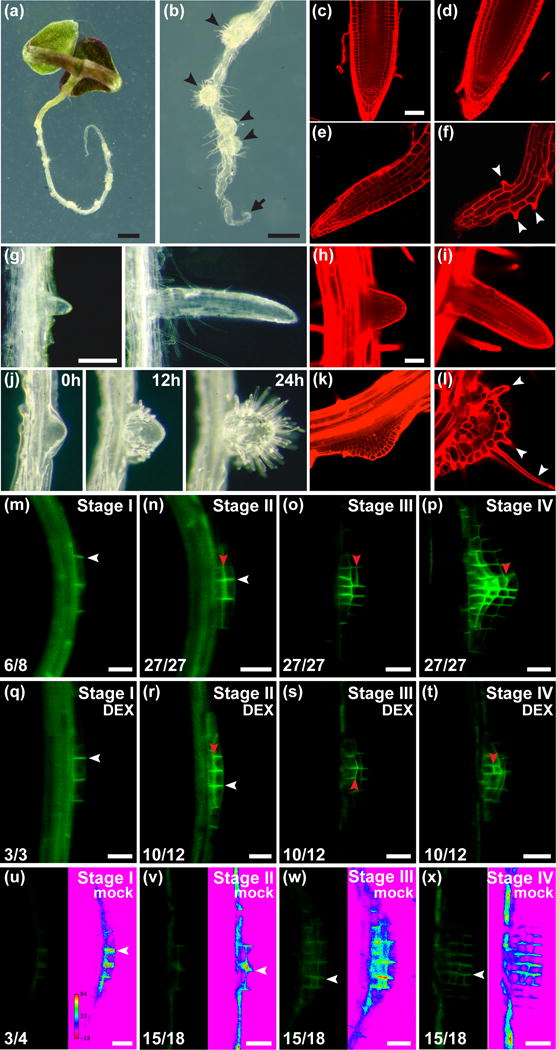
The role of MONOPTEROS (MP) in root apical meristem (RAM) maintenance and lateral root patterning in Arabidopsis.
(a,b) 9d after germination (DAG) mp nph4 MP-GRer seedlings grown in the absence of dexamethasone (DEX). Disorganized lateral roots (arrowheads) and inactive primary root tip (arrow) are indicated.
(c–f) Confocal images of primary roots. (c) 4DAG mp nph4 MP-GRer grown on 15μM DEX. RAM is normal and resembles 4DAG nph4 root (d). (e) 4DAG and (f) 6DAG mp nph4 MP-GRer RAMs grown in the absence of DEX. Arrowheads indicate epidermal root hair cells.
(g) Lateral roots from mp nph4 MP-GRer seedlings grown on 15μM DEX.
(h,i) Confocal images of nph4 lateral roots showing normal RAM.
(j–l) Time course (j) and confocal images (k,l) of lateral root primordia (LRPs) from mp nph4 MP-GRer seedlings grown in the absence of DEX. Arrowheads denote root hairs.
(m–x) PIN1-GFP in LRPs (distal tip to the right). (m–p) Wild type. (q–x) mp nph4 MP-GRer germinated and grown continuously with (q–t) or without (u–x) 15μM DEX. Transverse and lateral cell membranes are denoted by white and red arrowheads, respectively. Ratios of LRPs exhibiting the depicted PIN1-GFP distribution and intensity are shown. LRPs in (u–x) are also depicted as rainbow spectrum look-up-tables (LUTs) to show relative PIN1-GFP expression. LRP stages are according to Malamy & Benfey (1997).
Bars: (a) 1mm; (b) 0.5mm; (c–f,h,i,k,l) 50μm; (g,j) 0.2mm; (m–x) 20μm.
Role of MP in RAM Function
The formation of a primary RAM and positions of new lateral root primordia (LRPs) are dictated by local auxin response maxima (Sabatini et al., 1999; Benkova et al., 2003). The mp nph4 MP-GRer phenotype demonstrates the critical impact of MP regulatory potential on postembryonic de novo root organization and on maintenance of the RAM, which would not have been evident in mp single mutants (Supporting Information Fig. S1a,b). In untreated mp nph4 MP-GRer primary roots, initial elongation is invariably followed by growth cessation after 4 days postgermination (Fig. 1a,b; Supporting Information Fig. S2a). This is accompanied by the gradual disintegration of the RAM, as cell file numbers decrease and terminal differentiation occurs, evidenced by increased cell size and root hair production close to the root tip (Fig. 1e,f). Conversely, continuous DEX treatment maintains normal RAM organization and function (Fig. 1c). Furthermore, DEX exposure establishes a concentration-dependent memory effect, as duration of mp nph4 MP-GRer root growth in the absence of DEX directly correlates with the extent of prior treatment (Supporting Information Fig. S2a). Our findings also implicate MP activity in gravitropic responses, as untreated mp nph4 MP-GRer roots are agravitropic, a defect that can be rescued by DEX application (Supporting Information Fig. S1c).
ARF19 and ARF7/NPH4 have been shown to act redundantly in the initiation of LRPs (Okushima et al., 2005), a developmental process that is retained in mp nph4 MP-GRer seedlings. Under continuous DEX exposure, mp nph4 MP-GRer lateral roots exhibit proper patterning and outgrowth similar to nph4 mutants (Fig. 1g–i). By contrast, lateral organs of untreated mp nph4 MP-GRer roots display gross morphological abnormalities not previously seen in other mutant backgrounds (Fig. 1b,j), apparently reflecting MP function in LRP development (De Smet et al., 2010). Initially, an increased number of cells are recruited into LRPs of these mutants, resulting in broader outgrowths (Fig. 1j,k). Upon further development, the excessively wide primordia fail to specify distal cell identities and hence a functional RAM. Instead, these cell masses typically arrest and become covered with epidermal root hair cells, suggesting that they comprise only unspecified or proximal cell identities and fail to maintain a stem cell niche (Fig. 1j,l).
Aspects of root growth affected in untreated mp nph4 MP-GRer seedlings are reminiscent of roots compromised in PIN-mediated auxin transport, including an inability to pattern and maintain a RAM, agravitropism and widened LRPs (Benkova et al., 2003; Geldner et al., 2001; Geldner et al., 2004). Therefore, we investigated whether PIN1::PIN1-GFP expression is altered in mp nph4 MP-GRer. In wild type and DEX-treated mp nph4 MP-GRer, PIN1-GFP localizes to the transverse sides of LRP initial cells (Fig. 1m,q). Subsequent LRP development is characterized by a prominent shift in PIN1-GFP localization to lateral sides of cells (Fig. 1n–p,r–t), which is associated with the focusing of auxin transport to the tip of growing primordia (Benkova et al., 2003). In LRPs of untreated mp nph4 MP-GRer roots, intensity of PIN1-GFP expression was dramatically reduced in all stages, while prominent relocalization of PIN1 to lateral sides of cells was not apparent (Fig. 1u–x). This suggests that abnormalities in LRP patterning in mp nph4 are due to defects in both the expression and subcellular relocalization of PIN1.
We sought to determine whether reactivation of MP in mp nph4 MP-GRer roots was sufficient to restore PIN1-GFP levels and to rescue RAM patterning and function. Upon transfer of mp nph4 MP-GRer seedlings to DEX media, almost half of early LRPs analyzed showed an extremely rapid increase in PIN1-GFP levels and exhibited correct relocalization of PIN1 to lateral cell surfaces (Fig. 2d–f). In contrast, PIN1-GFP in wild-type roots treated with DEX showed no change or a decrease in expression level, while subcellular localization remained normal (Fig. 2a–c). Strikingly, when older LRPs from untreated mp nph4 MP-GRer seedlings were transferred to DEX, lateral roots with normalized RAMs emerged from within the disorganized cell masses (Fig. 2g–i). This de novo RAM emergence was associated with the initiation of PIN1-GFP expression foci within the interior cells (Fig. 2j). These results implicate PIN1 as a major target of MP in the control of root patterning. Finally, the primary roots of older untreated mp nph4 MP-GRer seedlings were incapable of reinitiating growth upon transfer to DEX (Fig. 2g,h), possibly due to the absence of reversible cell states among the very few cells in these locations (Fig. 1f).
Figure 2.
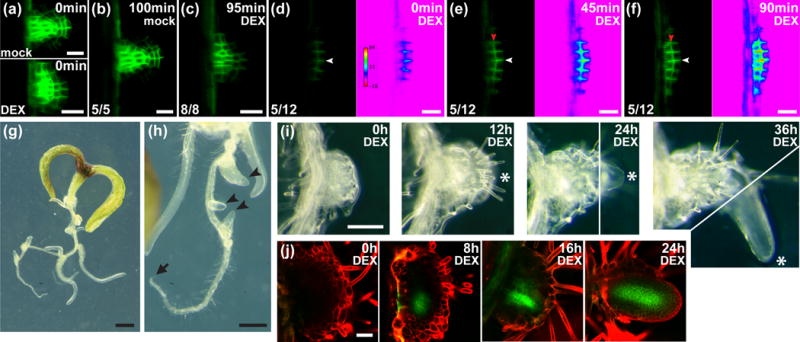
Effect of MONOPTEROS (MP) on lateral root normalization and PIN FORMED1 (PIN1) activation in Arabidopsis.
(a–f) PIN1-GFP in lateral root primordia (LRPs) of 4-to-5d after germination (DAG) seedlings (distal tip to the right). (a–c) Wild type and (d–f) mp nph4 MP-GRer LRPs (with rainbow spectrum look-up-tables) transferred to 15μM dexamethasone (DEX) at 0min. Ratios of LRPs exhibiting the depicted PIN1-GFP distribution and intensity are shown. LRPs that differed from images depicted in (d–f) did not show an increase in PIN1-GFP expression. Transverse and lateral cell membranes are denoted by white and red arrowheads, respectively.
(g,h) 9DAG seedlings germinated and grown without DEX for 6d, then transferred to 15μM DEX for 3d. When transferred at 6DAG, the primary root tip (arrow) had ceased growth and disorganized lateral outgrowths had initiated. Arrowheads point to emerging lateral roots.
(i) Time course of mp nph4 MP-GRer lateral root outgrowth transferred to 30μM DEX at 0h. Asterisks denote an emerging lateral root. 24h and 36h pictures are composites of two images (separated by white lines) at different focal planes.
(j) PIN1-GFP expression in mp nph4 MP-GRer lateral root outgrowths of 8DAG seedlings transferred to 30μM DEX at 0h.
Bars: (a–f) 20μm; (g) 1mm; (h) 0.5mm; (i) 0.2mm; (j) 50μm.
Role of MP in SAM Function
The initiation sites of shoot lateral organs are defined by areas of high PIN1 expression (with PIN1 protein localization suggestive of auxin transport towards convergence points), and in mathematical models of organ positioning, auxin signal transduction and transport constitute key parameters (Sassi & Vernoux, 2013). Consistent with this, both mp and pin1 mutants display severe distortions in this positioning process (Okada et al., 1991; Przemeck et al., 1996), and mp nph4 double mutants fail to form such organs at all (Hardtke et al., 2004). By controllably activating MP in mp nph4 MP-GRer seedlings, we observed that continuous MP activity was required for shoot organ formation, and that the intensity and duration of MP induction correlated with the extent of organ production (Supporting Information Fig. S2b). These findings reveal the sensitivity of the self-organizing patterning process, which appears continuously reliant on MP activity.
In the absence of MP and NPH4 activity, SAMs do not show signs of lateral organ outgrowth for 2–3 weeks (Fig. 3a,b,l,o), after which they generate lateral bulges that vary in their spatial arrangement depending on whether cotyledons are present (Fig. 3c–f). If cotyledons are absent (as in mp nph4 MP-GR), the primary SAM turns into a rotationally symmetrical, grossly oversized apical mound (Fig. 3a,b) that goes on to initiate lateral bulges evenly spaced over its entire surface (Fig. 3c,d). In contrast, if cotyledons are present (as in mp nph4 MP-GRer), SAMs become more elliptical in shape (Fig. 3l,o) and later initiate equally spaced bulges along a straight line perpendicular to the axis connecting the cotyledons (Fig. 3e,f). These findings suggest that under conditions of highly diminished ARF activity, position-defining auxin focusing, as described for normal SAMs (Sassi & Vernoux, 2013), is extremely delayed but qualitatively unchanged. In the absence of cotyledons, the previously postulated auxin-based lateral inhibition model (Reinhardt et al., 2003) would predict new initiation positions to be largely random but separated as far as possible from one other (Fig. 3c,d). In the presence of cotyledons, the same principles, in combination with the postulated inhibitory influence of cotyledons, would also be consistent with the observed distribution of primordia (Fig. 3e,f).
Figure 3.
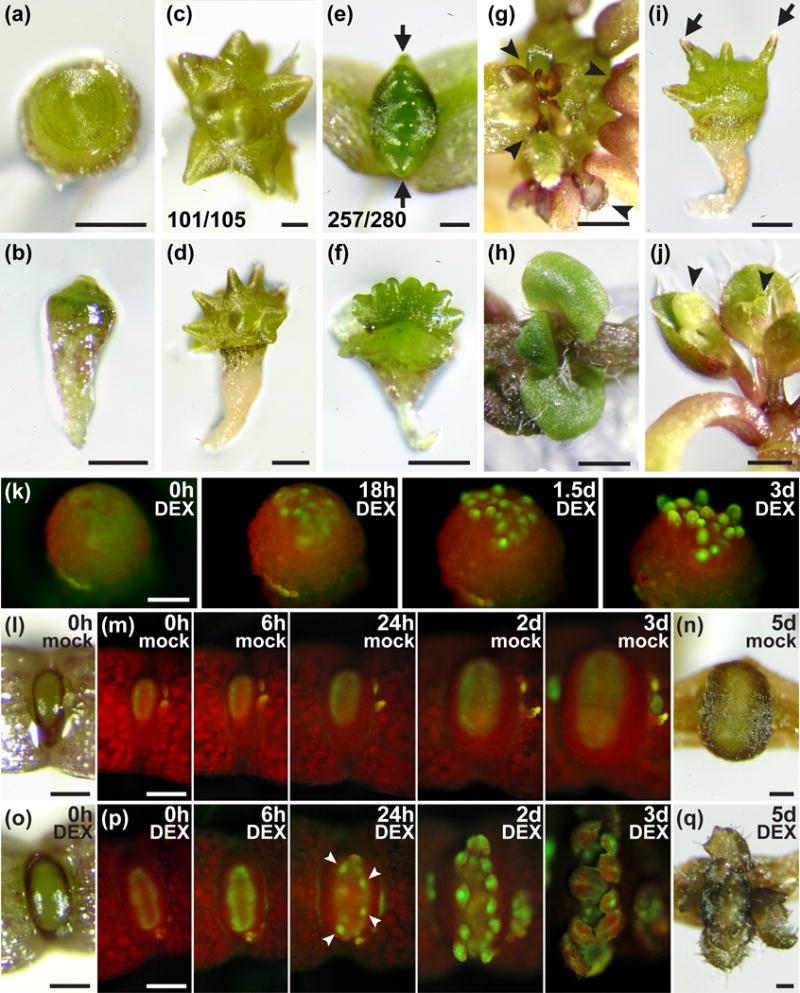
Effect of MONOPTEROS (MP) activity on shoot organ initiation and PIN FORMED1 (PIN1) expression in Arabidopsis.
(a) Apical and (b) lateral views of 17d after germination (DAG) mp nph4 MP-GR seedlings grown in the absence of dexamethasone (DEX).
(c–f) Apical and lateral views of mp nph4 MP-GR (c,d) and mp nph4 MP-GRer (e,f) seedlings (24DAG) grown in the absence of DEX. The fractions of mp nph4 MP-GR individuals with randomly arranged lateral bulges (c) and of mp nph4 MP-GRer seedlings with lateral bulges arranged in a linear plane perpendicular to cotyledons (arrows, e) are given. Cotyledons have been removed in (f) to allow visualization of bulges.
(g) Clusters of leaf-like organs (arrowheads) initiated from lateral bulges of mp nph4 MP-GR seedlings transferred to 15μM DEX at 17DAG and grown for an additional 11d.
(h) Apical view of 17DAG mp nph4 MP-GRer seedling transferred to 15μM DEX at 10DAG.
(i) mp nph4 MP-GR seedling (28DAG) grown in the absence of DEX. Lateral bulges have transitioned to tapered, pin-like structures (arrows).
(j) Flowers (arrowheads) initiated from pin-like SAM outgrowths of 28DAG mp nph4 MP-GR seedlings transferred to 15μM DEX at 17DAG.
(k) PIN1-GFP expression in the shoot apical meristem (SAM) of an mp nph4 MP-GR seedling (13DAG) lacking cotyledons, transferred to 30μM DEX at 0h. Foci of PIN1-GFP expression at 18h presage lateral organ positions.
(l–q) PIN1-GFP expression in dicotyledonous mp nph4 MP-GRer seedlings (10DAG) transferred to mock (l–n) or 30μM DEX (o–q) at 0h. Appearances of the SAMs at the start (l,o) and end (n,q) of each time course are shown. A uniform ring of PIN1-GFP shows increased expression by 6h of DEX treatment (p). PIN1-GFP foci (arrowheads at 24h) presage lateral organ formation.
Bars: (a,b,d,f,g,i,j) 0.5mm; (c,e) 0.2mm; (h) 1mm; (k–q) 0.2mm.
The lateral bulges formed in the absence of DEX are meristematic in identity, as transfer to DEX shortly after their formation results in each bulge initiating an array of leaf-shaped organs (Fig. 3g). The ability of MP to promote leaf formation is also reflected in the immediate generation of leaves (as opposed to later-forming bulges) in mp nph4 MP-GR and mp nph4 MP-GRer primary SAMs treated with DEX shortly after germination (Fig. 3h). In contrast, if the bulges develop in the continued absence of DEX, they enter reproductive growth as evidenced by their transition into pin-like inflorescence stems (Fig. 3i), which initiate flowers when finally exposed to DEX (Fig. 3j). These findings indicate that MP is continuously required for the formation of leaves but has no influence on the transition to reproductive development.
Because PIN1 is postulated to have an instrumental role in primordia initiation and positioning in the SAM (Benkova et al., 2003; Reinhardt et al., 2003), abnormalities displayed by mp nph4 SAMs may be due to reduced PIN1 expression. Visualization of PIN1-GFP supports this interpretation, as PIN1 expression in SAMs of mp nph4 MP-GR seedlings (which lack cotyledons) is not apparent even after two weeks postgermination (Fig. 3k, left). Expression of PIN1-GFP is only faintly visible in dicotyledonous mp nph4 MP-GRer seedlings, where it forms a stable ring of homogeneous intensity in the peripheral zone of the narrow, oval SAM (Fig. 3m,p). The effect of MP-GR activation on PIN1 expression in both seedling types is immediate and dramatic. Upon DEX application to mp nph4 MP-GR SAMs, distinct spots of strong PIN1-GFP expression become visible within 18h, then further intensify to eventually become associated with outgrowing primordia (Fig. 3k). In mp nph4 MP-GRer SAMs, DEX application increases ring-shaped PIN1 expression after 6h (Fig. 3p). Together with increased intensity, PIN1-GFP expression becomes uneven by 24h, narrowing into bright foci that precede organ primordia. As the SAM grows for the next two days, more PIN1 expression foci emerge, which also mark organ initiation sites (Fig. 3p).
In summary, our analysis of the SAM identifies ARF activity as the transcriptional driving force underlying dynamic PIN1 expression, auxin distribution and eventually organ initiation and phyllotaxis. In the near-absence of ARF activity, the generation of PIN1 expression foci and lateral organs is strongly attenuated and shoot development operates in vastly different dimensions and time scales. Intriguingly, MP remains sufficient to restore organ production from these abnormal conditions. As a genetic tool, this ability of a single gene to controllably reset the self-organizing process of organ initiation under various experimentally designed conditions, including those with or without positional references, affords a tremendous opportunity to interrogate the underlying principles and mechanisms of auxin-based patterning.
Direct regulation of PIN genes by MP
The rapid response of PIN1-GFP expression levels to MP activity (Fig. 2d–f; Fig. 3p) suggests direct regulation of PIN1 by MP. To test this, we quantified PIN1 transcript levels by RT-PCR on isolated mp nph4 MP-GRer roots and shoots treated with varying combinations of DEX, auxin, and the translational inhibitor cycloheximide (Fig. 4a,b). In cycloheximide-treated root tissue, DEX-mediated MP activation resulted in over 5-fold induction of PIN1 expression in the presence of auxin (Fig. 4a). We further monitored expression of PIN3 and PIN7 and found both to be responsive to MP activity under conditions of translational inhibition (Fig. 4a). In auxin-treated mp nph4 MP-GRer SAMs, all three tested PIN genes were also reproducibly upregulated by MP activation in the presence of cycloheximide (Fig. 4b). Consistent with these observations, PIN1, PIN3 and PIN7 show auxin inducible expression (Supporting Information Fig. S3a) (Vieten et al., 2005) and have been implicated in RAM patterning (Blilou et al., 2005). Furthermore, the spatio-temporal expression profiles of all three PIN genes show significant overlap with MP in a variety of developmental contexts (Supporting Information Fig. S3b). Collectively, these results indicate that PIN1, PIN3 and PIN7 are direct transcriptional targets of MP.
Figure 4.
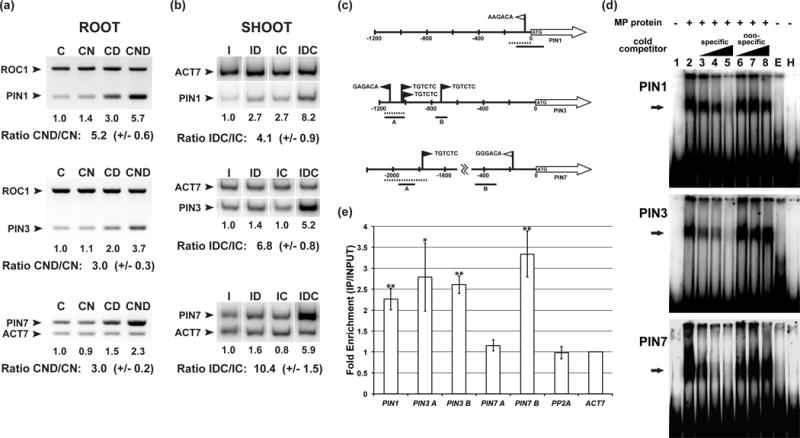
Binding of MONOPTEROS (MP) to PIN FORMED (PIN) upstream regulatory regions.
(a) Reverse transcription-polymerase chain reaction (RT-PCR) on dissected mp nph4 MP-GRer roots (8-to-9d after germination (DAG)) treated for 4h with 30μM cycloheximide (‘C’) and varying combinations of 5μM α-naphthaleneacetic acid (‘N’) and 30μM dexamethasone (‘D’). ROC1 or ACT7 genes are internal controls used for normalization. Normalized signal in lane (‘C’) was arbitrarily set to 1.0. Mean +/− s.e.m of the normalized ‘CND’/‘CN’ ratios for three independent biological replicates are given.
(b) RT-PCR on dissected mp nph4 MP-GRer SAMs (21DAG) treated for 4h with 10μM indole-3-acetic acid (‘I’) and varying combinations of 30μM dexamethasone (‘D’) and 30μM cycloheximide (‘C’). ACT7 is an internal control used for normalization. Normalized signal in lane (‘I’) was arbitrarily set to 1.0. Mean +/− s.e.m. of the normalized ‘IDC’/‘IC’ ratios for three independent biological replicates are given.
(c) Schematics of PIN promoters. Arrows depict open reading frames (ORFs), black flags mark consensus Auxin Response Elements (AuxREs), and white flags denote near-perfect AuxREs. Dashed bars designate regions used as electrophoretic mobility shift assay (EMSA) probes in (d), while solid bars delineate regions tested by chromatin immunoprecipitation (ChIP) PCR in (e).
(d) EMSAs using PIN DNA probes and recombinant His-MP(432) protein. Arrows indicate positions of MP-probe complexes. Lanes 3–5 and lanes 6–8 contain increasing amounts (10X, 50X, 100X) of specific unlabeled competitor DNA and nonspecific unlabeled DNA lacking consensus AuxREs, respectively. Lane ‘E’ contains protein from an empty vector control purification. Lane ‘H’ contains an unrelated prokaryotic protein with the same amino-terminal His-tag as the MP protein.
(e) Anti-GFP ChIP of MP::MP-GFP tissue showing fold enrichment of PIN promoter regions depicted in (c). Mean fold enrichments +/− s.e.m. for three independent biological replicates are shown. Student’s t-test was used to determine the significance of target enrichment relative to enrichment of PP2A control (*, P < 0.05, **, P < 0.01).
To delineate which PIN cis-regulatory regions MP directly targets, we performed in vitro and in vivo binding assays on PIN1, PIN3 and PIN7 promoters, each of which contain canonical or near-canonical Auxin Response Elements (AuxREs) (Fig. 4c). MP specifically bound AuxRE-containing promoter fragments of all three tested PINs in electrophoretic mobility shift assays (EMSAs) (Fig. 4d). Chromatin immunoprecipitation (ChIP) was performed on MP::MP-GFP to test these interactions in planta, and at least for PIN1 and PIN3, MP bound the same promoter regions identified by EMSA (Fig. 4e). Interestingly, this tested region of PIN3 was recently identified as a binding site for ARF7, and the mutation of its three canonical AuxREs reduces the auxin inducibility of PIN3 (Chen et al., 2015). Downstream of this region, another canonical AuxRE exists (Fig. 4c), the position of which also showed enrichment in MP ChIP analyses (Fig. 4e). In the case of PIN7, ChIP did not show enrichment of the promoter fragment bound by MP in vitro (Fig. 4e). We therefore scanned the PIN7 promoter for near-consensus AuxREs and found one such element at a more proximal position (Fig. 4c). Our MP::MP-GFP ChIP analysis showed enrichment in the vicinity of this proximal position (Fig. 4e). Together, our results demonstrate that MP binds to AuxRE-containing regulatory regions in the promoters of PIN1, 3 and 7 in planta, and that through this binding, MP is strictly required for RAM maintenance, LRP patterning and SAM lateral organ initiation.
A Central Molecular Link between Auxin Signaling and Auxin Transport
Like other key processes in plant development, the formation of new organs in roots and shoots has been attributed to the dynamic, self-organizing interplay between auxin signal transduction and auxin transport. The regulatory relationships between critical parameters of this interplay have remained subject to mathematical modeling (Kuhlemeier, 2007), and many of the corresponding cellular mechanisms have yet to be unravelled. Surprisingly, however, one central tenet in most mathematical models, a positive regulation of auxin transport by auxin, has not been adequately explained at the molecular level. In most models, disruption of this regulation should have the most dramatic consequences on respective patterning processes (Wabnik et al., 2013), but hitherto no genes with correspondingly severe mutant phenotypes have been implicated. In this study, we have demonstrated that mp, in an appropriate multiple mutant background, displays the severe patterning defects expected for a critical connector between auxin and auxin transport. In this capacity, MP serves as a nearly perfect on-off switch regulating PIN expression and organ formation in both root and shoot. This regulation occurs through direct binding of MP to distinct promoter elements in multiple PIN genes, and the possibility to control the entire process through the nuclear entry of a single transcription factor provides vast opportunities to interrogate the systems properties of auxin’s self-organizing regulation.
Supplementary Material
Fig. S1 Arabidopsis root gravitropic responses.
Fig. S2 mp nph4 MP-GRer root and shoot apical meristem activity depends directly on dexamethasone (DEX) dosage in Arabidopsis.
Fig. S3 Arabidopsis PIN FORMED (PIN)::β-glucuronidase (GUS) expression patterns.
Table S1 Reverse transcription-polymerase chain reaction (RT-PCR) primer sequences.
Table S2 Primer sequences used to create electrophoretic mobility shift assay (EMSA) probes.
Table S3 Chromatin immunoprecipitation (ChIP) primer sequences.
Table S4 (a) β-Glucuronidase (GUS)-staining conditions of auxin-induced Arabidopsis tissues; (b) β-Glucuronidase (GUS)-staining conditions of Arabidopsis tissues not auxin treated.
Acknowledgments
We would like to thank our colleagues J. Friml and D. Weijers for PIN1::PIN1-GFP and MP::MP-GFP Arabidopsis seeds, respectively. This work was supported by a Natural Sciences and Engineering Research Council of Canada discovery grant to T.B. and with support from the Center for Analysis of Genome Evolution and Function (CAGEF). Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R15GM114733 to N.T.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article.
AUTHOR CONTRIBUTIONS
N.T.K. and T.B. planned and designed the research and wrote the manuscript. N.T.K., D.M. and A.I.W. performed the experiments.
References
- Adamowski M, Friml J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml F. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Chapman E, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev of Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chen Q, Liu Y, Maere S, Lee E, Van Isterdael G, Xie Z, Xuan W, Lucas J, Vassileva V, Kitakura S, et al. A coherent feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nat Commun. 2015;6:8821. doi: 10.1038/ncomms9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136:1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 2013;9:e1003759. doi: 10.1371/journal.pgen.1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Ortiz-Moreno E, de la Paz Sanchez M, Murphy AS, Garcia-Ponce B, Marsch-Martinez N, de Folter S, Corvera-Poire A, Jaimes-Miranda F, Pacheco-Escobedo MA, et al. The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO J. 2013;32:2884–2895. doi: 10.1038/emboj.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, Jurgens G. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development. 2004;131:389–400. doi: 10.1242/dev.00926. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1404–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the Auxin Response Factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139:4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Yin X, Ckurshumova W, Berleth T. Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. New Phytol. 2014;204:474–483. doi: 10.1111/nph.12994. [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C. Phyllotaxis. Trends Plant Sci. 2007;12:143–150. doi: 10.1016/j.tplants.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okishuma Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Sabitini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sassi M, Vernoux T. Auxin and self-organization at the shoot apical meristem. J Exp Bot. 2013;64:2579–2592. doi: 10.1093/jxb/ert101. [DOI] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wisniewska J, Reinohl V, Friml J, Benkova E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20:2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Simaskova M, O’Brien JA, Khan M, Van Noorden G, Otvos K, Vieten A, De Clercq I, Van Haperen JM, Cuesta C, Hoyerova K, et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat Commun. 2015;6:8717. doi: 10.1038/ncomms9717. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development. 2007;134:2561–2567. doi: 10.1242/dev.006759. [DOI] [PubMed] [Google Scholar]
- Wabnik K, Robert HS, Smith RS, Friml J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr Biol. 2013;23:2513–2518. doi: 10.1016/j.cub.2013.10.038. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Yang KZ, Zou JJ, Zhu LL, Xie ZD, Morita MT, Tasaka M, Friml J, Grotewold E, Beeckman T, et al. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat Commun. 2015;6:8822. doi: 10.1038/ncomms9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Arabidopsis root gravitropic responses.
Fig. S2 mp nph4 MP-GRer root and shoot apical meristem activity depends directly on dexamethasone (DEX) dosage in Arabidopsis.
Fig. S3 Arabidopsis PIN FORMED (PIN)::β-glucuronidase (GUS) expression patterns.
Table S1 Reverse transcription-polymerase chain reaction (RT-PCR) primer sequences.
Table S2 Primer sequences used to create electrophoretic mobility shift assay (EMSA) probes.
Table S3 Chromatin immunoprecipitation (ChIP) primer sequences.
Table S4 (a) β-Glucuronidase (GUS)-staining conditions of auxin-induced Arabidopsis tissues; (b) β-Glucuronidase (GUS)-staining conditions of Arabidopsis tissues not auxin treated.


