1 Abstract
Recombinant adeno-associated viral (AAV) vectors are one of the most promising therapeutic delivery systems for gene therapy to the central nervous system (CNS). Preclinical testing of novel gene therapies requires the careful design and production of AAV vectors and their successful application in a model of CNS injury. One major limitation of AAV vectors is their limited packaging capacity (<5 kb) making the co-expression of two genes (e.g., from two promoters) difficult. An Internal Ribosomal Entry Sequence (IRES) has been used to express two genes: However, the second transgene is often expressed at lower levels than the first. In addition to this, achieving high levels of transduction in the CNS can be challenging. In this chapter we describe the cloning of a bicistronic AAV vector that uses the foot and mouth disease virus 2A sequence to efficiently express two genes from a single promoter. Bicistronic expression of a therapeutic gene and a reporter gene is desirable so that the axons from transduced neurons can be tracked and, after CNS injury, the amount of axonal sprouting or regeneration quantified. We go on to describe how to perform a pyramidotomy model of CNS injury and the injection of AAV vectors into the sensorimotor cortex to provide efficient transduction and bicistronic gene expression in cortical neurons such that transduced axons are detectable in the dorsal columns of the spinal cord.
Keywords: AAV, CNS injury, regeneration associated gene, bicistronic vector, 2A sequence, ribosomal skipping, self-cleavage, intracortical injection, pyramidotomy
2. Introduction
Following an injury to the central nervous system (CNS) neurons undergo collateral sprouting but very limited long distance regeneration which results in their failure to form functional connections with their original targets. This is due in part to the reduced intrinsic growth state of mature CNS neurons, which is characterised by their failure to express regeneration associated genes (RAGs) after an injury (1–5). Steady advancements in molecular neurobiology have elevated gene therapy into a promising therapeutic strategy for increasing the expression of RAGs and improving regeneration after a CNS injury (6–9).
To test a potential gene therapy an in vivo model of CNS injury is required. The corticospinal tract (CST) is present in all mammals and originates from the pyramidal corticospinal neurons (CSNs) in layer V of the cerebral cortex that project through the brainstem to the spinal cord. The CST fibres descend through the ventral brainstem in the medullary pyramids after which the main component of the CST decussates at the spinomedullary junction to form the dorsal CST which runs in the ventral portion of the contralateral dorsal funiculus in the adult rat (10). The CST is primarily a descending motor pathway that controls locomotion, posture and voluntary skilled movements (11). Following injury to the rodent CST, persistent deficits in sensorimotor function are observed and restorative strategies are often assessed using behavioural tests of sensorimotor and skilled motor function (11–14). The CST can be lesioned by performing a unilateral pyramidotomy (i.e. one of the medullary pyramids is cut) (15). This leads to the anterograde degeneration of the injured axons and functional deficits on the contralateral side of the body. These deficits can be overcome by enhancing the growth state of the uninjured CSNs, allowing them to sprout their axons across the midline and reinnervate the area denervated by the lesion. The pyramidotomy model has been extensively used to assess potential therapies to enhance neuronal regeneration, sprouting and functional recovery following CNS injury (8, 16). However, efficient, wide-spread transduction of the CSNs in the cortex is challenging and has only been achieved when high titres (1x1014 Genome Copies (GC)/ml) and slow infusion rates were used (6, 8, 17). Here we outline a procedure to inject AAVs into the cortex to transduce CSNs and then perform a pyramidotomy injury.
Recombinant adeno-associated viral (AAV) vectors represent one of the most attractive gene delivery systems for the CNS due to their ability to efficiently transduce neurons and to provide long-term expression with only a minimal immune response (18, 19). AAV vectors have one of the best characterised safety profiles and have been taken to clinical trial for a number of neurodegenerative diseases (20, 21). In addition to this, Glybera (an AAV vector encoding lipoprotein lipase) has recently been approved for use in the EU to reduce incidence of pancreatitis in people with inherited lipoprotein lipase deficiency (22) and paves the way for further AAV based gene therapies for other conditions. Each AAV serotype expresses different capsid proteins that determine which receptors the AAV vector can bind to for cell entry and therefore establishes the AAV vector’s tropism (23). Pseudotyping AAV2 genome with the capsid from different AAV serotypes can generate vectors with different tissue and cellular tropisms, which can improve the efficiency and pattern of transduction in specific regions of the CNS (24–26). We have previously shown that AAV1 is the optimal serotype for transducing CSNs in adult rats (17) although other AAVs also work well; AAV5 (27–29) and AAV8 (6, 28, 30). Viral vector mediated over-expression of pro-regenerative genes, including retinoic acid receptor, beta (RARbeta2), neuronal calcium sensor 1 (NCS1) and Kruppel-like factor 7 (KLF-7) have been shown to enhance the intrinsic growth state of CSNs and facilitate axonal regeneration after injury (6–9). However, these studies have either used viral vectors with larger insert capacities such as lentiviral vectors or used two separate AAV vectors; one encoding a RAG and the other a reported gene. It would therefore be very valuable to have an AAV which can co-express a pro-regenerative gene together with a fluorescent reporter so that transduced axons can be distinguished from axons that are not transduced: this would increase our ability to identify useful pro-regenerative therapies.
A disadvantage of AAV vectors is their relatively low insert capacity which can be an issue if two genes are required to be expressed (e.g., from two promoters). One partial solution is to use a single promoter which drives two genes linked by an internal ribosomal entry site (IRES) (31, 32): typically the RAG would be expressed immediately downstream of the promoter whilst the fluorescent reporter gene is expressed downstream of the IRES. Although this works reasonably well, the IRES-dependent second gene is usually expressed at a significantly lower level, estimated at 10-fold less than the cap-dependent first gene (33, 34). To date we know of no reports where transduction of supraspinal neuronal cell bodies with such an AAV results in detectable labelling of their axons in the spinal cord.
This problem can be mitigated by using the 2A peptide from the Foot and Mouth Disease Virus (35). The 2A peptide sequence is only 24 amino acids long and can mediate efficient bicistronic expression of two gene products from a single promoter by undergoing ribosomal skipping (referred to as "self-cleavage") during protein translation. Theoretically, the two gene products are expressed in a 1:1 ratio and in practice each gene product is expressed at a high level. The 2A and 2A-like bicistronic systems have been shown to be highly effective in the CNS where high expression levels of both genes have been demonstrated (6, 34).
3. Materials
Chemicals and solutions
Plasmids
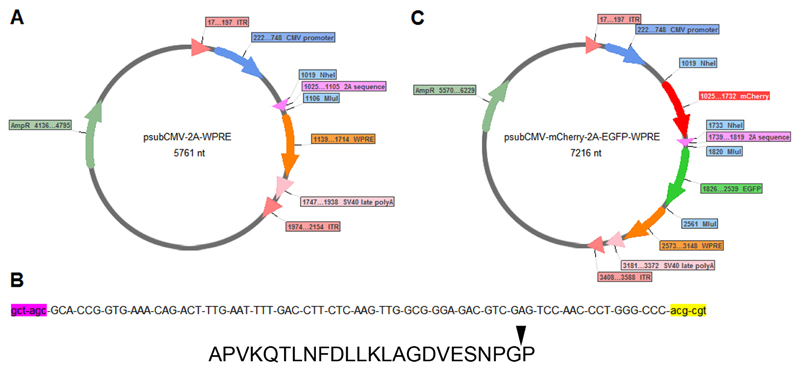
The psubCMV-2A-WPRE plasmid (Figure 1A) was a kind gift from Dr. Hansruedi Bueler, University of Kentucky. The plasmid contains a cytomegalovirus (CMV) promoter (to drive expression of the bicistron), the Foot-and-mouth disease virus (FMDV) 2A sequence and a Woodchuck Hepatitis Virus Post-transcriptional Regulatory Element (WPRE). The 2A sequence is immediately flanked by an upstream (i.e., 5’) NheI restriction site and a downstream (i.e., 3’) MluI restriction site (Figure 1B).
Figure 1.
(A) psubCMV-2A-WPRE plasmid map showing NheI site for cloning a transgene immediately downstream of the CMV promoter (and upstream of the 2A sequence) and an MluI site for cloning a transgene downstream of the 2A sequence. (B) Nucleotide sequence showing NheI restriction site (lower case, pink), the 2A sequence (upper case) and MluI restriction site (lower case, yellow). The 2A sequence encodes 24 amino acids APVKQTLNFDLLKLAGDVESNPGP, the arrowhead indicates the ribosome skipping ("self-cleavage") site. (C) psubCMV-mCherry-2A-EGFP-WPRE plasmid map showing mCherry cloned upstream and EGFP downstream of the 2A sequence.
Enzymes
NheI and MluI, NEBuffer 4 and BSA (NEB, UK). Alkaline phosphatase (Calf intestinal) (NEB, UK).
PCR reagents
Vent DNA polymerase (NEB, UK), 10X Vent DNA polymerase buffer, DMSO, 10 mM Nucleotide mix (Promega, UK), 100 mM MgSO4, 100 μM forward and reverse primers, 50 ng plasmids containing mCherry or EGFP, PCR-grade nucleotide free H2O.
Anaesthetic
0.5 mg/kg Domitor (medetomidine hydrochloride) and 100 mg/kg Vetalar (ketamine hydrochloride) or with a mixture of Isoflurane (1.5 %) and O2 (1 L/min).
Analgesia
5 mg/kg Carprofen was administered subcutaneously for post-operative analgesia.
Euthanasia solutions
400 mg/kg Lethabarb (sodium pentobarbital).
Cloning strategy and primer design
Our goal was to generate a series of plasmids with the generic structure (AAV-RAG-2A-EGFP). First, we cloned EGFP downstream of 2A and then we cloned mCherry upstream of 2A. The resulting plasmid (AAV-mCherry-2A-EGFP-WPRE) (Figure 1C) has been used in many of our experiments as a negative control (because mCherry is in place of a given RAG). It also enabled us to check easily that, indeed, both genes are expressed (Figures 2 and 3). Subsequently, we have cloned more than 10 different pro-regenerative transgenes in place of mCherry.
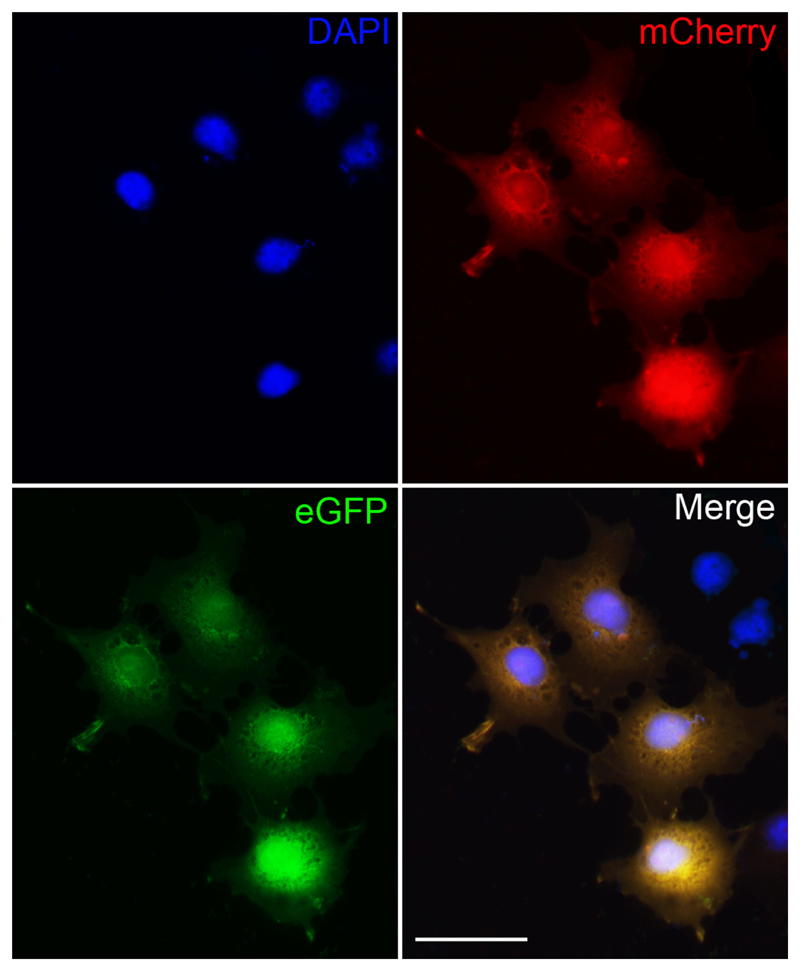
Figure 2.
COS-7 cells transfected with the psub-CMV-mCherry-2A-EGFP-WPRE plasmid efficiently expresses both mCherry and EGFP. Fluorescent images demonstrate that transfected cells express both mCherry and EGFP and appear yellow. Non-transfected cells do not express mCherry or EGFP but can be identified by DAPI staining. Scale bar - 25 µm.
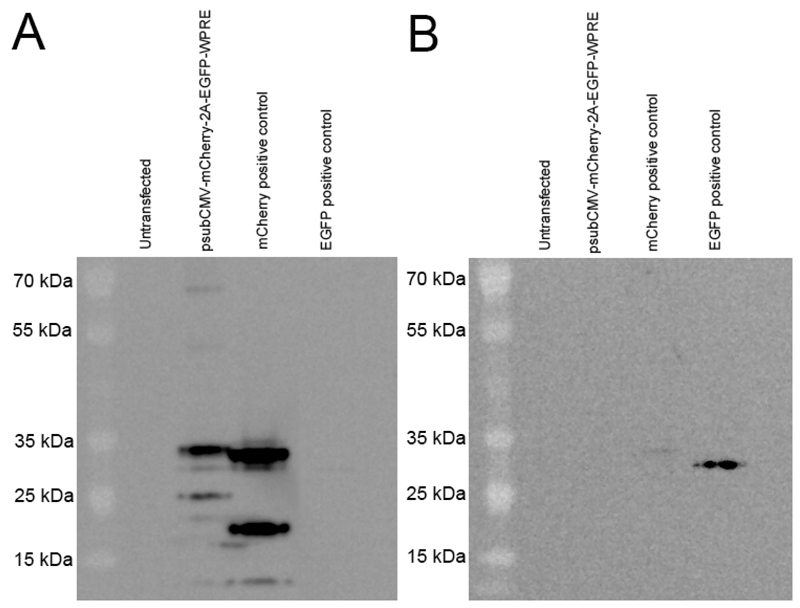
Figure 3.
Western blotting confirms efficient "self-cleavage" of the 2A sequence. COS-7 cells were transfected with the psub-CMV-mCherry-2A-EGFP-WPRE plasmid (Figure 3) or the mCherry- or EGFP-monocistronic plasmids. (A) Western blotting of 10 mg of protein lysate using an anti-mCherry antibody (Abcam - ab167453) detected a band at the expected size for mCherry protein (~30 kDa) and was a similar size as our mCherry positive control (mCherry expressed from a monocistronic plasmid). The mCherry protein from the 2A plasmid was at a slightly larger molecular weight than the control mCherry protein which was expected due to the addition of 23 amino acids at its C-terminus following "self-cleavage" of the 2A sequence. Only a very faint band was detected at ~60 kDa, as would occur if a mCherry-2A-EGFP fusion protein was produced, demonstrating efficient 2A "self-cleavage" of the two proteins. Several unidentified bands were detected at smaller molecular weights for both the 2A plasmid and the mCherry positive control. (B) We were unable to detect EGFP protein expression from the 2A plasmid via western blotting using several different anti-EGFP antibodies (Lifespan Bioscience - C183899, Clontech - 632569, Abcam - ab13970, Sigma - G1546), see Note 9. However we did detect a band at the correct size for EGFP (~30 kDa) in our EGFP positive control (EGFP expressed from a monocistronic plasmid).
Forward and reverse primers were designed for EGFP (Table 1) and mCherry (Table 2) to ensure that the coding region of the plasmid produced one single open reading frame encoding all of the following: a start codon, the upstream cDNA (mCherry in this case), the 2A sequence, and the downstream cDNA (EGFP in this case), and a stop codon.
Table 1. Primers used for cloning EGFP downstream of 2A.
| Forward:5′-GCG-CGC-ACG-CGT-GTG-AGC-AAG-GGC-GAG-GAG-CTG-TTC-ACC-G-3′ |
| Reverse:5′-GCG-CGC-ACG-CGT-CTA-GTA-GGA-TCT-GAG-TCC-GGA-CTT-GTA-C-3′ |
Table 2. Primers used for cloning mCherry upstream of 2A.
| Forward:5′-GCG-CGC-GCT-AGC-ATG-GTG-AGC-AAG-GGC-GAG-GAG-GAT-AAC-A-3′ |
| Reverse:5′-GCG-CGC-GCT-AGC-CTT-GTA-CAG-CTC-GTC-CAT-GCC-GCC-GGT-G-3′ |
For cloning EGFP downstream of the 2A sequence, the forward primer was designed to contain a hexamer of GC-repeats (blue) for stability of the PCR product and efficient restriction enzyme cleavage, an MluI restriction site (yellow), and the first 28 bases of the EGFP cDNA excluding the ATG start codon. The reverse primer contained a hexamer of GC-repeats (blue), an MluI restriction site (yellow), and the reverse complement of the last 28 bases of the EGFP cDNA (green) including the stop codon (underlined) to ensure that translation ends after the synthesis of the bicistron.
For cloning mCherry upstream of the 2A, the forward primer was designed to contain a hexamer of GC-repeats (blue) for stability of the PCR product and efficient restriction enzyme cleavage, a NheI restriction site (pink), and the first 28 bases of the mCherry cDNA (red) including the start codon (ATG; underlined). The reverse primer contained the GC-repeat hexamer (blue), a NheI restriction site (pink), and the reverse complement of the last 28 bases of the mCherry cDNA with the stop codon excluded (red).
Primers were synthesised at 50 nmoles with desalting but no other purification (Sigma, UK). We always check the resulting AAV plasmids by DNA sequencing to ensure that sequences are correct.
Sequencing primer used to check the success of cloning a gene upstream of 2A: 5'-AGC-TGC-GGA-ATT-GTA-CCC-GC-3'
Sequencing primer used to check the success of cloning a gene downstream of 2A: 5'-AAG-GCA-TTA-AAG-CAG-CGT-ATC-CAC-A-3'
Polymerase chain reaction reagents
The reagents included in our standard PCR were:
| Volume (µl) | Final concentration | |
|---|---|---|
| DNA template | 1 | Determined by user; we used 50 ng |
| Forward primer (100 µM) | 1 | 1 µM |
| Reverse primer (100 µM) | 1 | 1 µM |
| ThermoPol Reaction Buffer (10x) | 10 | 1x |
| Vent polymerase (2 units/µL) | 1 | 2 units |
| dNTPs (pool of 10 mM of each) | 2 | 200 µM |
| DMSO | 5 | 5% |
| MgSO4 (100 mM) | 4 | 4 mM |
| Nuclease-free water | To a final volume of 100 µl | |
Viral Vector production and titring
AAV vectors were generated by the Miami Project Viral Vector Core using fast protein liquid chromatography (FPLC)-based purification and pseudotyped with AAV capsid serotype 1. AAV vectors were suspended in HBSS at 2.9x1013 GC/ml.
Animals and surgery
Animals
Female Lister Hooded rats weighing 200-220 g (Charles River, UK) were used for these experiments. Rats were maintained under standard animal care conditions (12:12 hr light/dark cycle), with food and water ad libitum. All procedures were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 and approved by the local veterinarian and ethical committee.
Brain atlas for stereotaxic coordinates
Holes were drilled through the skull above the sensorimotor cortex using coordinates reported in a microstimulation mapping study relative to Bregma, midline and the brain surface (36, 37) (see methods section for coordinates).
Stereotaxic apparatus
Stereotaxic frame for rats (World precision instruments, FL, USA) which allowed the attachment of a microdrill (Power performance, UK) or ultra microsyringe pump with Micro4 control panel (World Precision Instruments, FL, USA).
Surgery materials
Eye lubricant (Viscotears, Germany), alcohol-based permanent marker, chlorhexidine disinfectant, sterile cotton buds, small bulldog clamps (World Precision Instruments # 14118), blunted scissors (Fine Science Tools, Fine Scissors – Tough Cut 14058-09), toothed forceps (Fine Science Tools, 11019-12), Long-toothed Alm retractors (Fine Science Tools, 17009-07), fine Dumont forceps (Fine Science Tools, 11251-10), dental hand drill, 0.7 mm drill bits (Fine Science Tools, 19007-07), Vannas Spring Scissors (Fine Science Tools, 15000-03), gelfoam (Equimedical, Gelatin Absorbable Haemostatic Sterile EQU705001), 3.0 Vicryl sutures (Ethicon), scalpel and #10 scalpel blades, needle holder, 27-G needles, 1 ml disposable syringes (for intraperitoneal (i.p.) injection of the anaesthetic), 10 μl Hamilton syringe (Hamilton, Reno, NV, USA), low magnification stereomicroscope (2-20x).
Histology, immunohistochemistry and microscopy
Fixative and buffers
4% paraformaldehyde in phosphate-buffered saline (PBS) pH 7 and cryoprotectant (30% sucrose in PBS).
Embedding and Sectioning
10% porcine gelatine in ddH2O or cryoprotectant embedding medium OCT. A freezing microtome (Leitz, Wetzlar, Germany) or cryostat (Bright, UK) was used to cut the tissue.
Microscope
Confocal microscope (Carl Zeiss LSM 710)
4. Methods
Plasmid design and cloning
- The coding sequences for mCherry and EGFP were amplified separately using primers and PCR reagents listed above at the cycling conditions recommended by the manufacturer for the Vent polymerase. The conditions for our typical PCR are shown below:
Temperature (ºC) Time 1 cycle Holding 95 2 minutes 25 cycles Denaturation 95 30 seconds Annealing 54 30 seconds Extension 73 3 minutes 1 cycle Final extension 73 11 minutes An aliquot of the amplicons (2.5 μl of the 100 μl reaction) was analysed using agarose gel electrophoresis to ensure the PCR products were of the expected sizes.
These blunt-ended amplicons were separately cloned into the pCR-BluntII-TOPO plasmid (Invitrogen, UK) according to manufacturer’s instructions, creating pCR-BluntII-TOPO-mCherry and pCR-BluntII-TOPO-EGFP. Competent E. coli were then transformed using the heat-shock protocol and 3 μl of the cloning reaction. Transformants were streaked out onto LB-agar plates containing kanamycin (10 μg/ml).
A day later, putative positive clones were inoculated into 5 ml of LB-kanamycin and DNA was purified using standard minipreps (Promega, UK).
- Clones were screened by restriction digests of the purified DNA using 0.5-1.0 μg of pCR-BluntII-TOPO-mCherry or pCR-BluntII-TOPO-EGFP digested by NheI or MluI, respectively. Our standard screening restriction digests were:
Reagent Volume DNA (Up to 1 µg) 1 µl 10x Buffer 4 2 µl 100x BSA 0.2 µl Restriction enzyme (NheI or MluI) 0.5 µl (2-10 units) Nuclease-free water To a final volume of 20 µl Clones were screened by performing an MluI or NheI restriction digest and visualised using agarose gel electrophoresis to ensure the cDNA inserts were present. Subsequently, these clones were sequenced using the SP6 and T7 primer sites that flank the "multiple cloning site" of pCR-Blunt II-TOPO to ensure the fidelity of the PCR amplification.
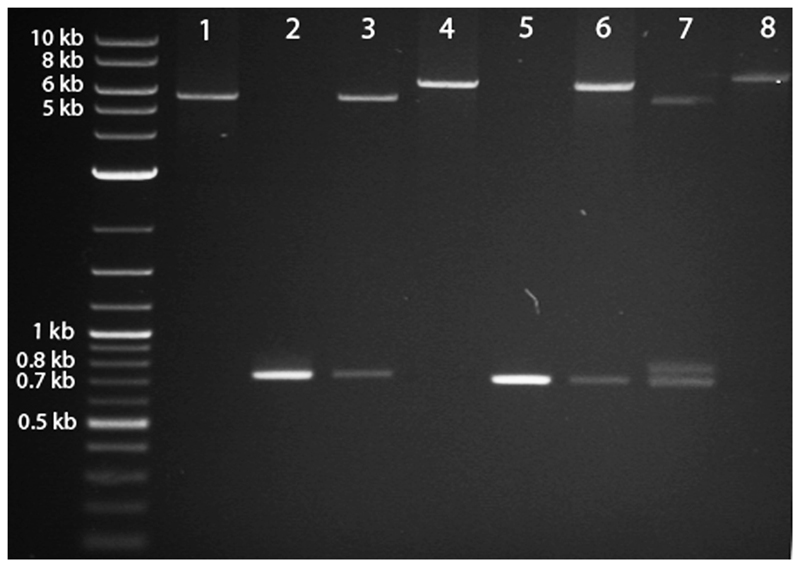
At least 10 μg of the psub-CMV-2A-WPRE vector was digested with MluI and then treated with alkaline phosphatase to prevent re-circularization. Similarly, 10 μg of pCR-BluntII-TOPO-EGFP was also digested with MluI to release the EGFP cDNA. Both the vector (Figure 4, lane 1) and the insert (Figure 4, lane 2) were then purified using gel extraction.
The EGFP cDNA insert was then ligated into the vector using T4 DNA ligase, following the manufacturer's protocol, to create psub-CMV-2A-EGFP-WPRE.
This new plasmid was transformed into E. coli and clones were screened for the presence of the insert using an MluI restriction digest (as described above) (Figure 4, lane 3) and then sequenced to ensure the correct orientation of the EGFP insert.
Next, to clone in mCherry upstream of the 2A-EGFP sequence, psub-CMV-2A-EGFP-WPRE was digested with NheI and treated with alkaline phosphatase. The pCR-BluntII-TOPO-mCherry was also digested with NheI to release the mCherry cDNA.
The linearised psub-CMV-2A-EGFP-WPRE (Figure 4, lane 4) and the mCherry cDNA insert (Figure 4, lane 5) were gel purified and then ligated using T4 DNA ligase, to create psub-CMV-mCherry-2A-EGFP-WPRE, and transformed into E. coli. The following day, DNA minipreps were obtained for 10 to 20 colonies.
Clones were screened for the presence of the insert via NheI restriction digest (Figure 4, lane 6) and then sequenced to ensure the correct orientation of mCherry.
psub-CMV-mCherry-2A-EGFP-WPRE digested with MluI and NheI to show presence of both EGFP and mCherry (Figure 4, lane 7).
psub-CMV-mCherry-2A-EGFP-WPRE was digested with XhoI to illustrate the increased size of the linearised plasmid (Figure 4, lane 8) relative to lanes 1 and 4.
Figure 4.
Agarose gel electrophoresis of restriction-digested plasmid DNA depicts the expected products at each step when cloning mCherry and EGFP into the psub-CMV-2A-WPRE plasmid. See text for a detailed explanation of the steps illustrated in the figure.
Electroporation of plasmids
Viral vector production and titring
AAV vectors were generated by the Miami Project Viral Vector Core using FPLC-based purification.
The AAV vector transgene cassette was flanked by non-coding inverted terminal repeats (ITRs), which were the only wild-type viral sequences present. The AAV vectors were pseudotyped with AAV capsid serotype 1, Following production, the AAV vectors were titred using quantitative PCR to determine the number of genomic copies and an infection assay on 293T cells was conducted to verify transduction efficiency.
In vivo experiments
In this section we describe methods for the injection of AAVs into the cortex of adult rats. We also describe methods for performing a pyramidotomy. Depending on the goal of the experiment, AAVs can be injected prior or subsequent to the CNS injury.
Intracortical injections
Female adult Lister Hooded rats (Charles River, UK) weighing 200-220 g were anesthetised with an intraperitoneal injection of 0.5 mg/kg Domitor (medetomidine hydrochloride) and 100 mg/kg Vetalar (ketamine hydrochloride).
Once anesthetised, the head was shaved and sterilised using chlorhexidine disinfectant.
Eye lubricant was applied to the eyes and throughout the surgery, the animals’ body temperature was maintained at 37 °C with a thermal heating blanket connected to a rectal temperature probe.
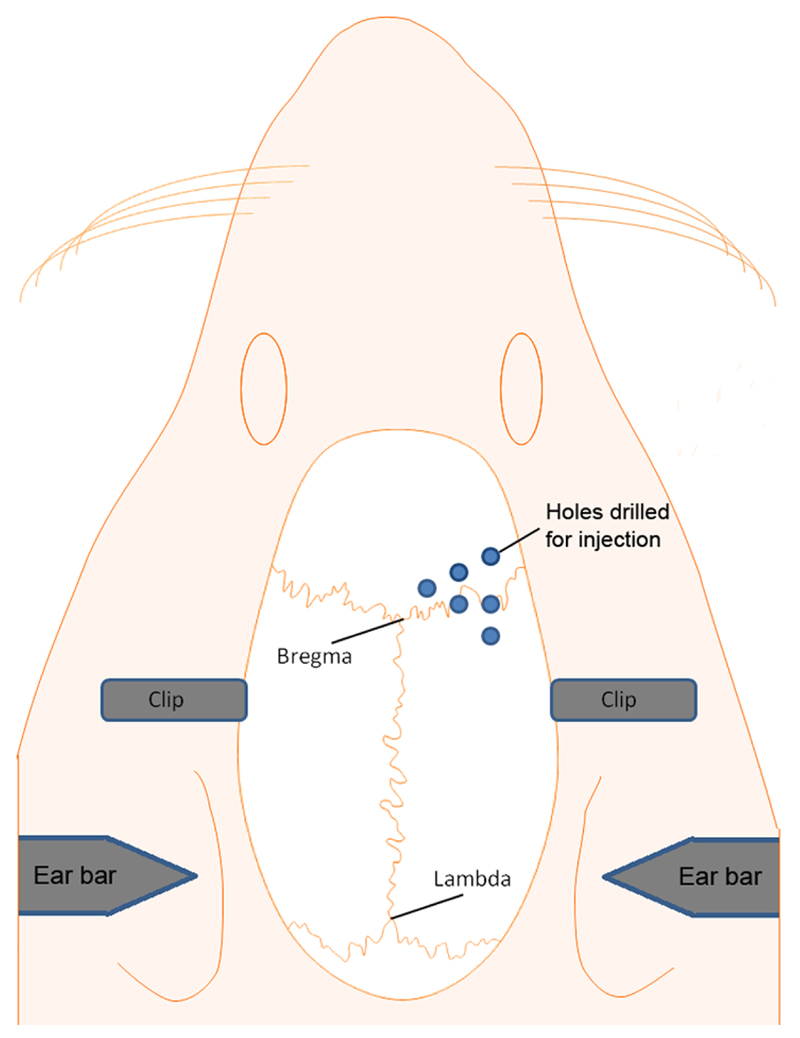
The rats' head was placed into the stereotaxic frame, the incisor bar was lowered 2 mm below the horizontal plane so that the heights of Lambda and Bregma were equal and the surface of the skull was flat (Figure 5A).
Following a midline incision, the skin was retracted using small bulldog clamps and the periosteum was removed with a scalpel blade to expose the skull surface.
- The microdrill was fixed to the stereotaxic frame holder and the drill burr centred over Bregma. Six holes were then drilled through the skull above the left or right sensorimotor cortex using coordinates reported in a microstimulation mapping study relative to Bregma, midline and the brain surface (Figure 5B). Defined as anterior–posterior (AP), medial–lateral (ML), dorsal–ventral (DV):
-
1st: 3.5mm ML, -0.5mm AP, DV -1.5mm2nd: 3.5mm ML, +0.5mm AP, DV -1.5mm3rd: 3.5mm ML, +2mm AP, DV -1.5mm4th: 2.5mm ML, +1.5mm AP, DV -1.5mm5th: 2.5mm ML, +0.5mm AP, DV -1.5mm6th: 1.5mm ML, +1mm AP, DV -1.5mm
-
A 10 μl Hamilton syringe was loaded with 3 μl of viral vector and fixed to the stereotaxic frame holder.
The tip of the micropipette was positioned over the first hole and lowered onto the brain surface. Once on the brain surface it was lowered a further 1.5 mm into the sensorimotor cortex.
0.5 μl of AAV vector was infused into the brain using a microsyringe pump. The infusion rate was set to 200 nl/min on the Micro4 control panel.
Once the pump infusion was finished the microsyringe was left in place for 3 minutes to allow the AAV vector to diffuse from the injection site.
The micropipette was then removed, positioned at the second coordinate to inject the next 0.5 μl of AAV vector. This process was repeated until all six injections were complete.
After the last injection the skin was closed using 3.0 vicryl sutures.
The rats were then placed into an incubator at 37°C and 1 mg/kg Antisedan (atipamezole hydrochloride) was administered intramuscularly. Once awake, 5 mg/kg Carprofen was administered subcutaneously for post-operative analgesia.
Figure 5.
Schematic showing the procedure for exposing the brain and injection of AAV vectors into the sensorimotor cortex. (A) A stereotaxic frame is used for both drilling and AAV vector injection (B) Location of the holes over the sensorimotor cortex used for AAV vector injection. See text for a detailed explanation of the steps illustrated in the figure.
Pyramidotomy injury model
Rats were anesthetised using 5% isoflurane in O2 (flowrate 1.5 L/min) and maintained with 1-2% isoflurane in O2. Throughout the surgery the animals’ body temperature was maintained at 37 °C with a thermal heating blanket connected to a rectal temperature probe.
The ventral neck region was shaved and sterilised using chlorhexidine disinfectant.
A midline incision is made from the chin to the rostral end of the sternum and the outer layers of gland and muscle tissue is blunt dissected until the trachea is exposed.
The trachea is displaced to one side and the underlying muscle is dissected rostrally until the basioccipital skull is reached.
Surgical long-teethed retractors are inserted to allow access to the basioccipital bone.
A craniotomy of the ventral occipital bone was performed using a microdrill. This allows access to the medullary pyramids, which are located medially.
The left or right pyramid was cut using Vannas microsissors making sure the medial boundary close to the basilar artery is lesioned as well.
Any bleeding is stopped and the skin is sutured using 3.0 vicryl sutures.
The rats were placed in an incubator at 37°C until fully awake. 5 mg/kg Carprofen was administered subcutaneously for post-operative analgesia
Histology
Ten weeks post-injection, the rats were terminally anaesthetised by intraperitoneal injection of 400 mg/kg Lethabarb (sodium pentobarbital) and transcardially perfused with 200 ml PBS (pH 7.4) to remove the blood, followed by 200 ml 4% paraformaldehyde in PBS (pH 7.4).
Immediately following perfusion the brain and spinal cord were dissected, post-fixed in 4% paraformaldehyde in PBS for 2 hours at 4°C.
The tissue was then cryoprotected in 30% sucrose with 0.1% sodium azide for 5 days at 4°C.
The brain and spinal cord tissue were placed in small plastic boxes and blocked in porcine gelatin.
Once the gelatin had set, they were placed overnight in 4% paraformaldehyde in PBS at 4°C to harden.
To allow tissue from each brain to undergo different immunohistological analysis the brains were cut rostro-caudally into 50 μm coronal serial sections using a freezing microtome and collected into ten series in a 24-well plate containing PBS and 0.1% sodium azide. The spinal cords were cut rostro-caudally into 30 μm transverse serial sections and stored in the same manner.
The tissue sections were then mounted onto microscope slides using Mowiol and kept in the dark.
Microscopy
Slides were imaged using a confocal microscope.
5. Notes
The cDNA to be inserted must be checked to ensure that none of the restriction sites used for cloning (MluI and NheI) are present within the coding region. For example one of our RAGs contained an NheI site within its coding sequence. We replaced the NheI restriction site of the upstream and downstream primers with an SpeI restriction site that has an identical overhang to NheI. However, a ligation of DNA products digested by NheI and SpeI lose the restriction site for both enzymes and it cannot be subsequently restriction digested by either enzyme.
The extension time of the PCR cycle was increased in instances where the cDNA was significantly larger. Some of the RAGs we cloned were over 3,000 bp long and required an increase in the extension time to 6 minutes.
The cloning capacity of AAVs is approximately 5 kB from ITR to ITR and longer sequences may lead to poor packaging and reduced titres. When using psubCMV-2A-EGFP-WPRE, only about 2.1 kb is available for a transgene. However, if longer transgenes need to be expressed, then WPRE could be deleted to make additional space.
It is important to choose the correct AAV serotype for the cell type you wish to transduce. We and others have previously shown that AAV1 and 5 are the optimal serotypes for transducing cortical neurons.
An important step in achieving high levels of transduction and a large area of transduction is the use of a micropump to slowly inject the viral vector over several minutes. In addition once the injection is complete it is also important to leave the needle in place for at least a couple of minutes to allow viral vector diffusion away from the injection sites
Using high-titre AAV vectors is a crucial factor for achieving high level transduction, ideally 1x1013 GC/ml or higher.
Note that "self-cleavage" of the 2A plasmid results in the upstream gene product having an additional 23 amino acids attached to the C-terminus and the downstream gene product having one additional amino acid attached to the N-terminus. The consequences of this additional sequence needs to be verified empirically on a case-by-case basis but in our experience, most pro-regenerative genes continue to show pro-regenerative effects in vitro after being shuttled from an original plasmid into the psub-CMV-2A-EGFP-WPRE plasmid.
EGFP protein could not be detected from the 2A plasmids via western blotting. This is a highly surprising result considering that the transfected COS-7 cells strongly expressed EGFP (Figure 3) and our EGFP positive control (EGFP expressed from a monocistronic plasmid) worked. Possible explanations for this may be because part of the "self-cleaved" 2A sequence remains attached to the N-terminus of EGFP and prevents binding to the anti-EGFP antibodies. However this is unlikely as we have tested several different antibodies (Lifespan Bioscience - C183899, Clontech - 632569, Abcam - ab13970, Sigma - G1546) raised against different regions of the EGFP protein including the N- and C-terminus, and the C-terminus of the downstream gene should not be affected by "self-cleavage" of the 2A sequence.
Note that our intermediate plasmid (psub-CMV-2A-EGFP) will not express EGFP in vitro or in vivo unless a transgene (containing an appropriate start codon) is cloned upstream of 2A.
The pyramidotomy surgery requires practice. High mortality rates are associated with the pyramidotomy surgery. (see Steward et al 2012 (39) and Kathe et al in preparation (40)).
Displacement of the trachea can cause breathing difficulties or apnea. Giving Dopram (Dopram V-Drops, 20 mg/ml), a respiratory stimulant, during the surgery can increase breathing activity. Furthermore, loosening of the retractors multiple times during the surgery can stabilise the animal.
When the pyramids are lesioned the basilar artery should be avoided. In addition to this, if the cut is made too deep it can affect the midbrain nuclei that are part of the breathing and cardiovascular centres.
Lesions are often incomplete. Repeating the cut with a 26 gauge needle including the fibres close to the basilar artery will prevent this to some extent.
Immunohistochemical staining for PKCγ can be use to verify completeness of the pyramidotomy lesion.
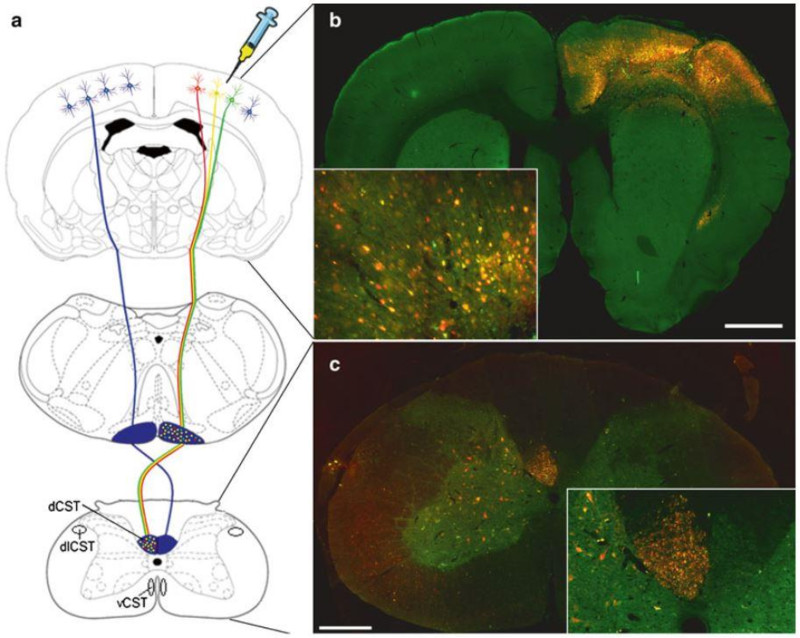
Figure 6.
(A) Schematic representation showing the rodent CST originating from the pyramidal CSNs in layer V of the sensorimotor cortex, decussating at the spinomedullary junction, forming the main dCST and the minor dlCST and vCST. (B) Image showing widespread transduction of the cortex, the majority of cortical neurons including CSNs in layer V are expressing both mCherry and EGFP (cells appear yellow). (C) Image showing numerous mCherry and EGFP positive CST axons (axons appear yellow) in the contralateral dCST of the cervical spinal cord. Unexpectedly, some fibres in the dorsal columns appear to be just mCherry or EGFP positive. We are unsure why this is, but it may reflect differences in the vesicular transport of the two proteins.
Acknowledgments
We would like to thank Dr. Vance Lemmon and the Miami Vector Core for production of the AAV vectors. We would also like to acknowledge support from a Research Councils UK Academic Fellowship (L.M.), the British Pharmacological Society’s Integrative Pharmacology Fund (L.M), Friends of Guy’s Hospital Research Grants (L.M.) and a grant from the Henry Smith Charity (L.M. and T.H.).
References
- 1.Chen DF, Jhaveri S, Schneider GE. Intrinsic changes in developing retinal neurons result in regenerative failure of their axons. Proceedings of the National Academy of Sciences U.S.A. 1995;92:7287–7291. doi: 10.1073/pnas.92.16.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholls JG, Saunders N. Regeneration of immature mammalian spinal cord after injury. Trends in neurosciences. 1996;19:229–234. doi: 10.1016/0166-2236(96)10021-7. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg JL. Intrinsic neuronal regulation of axon and dendrite growth. Curr Opin Neurobiol. 2004;14:551–557. doi: 10.1016/j.conb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg JL, Klassen MP, Hua Y, et al. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science (New York, N.Y. 2002;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- 5.Afshari FT, Kappagantula S, Fawcett JW. Extrinsic and intrinsic factors controlling axonal regeneration after spinal cord injury. Expert reviews in molecular medicine. 2009;11:e37. doi: 10.1017/S1462399409001288. [DOI] [PubMed] [Google Scholar]
- 6.Blackmore MG, Wang Z, Lerch JK, et al. Kruppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7517–7522. doi: 10.1073/pnas.1120684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yip PK, Wong LF, Pattinson D, et al. Lentiviral vector expressing retinoic acid receptor {beta}2 promotes recovery of function after corticospinal tract injury in the adult rat spinal cord. Hum Mol Genet. 2006;15:3107–3118. doi: 10.1093/hmg/ddl251. [DOI] [PubMed] [Google Scholar]
- 8.Yip PK, Wong LF, Sears TA, et al. Cortical overexpression of neuronal calcium sensor-1 induces functional plasticity in spinal cord following unilateral pyramidal tract injury in rat. PLoS biology. 2010;8:e1000399. doi: 10.1371/journal.pbio.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore DL, Blackmore MG, Hu Y, et al. KLF family members regulate intrinsic axon regeneration ability. Science (New York, N.Y. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tracey DJ. The Rat Nervous System. Second Edition. 1995. Ascending and Descending Pathways in the Spinal Cord; pp. 67–77. [Google Scholar]
- 11.Lemon RN. Descending pathways in motor control. Annual review of neuroscience. 2008;31:195–218. doi: 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- 12.Whishaw IQ, Gorny B, Sarna J. Paw and limb use in skilled and spontaneous reaching after pyramidal tract, red nucleus and combined lesions in the rat: behavioral and anatomical dissociations. Behavioural brain research. 1998;93:167–183. doi: 10.1016/s0166-4328(97)00152-6. [DOI] [PubMed] [Google Scholar]
- 13.Whishaw IQ, Metz GA. Absence of impairments or recovery mediated by the uncrossed pyramidal tract in the rat versus enduring deficits produced by the crossed pyramidal tract. Behavioural brain research. 2002;134:323–336. doi: 10.1016/s0166-4328(02)00051-7. [DOI] [PubMed] [Google Scholar]
- 14.Starkey ML, Barritt AW, Yip PK, et al. Assessing behavioural function following a pyramidotomy lesion of the corticospinal tract in adult mice. Experimental neurology. 2005;195:524–539. doi: 10.1016/j.expneurol.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Thallmair M, Metz GAS, Z'Graggen WJ, et al. Neurite growth inhibitors restrict plasticity and functional recovery following corticospinal tract lesions. Nature neuroscience. 1998;1:124–131. doi: 10.1038/373. [DOI] [PubMed] [Google Scholar]
- 16.Cafferty WB, Strittmatter SM. The Nogo-Nogo receptor pathway limits a spectrum of adult CNS axonal growth. J Neurosci. 2006;26:12242–12250. doi: 10.1523/JNEUROSCI.3827-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutson TH, Verhaagen J, Yanez-Munoz RJ, et al. Corticospinal tract transduction: a comparison of seven adeno-associated viral vector serotypes and a non-integrating lentiviral vector. Gene Ther. 2012;19:49–60. doi: 10.1038/gt.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCown TJ. Adeno-associated virus (AAV) vectors in the CNS. Current gene therapy. 2005;5:333–338. doi: 10.2174/1566523054064995. [DOI] [PubMed] [Google Scholar]
- 19.Papale A, Cerovic M, Brambilla R. Viral vector approaches to modify gene expression in the brain. Journal of neuroscience methods. 2009;185:1–14. doi: 10.1016/j.jneumeth.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Kaplitt MG, Feigin A, Tang C, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 21.Lim ST, Airavaara M, Harvey BK. Viral vectors for neurotrophic factor delivery: a gene therapy approach for neurodegenerative diseases of the CNS. Pharmacol Res. 2009;61:14–26. doi: 10.1016/j.phrs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yla-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandenberghe LH, Wilson JM, Gao G. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 2009;16:311–319. doi: 10.1038/gt.2008.170. [DOI] [PubMed] [Google Scholar]
- 24.Burger C, Nash K, Mandel RJ. Recombinant adeno-associated viral vectors in the nervous system. Human gene therapy. 2005;16:781–791. doi: 10.1089/hum.2005.16.781. [DOI] [PubMed] [Google Scholar]
- 25.Cearley CN, Wolfe JH. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Rabinowitz JE, Rolling F, Li C, et al. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. Journal of virology. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foust KD, Flotte TR, Reier PJ, et al. Recombinant adeno-associated virus-mediated global anterograde delivery of glial cell line-derived neurotrophic factor to the spinal cord: comparison of rubrospinal and corticospinal tracts in the rat. Human gene therapy. 2008;19:71–82. doi: 10.1089/hum.2007.104. [DOI] [PubMed] [Google Scholar]
- 28.Blits B, Derks S, Twisk J, et al. Adeno-associated viral vector (AAV)-mediated gene transfer in the red nucleus of the adult rat brain: comparative analysis of the transduction properties of seven AAV serotypes and lentiviral vectors. Journal of neuroscience methods. 2010;185:257–263. doi: 10.1016/j.jneumeth.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Burger C, Gorbatyuk OS, Velardo MJ, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Taymans JM, Vandenberghe LH, Haute CV, et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Human gene therapy. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- 31.Klein RL, Lewis MH, Muzyczka N, et al. Prevention of 6-hydroxydopamine-induced rotational behavior by BDNF somatic gene transfer. Brain Res. 1999;847:314–320. doi: 10.1016/s0006-8993(99)02116-2. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Current opinion in biotechnology. 1999;10:458–464. doi: 10.1016/s0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi H, Xu Z, Ishii-Watabe A, et al. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 34.Furler S, Paterna JC, Weibel M, et al. Recombinant AAV vectors containing the foot and mouth disease virus 2A sequence confer efficient bicistronic gene expression in cultured cells and rat substantia nigra neurons. Gene Ther. 2011;8:864–873. doi: 10.1038/sj.gt.3301469. [DOI] [PubMed] [Google Scholar]
- 35.Minskaia E, Nicholson J, Ryan MD. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC biotechnology. 2013;13:67. doi: 10.1186/1472-6750-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Atlas. 1982 doi: 10.1016/0165-0270(80)90021-7. [DOI] [PubMed] [Google Scholar]
- 37.Neafsey EJ, Bold EL, Haas G, et al. The organization of the rat motor cortex: a microstimulation mapping study. Brain Res. 1986;396:77–96. doi: 10.1016/s0006-8993(86)80191-3. [DOI] [PubMed] [Google Scholar]
- 38.Hutson TH, Buchser WJ, Bixby JL, et al. Optimization of a 96-Well Electroporation Assay for Postnatal Rat CNS Neurons Suitable for Cost-Effective Medium-Throughput Screening of Genes that Promote Neurite Outgrowth. Frontiers in molecular neuroscience. 2011;4:55. doi: 10.3389/fnmol.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steward O, Sharp K, Yee KM. A re-assessment of the effects of intracortical delivery of inosine on transmidline growth of corticospinal tract axons after unilateral lesions of the medullary pyramid. Experimental neurology. 2012;233:662–673. doi: 10.1016/j.expneurol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kathe C, McMahon S, Moon LDF. Unilateral pyramidotomy of the corticospinal tract in rats for assessment of neuroregenerative therapies. JoVe. doi: 10.3791/51843. (In preperation) [DOI] [PMC free article] [PubMed] [Google Scholar]