Abstract
Objectives
Breast milk is a complex bioactive fluid that varies across numerous maternal and environmental conditions. While breastfeeding is known to impact neonatal gut microbiome, the milk components responsible for this effect are not well-characterized. Given the wide range of immunological activity breast milk cytokines engage in, we investigated three essential breast milk cytokines and their association with early life gut microbiota.
Methods
A total of 52 maternal-child pairs were drawn from a racially diverse birth cohort based in Detroit, Michigan. Breast milk and neonatal stool specimens were collected at 1-month postpartum. Breast milk TGFβ1, TGFβ2, and IL-10 were assayed using ELISAs, while neonatal gut microbiome was profiled using 16S rRNA sequencing.
Results
Individually, immunomodulators TGFβ1 and TGFβ2 were significantly associated with neonatal gut microbial composition (R2=0.024, p=0.041; R2=0.026, p=0.012, respectively) and increased richness, evenness, and diversity, but IL-10 was not. However, the effects of TGFβ1 and TGFβ2 were not independent of one another, and the effect of TGFβ2 was stronger than that of TGFβ1. Higher levels of TGFβ2 was associated with the increased relative abundance of several bacteria, including members of Streptococcaceae and Ruminococcaceae, and lower relative abundance of distinct Staphylococcaceae taxa.
Conclusions
Breast milk TGFβ concentration explains a portion of variability in gut bacterial microbiota composition among breastfed neonates. Whether TGFβ acts in isolation or jointly with other bioactive components to alter bacterial composition requires further investigation. These findings contribute to an increased understanding of how breastfeeding affects the gut microbiome—and potentially immune development—in early life.
Keywords: breastfeeding, cytokines, microbiome
Introduction
Microbes residing in the human gastrointestinal tract are being increasingly recognized for their essential role in defining human health. In addition to nutrient and energy extraction, gut microbes influence appropriate physiological and immune development and defend against pathogenic invasions (1–3). After birth, a rapid expansion of intestinal bacterial diversity occurs in the first year of life before reaching a relatively stable state, which is typically maintained throughout healthy adulthood (4). Microbiome development in early life occur in parallel with immune maturation, and are thought to be foundational to health later in life. Indeed, dysbiotic patterns in early life are associated with the increased risk of several immunological diseases, including allergy and asthma (4–6). Though colonization patterns occur in a highly host-specific manner, early life exposures are key factors in the shaping of infant gut microbial communities, many of which involve maternal-offspring interactions and exchanges (7, 8). Specifically, in addition to delivery mode, breastfeeding has been shown to be one of the strongest determinants of early life gut microbial composition (2, 8–10). However, breast milk is a complex mixture that, in addition to nutritional factors, contains a variety of bioactive components, including immunoglobulins, fatty acids, hormones, cytokines, oligosaccharides, and bacteria. Further, its constitution has been shown to vary depending on several maternal and environmental factors, including maternal age, diet, and country of origin (11, 12). Therefore, though gut microbial composition of breastfed children is compositionally distinct from that of formula-fed children (2, 8–10), it is likely that the heterogeneity of breast milk components leads to differential gut microbiota development within breastfed offspring; however, little is known of these relationships.
Cytokines are important constituents of breast milk with a wide range of immunological activity. They can cross the intestinal barrier and communicate with cells to influence immune development and maturation (13). Although a number of different interleukins (ILs) have been detected in human breast milk, their levels are typically low (14). Nonetheless, IL-10 has often been detected in substantial quantities, and is thought to inhibit Th1-type inflammation as well as facilitate immune tolerance (13, 14). IL-10 has also been shown to play an important role in maintaining intestinal homeostasis in both murine models and humans (15, 16). The transforming growth factor beta (TGFβ) family is the most abundant family of cytokines in human breast milk and is expressed by many cell types, with TGFβ2 being predominant (13). Breast milk TGFβ not only maintains immune homeostasis in the infant gut, but also regulates inflammation, promotes IgA isotype switching to both IgA1 and IgA2 (17), initiates IgA production in newborn infants (18), and plays a major role in the induction of oral tolerance (14, 19). Given the expansive immunological role these breast milk cytokines play in early life and their ability to alter the gastrointestinal environment, we hypothesized that they would be associated with gut bacterial composition among breastfed neonates. We examined these associations among maternal-child pairs drawn from a metropolitan Detroit birth cohort, where participants were selected based on geographic area of residence and not on any parental disease history (20).
Methods
Study Population
The study population consisted of maternal-child pairs from the Wayne County Health Environment Allergy and Asthma Longitudinal Study (WHEALS), a general-risk, prospective birth cohort with racially and socioeconomically diverse participants. Briefly, pregnant women ages 21 to 49 receiving care at Henry Ford Health System (HFHS) obstetrics clinics in urban and suburban Detroit were recruited from 2003–2007. All women were living in a predefined geographic area in Wayne and Oakland counties that included the western portion of the city of Detroit as well as the suburban areas immediately surrounding the city. All participants provided written, informed consent and the study was performed in accordance with protocol guidelines approved by the Institutional Review Board at HFHS. Additional details about the cohort have been previously published (20).
Inclusion/Exclusion Criteria
A subset of children was selected from the WHEALS cohort for inclusion in the Microbes, Allergy, Asthma, and Pets (MAAP) study if they completed a 24-month study-specific clinic visit and had early life dust samples collected from their homes at the same time as their stool collection (N=308). Early life stool samples were successfully sequenced from 298 infants (130 with 1-month stool samples and 168 with 6-month stool samples). Because breast milk samples were collected at the 1-month home visit (from mothers who were currently—but not necessarily exclusively—breastfeeding), only 1-month stool was analyzed for the current study so that breast milk and stool sample collection was concurrent. Among the 130 maternal-child pairs with 1-month stool samples, 52 also had a breast milk sample collected and analyzed for cytokine concentrations, which comprised our final analytic sample. Given the inclusion criteria and breastfeeding requirement, participants in the analytic sample (N=52) differed from WHEALS participants excluded (N=1206), as they were more likely to be married (p=0.019), Caucasian (p=0.004), have higher education (p<0.001), and have higher incomes (p=0.017).
Breast milk and Measurement of Cytokines
Two ounces of breast milk were collected from participants using a sterilized disposable breast pump kit (Medela Classic). Using a sterile pipette, 1 ml aliquots of whole breast milk were transferred into sterile cryovials. Breast milk samples were placed on ice until delivery to the Department of Public Health Sciences lab where the remaining breast milk was centrifuged at 4°C to separate the lipid and aqueous fractions. After removal of the lipid layer, 1 ml aliquots of the aqueous fraction were transferred into cryovials and the whole milk, lipid layer and aqueous fractions were stored at −80°C. The aqueous fraction was utilized for assay of cytokines. TGFβ1, TGFβ2, and IL-10 were assayed using commercial ELISAs (R&D Systems, Minneapolis, MN) according to manufacturer instructions, with resulting concentrations expressed as pg/mL. These specific cytokines were chosen because they were thought to be associated with risk of allergic disease, the focus of the WHEALS cohort. The minimum detectable concentrations were 4.6, 7.0, and 3.9 pg/mL for TGFβ1, TGFβ2 and IL-10, respectively. Samples below the minimum detectable concentration of IL-10 were re-assayed with a high sensitivity IL-10 ELISA (R&D Systems, Minneapolis, MN). Values below the minimum detectable concentration for the high sensitivity kit (0.09 pg/mL) were reported as half the minimum detectable concentration (0.045 pg/mL).
Stool Specimens and Gut Microbiota Sequencing
A detailed description of processing and sequencing protocols has been previously published (6). Briefly, stool specimens were collected by field staff during the home visit, placed into cryovials, and stored at −80°C until stool aliquots were shipped to University of California, San Francisco on dry ice, where they were again stored at −80 °C until processed. Stool samples were extracted using a modified cetyltrimethylammonium bromide (CTAB) buffer based protocol, as previously described (6). Bacterial communities were characterized by sequencing the V4 region of the 16S rRNA gene, using the Illumina MiSeq sequencing platform (21). Sequence processing and quality assurance was conducted using the QIIME software version 1.8.0 (22). The data were then rarefied to the minimum read depth of 202,367 sequences per sample, using a representative rarefying approach described previously (6).
Statistical Analysis
For descriptive purposes, maternal and baseline covariates were examined for associations with raw breast milk cytokine concentrations using standard non-parametric statistical tests (Wilcoxon rank sum, Kruskal-Wallis, or Spearman correlation), which account for the skewed nature of raw cytokine distributions. However, breast milk cytokine concentrations were natural log-transformed prior to all microbiome-related analyses. Gut microbial alpha diversity metrics (richness, Pielou’s evenness, and Faith’s phylogenetic diversity) were calculated using QIIME (22) and the R vegan package (23), and their associations with breast milk cytokines were assessed using Spearman correlations. Compositional differences (i.e., beta diversity) in the gut microbiome were captured using an Unweighted UniFrac distance matrix (24), a measure of between-sample phylogenetic similarity in terms of bacteria present in the gut. These distances were tested for compositional differences by breast milk cytokine levels using (unadjusted and adjusted) permutational multivariate analysis of variance (PERMANOVA) with 10,000 permutations, using the “adonis” function in the R vegan package (23). Compositional differences were visualized using non-metric multidimensional scaling (nMDS)—which attempts to group samples based on relative similarity—with the “metaMDS” function in the R vegan package. Contour lines were overlaid onto the nMDS plot using generalized additive models (GAMs) with the “ordisurf” function in the vegan package.
Tests of differential taxon relative abundance were performed on all taxa with at least 5 total sequences using zero-inflated negative binomial regression, or standard negative binomial regression in cases where the model failed to converge. Tests were then corrected for multiple comparisons using the Benjamini & Hochberg false discovery rate (FDR) adjustment (25), with FDR adjusted p-value<0.05 considered significant. Phylogenetic trees of significant taxa were built using Interactive Tree Of Life (iTOL) (26). As a means to determine the functional implications of significant taxa, their metagenomic function was predicted using the Phylogenetic Reconstruction of Unobserved States (PICRUSt) algorithm (27). Predicted pathways were then tested for differences using the standard negative binomial model and FDR corrected, as described previously. All analyses were performed in R (Version 3.1.2) and SAS Software (Version 9.4).
Results
Among the 52 maternal-child pairs included in the analysis, home visits where both milk and stool were collected successfully targeted 1-month of age (mean=36 days, IQR=17 days). All but 12 (23%) of the paired milk and stool samples were collected on the same day; otherwise, paired samples were collected within 7 days of each other. A total of 13 (25%) mothers reported exclusively breastfeeding at the study visit while the remainder were partially breastfeeding. IL-10 concentrations were detectable in 30 (58%) breast milk samples, while TGFβ1 and TGFβ2 were detectable in all samples (median concentrations: 0.2, 1805.6, 2963.0 pg/mL, respectively).
Maternal marital status, urban residence, maternal race, maternal atopy, maternal age, and days postpartum at milk collection were associated with at least one of the three breast milk cytokines (all p<0.05, Table 1). Specifically, maternal atopy was associated with significantly higher breast milk IL-10 concentrations (p=0.002), and concentrations of this cytokine were significantly lower in breast milk samples collected later in the postpartum period (p=0.005). African American and urban-dwelling (which are highly related in this cohort, chi-square p=0.008) mothers had significantly higher concentrations of TGFβ1, and unmarried mothers had significantly higher concentrations of both TGFβ1 and TGFβ2. Lastly, maternal age was significantly associated with TGFβ2, with older mothers having lower concentrations.
Table 1.
Association between baseline and maternal factors and breast milk cytokines.
| Breast Milk Cytokines (pg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| IL-10 | TGFβ1 | TGFβ2 | ||||||
|
|
||||||||
| Variable | Level | N | Median | p-value* | Median | p-value | Median | p-value |
| Demographics | ||||||||
|
| ||||||||
| Maternal Education | Some college or less | 26 | 0.21 | 0.89 | 1958.5 | 0.17 | 3600.3 | 0.15 |
| Bachelor’s degree or more | 26 | 0.21 | 1648.0 | 2093.6 | ||||
|
| ||||||||
| Household Income | <80K | 26 | 0.16 | 0.22 | 1958.5 | 0.33 | 3339.3 | 0.21 |
| >80K | 20 | 0.54 | 1611.1 | 1872.1 | ||||
| Refused to answer | 6 | 0.05 | 1659.1 | 2923.6 | ||||
|
| ||||||||
| Married | No | 12 | 0.41 | 0.20 | 2820.2 | 0.009 | 10383.5 | 0.012 |
| Yes | 40 | 0.14 | 1605.8 | 2365.7 | ||||
|
| ||||||||
| Urban Residence | No | 32 | 0.29 | 0.30 | 1513.5 | 0.037 | 2294.5 | 0.16 |
| Yes | 20 | 0.08 | 2086.4 | 3827.3 | ||||
|
| ||||||||
| Maternal Race | Caucasian/Other | 29 | 0.21 | 0.63 | 1347.9 | 0.002 | 2248.2 | 0.11 |
| African American | 23 | 0.21 | 2123.3 | 4290.7 | ||||
|
| ||||||||
| Medical History | ||||||||
|
| ||||||||
| Maternal Atopy | No | 24 | 0.05 | 0.002 | 1472.5 | 0.17 | 3079.3 | 0.87 |
| Yes | 28 | 0.62 | 1893.9 | 2011.4 | ||||
|
| ||||||||
| Prenatal Antibiotic Use | No | 28 | 0.19 | 0.73 | 1605.8 | 0.089 | 2549.6 | 0.12 |
| Yes | 22 | 0.21 | 1990.7 | 3985.0 | ||||
|
| ||||||||
| Prenatal Antifungal Use | No | 41 | 0.21 | 0.47 | 1913.9 | 0.28 | 3083.5 | 0.24 |
| Yes | 9 | 0.21 | 1164.9 | 1378.7 | ||||
|
| ||||||||
| Pregnancy and Delivery | ||||||||
|
| ||||||||
| Season of Birth | Winter | 17 | 0.16 | 0.45 | 1970.3 | 0.42 | 2851.0 | 0.44 |
| Spring | 14 | 0.05 | 1282.0 | 2021.9 | ||||
| Summer | 11 | 0.21 | 2123.3 | 3388.9 | ||||
| Fall | 10 | 0.72 | 1758.1 | 4034.8 | ||||
|
| ||||||||
| Mode of Delivery | Vaginal | 34 | 0.21 | 0.84 | 1805.6 | 0.82 | 2667.1 | 0.95 |
| C-Section | 18 | 0.21 | 1788.5 | 3376.4 | ||||
|
| ||||||||
| Gender | Male | 27 | 0.21 | 0.89 | 1604.9 | 0.090 | 2248.2 | 0.34 |
| Female | 25 | 0.21 | 2049.6 | 3083.5 | ||||
|
| ||||||||
| First Born Child | No | 29 | 0.21 | 0.64 | 2003.1 | 0.23 | 3289.7 | 0.35 |
| Yes | 23 | 0.21 | 1606.8 | 1959.0 | ||||
|
| ||||||||
| Maternal Age at BirthƗ | --- | 52 | −0.18 | 0.20 | −0.25 | 0.075 | −0.35 | 0.012 |
|
| ||||||||
| Gestational AgeƗ | --- | 52 | 0.05 | 0.71 | −0.16 | 0.26 | −0.19 | 0.18 |
|
| ||||||||
| Birthweight Z-ScoreƗ | --- | 49 | 0.03 | 0.86 | −0.08 | 0.61 | −0.07 | 0.62 |
|
| ||||||||
| Environment | ||||||||
|
| ||||||||
| Prenatal Household Smoke Exposure | No | 43 | 0.21 | 0.89 | 1786.1 | 0.21 | 2248.2 | 0.093 |
| Yes | 9 | 0.21 | 2107.4 | 5535.8 | ||||
|
| ||||||||
| Prenatal Indoor Pets | No | 26 | 0.21 | 0.68 | 1849.6 | 0.53 | 3079.3 | 0.42 |
| Yes | 26 | 0.19 | 1738.5 | 2457.4 | ||||
|
| ||||||||
| Breast Milk and Feeding | ||||||||
|
| ||||||||
| Exclusive Breastfeeding at 1-Month Interview | No | 39 | 0.21 | 0.40 | 1913.9 | 0.10 | 3289.7 | 0.34 |
| Yes | 13 | 0.16 | 1522.9 | 2105.7 | ||||
|
| ||||||||
| Days Postpartum at Milk CollectionƗ | --- | 52 | −0.38 | 0.005 | 0.13 | 0.38 | 0.15 | 0.30 |
Wilcoxon rank sum, Kruskal-Wallis, or Spearman correlation p-value, as appropriate.
Continuous variable: spearman correlation rather than median.
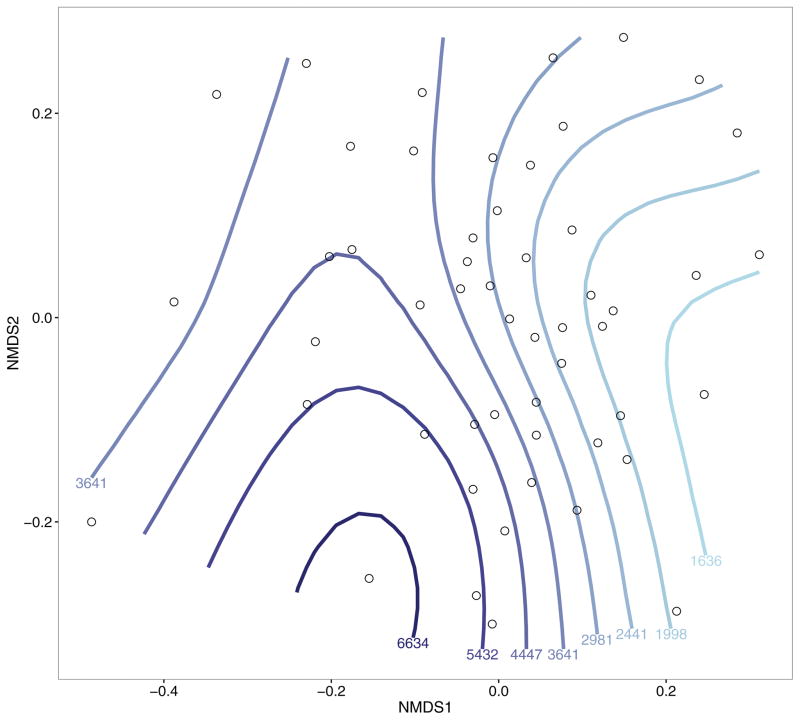
Breast milk IL-10 was not univariately (Unweighted UniFrac: R2=0.017, p-value=0.84) nor multivariately associated with neonatal gut composition (Table 2). However, individually, concentrations in breast milk of both TGFβ1 and TGFβ2 were significantly associated with gut microbial composition in breastfed neonates, with TGFβ1 explaining 2.4% of the microbial variability (p=0.041) and TGFβ2 explaining 2.6% (p=0.012). An nMDS ordination of Unweighted UniFrac distances with fitted smooth curves for TGFβ2 provided further support for this association (Fig. 1), which was primarily explained by the first axis. Specifically, mothers of neonates positively loaded on the first axis tended to have lower levels of breast milk TGFβ2 (Spearman r=−0.38; p=0.005), as reflected by the vertical smooth curves in Figure 1. When both TGFβ1 and TGFβ2 were included in a multivariate PERMANOVA model, each individual effect was no longer significant, demonstrating that their effects are not independent of one another. This result was somewhat expected, given that these two cytokines are highly correlated with one another (Spearman r=0.80, p<0.001). After adjusting for several potential confounders (Table 2), the association between TGFβ1 and neonatal gut composition was diminished, with the association being most influenced by maternal race (13% drop in R2) and marital status (8% drop in R2). However, the association with TGFβ2 was not substantially affected by covariate adjustment.
Table 2.
Association between breast milk cytokines and neonatal gut microbiome composition.
| Adjustment Variable | IL-10 | TGFβ1 | TGFβ2 |
|---|---|---|---|
|
| |||
| PERMANOVAR2 (p-value) | |||
|
|
|||
| (Unadjusted)* | 0.017 (0.84) | 0.024 (0.041)‡ | 0.026 (0.012) |
|
| |||
| Maternal EducationƗ | 0.017 (0.86) | 0.024 (0.040) | 0.026 (0.014) |
|
| |||
| Household IncomeƗ | 0.016 (0.97) | 0.024 (0.041) | 0.026 (0.009) |
|
| |||
| MarriedƗ | 0.018 (0.65) | 0.022 (0.11) | 0.025 (0.023) |
|
| |||
| Urban ResidenceƗ | 0.017 (0.85) | 0.023 (0.10) | 0.025 (0.026) |
|
| |||
| Maternal RaceƗ | 0.018 (0.81) | 0.021 (0.25) | 0.024 (0.043) |
|
| |||
| Maternal AtopyƗ | 0.017 (0.93) | 0.024 (0.034) | 0.026 (0.014) |
|
| |||
| Prenatal Antibiotic UseƗ | 0.018 (0.78) | 0.024 (0.079) | 0.025 (0.040) |
|
| |||
| Prenatal Antifungal UseƗ | 0.019 (0.76) | 0.023 (0.13) | 0.025 (0.043) |
|
| |||
| Season of BirthƗ | 0.017 (0.90) | 0.023 (0.071) | 0.025 (0.021) |
|
| |||
| Mode of DeliveryƗ | 0.018 (0.78) | 0.024 (0.031) | 0.026 (0.010) |
|
| |||
| GenderƗ | 0.017 (0.84) | 0.023 (0.057) | 0.025 (0.018) |
|
| |||
| First Born ChildƗ | 0.017 (0.91) | 0.024 (0.044) | 0.026 (0.014) |
|
| |||
| Prenatal Household Smoke ExposureƗ | 0.017 (0.85) | 0.023 (0.063) | 0.025 (0.026) |
|
| |||
| Prenatal Indoor PetsƗ | 0.018 (0.70) | 0.025 (0.025) | 0.026 (0.008) |
|
| |||
| Exclusive Breastfeeding at 1-Month InterviewƗ | 0.018 (0.72) | 0.022 (0.16) | 0.024 (0.046) |
|
| |||
| Maternal Age at BirthƗ | 0.018 (0.72) | 0.023 (0.078) | 0.025 (0.026) |
|
| |||
| Gestational AgeƗ | 0.017 (0.86) | 0.024 (0.051) | 0.025 (0.017) |
|
| |||
| Birthweight Z-ScoreƗ | 0.02 (0.61) | 0.026 (0.035) | 0.028 (0.014) |
|
| |||
| Days Postpartum at Milk Collection | 0.018 (0.68) | 0.024 (0.052) | 0.025 (0.015) |
Association between each breast milk cytokine and gut microbiome composition, unadjusted
Association between each breast milk cytokine and gut microbiome composition, adjusted for specified covariate
p<0.05 in bold
Figure 1.
Non-metric multidimensional scaling (nMDS) plot of Unweighted UniFrac distances, with contour lines showing breast milk TGFβ2 levels. Values below contour lines show fitted TGFβ2 concentrations (pg/mL) for each line, and lines are colored by these values (lighter blue: lower TGFβ2 levels; darker blue: higher TGFβ2 levels).
Examining the association between breast milk cytokines and gut microbial alpha diversity metrics (Supplemental Table 1), higher concentrations of TGFβ2 were consistently associated with higher gut richness (number of taxa detected; r=0.35, p-value=0.011), evenness (taxon distribution; r=0.36, p-value=0.008), and phylogenetic diversity (r=0.33, p-value=0.016). Similar associations were found for TGFβ1, though effect sizes were smaller and often diminished after covariate adjustment. IL-10 was not associated with these metrics in either unadjusted or adjusted models.
Given the low rates of exclusive breastfeeding in this socioeconomically and racially diverse population (25% in the analytic sample), it was of interest to determine if the association between breast milk cytokines and neonatal gut microbial composition was dependent upon frequency of breastfeeding. For each of the three breast milk cytokines, interactions were non-significant (all p>0.44). Though these differences were not statistically significant, effect sizes were consistent with a dose-response relationship, where IL-10, TGFβ1, and TGFβ2 explained a greater proportion of the variation among exclusively breastfed neonates compared to partially breastfed neonates (among exclusively breastfed neonates: R2=0.086, 0.067, 0.083; among partially breastfed neonates: R2=0.024, 0.030, 0.030, respectively).
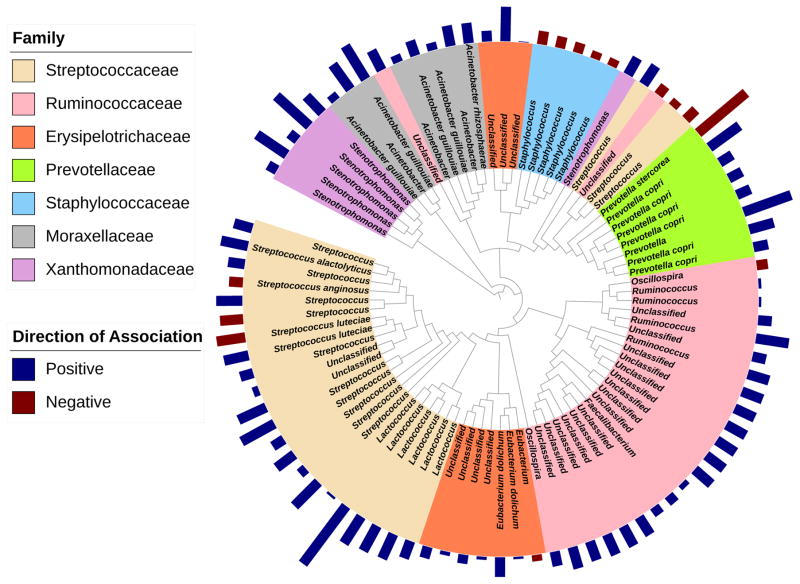
Given that breast milk TGFβ2 appeared to be more strongly associated with neonatal gut microbial composition than TGFβ1 (both in terms of effect size and persistence after covariate adjustment) and because the effects of TGFβ1 and TGFβ2 were not independent, taxa-specific tests were conducted to identify those associated with TGFβ2. We identified a total of 700 taxa significantly associated with breast milk TGFβ2, the majority of which were positively associated (428 (61%)). Neonates of mothers with higher concentrations of TGFβ2 were enriched with members of Ruminococcaceae (22 positive of 24 significant=22/24), including Ruminococcus and Faecalibacterium, Streptococcaceae (20/25), including Streptococcus and Lactococcus, Moraxellaceae (8/8), including Acinetobacter, Erysipelotrichaceae (9/10), Xanthomonadaceae (6/6), and Prevotellaceae (7/8), and were depleted of multiple Staphylococcus taxa (5 negative of 5 significant=5/5; Fig. 2, Supplemental Table 2). Using PICRUSt, several metabolic functions were predicted to be enriched in TGFβ2-associated microbiota, including pathways involved in fatty acid elongation, as well as steroid and flavonoid biosynthesis, among others (Supplemental Table 3).
Figure 2.
Taxa of select families significantly associated with TGFβ2 (FDR adjusted p-value<0.05). Bar lengths correspond to regression model coefficients, colored by direction of association, and represent the effect size of the association between each taxon and TGFβ2.
Discussion
Though it is well-established that gut microbiota of breastfed neonates are distinct from gut microbiota of formula-fed neonates (2, 9, 10), it is less clear if bioactive components of breast milk further differentiate neonatal gut microbial composition amongst breastfed neonates. In the current study, we demonstrated that breast milk TGFβ2 was significantly associated with neonatal gut bacterial composition among breastfed neonates and explained 2.6% of its variation. This is a fairly large effect size considering that neonatal composition is highly host-specific, and that current breastfeeding alone explains 2.1% of the variation among our larger cohort of 130 neonates (from which our analytic sample of 52 was drawn), using the same metric and statistical testing procedure (8). However, only 25% of mothers included were exclusively breastfeeding, which is particularly relevant to understanding the true magnitude of the effect of TGFβ2 on gut microbiota.
There are several factors that complicate our ability to analyze the “dose” of TGFβ among partially breastfed neonates. We did not collect information on the total daily intake of formula per day and did not attempt to assess the quantity of breast milk consumed per day. The quantity of biologically active TGFβ in formula consumed by each infant was also not measured. However, studies have suggested that bovine milk (which is most commonly used in infant formulas) is not identical to human milk in terms of TGFβ composition. Though TGFβ1 and 2 are present in fairly high levels in both humans and cows, the dominant isoform varies between the two species, and most TGFβ in bovine colostrum is latent, while approximately half of TGFβ in human colostrum is active (28). Whether TGFβ is latent or active is particularly important in the context of neonates, as newborn guts are generally not acidic enough to cause activation (28). Additionally, manufacturing formula from bovine milk can affect the retention and stability of bioactive molecules (29), including TGFβ, which some studies have failed to detect in infant formula (30). Further, a study comparing bovine colostrum to bovine infant formula demonstrated that TGFβ2 was abundant in colostrum but negligible in infant formula, and these differences were also reflected in the guts of preterm pigs fed these two diets (31). Due to these dissimilarities, we performed an additional analysis stratified by exclusive versus partial breastfeeding, and found that TGFβ2 explained more variation in gut microbiota composition among exclusively breastfed neonates compared to partially breastfed neonates, though these differences were not statistically significant. Future studies seeking to disentangle these more detailed aspects of exclusive versus partial breastfeeding should ensure statistical power is adequate for subgroup analyses.
A major strength of our study is that conclusions are drawn from a population-based, racially diverse birth cohort representative of the general population living in and around Detroit (e.g., not selected based on parental history of disease). Additionally, adjusted analyses were able to be performed with a wide range of sociocultural factors that may be associated with maternal breast milk constitution, which determined that the TGFβ2/neonatal gut composition was independent of these factors. Notably, though previous studies have reported a decrease in TGFβ levels over time (28), we failed to detect an association between time of milk collection and TGFβ levels, and adjusting for time did not meaningfully alter results. However, this may be due to the relatively restricted age range we observed (mean=36 days, IQR=17 days). Nonetheless, additional information would be gained from studies examining longitudinal breast milk samples with respect to infant gut microbiota.
A natural next step in understanding the association between breast milk TGFβ2 and neonatal gut microbial composition is the mechanism, or more specifically, whether TGFβ2 acts independently to impact gut microbial composition, jointly with other bioactive components, or is a biomarker correlated with an underlying causal breast milk constituent, such as breast milk bacteria or specific oligosaccharides. For example, the fact that Streptococcus are some of the most abundant bacteria in colostrum and mature milk (32) makes it difficult to determine in an observational setting if breast milk TGFβ2 is a marker for breast milk Streptococcus that are transferred to the neonatal gut, or if it alters neonatal gut Streptococcus abundance independently. However, to our knowledge, no studies have detected Ruminococcus in breast milk, making a simple bacterial transfer hypothesis unlikely in this particular case. However, a plausible alternative hypothesis is that breast milk TGFβ2 correlates with breast milk oligosaccharides, which have been shown to affect the presence of Ruminococcaceae in guts of breastfed infants (33). Given the limited number of breast milk components measured in the current analysis, further studies are needed to determine how other breast milk cytokines (TNFα and IFNγ, for example) (13), as well as a broader array of milk components (oligosaccharides, lactoferrin, microbes, etc.), interact with one another and offspring gut microbiota.
TGFβ2 may also play more of a causative role in the association with neonatal gut composition by facilitating the creation of a non-inflammatory environment that favors particular microbes; TGFβ2 can directly inhibit inflammation in immature intestinal epithelia (34). TGFβ has also been shown to decrease gut permeability by increasing the expression of tight junction proteins (35), which may improve the epithelial colonization landscape in the developing gut for specific bacteria (36). Further, TGFβ2 has been shown to act in synergy with lipopolysaccharide to stimulate epithelial repair mechanisms in pig intestines that are necessary to maintain homeostasis during gut colonization (31). Additionally, intestinal homeostasis could be induced and maintained by TGFβ2 induction of intestinal regulatory T cells (1). So, through direct or indirect mechanisms of TGFβ2, a non-inflammatory environment could be established in the infant gut that creates and maintains a gut microbiome that is more balanced and diverse (37). Further multidisciplinary investigations are required to explore these complex relationships in order to understand biological mechanisms.
Despite the observational nature of our study, we found that microbial composition in very early life varies by levels of TGFβ1 and TGFβ2, during the critical period when microbial succession occurs in parallel with immune system education and metabolic programming. Further, TGFβ2-associated bacteria were involved in a wide range of metabolic functions, including steroid and flavonoid biosynthesis, which may contribute to metabolic profile differentiation induced by breastfeeding compared to formula-feeding (38). Additionally, given that several studies have found a protective effect of TGFβ1 and TGFβ2 against atopic disorders (19)—consistent with what we have previously found in this cohort (39)—it is conceivable that neonatal gut microbiota may partially mediate this association. Interestingly, a recent case-control study found that Streptococcus and Prevotella were increased in abundance in the guts of healthy controls compared to food-sensitized children (40). Further, a recent population-based birth cohort study found that Ruminococcaceae were protective against food sensitization at 1 year of age (41). In this cohort, we observed that Faecalibacterium, Lactococcus and Acinetobacter were enriched in healthy neonates at significantly reduced risk of developing childhood allergic sensitization and asthma (6). Given our findings that breast milk TGFβ2 is associated with the increased abundance of these specific taxa in the neonatal gut, this provides further support for a mediation hypothesis. Finally, because we observed that one aspect of breast milk constitution has a significant association with early life microbial colonization patterns, which have been shown to alter the risk of immunological diseases later in life, our findings suggest a possible explanation for the inconsistent epidemiological reports regarding the effect of breastfeeding on atopic disorders later in life (42), and highlights the need to better sub-classify breastfed neonates based on maternal milk content.
In summary, breast milk TGFβ2 is associated with bacterial composition within the breastfed neonatal gastrointestinal tract, indicating that further gut microbiome variability can be explained beyond breastfeeding status through the examination of maternal breast milk constitution. Though there is a conceivable direct link between breast milk TGFβ and intestinal microbes, additional studies are needed to understand the underlying mechanisms. These findings may contribute to an improved comprehension of how breastfeeding affects immune development in early life, and adds valuable information in terms of characterizing the composition of neonatal gut microbial communities.
Supplementary Material
Correlation between breast milk cytokines and neonatal gut microbiome alpha diversity measures.
Taxa significantly associated with TGFβ2 (FDR adjusted p-value<0.05). Taxa are ordered by direction of association (red: negative, blue: positive) and significance, within family.
PICRUSt-predicted KEGG pathways significantly associated with TGFβ2 (FDR adjusted p-value<0.05). Pathways are ordered by direction of association and effect size.
What is Known
Gastrointestinal microbiome composition in early life is foundational to human health
Breastfeeding is one of the most influential factors affecting early life gut microbiome development
What is New
Breast milk cytokines TGFβ1 and TGFβ2 partially explain variation in gut microbial composition among breastfed neonates
High concentrations of TGFβ2 are associated with a more rich, even, and diverse microbiome, and the increased abundance of several bacteria: primarily members of Streptococcaceae and Ruminococcaceae
Acknowledgments
Source of Funding: The MAAP study and funding for breast milk cytokine analysis were both supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (P01 AI089473; A1080066-A101, respectively). The WHEALS study was supported by the National Institutes of Health (R01 AI050681; R01 HL-113010). This work was also supported by the Fund for Henry Ford Hospital.
We would like to acknowledge the continued dedicated participation of the WHEALS families in this long-standing birth cohort study. The MAAP study and funding for breast milk cytokine analysis were both supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (P01 AI089473; A1080066-A101, respectively). The WHEALS study was supported by the National Institutes of Health (R01 AI050681; R01 HL-113010). This work was also supported by the Fund for Henry Ford Hospital.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Author Contributions
A.R.S. performed statistical analyses and wrote the manuscript. A.R.S., S.L.H., and C.C.J. conceived the research. K.E.F. and S.V.L. performed 16S rRNA gene sequencing. K.R.B. and K.J.W. performed cytokine assays. K.E.F., S.V.L., K.R.B., and K.J.W. assisted in interpretation of results. D.R.O., G.W., S.L.H., E.M.Z., C.L.M.J., and C.C.J. were responsible for the conduct and design of the parent studies. H.K., E.M.Z., A.M.L., and G.W. provided conceptual advice. All authors critically reviewed and edited the manuscript. C.C.J. guided the project.
Data and Materials Availability
16S sequence reads were deposited to the European Bioinformatics Institute (EBI) with accession number PRJEB13896 (http://www.ebi.ac.uk/ena/data/view/PRJEB13896).
References
- 1.Maynard CL, Elson CO, Hatton RD, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–41. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185(5):385–94. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature Reviews Immunology. 2004;4(6):478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 4.Wopereis H, Oozeer R, Knipping K, et al. The first thousand days–intestinal microbiology of early life: establishing a symbiosis. Pediatric Allergy and Immunology. 2014;25(5):428–38. doi: 10.1111/pai.12232. [DOI] [PubMed] [Google Scholar]
- 5.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nature Medicine. 2016 doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller NT, Bakacs E, Combellick J, et al. The infant microbiome development: mom matters. Trends in molecular medicine. 2015;21(2):109–17. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin AM, Sitarik AR, Havstad SL, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Scientific Reports. 2016;6(31775) doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 10.Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51(1):77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 11.Andreas NJ, Kampmann B, Le-Doare KM. Human breast milk: A review on its composition and bioactivity. Early human development. 2015;91(11):629–35. doi: 10.1016/j.earlhumdev.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Munblit D, Sheth S, Abrol P, et al. Exposures influencing total IgA level in colostrum. Journal of developmental origins of health and disease. 2016;7(01):61–67. doi: 10.1017/S2040174415001476. [DOI] [PubMed] [Google Scholar]
- 13.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatric Clinics of North America. 2013;60(1):49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munblit D, Boyle R, Warner J. Factors affecting breast milk composition and potential consequences for development of the allergic phenotype. Clinical & Experimental Allergy. 2015;45(3):583–601. doi: 10.1111/cea.12381. [DOI] [PubMed] [Google Scholar]
- 15.Kühn R, Löhler J, Rennick D, et al. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 16.Glocker EO, Kotlarz D, Klein C, et al. IL-10 and IL-10 receptor defects in humans. Annals of the New York Academy of Sciences. 2011;1246(1):102–07. doi: 10.1111/j.1749-6632.2011.06339.x. [DOI] [PubMed] [Google Scholar]
- 17.Banchereau J, Pascual V, O’Garra A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nature immunology. 2012;13(10):925–31. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa J, Sasahara A, Yoshida T, et al. Role of transforming growth factor-β in breast milk for initiation of IgA production in newborn infants. Early human development. 2004;77(1):67–75. doi: 10.1016/j.earlhumdev.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Oddy WH, Rosales F. A systematic review of the importance of milk TGF-β on immunological outcomes in the infant and young child. Pediatric Allergy and Immunology. 2010;21(1-Part-I):47–59. doi: 10.1111/j.1399-3038.2009.00913.x. [DOI] [PubMed] [Google Scholar]
- 20.Aichbhaumik N, Zoratti EM, Strickler R, et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin Exp Allergy. 2008;38(11):1787–94. doi: 10.1111/j.1365-2222.2008.03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–24. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.vegan: Community Ecology Package [Computer Program]. Version R package version 2.0–10. 2013.
- 24.Lozupone C, Lladser ME, Knights D, et al. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- 26.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016:gkw290. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31(9):814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CHUN S-K, NAM M-S, GOH J-S, et al. Kinetics and biological function of transforming growth factor-β isoforms in bovine and human colostrum. Journal of microbiology and biotechnology. 2004;14(6):1267–74. [Google Scholar]
- 29.Li Y, Jensen ML, Chatterton DE, et al. Raw bovine milk improves gut responses to feeding relative to infant formula in preterm piglets. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2014;306(1):G81–G90. doi: 10.1152/ajpgi.00255.2013. [DOI] [PubMed] [Google Scholar]
- 30.Penttila I. Effects of transforming growth factor-beta and formula feeding on systemic immune responses to dietary β-lactoglobulin in allergy-prone rats. Pediatric research. 2006;59(5):650–55. doi: 10.1203/01.pdr.0000203149.75465.74. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen DN, Sangild PT, Østergaard MV, et al. Transforming growth factor-β2 and endotoxin interact to regulate homeostasis via interleukin-8 levels in the immature intestine. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2014;307(7):G689–G99. doi: 10.1152/ajpgi.00193.2014. [DOI] [PubMed] [Google Scholar]
- 32.Fernández L, Langa S, Martín V, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacological Research. 2013;69(1):1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Coppa GV, Gabrielli O, Zampini L, et al. Oligosaccharides in 4 different milk groups, Bifidobacteria, and Ruminococcus obeum. Journal of pediatric gastroenterology and nutrition. 2011;53(1):80–87. doi: 10.1097/MPG.0b013e3182073103. [DOI] [PubMed] [Google Scholar]
- 34.Rautava S, Nanthakumar NN, Dubert-Ferrandon A, et al. Breast milk-transforming growth factor-β2 specifically attenuates IL-1β-induced inflammatory responses in the immature human intestine via an SMAD6-and ERK-dependent mechanism. Neonatology. 2011;99(3):192–201. doi: 10.1159/000314109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hering NA, Andres S, Fromm A, et al. Transforming growth factor-β, a whey protein component, strengthens the intestinal barrier by upregulating claudin-4 in HT-29/B6 cells. The Journal of nutrition. 2011;141(5):783–89. doi: 10.3945/jn.110.137588. [DOI] [PubMed] [Google Scholar]
- 36.Arrieta M, Bistritz L, Meddings J. Alterations in intestinal permeability. Gut. 2006;55(10):1512–20. doi: 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamoto S, Maruya M, Kato LM, et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41(1):152–65. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Hellmuth C, Uhl O, Kirchberg FF, et al. Preventive Aspects of Early Nutrition. Karger Publishers; 2016. Effects of Early Nutrition on the Infant Metabolome; pp. 89–100. [DOI] [PubMed] [Google Scholar]
- 39.Joseph CL, Havstad S, Bobbitt K, et al. Transforming growth factor beta (TGFbeta1) in breast milk and indicators of infant atopy in a birth cohort. Pediatr Allergy Immunol. 2014;25(3):257–63. doi: 10.1111/pai.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CC, Chen KJ, Kong MS, et al. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatric Allergy and Immunology. 2016 doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 41.Azad MB, Konya T, Guttman D, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clinical & Experimental Allergy. 2015;45(3):632–43. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 42.Matheson M, Allen K, Tang M. Understanding the evidence for and against the role of breastfeeding in allergy prevention. Clinical & Experimental Allergy. 2012;42(6):827–51. doi: 10.1111/j.1365-2222.2011.03925.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between breast milk cytokines and neonatal gut microbiome alpha diversity measures.
Taxa significantly associated with TGFβ2 (FDR adjusted p-value<0.05). Taxa are ordered by direction of association (red: negative, blue: positive) and significance, within family.
PICRUSt-predicted KEGG pathways significantly associated with TGFβ2 (FDR adjusted p-value<0.05). Pathways are ordered by direction of association and effect size.