Abstract
Hyperoxia exposure of newborn rodents has served as a model for bronchopulmonary dysplasia (BPD) phenotypes found in a sub-population of human premature infants. We previously demonstrated that Nrf2 modulates molecular events during saccular-to-alveolar lung maturation and also has a protective role in the pathogenesis of hyperoxia-induced acute lung injury, mortality, arrest of saccular-to-alveolar transition, and lung injury, using Nrf2-deficient and wild-type neonate mice. In this review, we describe how whole-genome transcriptome analyses can identify the means through which Nrf2 transcriptionally modulates organ injury and morphology, cellular growth/proliferation, vasculature development, and immune response during BPD-like pathogenesis. We illustrate how recently developed bioinformatics tools can be used to identify sets of Nrf2-dependently modulated genes in the BPD model, and elucidate direct Nrf2 downstream targets and chemicals/drugs that may act on them. These approaches will provide significant insights into promising therapeutic agents for Nrf2-dependent treatments of complications of preterm birth like BPD.
Keywords: Bronchopulmonary dysplasia, acute lung injury, prematurity, mRNA expression, pathway analysis, canonical, LINCS1000CDS2 search engine, inflammation, antioxidant
Introduction
Respiratory distress syndrome is a breathing disorder that affects a subpopulation of infants with preterm birth (prior to 37 weeks of completed gestation). Premature infant lungs are underdeveloped (saccular phase at 24–36 weeks of gestation) and are unable to make enough surfactant to maintain alveolar ventilation of blood oxygenation until they enter into the alveolar phase. Critical morphologic processes of the saccular phase include expansion of distal airways for subsequent alveolar formation, differentiation of type 1 and 2 pneumocytes, and thinning of the air-blood barrier. Treatments for lung prematurity include surfactant replacement therapy, mechanical ventilation, oxygen, and other strategies to reduce further lung injury, provide nutrition and other support for lung growth and recovery, and prevent lung infections. If premature infants still require oxygen therapy by the time they reach normal delivery dates, they’re diagnosed with bronchopulmonary dysplasia (BPD). Although BPD has multifactorial causes, it has been recently defined as a result from aberrant development of immature lungs exposed to the ex-uterine milieu [1] because the disease is mostly concentrated in very-low-birth weight (< 1,000 g) premature infants [2]. The risk of BPD is indeed inversely proportional to the gestational age at birth [3]. Unfortunately, permanent arrest of lung growth in BPD may cause lifelong functional abnormalities [4].
Due to lack of a safe and effective treatment, BPD remains the most significant pulmonary complication of preterm birth. Excessive oxygen use can paradoxically increase the risk of BPD because it can lead to oxidant injury while low oxygen saturation targets may increase the likelihood of death [5]. In addition, studies with high frequency ventilation have led to potentially less invasive care strategies [6]. Inhaled nitric oxide (iNO) has been approved clinically for more than a decade while efficacy to prevent or treat BPD is controversial [7]. Moreover, the National Institutes of Health Consensus Development Conference (https://consensus.nih.gov/2010/inofinalstatement.htm) concluded with avoidance of routine iNO use for premature infants. In addition to clinical investigations, experimental BPD models in rodents have been widely examined for pathogenesis and therapeutic intervention because neonatal rodents are normally born with premature lungs and develop pulmonary changes similar to BPD after hyperoxia exposure. More recent studies with BPD models demonstrated a novel stem cells therapeutic option. These studies demonstrated that prophylactic administration of mesenchymal stromal cells before hyperoxia exposure showed marked mitigation of alveolar injury, inflammation, pulmonary hypertension, and lung compliance, while administration of these stem cells after hyperoxia exposure did not provide convincing results (see review [8]).
In utero expression of airway antioxidant enzymes are known to increase toward term gestation to prepare for birth into an oxygen (O2)-rich (from 3% to 21% O2) environment [9]. Therefore, preterm infants with low birth weight are not only more sensitive to increased oxygen concentrations compared to adults [10], but they also have diminished/compromised endogenous antioxidant activity relative to full term infants [9], which is thought to contribute to the critical consequence of hyperoxic insult in BPD pathogenesis. While multiple studies are under investigation [11], overall clinical trials of antioxidant therapies [e.g., superoxide dismutases (SODs), vitamins A and E, N-acetylcysteine, metalloporphyrin] in management of BPD have remained inconclusive although recombinant human SOD has shown marginal efficacy (see review [12]). Nuclear factor, erythroid-derived 2, like 2 (Nfe2l2) or NF-E2-related factor 2 (Nrf2) is an indispensable transcriptional regulator for diverse antioxidant enzyme and host defense protein genes bearing antioxidant response elements (AREs) that bind Nrf2. Critical roles of Nrf2 function have been well defined in vivo using adult mice with genetically engineered Nrf2 and its cytoplasmic inhibitor Kelch-like ECH-associated protein (Keap1) in acute lung injury (ALI), emphysema, allergy and asthma, pulmonary fibrosis, lung tumor, and respiratory syncytial virus disease [13]. Roles for Nrf2 in organ development and neonatal disease have also recently been identified in murine experimental BPD. Augmented lethality, lung injury, and arrested saccular-to-alveolar transition after hyperoxia exposure were found in neonatal mice deficient in Nrf2 (Nrf2−/−) mice compared to wild type controls [14,15]. The surviving juvenile Nrf2−/− mice had decreased surfactant-producing type 2 cells [15], which indicated long term pulmonary outcome and the predisposition potential for oxidative pulmonary disease in adults or adolescents. Data also suggested a therapeutic potential to enhance Nrf2-mediated pulmonary responses in BPD pathogenesis. Supporting the role for Nrf2-ARE responses, increased BPD risk was associated with functional polymorphisms in NRF2 and in ARE-responsive glutathione-s-transferase (GSTP1), NAD(P)H:quinone oxidoreductase 1 (NQO1), and UDP glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1) in several human cohorts [16–18].
In order to further elucidate the gene transcripts affected by Nrf2 in healthy and diseased lungs we performed genome-wide lung cDNA microarray analyses in the murine model of neonatal ALI. The analyses demonstrated that Nrf2-mediated protection against hyperoxia-induced ALI phentoypes may be through transcriptional regulation of genes associated with DNA replication and cell cycle, various metabolism and small molecular processes, and cell-cell interaction as well as redox homeostasis in the saccular phase lungs of newborn mice [14,19]. We also found altered lung transcriptomes for sustaining lung morphogenesis, cell growth machinery, and lymphocyte immunity during saccular lung maturation in hyperoxia-susceptible Nrf2−/− neonates [14], suggesting their roles in predisposing the immature lung to oxidant-induced disorders. Among the key Nrf2 effectors contributing to the protection against BPD-like disorders are glutathione peroxidase 2 (Gpx2) and macrophage receptor with collagenase structure (Marco) [14].
In the current review, we revisited selected sets of our neonatal lung microarray data which were differentially regulated by Nrf2 basally, and during hyperoxia exposure, in order to demonstrate the value of transcriptomics and bioinformatic approaches for identifying novel gene transcripts and gene networks that may contribute to normal lung development and ALI pathogenesis [14]. We elucidated direct Nrf2 downstream target genes by searching genomic sequence near transcription start sites (TSS) for Nrf2/Maf binding sites (or AREs) using a position weight matrix (PWM) model [20] and mapping AREs into peaks of Nrf2 chromatin immunoprecipitation-sequencing (ChIP-seq) [21–24]. We also determined chemicals and drugs that are predicted to perturb the mode of gene expression (higher or lower in Nrf2−/− compared to Nrf2+/+ at baseline or after hyperoxia) by The Library of Integrated Cellular Signatures (LINCS) analysis. The results from these recently developed bioinformatics tools provide informative clues for promising intervention compounds in BPD protection or treatment.
Transcriptome data for bioinformatics analyses
The microarray data were deposited in Gene Expression Omnibus (GEO, accession number GSE29632) and in NIEHS Chemical Effects in Biological Systems (CEBS, accession number: 005-00003-0012-000-0). We reanalyzed the previously profiled data sets with a newer version of GeneSpring 12.6 (Agilent Technologies, Inc., Santa Clara, CA), to generate gene lists for further analyses. A number of bioinformatics tools have been developed to query transcription profiles for gene networks, canonical pathways, and co-expressed genes [e.g. Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, CA), Genomic Regions Enrichment of Annotations Tool (GREAT, http://bejerano.stanford.edu/great/public/html/), Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), Extracting Patterns and Identifying co-expressed Genes (EPIG, http://www.niehs.nih.gov/research/resources/software/biostatistics/epig/index.cfm)]. In order to provide the most recent updates in gene identity and networks, we focused first on Nrf2-dependently varied gene transcripts (> 2-fold at least one time point, n=396) from the saccular stage (post-natal days P1-P3, characterized by simple, poorly septated saccules) and the more mature late saccular/early alveolar phase (P4, bearing branched septa and multilobular alveoli). While developmental gene variation was highest between P1 and P4 in Nrf2+/+ mice, the greatest genotype effects were found at days P2-P3. Our previous visual profile analysis also revealed distinct sets of genes which were markedly overexpressed at P2-P3 of Nrf2−/− mice [14], so we used analysis by similar entities (GeneSpring, Agilent technologies, Santa Clara, CA) to identify 141 correlated transcripts (Figure 1-A). Using IPA we found that among the canonical pathways affected in this Nrf2-dependent transcript set was T cell receptor signaling (Figure 1-B), which has not been previously implicated in studies of this transcription factor. Following similar approaches, we also analyzed gene transcript expression changes induced by postnatal hyperoxia exposure (1–3 days starting from P1) and were Nrf2-dependent (p<0.01, >2-fold n=437). Description of these Nrf2-dependently modulated neonatal lung genes and their expression profiles as well as the associated functions and molecular networks have been published previously [14].
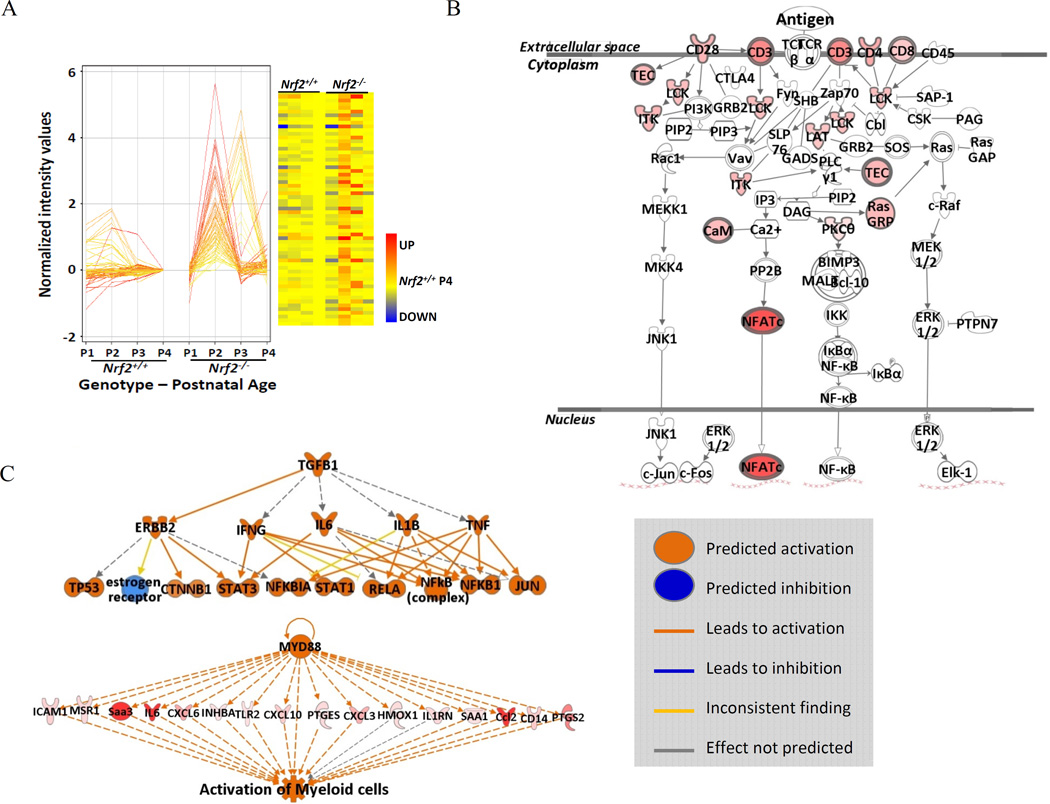
Figure 1. Transcriptomics and molecular events in Nrf2 deficient murine lungs during saccular-to alveolar transition and bronchopulmonary dysplasia (BPD)-like pathogenesis.
(A) cDNA microarray and profile analysis elucidated a set (n=141) of markedly elevated genes during postnatal days 2 and 3 (P2-P3) in Nrf2-deficient (Nrf2−/−) mice compared to their wild type controls (Nrf2+/+). Data were normalized to the average expression level of Nrf2+/+ mice at P4. The expression profiles of these genes were depicted as a heat map. Color bar indicates relative expression intensity. (B) Function analysis determined T cell receptor signaling as one of the most predominant molecular events in Nrf2-deficient saccular phase lungs. The intensity of the red highlight for the affected genes in the pathway indicates the level of significance (p value). (C) Pathway analysis demonstrated that transforming growth factor beta 1 (TGF-β1) is one of the key upstream regulators for severe BPD-like disease development in Nrf2−/− neonates and myeloid differentiation primary response gene 88 (Myd88) and related innate immune signaling pathway genes are essential in the heightened inflammatory responses in Nrf2−/− neonates of the current model. Analyses were done using GeneSpring software and Ingenuity Pathway Analysis.
Putative ARE search for potential Nrf2 downstream effectors
The putative Nrf2 binding sites (AREs) within 5 kb of TSS were predicted using a PWM model [20], and they were identified as functional AREs only if they were under ChIP-seq peaks [21,25–27]. About 15% (62 out of 424) or 19% (73 out of 377) of transcripts bearing potential AREs in the Nrf2-dependent neonatal lung genes at baseline or after hyperoxia, respectively (Table 1, Appendices A, B) had nearby putative AREs. For the lung genes basally heightened in Nrf2−/− neonates at P2-P3, a motif discovery analysis using MEM-ChIP [28] elucidated two enriched motifs for DNA binding proteins, Ets1 (the significance value, e = 2.3e-9) and Klf4 (e = 1.4e-2) in the TSS +/−1 kb regions and others including Etv6 (e = 1.1e−7), Nrf1 (e = 3.3e−5), E2f4 (e = 7.6e−5), Tcfl5(e = 1.4e-4), and Sp2 (e = 5.3e−4) in TSS +/−5 kb regions. Ets family genes are known to associate with cancer progression and studies using gene deficient mice demonstrated that Ets1 regulate hematopoietic cell (T cell, B cell, and natural killer cell) and Etv6 also play roles in hematopoiesis and embryonic angiogenesis [29]. Nrf1, similar to Nrf2, is known to regulate ARE-bearing antioxidants but it also modulates inflammatory responses [30]. Klf4 (Krüppel-like factor 4) is a pluripotent transcription factor known to be involved in carcinogenesis or anti-carcinogenesis by modulating various genes (e.g., p21, cyclins, Bcl2) in cell cycle and survival [31]. It corresponds to the pathway analysis (http://www.ingenuity.com/products/ipa) in which these genes were highly linked to the T cell development and T cell receptor signaling (Figure 1-B) with interleukin 7 and TCF3 as predicted upstream regulators. It is therefore postulated that Nrf2-deficieny in immature lung activates compensatory mechanism to evoke acute immune response for host defense. It is also suggested that these compensatory immune responses in Nrf2-deficient saccular lung may predispose the hyperoxia-induced BPD-like disorder in which transforming growth factor beta 1 (TGF-β1) is one of the key upstream regulator and myeloid differentiation primary response gene 88 (Myd88) plays a key role in inflammatory cell infiltration (Figure 1-C)
Table 1.
Potentially functional antioxidant response elements (ARE) on Nrf2-dependent acute lung injury (ALI) genes determined in a neonate mouse model.
| Gene Symbol |
Gene Title | PWM | MS | Distance to Transcription Start Site |
*Data Source |
† FD (Baseline) or Trend/p value (Hyperoxia) |
|---|---|---|---|---|---|---|
| Akr1b8 | aldo-keto reductase family 1, member B8 | 19.4, 13.7 |
0.922, 0..934 | −3742, −4012 | P2 | −2.0 |
| Days 1–2 | Low (0.001/ 0.002) | |||||
| Aox1 | aldehyde oxidase 1 | 15, 12.6 | 0.943, 0.836 | −323, −225 | P1-P4 | −2.3/−2.7/−2.4/−2.1 |
| Day 3 | Low (0.001) | |||||
| Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide |
15.3, 12.1 | 0.917, 0.891 | −972, −817 | P3 | 2.4 |
| Birc6 | baculoviral IAP repeat-containing 6 | 13.9 | 0.877 | −800 | P2 | −2.0 |
| Bscl2 | Bernardinelli-Seip congenital lipodystrophy 2 homolog |
18.7, 11.7 | 0.947, 0.843 | 204, −4499 | Day 3 | Low (0.009) |
| Cbr3 | carbonyl reductase 3 | 14.1 | 0.875 | −736 | Day 3 | Low (0.003) |
| Cdca7 | cell division cycle associated 7 | 11 | 0.795 | −2462 | P3 | −2.3 |
| Cltc | clathrin, heavy polypeptide (Hc) | 15.2–10.9 | 0.935–0.797 | −1984, −1307, −1795, −542 | Day 1 | Low (0.017) |
| Cyb5a | cytochrome b-5 | 18.5–13 | 0.952–0.895 | −2308, −2742, −1836 | Day 3 | Low (0.007) |
| Egr1 | early growth response 1 | 11.9 | 0.839 | −3334 | P3 | −2.7 |
| Ehd1 | EH-domain containing 1 | 11.5 | 0.89 | −3075 | Day 2 | High (0.001) |
| Eif4g2 | eukaryotic translation initiation factor 4, gamma 2 |
13.7 | 0.893 | −644 | Day 2 | High (0.001) |
| Fgfbp1 | fibroblast growth factor binding protein 1 | 14.5 | 0.942 | −1238 | Day 3 | Low (0.003) |
| Fkbp5 | FK506 binding protein 5 | 15 | 0.931 | −4611 | P3 | 2.2 |
| Ftl1 | ferritin light chain 1 | 20.7 | 0.981 | −1087 | Day 2 | Low (0.002) |
| Gclc | glutamate-cysteine ligase, catalytic | 21.6 | 0.99 | −3795 | Days 1–2 | Low (0.005/0.004) |
| Glipr1 | subunit GLI pathogenesis-related 1 (glioma) | 12.5 | 0.884 | −3418 | Day 2 | Low (0.003) |
| Gsta3 | glutathione S-transferase, alpha 3 | 12.9 | 0.86 | −126 | P1, P3, P4 | −4.9/−4.8/−5.3 |
| Hexa | hexosaminidase A | 13, 10.5 | 0.836, 0.81 | −2057, −1779 | Day 2 | Low (0.01) |
| Hmox1 | heme oxygenase (decycling) 1 | 19.3–10.8 | 0.968–0.862 | −3873, −3925, −3201 | Day 2 | Low (0.002) |
| Hsp90aa1 | heat shock protein 90, alpha (cytosolic), class A member 1 |
16.3, 10.3 | 0.947, 0.867 | −2986, −4958 | Day 2 | High (0.005) |
| Hspa8 | heat shock protein 8 | 13 | 0.904 | −1158 | Day 1 | High (0.027) |
| Htatip2 | HIV-1 tat interactive protein 2, homolog | 16.4 | 0.954 | −54 | Day 3 | Low (0.003) |
| Ift80 | intraflagellar transport 80 homolog | 10.8 | 0.851 | −3728 | P3 | −2.6 |
| Day 2 | Low (0.003) | |||||
| Il6 | interleukin 6 | 18.3 | 0.965 | −256 | Day 2 | Low (0.007) |
| Kif5b | kinesin family member 5B | 10.6 | 0.907 | −1043 | P1-P4 | 2.6/2.9/2.8/2.7 |
| Malat1 | metastasis associated lung | 15.3 | 0.904 | −4261 | P3 | −2.0 |
| Mcm5 | minichromosome maintenance deficient 5, cell division cycle 46 |
10.2 18 | 0.829 | −750 | P3 | −2.4 |
| Me1 | malic enzyme 1, NADP(+)-dependent, cytosolic |
18 | 0.944 | 8 | Days1, 3 | Low (0.001/0.003) |
| Mki67 | antigen identified by monoclonal | 11.8 | 0.849 | −740 | P3 | −2.3 |
| Nek6 | NIMA (never in mitosis gene a)-related expressed kinase 6 |
18 | 0.967 | −4345 | Day 2 | High (0.004) |
| Nfat5 | Nuclear factor of activated T-cells 5 (Nfat5), transcript variant a, mRNA |
10.3 | 0.896 | −928 | P2 | −2.2 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 16.4 | 0.87 | −422 | Day 3 | Low (0.003) |
| Oasl2 | 2’-5’ oligoadenylate synthetase-like 2 | 10.7 | 0.883 | −61 | P2 | 2.3 |
| Pgd | phosphogluconate dehydrogenase | 18.6, 11.7 | 0.971, 0.808 | −4145, −4177 | Day 1 | Low (0.03) |
| Pir | pirin | 20, 12.6 | 0.958, 0.909 | 118, −219 | Day 1 | Low (0.007) |
| Pola1 | polymerase (DNA directed), alpha 1 | 12.5 | 0.872 | −3001 | P3 | −2.5 |
| Prdx1 | peroxiredoxin 1 | 15.7–14.9 | 0.923–0.937 | −138, −2451, −4889 | Day 2 | Low (0.001) |
| Rrm2 | ribonucleotide reductase M2 | 16.2 | 0.947 | −3190 | P3 | −2.4 |
| Slc7a11 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
14.1 | 0.843 | −99 | Day 3 | Low (0.007) |
| Sord | sorbitol dehydrogenase | 11.8 | 0.899 | −4245 | P1 | −2.0 |
| Srxn1 | sulfiredoxin 1 homolog | 12.7 | 0.862 | −50 | Days 2–3 | Low (0.004/0.004) |
| Timp1 | tissue inhibitor of metalloproteinase 1 | 13 | 0.877 | −1957 | Day 1 | Low (0.04) |
| Tlcd2 | TLC domain containing 2 | 13.7 | 0.877 | −4977 | P1 | 2.1 |
| Day 3 | High (0.002) | |||||
| Tomm20 | Translocase of outer mitochondrial membrane 20 homolog |
10.3 | 0.915 | −131 | Day 2 | Low (0.002) |
| Txnl1 | thioredoxin-like 1 | 13.1 | 0.9 | 45 | Day 2 | Low (0.001) |
| Txnrd1 | thioredoxin reductase 1 | 17.9, 10.6 | 0.928, 0.84 | −97, −403 | Days 1–3 | Low (0.005/0.001/0) |
| Ubc | ubiquitin C | 12.7, 10.6 | 0.907, 0.871 | −1794, −254 | Days 1, 3 | Low (0.009/0.003) |
| Wac | WW domain containing adaptor with Coiled coil |
16.8 | 0.873 | −3912 | P2 | −2.2 |
ARE, antioxidant response element. PWM, position weight matrix. MS, matrix significance.
Selected bound AREs determined by mouse ChIP-seq analyses [21–24] with PWM greater than 10 in 5’-untranslated region (UTR) and 5 kb promoter of the genes are shown. Expanded data for full gene list and ARE search in Appendices A and B. †Fold difference at baseline (postnatal days P1, P2, P3, or P4) or expression trend and p value during hyperoxia exposure (Days 1, 2, or 3) in Nrf2−/− neonates compared to Nrf2+/+ neonates.
Microarray analyses data sources of the Nrf2-dependent genes - baseline (P1-P4) or during hyperoxia (Days 1–3).
LINCS analysis for chemical targets
LINCS analysis was performed on the gene signatures obtained from our microarray gene expression experiment on the Nrf2-dependently modulated neonatal lung gene sets using the L1000CDS2 search engine (http://amp.pharm.mssm.edu/L1000CDS2/#/index). We took gene transcripts significantly higher or lower (>2-fold) in Nrf2−/− mice compared to the same exposure condition or postnatal age, chose “reverse” mode for small molecule signatures that reversed our input, and allowed the small molecular combination [32]. For the gene sets markedly high only in Nrf2−/− at baseline, we used the signatures and their relative expression ratio between Nrf2+/+ and Nrf2−/− as input for the analysis. For the top 50 search results ranked by the “search score” obtained from the L1000CDS2 analysis (when up and down lists were available) or the largest cosine distance (Nrf2−/− baseline), we performed additional statistical tests for significance. To do so, we downloaded the original L1000 gene expression signatures computed using the characteristic direction signature method [33] for each chemical compound perturbation and the respective cell line landmark gene signatures and all ∼22,000 L1000 genes. We then implemented the hypergeometric test that produced a p value for each enrichment result. All gene signatures were annotated with Entrez symbols and duplicate gene entries were filtered prior to the test.
We next used LINCS to identify via signature overlap assessment, transcript profiles from cell-based drug perturbation experiments that overlap with transcript profiles generated for hyperoxia exposure responses (Table 2, Appendix C). At baseline, significant overlaps with perturbation-induced profiles were determined only at P4 for the Nrf2-dependent transcripts and they included anti-inflammatory (e.g., Zolantidine, Meclocycline, Fenbufen) and chemotherapeutic (e.g., Tipifarnib) agents (Table S3). Significantly high overlaps were also found between our differential transcript lists at all three time points. The minimum overlap ratios (the input differential transcripts and the signature differential transcripts divided by the effective input) ranged from 0.0349 to 0.0581 across the time points.
Table 2.
Representative list of drugs identified using the L1000CDS2 search engine of the Library of Integrated Cellular Signature (LINCS) analysis to be predicted to act on Nrf2-dependent lung transcriptome during the bronchopulmonary dysplasia pathogenesis.
| Hyperoxia exposure day |
Drug name | Activity of compound* | Disease treatment | Minimum overlap ratio |
Corresponding p value |
|---|---|---|---|---|---|
| day 1 | Elesclomol | Induction of cancer cell death by oxidative stress |
Anti-cancer activity | 0.1765 | 3.060 × 10−6 |
| 1 | Isoliquiritigenin | Inhibits NLRP3 inflammasome |
Treatment of inflammatory diseases |
0.1765 | 7.170 × 10−7 |
| 1/3 | Parthenolide | Suppression of apoptotic genes | Anti-cancer and anti- inflammatory activities |
0.2059/0.2051 | 6.280 × 10−6 /9.929 × 10−4 |
| 1/3 | L-sulforaphane | Induction of Nrf2 and prevent NF-κB binding |
Induce detoxification and xenobiotic enzymes |
0.1765/0.2051 | 3.100 × 10−10 /7.32 × 10−5 |
| 1/3 | Arachidonyl trifluoro- methyl ketone |
Inhibits PLA2 | Neuroprotective and treatment of multiple |
0.1765/0.2564 | 6.280 × 10−6 7.650 × 10−4 |
| 1/3 | Parthenolide | Binds directly to and inhibit IKKβ |
Anti-inflammatory and anti- hyperalgesic activities |
0.2059/0.2051 | 6.28 × 10−6 /9.929 × 10−4 |
| 3 | Celastrol | Inhibits NF-κB | Antioxidant, anti- inflammatory, and anti-cancer |
0.2308 | 4.390 × 10−5 |
| 3 | Canertinib | Tyrosine kinase activity | activities Anti-cancer activity | 0.2051 | 9.010 × 10−6 |
| 3 | Afatinib | Inhibits EGFR and HER2 | Anti-cancer (NSCLC) activity |
0.2051 | 3.042 × 10−4 |
| 3 | 15-delta prostaglandin J2 |
PPAR-γ agonist | Antioxidant and anti- inflammatory activities |
0.2051 | 1.210 × 10−10 |
Drugs and chemicals were identified using the L1000CDS2 search engine for which there are statistically significant overlap ratios of transcript profiles from cell-based perturbation experiments and Nrf2-dependent lung transcriptome analysis in neonatal mice exposed to hyperoxia. Details for L1000CDS2 analyses are described elsewhere [41]. Full list of the drugs and chemicals in Appendix 3.
EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor receptor 2; IKKβ, I kappa B kinase beta; NLRP3, NLR Family, Pyrin Domain Containing 3; NSCLC, non-small cell lung cancer; NF-κB; nuclear factor kappa-light-chain-enhancer of activated B cells PLA2, phospholipase 2; PPAR-γ, peroxisome proliferator-activated receptor gamma.
Among many perturbing agents (Table 2, Appendix C), L-sulforaphane and 15-delta prostaglandin J2 (15d-PGJ2) were predicted to reverse the Nrf2-dependent transcriptome changes during the BPD-like pathogenesis. These finding are consistent with the current concepts of Nrf2 regulation. 15d PGJ2 is not only an Nrf2 activator acting through Keap1 binding [34] but is also a ligand for peroxisome proliferator-activated receptor gamma (PPAR-γ) which bears functional AREs for Nrf2 modulation [35]. Through nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) inhibition, it is known as a key regulator of inflammation [36,37]. We also demonstrated Nrf2-dependent protection against ALI by 15d-PGJ2 in mice [35]. Sulforaphane is a powerful phytochemical Nrf2 agonist, and its role in anti-carcinogenesis and pulmonary protection against bacterial infection following emphysema, respiratory syncytial virus infection, and inhaled arsenic has been demonstrated in rodent studies (see review [13]). Moreover, controlled human studies recently reported the therapeutic potential of sulforaphane in reducing adverse effects of airway toxicants [38–40]. This LINCS-based approach to investigation of transcript profiles has the potential to identify other drugs or chemicals that behave similar to 15d-PGJ2 and sulphoraphane, and provide additional means to upregulate anti-oxidant and other lung cellular defense mechanisms.
Conclusions
We have described new and emerging bioinformatics-based approaches that may be used to better understand direct Nrf2 downstream effector genes and drugs/chemicals that may act on the Nrf2-ARE pathways for BPD prevention or treatment. Nrf2 effectors bearing AREs determined by functional bioinformatics in the upstream region as well as in the vicinity of the genes (Table 1, Appendices A, B) have broad spectrum of activity including lung morphogenesis, cell growth machinery, and lymphocyte immunity (Summary in Figure 2). We believe that continued development of analytical tools to investigate global transcriptome changes in the developing lung will provide new insight to potential therapeutic strategies for lung diseases of prematurity, as well as other diseases where Nrf2 has an important role in pathogenesis.
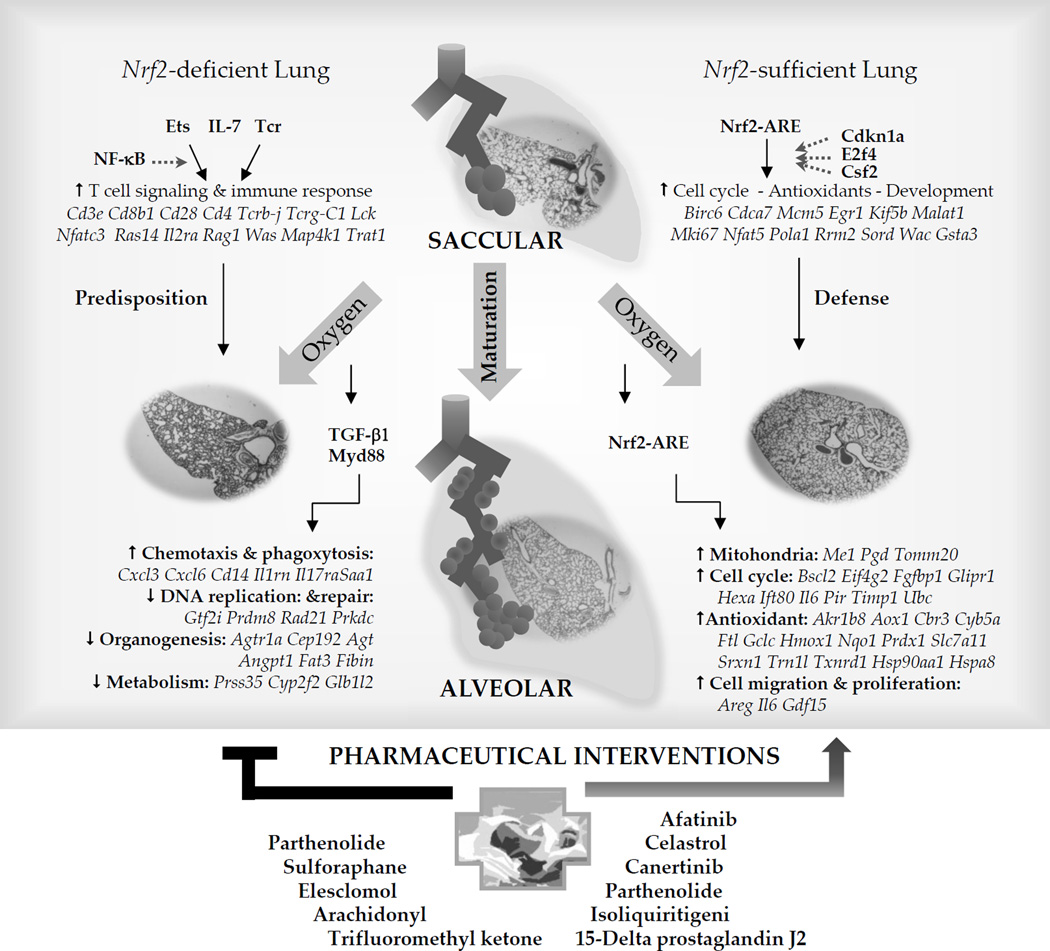
Figure 2. Proposed Nrf2-mediated regulatory mechanisms of saccular-to-alveolar transition and bronchopulmonary dysplasia (BPD)-like pathogenesis and potential therapeutic intervention.
Comparative microarray analysis of Nrf2+/+ and Nrf2−/− neonates followed by bioinformatics suggested downstream Nrf2 target genes bearing functional AREs in the lung undergoing saccular-to-alveolar transition and BPD-like pathogenesis. Molecules such as cell cycle dependent kinase 1a (Cdkn1a), E2F transcription factor 4 (F2f4), or colony stimulating factor 2 (Csf2) may participate in the Nrf2-ARE pathway during normal saccular-to-alveolar transition. Hyperoxia exposure to the premature lung at saccular phase with simple, poorly septated saccules causes lung injury and interrupts normal alveolar formation. Nrf2-mediated protection is predicted to be through transcriptional activation of antioxidant, cell cycle and DNA repair, and mitochondria-related metabolism. Nrf2 deficiency in saccular phase lung suppressed these cellular processes and aberrantly activated upstream molecules including transforming growth factor beta 1 (TGF-β1) and myeloid differentiation primary response gene 88 (Myd88) may drive inflammatory cell infiltration and attenuate DNA repair and cell cycle, metabolism, and morphogenesis signaling. Augmented transcriptome of T lymphocyte signaling and related acquired immunity during the saccular-to-alveolar transition Nrf2−/− neonate lung may predispose the BPD-like phenotypes. Ets family transcription factor, interleukin 7 (IL-7), or T cell receptor (Tcr) in association with nuclear factor kappa-light-chain enhancer of activated B cells (NF-kB) may play roles in the upstream of this process. Drugs or chemicals acting on redox balance (e.g., sulforaphane, Celastrol), anti-inflammation (e.g., 15-delta prostaglandin J2, Isoliquiritigenin, Pathenolide, Celastrol), or anti-cancer (e.g., Elesclomol, Pathenolide, Canertinib, Afatinib) mechanisms are suggested as pharmaceutical interventions for BPD pathogenesis.
Supplementary Material
HIGHLIGHTS.
Transcriptome analysis provided molecular aspects of BPD pathogenesis.
Pathway analysis indicated roles for Nrf2 in neonatal lung disease.
Bioinformatic analyses suggested potential drug targets and therapeutic compounds for BPD.
Acknowledgments
This research was supported by the Intramural Research Program of the NIEHS, National Institutes of Health (NIH), Department of Health and Human Services. Drs. Donald Cook and Michael Fessler of the NIEHS provided excellent critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR DISCLOSURE STATEMENT
The authors declare that no competing financial interests exist.
References and recommended reading
- 1.Baraldi E, Filippone M. Chronic lung disease after premature birth. N Engl J Med. 2007;357:1946–1955. doi: 10.1056/NEJMra067279. [DOI] [PubMed] [Google Scholar]
- 2.Fanaroff AA, Stoll BJ, Wright LL, Carlo WA, Ehrenkranz RA, Stark AR, Bauer CR, Donovan EF, Korones SB, Laptook AR, et al. Trends in neonatal morbidity and mortality for very low birthweight infants. Am J Obstet Gynecol. 2007;196:147. doi: 10.1016/j.ajog.2006.09.014. e141–148. [DOI] [PubMed] [Google Scholar]
- 3.Jobe AH. The new bronchopulmonary dysplasia. Curr Opin Pediatr. 2011;23:167–172. doi: 10.1097/MOP.0b013e3283423e6b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northway WH, Jr, Moss RB, Carlisle KB, Parker BR, Popp RL, Pitlick PT, Eichler I, Lamm RL, Brown BW., Jr Late pulmonary sequelae of bronchopulmonary dysplasia. N Engl J Med. 1990;323:1793–1799. doi: 10.1056/NEJM199012273232603. [DOI] [PubMed] [Google Scholar]
- 5.Vento M, Moro M, Escrig R, Arruza L, Villar G, Izquierdo I, Roberts LJ, 2nd, Arduini A, Escobar JJ, Sastre J, et al. Preterm resuscitation with low oxygen causes less oxidative stress, inflammation, and chronic lung disease. Pediatrics. 2009;124:e439–e449. doi: 10.1542/peds.2009-0434. [DOI] [PubMed] [Google Scholar]
- 6.Cools F, Askie LM, Offringa M, Asselin JM, Calvert SA, Courtney SE, Dani C, Durand DJ, Gerstmann DR, Henderson-Smart DJ, et al. Elective high-frequency oscillatory versus conventional ventilation in preterm infants: a systematic review and meta-analysis of individual patients’ data. Lancet. 2010;375:2082–2091. doi: 10.1016/S0140-6736(10)60278-4. [DOI] [PubMed] [Google Scholar]
- 7.Raffay TM, Martin RJ, Reynolds JD. Can nitric oxide-based therapy prevent bronchopulmonary dysplasia? Clin Perinatol. 2012;39:613–638. doi: 10.1016/j.clp.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pierro M, Ciarmoli E, Thebaud B. Bronchopulmonary Dysplasia and Chronic Lung Disease: Stem Cell Therapy. Clin Perinatol. 2015;42:889–910. doi: 10.1016/j.clp.2015.08.013. With absence of efficient therapy for bronchopulmonary dysplasia, one of novel approach for this significant burden in extreme prematurity may be stem cells which play a crucial role in organogenesis and organ repair. This review reported up-to-date potential of mesenchymal stem cell therapy by presenting prophylactic or rescue administration cases of rodent preclinical studies
- 9.Rickett GM, Kelly FJ. Developmental expression of antioxidant enzymes in guinea pig lung and liver. Development. 1990;108:331–336. doi: 10.1242/dev.108.2.331. [DOI] [PubMed] [Google Scholar]
- 10.Chessex P, Watson C, Kaczala GW, Rouleau T, Lavoie ME, Friel J, Lavoie JC. Determinants of oxidant stress in extremely low birth weight premature infants. Free Radic Biol Med. 2010;49:1380–1386. doi: 10.1016/j.freeradbiomed.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Lee JW, Davis JM. Future applications of antioxidants in premature infants. Curr Opin Pediatr. 2011;23:161–166. doi: 10.1097/MOP.0b013e3283423e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poggi C, Dani C. Antioxidant strategies and respiratory disease of the preterm newborn: an update. Oxid Med Cell Longev. 2014;2014:721043. doi: 10.1155/2014/721043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho HY, Kleeberger SR. Association of Nrf2 with airway pathogenesis: lessons learned from genetic mouse models. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho HY, van Houten B, Wang X, Miller-Degraff L, Fostel J, Gladwell W, Perrow L, Panduri V, Kobzik L, Yamamoto M, et al. Targeted Deletion of Nrf2 Impairs Lung Development and Oxidant Injury in Neonatal Mice. Antioxidants & redox signaling. 2012 doi: 10.1089/ars.2011.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGrath-Morrow S, Lauer T, Yee M, Neptune E, Podowski M, Thimmulappa RK, O’Reilly M, Biswal S. Nrf2 increases survival and attenuates alveolar growth inhibition in neonatal mice exposed to hyperoxia. Am J Physiol Lung Cell Mol Physiol. 2009;296:L565–L573. doi: 10.1152/ajplung.90487.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manar MH, Brown MR, Gauthier TW, Brown LA. Association of glutathione-S-transferase-P1 (GST-P1) polymorphisms with bronchopulmonary dysplasia. J Perinatol. 2004;24:30–35. doi: 10.1038/sj.jp.7211020. [DOI] [PubMed] [Google Scholar]
- 17.Sampath V, Garland JS, Helbling D, Dimmock D, Mulrooney NP, Simpson PM, Murray JC, Dagle JM. Antioxidant response genes sequence variants and BPD susceptibility in VLBW infants. Pediatr Res. 2015;77:477–483. doi: 10.1038/pr.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen JP, Ebbesen F, Hollegaard MV, Andersson S, Hougaard DM, Thorlacius-Ussing O, Henriksen TB. UGT1A1*28 Genotypes and Respiratory Disease in Very Preterm Infants: A Cohort Study. Neonatology. 2016;109:124–129. doi: 10.1159/000442042. [DOI] [PubMed] [Google Scholar]
- 19.McGrath-Morrow SA, Lauer T, Collaco JM, Lopez A, Malhotra D, Alekseyev YO, Neptune E, Wise R, Biswal S. Transcriptional responses of neonatal mouse lung to hyperoxia by Nrf2 status. Cytokine. 2014;65:4–9. doi: 10.1016/j.cyto.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med. 2016;91:45–57. doi: 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Uruno A, Yagishita Y, Katsuoka F, Kitajima Y, Nunomiya A, Nagatomi R, Pi J, Biswal SS, Yamamoto M. Nrf2-Mediated Regulation of Skeletal Muscle Glycogen Metabolism. Mol Cell Biol. 2016;36:1655–1672. doi: 10.1128/MCB.01095-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. HT-ChiP method leverages the next generation sequencing technology and applies in immunoprecipitation approach. The Transcription Factor (TF) binding motif was identified and the binding was revealed by the peak calling. The overall relationship between the gene regulation machinery and genomic responses were validated. This research reveals the mechanism and dynamic of such molecular activity, and it also lays out the ground work for upcoming research pursuits
- 25. Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. The Mouse ENCODE Consortium published an invaluable resource for researchers using mouse-based systems who pursue mechanisms of gene regulation and interaction
- 26.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic acids research. 2012 doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell MR, Chorley B, Wang X, Cho HY, Kleeberger SR, Bell DA. Discovery of novel genomic targets in the NRF2-mediated antioxidant response pathway by ChIP-on-Chip and ChIP-seq. Cancer Prev Res. 2010;3(1 Supplement):B51. [Google Scholar]
- 28.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HM, Han JW, Chan JY. Nuclear Factor Erythroid-2 Like 1 (NFE2L1): Structure, function and regulation. Gene. 2016;584:17–25. doi: 10.1016/j.gene.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park CS, Shen Y, Lewis A, Lacorazza HD. Role of the reprogramming factor KLF4 in blood formation. J Leukoc Biol. 2016;99:673–685. doi: 10.1189/jlb.1RU1215-539R. [DOI] [PubMed] [Google Scholar]
- 32.Ciencewicki JM, Verhein KC, Gerrish KE, McCaw ZR, Li J, Bushel PR, Kleeberger SR. Effects of mannose-binding lectin on pulmonary gene expression and innate immune inflammatory response to ozone. Am J Physiol Lung Cell Mol Physiol. 2016 doi: 10.1152/ajplung.00205.2015. ajplung 00205 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clark NR, Hu KS, Feldmann AS, Kou Y, Chen EY, Duan Q, Ma’ayan A. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics. 2014;15:79. doi: 10.1186/1471-2105-15-79. A novel geometrical multivariate method successfully overcomes a common limitation that microarray and RNA-seq data face. It creates a hyper-plane and quantifies the relative contribution of each gene to the overall differential expression. By incorporating other statistical tactics, this “characteristic direction” method outperforms existing methods and produces accurate differential expressed genes with high sensitivity and specificity
- 34. Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. This study provides a thorough examination of Nrf2 activators and specific targets that increased our understanding of cellular defense systems and responses to diverse chemicals
- 35.Cho HY, Gladwell W, Wang X, Chorley B, Bell D, Reddy SP, Kleeberger SR. Nrf2-regulated PPAR{gamma} expression is critical to protection against acute lung injury in mice. Am J Respir Crit Care Med. 2010;182:170–182. doi: 10.1164/rccm.200907-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 37.Ishii T. Close teamwork between Nrf2 and peroxiredoxins 1 and 6 for the regulation of prostaglandin D2 and E2 production in macrophages in acute inflammation. Free Radic Biol Med. 2015;88:189–198. doi: 10.1016/j.freeradbiomed.2015.04.034. [DOI] [PubMed] [Google Scholar]
- 38.Heber D, Li Z, Garcia-Lloret M, Wong AM, Lee TY, Thames G, Krak M, Zhang Y, Nel A. Sulforaphane-rich broccoli sprout extract attenuates nasal allergic response to diesel exhaust particles. Food Funct. 2014;5:35–41. doi: 10.1039/c3fo60277j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kensler TW, Egner PA, Agyeman AS, Visvanathan K, Groopman JD, Chen JG, Chen TY, Fahey JW, Talalay P. Keap1-nrf2 signaling: a target for cancer prevention by sulforaphane. Top Curr Chem. 2013;329:163–177. doi: 10.1007/128_2012_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Egner PA, Chen JG, Zarth AT, Ng DK, Wang JB, Kensler KH, Jacobson LP, Munoz A, Johnson JL, Groopman JD, et al. Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prev Res (Phila) 2014;7:813–823. doi: 10.1158/1940-6207.CAPR-14-0103. This population-based clinical trial demonstrated the efficacy of sulforaphane, a potent Nrf2 agonist from broccoli, in detoxification of airborn toxicants. Lowered toxicant excretion by genetic mutation in its metabolic enzyme (GSTT1) induced by Nrf2 and sustained detection of sulforaphane metabolites by daily intake supported the intervention with sulforapahne as a frugal means to attenuate long-term health risks
- 41. Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014;42:W449–W460. doi: 10.1093/nar/gku476. The Library of Integrated Network-based Cellular Signatures (LINCS) project founded by NIH was to investigate genomic responses via gene expression to the exposure to adverse environmental stress or chemical perturbation. It leverages the L1000 platform by measuring the land mark genes and extrapolating to the whole transcriptome repertoire, and stores experimental results in a scheme-less database. The web front-end browser allows user to query such database to extract useful and relevant information to customized experimental set up
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.