Abstract
Background
A small proportion of HIV-infected patients remain clinically and/or immunologically stable for years, including elite controllers (ECs) who have undetectable viremia (<50 copies/ml) and long-term nonprogressors (LTNPs) who maintain normal CD4+ T cell counts for prolonged periods (>10 years). However, the mechanism of nonprogression needs to be further resolved. In this study, a transcriptome meta-analysis was performed on nonprogressor and progressor microarray data to identify differential transcriptome pathways and potential biomarkers.
Methods
Using the INMEX (integrative meta-analysis of expression data) program, we performed the meta-analysis to identify consistently differentially expressed genes (DEGs) in nonprogressors and further performed functional interpretation (gene ontology analysis and pathway analysis) of the DEGs identified in the meta-analysis. Five microarray datasets (81 cases and 98 controls in total), including whole blood, CD4+ and CD8+ T cells, were collected for meta-analysis.
Results
We determined that nonprogressors have reduced expression of important interferon-stimulated genes (ISGs), CD38, lymphocyte activation gene 3 (LAG-3) in whole blood, CD4+ and CD8+ T cells. Gene ontology (GO) analysis showed a significant enrichment in DEGs that function in the type I interferon signaling pathway. Upregulated pathways, including the PI3K-Akt signaling pathway in whole blood, cytokine–cytokine receptor interaction in CD4+ T cells and the MAPK signaling pathway in CD8+ T cells, were identified in nonprogressors compared with progressors. In each metabolic functional category, the number of downregulated DEGs was more than the upregulated DEGs, and almost all genes were downregulated DEGs in the oxidative phosphorylation (OXPHOS) and tricarboxylic acid (TCA) cycle in the three types of samples.
Conclusions
Our transcriptomic meta-analysis provides a comprehensive evaluation of the gene expression profiles in major blood types of nonprogressors, providing new insights in the understanding of HIV pathogenesis and developing strategies to delay HIV disease progression.
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-017-1294-5) contains supplementary material, which is available to authorized users.
Keywords: HIV-1, LTNP, EC, Integrative transcriptome analyses, Microarray, Meta-analysis
Background
Disease progression in the absence of therapy varies significantly in HIV-infected individuals. Most patients experience progressive CD4+ T cell loss and develop AIDS [1]. However, a small proportion of HIV-infected patients remain clinically and/or immunologically stable for years, including long-term nonprogressors (LTNPs) who maintain normal CD4+ T cell counts for prolonged periods (>10 years) and elite controllers (ECs) who have undetectable viremia (<50 copies/ml) [2–4]. In past decades, numerous studies have been conducted searching for the cause for this lack of progression [5–7]. Among them, high-throughput techniques, such as microarray analysis, have contributed to the understanding of the complex host-virus interactions associated with the delayed disease progression in nonprogressors [8–12].
Using various microarray techniques and various types of samples, including whole blood, PBMC, CD4+ T cells, CD8+ T cells and monocytes, previous studies have provided valuable information on transcriptomic profiles in nonprogressors (including LTNPs and ECs, referred to as “nonprogressors” below). Several transcriptomic analyses of T cells have highlighted the role of reduced interferon-stimulated genes (ISGs) associated with the nonprogressing status in LTNPs and ECs [8, 10, 13]. T cell transcriptomic studies have also revealed the enhancing pathways in LTNPs, including the APK, WNT, and AKT pathways, contributing to cell survival and antiviral responses [14], cytokine–cytokine receptor interaction, a negative control of apoptosis or regulation of actin cytoskeleton [15]. A PBMC transcriptomic study demonstrated that cell death/proapoptotic genes were mostly downregulated and cell survival/antiapoptotic genes or genes belonging to the canonical Wnt/beta-catenin signaling pathway were upregulated in LTNPs [16]. A monocyte transcriptomic study revealed upregulation of Toll-like receptor (TLR) signaling with subsequent downregulation of MAPK, NF-kB, JAK-STAT, and the IRF cascades in LTNPs [17]. A transcriptomic study on whole blood from LTNPs and progressors identified a novel ISG gene, LY6E, which restrains the hyperactivation of monocytes during HIV-1 infection [11].
Although the accumulating transcriptome data provide useful information in studying the host protective immune responses in nonprogressors, the identification of key genes and pathways from these studies was restricted due to the limited sample size in the independent study. A transcriptome meta-analysis can incorporate high-throughput data from multiple independent studies and overcome the aforementioned limitations. In addition, different cells have unique functional activities in the immune responses of nonprogressors, but the differences in the transcriptomic profiles between the major cell types of nonprogressors have not been elucidated. In the present study, we performed a meta-analysis of five independent microarray datasets to identify differentially expressed genes (DEGs) in whole blood, CD4+ and CD8+ T cells from HIV-infected nonprogressors compared with progressors. Our study provides a comprehensive evaluation of the gene expression profiles in the major blood types of nonprogressors, which will provide new insights in the understanding of HIV pathogenesis and developing strategies to delay HIV-1 disease progression.
Methods
Microarray data collection
The gene expression microarray datasets with the keywords “long-term nonprogressor” and “elite controller” were downloaded from Gene Expression Omnibus (NCBI) database [18]. LTNPs or ECs were considered “case group” while progressors, including rapid progressors (RPs), chronic progressors (CPs) or normal progressors (NPs) were considered “control group”. Data sets with a sample source other than whole blood, CD4+ and CD8+ T cells from nonprogressors were excluded. Five independent microarray datasets with raw data were selected and the details about these datasets are outlined in Table 1. The following information was extracted from each of the studies that were selected: GEO accession; numbers of patients and controls; sample source; platform and gene expression data. Of the five datasets, three were conducted in Affymetrix HG U133 GeneChips while the others were performed in the Illumina beadchip platform. We compared microarray data for LTNP/ECs (n = 81) and progressors (n = 98) from the public database submissions. Two datasets included the transcriptome profiles of whole blood from 20 nonprogressors and 18 progressors. Two datasets included CD4+ T cells from 19 nonprogressors and 32 progressors. Three datasets included CD8+ T cells from 42 nonprogressors and 48 progressors.
Table 1.
Summary of transcriptome datasets used in this study
| Study | GEO accession | Sample size | Sample source | Platform | |
|---|---|---|---|---|---|
| LTNP case | Control | ||||
| 1 | GSE56837 | 15 | 11 | Whole blood | GPL6884, Illumina HumanWG-6 v3.0 expression beadchip |
| 2 | GSE57730 | 5 | 7 | Whole blood | GPL570, Affymetrix Human Genome U133 Plus 2.0 Array |
| 3 | GSE28128 | 14 | 27 | CD4+ T cell | GPL6884, Illumina HumanWG-6v3.0 expression beadchip |
| 4 | GSE28128 | 13 | 25 | CD8+ T cell | GPL6884, Illumina HumanWG-6v3.0 expression beadchip |
| 5 | GSE6740 | 5 | 5 | CD4+ T cell | GPL96, Affymetrix Human Genome U133A Array |
| 6 | GSE6740 | 5 | 5 | CD8+ T cell | GPL96, Affymetrix Human Genome U133A Array |
| 7 | GSE24081 | 24 | 18 | CD8+ T cell | GPL3921, Affymetrix HT Human Genome U133 Array |
Analysis of differential gene expression
To find DEGs in different types of samples between nonprogressors and progressors, the data collected from each eligible microarray study were imported to the INMEX (integrative meta-analysis of expression data) program (http://www.inmex.ca/INMEX/) [19] to carry out the meta-analysis. The data were annotated after converting the gene and probe IDs to the corresponding Entrez IDs. The intensity values for each probe set were log2 transformed then uploaded, processed, and annotated for data integrity. Batch effect correction option (ComBat) was used to reduce potential batch effect [20]. After a data integrity check, we carried out a meta-analysis using the combined rank orders method whose implementation is based on the RankProd package [21]. The whole process was repeated 100 times to compute the p value and false discovery rate (FDR) associated with each gene [19]. The selected genes are sorted based on the combined rank product (combined RP). The lower the combined RP value, the more significant the candidate gene is for heterologous expression [22]. Statistical analyses were performed using the INMEX program [19]. A p value <0.05 was considered statistically significant in the analysis.
Gene ontology terms and pathway enrichment
Functional interpretation (gene ontology analysis and pathway enrichment analysis) of the DEGs identified in this meta-analysis was performed with Database for Annotation, Visualization and Integrated Discovery (DAVID, Version 6.8 Beta), a web-based tool for Gene Ontology (GO) enrichment analysis (http://david.abcc.ncifcrf.gov/) [23]. Gene symbol lists were uploaded and analyzed using functional annotation chart for GO Biological Process Annotation (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway. Representative GO Biological Process terms and KEGG pathways selected from the top significantly enriched charts are reported in the figures. A Benjamini-corrected p value less than 0.05 was used to identify a statistically significant analysis.
Results
DEGs in whole blood, CD4+ and CD8+ T cells from nonprogressors
First, we identified the DEGs in whole blood from nonprogressors. According to the results of our meta-analysis, 1160 genes in whole blood were identified to be differentially expressed between nonprogressors and progressors. Among the 1160 DEGs, 215 genes were upregulated and 945 genes were downregulated. The downregulated genes included genes that have been reported to be associated with HIV-1 disease progression or pathogenesis, such as CD38 and some ISGs [11, 13, 24].
There were 854 DEGs in CD4+ T cells between nonprogressors and progressors across microarray datasets. Among the 854 DEGs, 310 genes were upregulated and 544 genes were downregulated. The downregulated genes included ISGs, such as IFI44L, MX1 and IFI27.
There were 1319 genes in CD8+ T cells identified to be differentially expressed between nonprogressors and progressors across microarray datasets. Among the 1319 DEGs, 524 genes were upregulated and 795 genes were downregulated. The downregulated genes also included CD38 and some ISGs. The top 20 most significantly upregulated and downregulated DEGs in different types of cells in the comparison of nonprogressors and progressors are shown in Additional file 1: Table S1A–C. The DEGs in different types of cells are shown in Additional file 2: Table S2A–C.
Second, we investigated whether some of the genes were shared in the comparison of different types of samples between nonprogressors and progressors. The Venn diagram showed that 175 DEGs were significantly altered in all three sample types. Among them, ten upregulated DEGs and 107 downregulated DEGs showed the same variation trend in all the three sample types (Fig. 1; Additional file 3: Table S3A and B). Ten upregulated DEGs and the top 20 most significantly downregulated DEGs shared in the three sample types are shown in Table 2. The upregulated DEGs included CCR7, and the downregulated DEGs involved some important ISGs, CD38 and co-inhibitory molecule LAG-3.
Fig. 1.
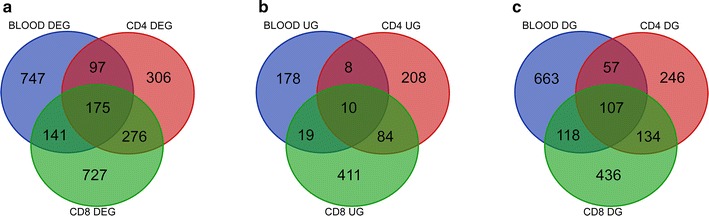
Venn diagram representing the number of DEGs in nonprogressors compared with progressors in whole blood (blue circle), CD4+ T cells (red circle) and CD8+ T cells (green circle). The overlapping area indicates DEGs identified in the three types of samples. UG upregulated genes. DG downregulated genes. a All the DEGs found in the three types of samples. b The upregulated genes found in the three types of samples. c The downregulated genes found in the three types of samples
Table 2.
The 10 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors of the three types of samples
| Entrez ID | Gene symbol | Combined rank product | Average log fold change |
|---|---|---|---|
| 10 most significantly up-regulated genes | |||
| 1236 | CCR7 | 693.97 | 0.087221 |
| 4753 | NELL2 | 1075.15 | 0.267438 |
| 23531 | MMD | 1092.11 | 0.317429 |
| 6653 | SORL1 | 1446.07 | 0.192771 |
| 54855 | FAM46C | 1718.18 | 0.210897 |
| 55544 | RBM38 | 1942.46 | 0.220304 |
| 7057 | THBS1 | 2070.14 | 0.187822 |
| 10320 | IKZF1 | 2090.63 | 0.055676 |
| 9882 | TBC1D4 | 2157.42 | 0.127719 |
| 55893 | ZNF395 | 2458.17 | 0.091132 |
| 20 most significantly down-regulated genes | |||
| 3429 | IFI27 | 35.61 | −0.948905 |
| 9636 | ISG15 | 259.82 | −0.534496 |
| 4061 | LY6E | 260.35 | −0.607156 |
| 26010 | SPATS2L | 278.23 | −0.519912 |
| 26519 | TIMM10 | 289.63 | −0.804127 |
| 10964 | IFI44L | 296.89 | −0.427484 |
| 9381 | OTOF | 360.09 | −0.464578 |
| 6036 | RNASE2 | 421.70 | −0.442343 |
| 952 | CD38 | 562.80 | −0.723494 |
| 3902 | LAG3 | 618.23 | −0.503929 |
| 644 | BLVRA | 623.86 | −0.523236 |
| 81030 | ZBP1 | 700.78 | −0.423206 |
| 3512 | IGJ | 704.39 | −0.522450 |
| 3665 | IRF7 | 762.54 | −0.566041 |
| 9997 | SCO2 | 982.28 | −0.652977 |
| 7298 | TYMS | 1192.70 | −0.491925 |
| 3965 | LGALS9 | 1449.29 | −0.420535 |
| 10549 | PRDX4 | 1557.82 | −0.435958 |
| 10287 | RGS19 | 1709.44 | −0.423876 |
| 3113 | HLA-DPA1 | 1992.11 | −0.496091 |
DEG functional classification and pathway assignment
Gene ontology analysis and KEGG pathway analysis were carried out for the functional investigation of DEGs. In the GO analysis, “type I interferon signaling pathway”, “interferon-gamma-mediated signaling pathway” and “defense response to virus” were significantly enriched for the DEGs in all three sample types (Fig. 2a–c). Additionally, the 10 most significantly enriched GO terms for DEGs in CD8+ T cells included “negative regulation of apoptotic process”, “inflammatory response” and “translational initiation” (Fig. 2c).
Fig. 2.
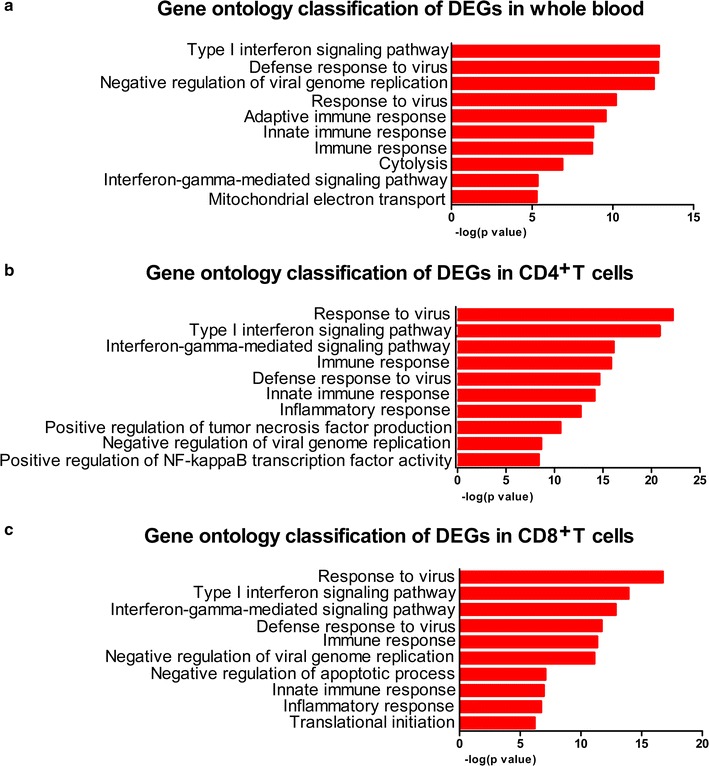
The gene ontology (GO) analysis of DEGs in nonprogressors. a The 10 most significantly enriched GO terms for whole blood. b The 10 most significantly enriched GO terms for CD4+ T cells. c The 10 most significantly enriched GO terms for CD8+ T cells. p < 0.05 and FDR < 0.01 were used as the threshold for GO analysis
In the pathway analysis, we identified the top 15 most significantly upregulated and downregulated pathways for DEGs in different sample types in the comparison between nonprogressors and progressors (Fig. 3a–f). Oxidative phosphorylation (OXPHOS) was significantly downregulated in whole blood (1 top, Fig. 3b), CD4+ T cells (10 top, Fig. 3d) and CD8+ T cells (53 top, data not shown). Antigen processing and presentation were significantly downregulated in both whole blood and CD4+ T cells. In addition, pathways including PI3K-Akt signaling pathway in whole blood, cytokine–cytokine receptor interaction and Jak-STAT signaling pathway in CD4+ T cells and MAPK signaling pathway in CD8+ T cells were identified.
Fig. 3.
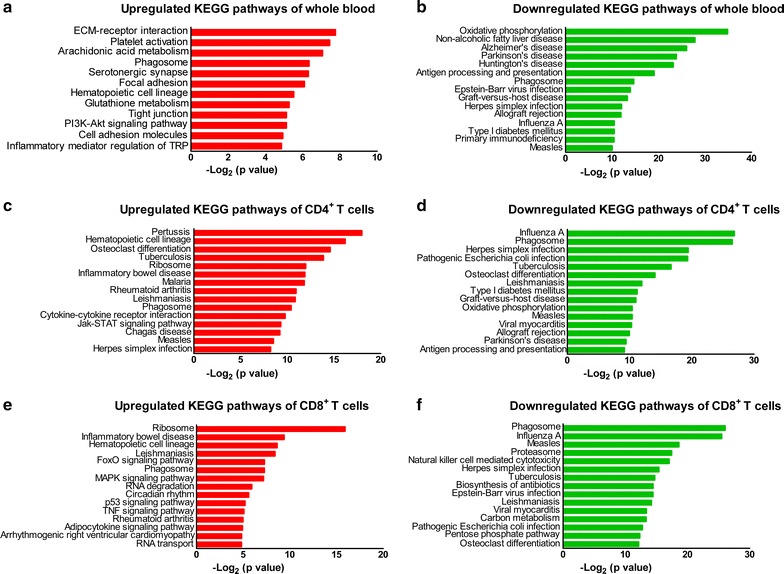
KEGG pathways of the upregulated and downregulated DEGs in nonprogressors. a, b The 15 most significantly upregulated pathways and 15 most significant downregulated pathways of whole blood. c, d The 15 most significantly upregulated and 15 most downregulated pathways of CD4+ T cells. e, f The 15 most significantly upregulated and 15 most downregulated pathways of CD8+ T cells. p < 0.05 and FDR < 0.01 were used as the threshold for KEGG pathway assignment
Metabolic pathways associated with HIV nonprogressors
It has become increasingly clear that immune function is intimately linked to metabolic programs [25, 26]. Metabolic pathways have been reported to play important roles in the pathogenesis of HIV [27, 28]. After we identified that OXPHOS was downregulated in all kinds of samples in nonprogressors compared with progressors, we then performed the GO metabolism analysis with the upregulated and downregulated DEGs to investigate the alterations of metabolic pathways in nonprogressors. We found that there are more downregulated DEGs than upregulated DEGs in “carbohydrate metabolic process”, “cellular amino acid metabolic process”, “lipid metabolic process”, “nucleobase-containing compound process”, “protein metabolic process”, “TCA cycle” and “OXPHOS” in whole blood, CD4+ and CD8+ T cells (Fig. 4a–c). Nearly all DEGs in OXPHOS and the TCA cycle were downregulated in the three kinds of samples in the comparison of nonprogressors and progressors.
Fig. 4.
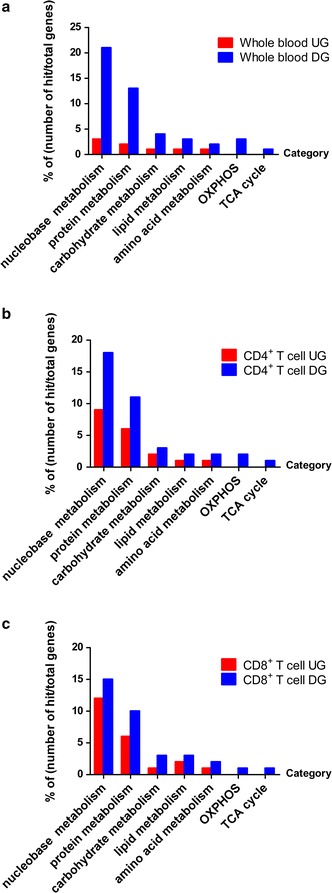
GO classification of DEGs specific to whole blood (a), CD4+ (b) and CD8+ T cells (c). A total of 1160 whole blood-specific, 854 CD4+ T cell-specific and 1319 CD8+ T cell-specific DEGs were categorized based on GO slim classification (p < 0.05). A bar represents each gene ontology category. The height of the bar represents the percentage of genes observed in each category. The percentage of genes per category is indicated upon the bars text. UG upregulated genes. DG downregulated genes
Discussion
Identification of the most relevant genes and pathways involved in HIV-infected nonprogressors is important in understanding the molecular and cellular processes determining the cause for the nonprogression status in these patients. In this study, we performed a meta-analysis of multiple public microarray datasets to investigate the transcriptomic profiles of HIV-infected nonprogressors, including LTNPs and ECs. Through transcriptomic meta-analysis, we identified DEGs in various types of samples in the comparison of nonprogressors and progressors, including whole blood, CD4+ and CD8+ T cells.
We found that the expression of multiple important ISG genes, CD38 and LAG-3 was significantly downregulated in HIV-infected nonprogressors compared with that of progressors in all the three sample types. The downregulation of important ISG genes, including LY6E, IFI27, ISG15 and IFI44L, was observed in our study. Type-1 interferons (IFNs) is of critical importance for its potent antiviral effects [29]. However, it is well established that type-1 IFNs exert their full antiviral effect at very low concentration and their expression is required locally [30, 31]. Emerging lines of evidence reveal that high level and sustained type-1 IFNs expression is associated with hyper-immune activation and disease progression in persistent infections [31–34]. Dynamics of type-1 IFNs distinguishes Simian immunodeficiency virus (SIV) infection of natural hosts, that do not develop AIDS, from pathogenic SIV infections [35–38]. Natural hosts rapidly mute their type-1 IFNs responses after acute SIV infection whereas disease-susceptible macaque species maintain type-1 IFNs signaling indefinitely, which triggers hyper-activation of immune system and contributes to provide an environment that favors progression to AIDS [29, 36–39]. As the primary producers of type-1 IFNs, plasmacytoid dendritic cells (pDCs) in natural hosts of SIV have attenuated recruitment to lymphoid tissues compared with non-natural host [39–41]. This reduces pDCs exposure to sites of high level virus replication, and limits the pathogenic T-cell activation and chronic inflammation driven by activated pDCs [39, 41, 42]. In HIV infected human, although pDCs are depleted in blood during chronic infection, they accumulate in lymph nodes [39, 41–43], which may directly contribute to triggering sustain pathogenic immune activation [41–45]. High levels of IFN-α in the serum of chronically HIV-infected patients, as well as in tonsils during both chronic and acute HIV infection were observed [46, 47], which positively correlated with markers of immune activation [48, 49]. The expression of ISGs is elevated in HIV-infected rapid progressors rather than in nonprogressors [9–11, 13, 14]. Our results indicated that reduced expression of ISGs, which leads to reduced activation of immune system, was a key factor affecting disease progression. This was confirmed by our further finding that CD38 was downregulated in HIV-infected nonprogressors. CD38 is an activation marker and a nicotinamide adenine dinucleotide consuming enzyme (NADases) [15, 50]. Previous studies showed that the elevated expression of CD38 has a strong relationship with activation and cell aging [15, 50]. Our results showed that nonprogressors have lower expression of CD38 in different types of cells, which was beneficial in maintaining the survival of the cell. In addition, we found a lower expression of LAG-3, an important coinhibitory molecule in the immune system, in HIV-infected nonprogressors. It has been reported that LAG-3 is associated with immune dysfunction/exhaustion of T cells [51]. Taken together, our data revealed that HIV-infected nonprogressors have lower expression of ISGs, CD38 and LAG-3, which was beneficial in maintaining a healthy status of the immune system and contributed to the nonprogression of the disease.
Gene ontology analysis and pathway enrichment analysis offered insight into the possible roles of DEGs in the pathogenesis of nonprogressors. Consistent with our data on DEGs, we found that the type I interferon signaling pathway was involved in the most significantly enriched terms shared by the three sample types. Furthermore, we found an upregulated PI3K-Akt signaling pathway in whole blood and MAPK pathway in CD8+ T cells in nonprogressors. Both pathways contribute to cell survival and a successful immune response depends upon the ability of T lymphocytes to respond to antigenic stimulation [14, 15, 52]. In addition, the MAPK pathway plays an important role in T cell homeostasis [53], which is required for the cytotoxic activity of most CD8+ T cells [54]. Wu et al. reported that the nonprogressing status in HIV-infected LTNPs was associated with the MAPK, WNT, and AKT pathways. Our results are consistent with their study, indicating that the upregulated signaling pathways in nonprogressors is helpful for cell survival and the development of effector functions of the immune system.
In the past several years, a wealth of evidence has emerged illustrating how metabolism supports many aspects of immune system biology [55]. The field of cellular immunometabolism has made big strides over the past decade, becoming one of the hottest areas of research in immunology [56]. Through KEGG Pathway analysis, we found OXPHOS was downregulated in all kinds of samples, including whole blood, CD4+ and CD8+ T cells. Through further analysis of metabolic pathways, we found the number of downregulated DEGs was more than upregulated DEGs in different metabolic processes, including carbohydrate metabolic process, cellular amino acid metabolic process, lipid metabolic process and protein metabolic process, and almost all the DEGs in OXPHOS and the TCA cycle were downregulated in the three sample types in nonprogressors compared with progressors. Previous studies have indicated that HIV itself and/or ART can damage the mitochondria, affecting the normal functioning of the cell [57–59]. Two recent studies showed that HIV infection caused the upregulation of components of OXPHOS, TCA cycle, amino acid metabolism, and fatty acid metabolism in human CD4+ T cell lines at the protein level, which may be compensation for the mitochondria dysfunction [60, 61]. Previous studies have demonstrated that the LTNPs had slighter mitochondrial impairment and lower frequencies of cells with decreased mitochondrial membrane potential; this correlates may result in suppression of spontaneous apoptosis and higher CD4+ T cells counts when compared to HIV infected individuals [44, 45]. Compared with viremic patients in the HAART (VIR) group, LTNP downregulated the OXPHOS pathway and the TCA cycle in CD4+ and CD8+ T cells [14, 62]. Our results revealed that nonprogressors have lower levels of the OXPHOS pathway and the TCA cycle, which may be due to the milder mitochondrial impairment in nonprogressors than progressors. Generating more effective adoptive cellular immunotherapies to rescue the abnormal metabolic profiles in progressors is valuable for delaying disease progression.
Conclusions
In summary, our meta-analysis of microarray studies provided an overview of the differential gene expression signatures in HIV-infected nonprogressors compared to progressors. We revealed that nonprogressors had reduced expression of type I interferon signaling pathway and associated genes. Downregulated DEGs in OXPHOS and the TCA cycle were also observed, which may be a compensation of mitochondrial dysfunction in HIV infection. Our meta-analysis will facilitate the understanding of unique transcriptomic profiles in HIV-infected nonprogressors and provide information for immune intervention of the disease progression of HIV.
Additional files
Additional file 1: Table S1A. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors. Table S2B. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors of CD4+ T cells. Table S3C. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors of CD8+ T cells.
Additional file 2: Table S2A. All the DEGs in nonprogressors of whole blood. Table S2B. All the DEGs in nonprogressors of CD4+ T cells. Table S3C. All the DEGs in nonprogressors of CD8+ T cells.
Additional file 3: Table S3A. All the DEGs in nonprogressors of the three types of samples. Table S3B. All the down-regulated DEGs in nonprogressors of the three types of samples.
Authors’ contributions
L-LZ and Z-NZ performed the majority of work described here. L-LZ, Z-NZ, and HS wrote and edited the manuscript. Z-NZ and HS supervised the project. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/geo/).
Consent for publication
All the authors have read and approved the paper and declare no potential conflicts of interest in the paper. If their paper is accepted, all the authors will observe the terms of the licence to publish.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by grants from the Mega-Projects of National Science Research for the 12th Five-Year Plan (2012ZX10001-006) and National Natural Science Foundation of China (81371884).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- HIV-1
human immunodeficiency virus 1
- AIDS
acquired immunodeficiency syndrome
- LTNP
long-term nonprogressor
- EC
elite controller
- NP
normal progressor
- CP
chronic progressor
- RP
rapid progressors
- ART
antiretroviral therapy
- DEGs
differentially expressed genes
- DAVID
database for annotation, visualization and integrated discovery
- GEO
gene expression omnibus
- GO
gene ontology
- KEGG
kyoto encyclopedia of genes and genomes
- LAG-3
lymphocyte activation gene 3
- ISG
interferon-stimulated gene
- ISG15
interferon-stimulated gene 15
- IFI44
interferon induced protein 44
- IFI44L
interferon induced protein 44 like
- OXPHOS
oxidative phosphorylation
- TCA
tricarboxylic acid
- IFNs
interferons
- SIV
Simian immunodeficiency virus
- pDCs
plasmacytoid dendritic cells
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-017-1294-5) contains supplementary material, which is available to authorized users.
Le-Le Zhang and Zi-Ning Zhang contributed equally to this work
Contributor Information
Le-Le Zhang, Email: le932560983@163.com.
Zi-Ning Zhang, Email: zi_ning101@hotmail.com.
Xian Wu, Email: wuxian891020@163.com.
Yong-Jun Jiang, Email: jiangjun55555@163.com.
Ya-Jing Fu, Email: fufu80s@sina.com.
Hong Shang, Phone: +86 (24) 8328-2634, Email: hongshang100@hotmail.com.
References
- 1.Carrington M, Walker BD. Immunogenetics of spontaneous control of HIV. Annu Rev Med. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27(3):406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Pantaleo G, Fauci AS. New concepts in the immunopathogenesis of HIV infection. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 4.Dyer WB, Zaunders JJ, Yuan FF, Wang B, Learmont JC, Geczy AF, et al. Mechanisms of HIV non-progression; robust and sustained CD4+ T-cell proliferative responses to p24 antigen correlate with control of viraemia and lack of disease progression after long-term transfusion-acquired HIV-1 infection. Retrovirology. 2008;5:112. doi: 10.1186/1742-4690-5-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez V, Costagliola D, Bonduelle O, N’go N, Schnuriger A, Theodorou I, et al. Combination of HIV-1-specific CD4 Th1 cell responses and IgG2 antibodies is the best predictor for persistence of long-term nonprogression. J Infect Dis. 2005;191(12):2053–2063. doi: 10.1086/430320. [DOI] [PubMed] [Google Scholar]
- 6.Petrovas C, Mueller YM, Katsikis PD. HIV-specific CD8+ T cells: serial killers condemned to die? Curr HIV Res. 2004;2(2):153–162. doi: 10.2174/1570162043484960. [DOI] [PubMed] [Google Scholar]
- 7.Pancre V, Delhem N, Yazdanpanah Y, Delanoye A, Delacre M, Depil S, et al. Presence of HIV-1 Nef specific CD4 T cell response is associated with non-progression in HIV-1 infection. Vaccine. 2007;25(31):5927–5937. doi: 10.1016/j.vaccine.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 8.Hyrcza MD, Kovacs C, Loutfy M, Halpenny R, Heisler L, Yang S, et al. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J Virol. 2007;81(7):3477–3486. doi: 10.1128/JVI.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16(10):1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, et al. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest. 2011;121(6):2391–2400. doi: 10.1172/JCI45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Qiu C, Zhu L, Huang J, Li L, Fu W, et al. IFN-stimulated gene LY6E in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the hyperactivation of monocytes during chronic HIV-1 infection. J Immunol. 2014;193(8):4125–4136. doi: 10.4049/jimmunol.1401249. [DOI] [PubMed] [Google Scholar]
- 12.Telenti A, Goldstein DB. Genomics meets HIV-1. Nat Rev Microbiol. 2006;4(11):865–873. doi: 10.1038/nrmicro1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, et al. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6(2):e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. Genome-wide analysis of primary CD4+ and CD8+ T cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology. 2011;8:18. doi: 10.1186/1742-4690-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salgado M, Lopez-Romero P, Callejas S, Lopez M, Labarga P, Dopazo A, et al. Characterization of host genetic expression patterns in HIV-infected individuals with divergent disease progression. Virology. 2011;411(1):103–112. doi: 10.1016/j.virol.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 16.Luque MC, Santos CC, Mairena EC, Wilkinson P, Boucher G, Segurado AC, et al. Gene expression profile in long-term non progressor HIV infected patients: in search of potential resistance factors. Mol Immunol. 2014;62(1):63–70. doi: 10.1016/j.molimm.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Wu JQ, Sasse TR, Wolkenstein G, Conceicao V, Saksena MM, Soedjono M, et al. Transcriptome analysis of primary monocytes shows global down-regulation of genetic networks in HIV viremic patients versus long-term non-progressors. Virology. 2013;435(2):308–319. doi: 10.1016/j.virol.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39(Database issue):D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia J, Fjell CD, Mayer ML, Pena OM, Wishart DS, Hancock RE. INMEX—a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res. 2013;41(Web Server issue):W63–W70. doi: 10.1093/nar/gkt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 21.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 22.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, RA Lempicki. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16(2):83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 25.O’Sullivan D, Pearce EL. Immunology. Expanding the role of metabolism in T cells. Science. 2015;348(6238):976–977. doi: 10.1126/science.aac4997. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mody A, Bartz S, Hornik CP, Kiyimba T, Bain J, Muehlbauer M, et al. Effects of HIV infection on the metabolic and hormonal status of children with severe acute malnutrition. PLoS ONE. 2014;9(7):e102233. doi: 10.1371/journal.pone.0102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassol E, Misra V, Holman A, Kamat A, Morgello S, Gabuzda D. Plasma metabolomics identifies lipid abnormalities linked to markers of inflammation, microbial translocation, and hepatic function in HIV patients receiving protease inhibitors. BMC Infect Dis. 2013;13:203. doi: 10.1186/1471-2334-13-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosinger SE, Utay NS. Type I interferon: understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep. 2015;12(1):41–53. doi: 10.1007/s11904-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 30.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murira A, Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbeuval JP, Shearer GM. HIV-1 immunopathogenesis: how good interferon turns bad. Clin Immunol. 2007;123(2):121–128. doi: 10.1016/j.clim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng L, Yu H, Li G, Li F, Ma J, Li J, et al. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV-1 infection. JCI Insight. 2017;2(12):e94366. [DOI] [PMC free article] [PubMed]
- 34.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32(6):737–742. doi: 10.1016/j.immuni.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science. 2012;335(6073):1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol. 2010;84(15):7886–7891. doi: 10.1128/JVI.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosinger SE, Sodora DL, Silvestri G. Generalized immune activation and innate immune responses in simian immunodeficiency virus infection. Curr Opin HIV AIDS. 2011;6(5):411–418. doi: 10.1097/COH.0b013e3283499cf6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwa S, Kannanganat S, Nigam P, Siddiqui M, Shetty RD, Armstrong W, et al. Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood. 2011;118(10):2763–2773. doi: 10.1182/blood-2011-02-339515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boasso A. HIV and DC: hate at first sight. Blood. 2010;116(19):3687–3689. doi: 10.1182/blood-2010-08-302331. [DOI] [PubMed] [Google Scholar]
- 42.Boasso A, Shearer GM. Chronic innate immune activation as a cause of HIV-1 immunopathogenesis. Clin Immunol. 2008;126(3):235–242. doi: 10.1016/j.clim.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien M, Manches O, Bhardwaj N. Plasmacytoid dendritic cells in HIV infection. Adv Exp Med Biol. 2013;762:71–107. doi: 10.1007/978-1-4614-4433-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boasso A, Hardy AW, Anderson SA, Dolan MJ, Shearer GM. HIV-induced type I interferon and tryptophan catabolism drive T cell dysfunction despite phenotypic activation. PLoS ONE. 2008;3(8):e2961. doi: 10.1371/journal.pone.0002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald-Bocarsly P, Jacobs ES. Plasmacytoid dendritic cells in HIV infection: striking a delicate balance. J Leukoc Biol. 2010;87(4):609–620. doi: 10.1189/jlb.0909635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbeuval JP, Nilsson J, Boasso A, Hardy AW, Kruhlak MJ, Anderson SA, et al. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc Natl Acad Sci USA. 2006;103(18):7000–7005. doi: 10.1073/pnas.0600363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tilton JC, Johnson AJ, Luskin MR, Manion MM, Yang J, Adelsberger JW, et al. Diminished production of monocyte proinflammatory cytokines during human immunodeficiency virus viremia is mediated by type I interferons. J Virol. 2006;80(23):11486–11497. doi: 10.1128/JVI.00324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinson JA, Montoya CJ, Usuga X, Ronquillo R, Landay AL, Desai SN. Chloroquine modulates HIV-1-induced plasmacytoid dendritic cell alpha interferon: implication for T-cell activation. Antimicrob Agents Chemother. 2010;54(2):871–881. doi: 10.1128/AAC.01246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, et al. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS ONE. 2013;8(2):e56527. doi: 10.1371/journal.pone.0056527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camacho-Pereira J, Tarrago MG, Chini CC, Nin V, Escande C, Warner GM, et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016;23(6):1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith KA, Lachman LB, Oppenheim JJ, Favata MF. The functional relationship of the interleukins. J Exp Med. 1980;151(6):1551–1556. doi: 10.1084/jem.151.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merritt C, Enslen H, Diehl N, Conze D, Davis RJ, Rincon M. Activation of p38 mitogen-activated protein kinase in vivo selectively induces apoptosis of CD8(+) but not CD4(+) T cells. Mol Cell Biol. 2000;20(3):936–946. doi: 10.1128/MCB.20.3.936-946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilic M, Kulig K, Messaoudi I, Remus K, Jankovic M, Nikolic-Zugic J, et al. CD8(+) T cell cytolytic activity independent of mitogen-activated protein kinase/extracellular regulatory kinase signaling (MAP kinase/ERK) Eur J Immunol. 1999;29(12):3971–3977. doi: 10.1002/(SICI)1521-4141(199912)29:12<3971::AID-IMMU3971>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 55.O’Sullivan D, Pearce EL. Targeting T cell metabolism for therapy. Trends Immunol. 2015;36(2):71–80. doi: 10.1016/j.it.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medzhitov R. Bringing Warburg to lymphocytes. Nat Rev Immunol. 2015;15(10):598. doi: 10.1038/nri3918. [DOI] [PubMed] [Google Scholar]
- 57.Cossarizza A, Mussini C, Mongiardo N, Borghi V, Sabbatini A, De Rienzo B, et al. Mitochondria alterations and dramatic tendency to undergo apoptosis in peripheral blood lymphocytes during acute HIV syndrome. AIDS. 1997;11(1):19–26. doi: 10.1097/00002030-199701000-00004. [DOI] [PubMed] [Google Scholar]
- 58.Feeney ER, Mallon PW. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr Pharm Des. 2010;16(30):3339–3351. doi: 10.2174/138161210793563482. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Matute P, Perez-Martinez L, Blanco JR, Oteo JA. Role of mitochondria in HIV infection and associated metabolic disorders: focus on nonalcoholic fatty liver disease and lipodystrophy syndrome. Oxid Med Cell Longev. 2013;2013:493413. doi: 10.1155/2013/493413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan EY, Qian WJ, Diamond DL, Liu T, Gritsenko MA, Monroe ME, et al. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J Virol. 2007;81(14):7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ringrose JH, Jeeninga RE, Berkhout B, Speijer D. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol. 2008;82(9):4320–4330. doi: 10.1128/JVI.01819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. Transcriptional profiles in CD8+ T cells from HIV+ progressors on HAART are characterized by coordinated up-regulation of oxidative phosphorylation enzymes and interferon responses. Virology. 2008;380(1):124–135. doi: 10.1016/j.virol.2008.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1A. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors. Table S2B. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors of CD4+ T cells. Table S3C. The 20 most significantly up-regulated and 20 most significantly down-regulated genes in nonprogressors of CD8+ T cells.
Additional file 2: Table S2A. All the DEGs in nonprogressors of whole blood. Table S2B. All the DEGs in nonprogressors of CD4+ T cells. Table S3C. All the DEGs in nonprogressors of CD8+ T cells.
Additional file 3: Table S3A. All the DEGs in nonprogressors of the three types of samples. Table S3B. All the down-regulated DEGs in nonprogressors of the three types of samples.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/geo/).


