Abstract
To characterize HIV-1 transmission dynamics in regions where the burden of HIV-1 is greatest, the “Phylogenetics and Networks for Generalised HIV Epidemics in Africa” consortium (PANGEA-HIV) is sequencing full-genome viral isolates from across sub-Saharan Africa. We report the first 3,985 PANGEA-HIV consensus sequences from four cohort sites (Rakai Community Cohort Study, n = 2,833; MRC/UVRI Uganda, n = 701; Mochudi Prevention Project, n = 359; Africa Health Research Institute Resistance Cohort, n = 92). Next-generation sequencing success rates varied: more than 80% of the viral genome from the gag to the nef genes could be determined for all sequences from South Africa, 75% of sequences from Mochudi, 60% of sequences from MRC/UVRI Uganda, and 22% of sequences from Rakai. Partial sequencing failure was primarily associated with low viral load, increased for amplicons closer to the 3′ end of the genome, was not associated with subtype diversity except HIV-1 subtype D, and remained significantly associated with sampling location after controlling for other factors. We assessed the impact of the missing data patterns in PANGEA-HIV sequences on phylogeny reconstruction in simulations. We found a threshold in terms of taxon sampling below which the patchy distribution of missing characters in next-generation sequences (NGS) has an excess negative impact on the accuracy of HIV-1 phylogeny reconstruction, which is attributable to tree reconstruction artifacts that accumulate when branches in viral trees are long. The large number of PANGEA-HIV sequences provides unprecedented opportunities for evaluating HIV-1 transmission dynamics across sub-Saharan Africa and identifying prevention opportunities. Molecular epidemiological analyses of these data must proceed cautiously because sequence sampling remains below the identified threshold and a considerable negative impact of missing characters on phylogeny reconstruction is expected.
Keywords: : human immunodeficiency virus, phylogenomics, phylodynamics, HIV-1 molecular epidemiology, sub-Saharan Africa, PANGEA
Introduction
Viral phylogenetic methods are proving effective in addressing central questions in HIV-1 epidemiology: from characterizing continued transmissions in vulnerable populations1,2 to quantifying their sources of transmission,3,4 and detecting HIV-1 outbreaks in near real time.5 In the past, these investigations were largely based on partial HIV-1 sequences of less than 1,500 nucleotides (nt) in length, obtained through Sanger sequencing.
To expand the utility of viral phylogenetic methods, several consortia are now generating HIV-1 sequence data sets that span the entire viral genome.6–9 The “Phylogenetics and Networks for Generalised HIV Epidemics in Africa” consortium (PANGEA-HIV) is in the process of providing more than 10,000 NGS from partnering cohort sites in sub-Saharan Africa for a comprehensive evaluation of current HIV-1 transmission dynamics.6
We report the first 3,985 PANGEA-HIV consensus sequences that were generated in high throughput at the Wellcome Trust Sanger Institute on the Illumina MiSeq platform, after automated extraction of viral RNA and amplification with a universal HIV-1 primer set.10 The sequences are from diverse settings in sub-Saharan Africa, including cohorts of the general population at various surveillance sites (Rakai Community Cohort Study,11 Mochudi Prevention Project,12,13 MRC/UVRI Uganda general population and fisherfolk cohorts14–16), a cohort of female sex-workers (MRC/UVRI Uganda Good Health for Women17), historical sequences from the 1980s, and a cohort of HIV-1 drug-resistant individuals from northern KwaZulu-Natal in South Africa (Africa Health Research Institute Resistance Cohort18).
Most PANGEA-HIV consensus sequences from Botswana, South Africa, and MRC/UVRI Uganda cover nearly the entire viral genome from the gag to nef genes. Sequencing success rates were considerably lower for samples from the Rakai Community Cohort and varied substantially across the genome. Potential reasons for variation in NGS success rates could be as follows: low viral RNA count at time of sampling; sample degradation before RNA extraction; failure to extract viral RNA from plasma or serum samples; failure to amplify extracted RNA with the universal HIV-1 primer set; and failure during sequencing or sequence assembly. Our investigations below indicate that a number of factors, and not only low serum/plasma HIV-1 RNA loads, were associated with partial sequencing failure.
Phylogenomic studies across the tree of life highlight that phylogenies can be accurately reconstructed from sequences with very high proportions of missing characters.19–25 This could also be the case for HIV-1 phylogenies. Longer sequences of HIV-1 genomes increase phylogenetic accuracy,26,27 because more nucleotide characters are available to resolve internal branches through characters that are uniquely shared among sets of sequences, and to infer multiple substitutions between convergent sequences.28
On the contrary, and similar to many other pathogens, the HIV-1 genome is short (9,719 nt for the reference strain HXB2, including 5′- and 3′-LTR sequences). Thus, the number of informative characters between full-genome HIV-1 sequences remains limited, and missing data act by reducing the number of shared informative characters disproportionally when any two sequences have missing characters at different alignment positions.
In addition, HIV-1 phylogenies from generalized epidemics in sub-Saharan Africa are broad and exhibit many long branches.12,29,30 Indeed, due to the sheer magnitude of generalized HIV-1 epidemics in sub-Saharan Africa, closely related sequences are often not available to break long branches in viral phylogenies,31 although other factors, including onward transmission months or years after infection, also contribute to the presence of long branches.32 When branches are long, more informative sites are required to correctly infer phylogenetic relationships, and missing data could in this context indirectly exacerbate tree reconstruction artifacts.31,33,34 These considerations suggest that missing data in HIV-1 sequences could have a substantial negative impact on tree reconstruction accuracy and subsequent molecular epidemiological studies, especially in sub-Saharan African settings where sequence sampling is limited.
To characterize the implications of missing nucleotide characters in PANGEA-HIV sequences on tree reconstruction accuracy, we conducted simulation studies. An individual-level transmission and prevention model was used to generate regional HIV-1 epidemics in populations of ∼80,000 individuals, as well as corresponding HIV-1 phylogenies and full-genome sequences.35 Each simulated sequence was paired at random with a PANGEA-HIV sequence, and missing nucleotide patterns of PANGEA-HIV sequences were superimposed onto the simulated sequences. We then tested several tree reconstruction tools in their ability to re-estimate known HIV-1 phylogenies from partially determined consensus sequences.
These analyses provide insight into the accuracy with which viral phylogenetic relationships can be reconstructed from NGS that have missing characters at different positions in the sequence alignment. Since all molecular epidemiological investigations rely on accurately reconstructed viral phylogenies, our findings are fundamental to PANGEA-HIV and analogous full-genome viral sequencing efforts.
Materials and Methods
Next-generation sequencing
Serum and plasma samples from PANGEA-HIV participating cohort sites in Uganda and Botswana were shipped to University College London Hospital, London, United Kingdom, for automated RNA sample extraction on QIAsymphony SP workstations with the QIAsymphony DSP Virus/Pathogen Kit (Cat. No. 937036, 937055; Qiagen, Hilden, Germany), followed by one-step reverse transcription polymerase chain reaction (RT-PCR) as described in Ref.10 Amplification was assessed through gel electrophoresis on a fraction of samples, and samples were shipped to the Wellcome Trust Sanger Institute, Hinxton, United Kingdom.
From plasma samples from the resistance cohort, RNA was extracted at the Africa Health Research Institute in Durban, South Africa, using the QIAamp Viral Mini kit (Cat. No 52906; Qiagen), followed by one-step RT-PCR as described in Ref.10 Amplicons were purified using the QIAquick Purification kit (Cat. No. 28106; Qiagen) and shipped to the Wellcome Trust Sanger Institute.
Next-generation sequencing was performed as described previously on the Illumina MiSeq platform in the DNA pipelines core facility at the Wellcome Trust Sanger Institute.36
HIV-1 consensus sequences
Next-generation sequencing output was assembled with the SHIVER sequence assembly pipeline.37 Briefly, short reads were mapped to a de novo reference constructed using contigs (that were assembled from the short reads with IVA38) and a set of standard whole-genome reference sequences.39 Using, where available, the contigs for mapping increased accuracy in the constructed consensus sequences compared to using standard reference sequences alone. Gaps between contigs in the reference sequence were filled with a “best guess” standard reference sequence, giving those short reads that failed to result in contigs a chance to be mapped and produce additional consensus sequence. The SHIVER pipeline thus combined de novo assembly and read mapping to maximize accuracy and the length of the genome that can be assembled.
The consensus sequence of mapped reads was determined by the most frequent read call at each site. To mitigate the effects of low-level contaminant reads, sites with less than 10 mapped reads were classified as undetermined. Consensus sequences were trimmed to the viral genome from HIV-1 gag (p17) to HIV-1 nef. This process yielded consensus sequences that were each aligned against a de novo reference sequence and a set of standard reference sequences, in this case the Los Alamos HIV-1 sequence compendium 2012.39
To construct alignments of HIV-1 consensus sequences, insertions in consensus sequences were excised if they were not present in the standard reference sequences. The resulting alignment was uncertain in that gap characters which flank missing data characters could represent a deletion or a missing nucleotide as in “AC-GT-??–?-ACGT.” These sites were set to missing nucleotide characters: “AC-GT???????ACGT.”
Statistical analysis of factors associated with partial sequencing failure
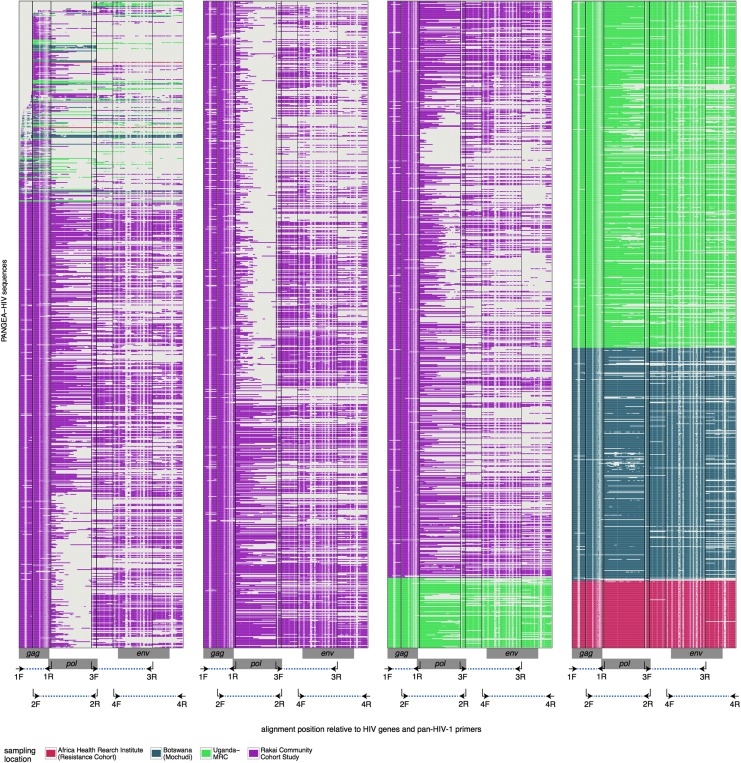
To evaluate factors associated with partial sequencing failure, we focused on four genomic regions in the unaligned consensus sequences that were amplified by exactly one of the four primer sets of the Gall protocol10: region start-2F between the start of the gag gene and the 2F primer on amplicon 1, region 1R-3F between the 1R and 3F primers on amplicon 2, region 2R-4F between the 2R and 4F primers on amplicon 3, and region 3R-end between the 3R primer and the end of the nef gene on amplicon 4 (“partial amplicon sequences,” see Fig. 1).
FIG. 1.
Alignment of the first PANGEA-HIV consensus sequences. Three thousand nine hundred eighty-five HIV-1 consensus sequences were generated from samples collected as part of the Mochudi Prevention Project (dark blue), the Rakai Community Cohort Study (purple), the Africa Health Research Institute Resistance Cohort (red), and the general population, fisherfolk, and female sex worker cohorts from MRC/UVRI Uganda (green). Locations of the HIV-1 gag, pol, and env genes are indicated on the x-axis, along with the primer sets of the Gall protocol that were used to amplify four overlapping genomic regions (arrows and blue dots). Vertical lines indicate the position of primers in the alignment. Missing data and gaps are shown in white. The total length of the alignment is 9,742 nt and covers the viral genome between HIV-1 gag and nef (length 8,628 nt in reference strain HXB2).
Based on PANGEA-HIV sequencing failure rates, partial amplicon sequences were classified into “undetermined” when more than 80% of nucleotide characters were missing, and “determined” when less than 60% of characters were missing. Ambiguous partial amplicon sequences with 20%–40% missing characters were not used in the analysis. Multivariate logistic regression analysis (gamlss,40 R version 3.2.0) was used to identify covariates that were significantly associated with undetermined partial amplicon sequences.
Simulations to assess impact of missing nucleotides in PANGEA-HIV sequences
Viral trees were generated under the regional PANGEA-HIV simulation model35 and captured disease dynamics in a regional population of ∼80,000 individuals from 1985 until 2020. Sequences of 6,807 nt were simulated along the viral trees with SeqGen version 1.3.2,41 using codon- and gene-specific evolutionary rates and relative substitution rate parameters that were estimated from HIV-1 subtype C sequences (see supplementary fig. S13 in Ref.35). The simulated sequences correspond to concatenated gag, pol, and env genes, excluding the gag stem loop and variable loops in the env gene.
To create alignments with missing data, missing nucleotide patterns of aligned PANGEA-HIV sequences were superimposed onto the simulated sequences. This step preserved the nonrandom distribution of missing data in PANGEA-HIV sequences. Other missing data patterns were also considered. Simulated sequence alignments systematically varied in the average proportion of missing characters per sequence in an alignment (0% to 60%), the distribution of missing characters [structured as in PANGEA-HIV sequences (“patchy” sequences), or in a single block after a given genomic position (“partial” sequences)], and sequence sampling coverage [1,600 (6%) to 9,629 (30%) of individuals living with HIV-1 in 2020 in the simulations].
Alignments and corresponding viral trees were indexed as described in Supplementary Table S1, and are available from https://doi.org/10.6084/m9.figshare.5056837.v1
Maximum-likelihood tree reconstruction
To ensure optimal deployment of existing phylogenetic inference tools, HIV-1 trees were reconstructed with IQ-TREE,42,43 PhyML,44 and RAxML45 by the respective software developers. To determine best program settings, the ‘true’ phylogeny, from which the sequences were simulated, was provided for one data set without missing nucleotides to the teams. Trees were also reconstructed with FastTree46 by the authors of this study. The command line options that were used for HIV-1 tree reconstructions are listed in Supplementary Data; Supplementary Data are available online at www.liebertpub.com/aid). Trees were subsequently dated and rooted with LSD version 0.3beta.47
Assessment of phylogeny reconstructions from sequences with missing data
Reconstructed trees were compared to true trees using several distance measures for tree topologies and HIV-1 transmission pairs. The central aim of PANGEA-HIV is to characterize recent transmission dynamics. For this reason, we focused on comparing the topology of phylogenetic clades that corresponded to transmission chains within the simulated regional population. This excluded deep splits in the true and inferred phylogenies from consideration. For each clade with at least four taxa, we calculated the proportion of unrooted, labeled subtrees of four taxa whose topologies differed between inferred and true clades (Quartet distance).48 In addition, we evaluated the Kendall-Colijn distance on the same clades.49 Tree distances typically scaled with clade size.50 We estimated average functional relationships between tree distance and clade size with polynomial regression techniques, and adjusted tree distances for differences in clade size.
To evaluate whether transmission pairs were accurately identified, we considered phylogenetically very close individuals as a proxy of transmission pairs and evaluated the proportion of false positives. The divergence cutoff was set deliberately at a low value of 1% substitutions per site,51,52 so that a high proportion of true transmission pairs was expected under baseline analyses from near complete sequences.
To evaluate whether transmission pairs were accurately dated, we considered for each sampled transmission pair (for whom both the transmitter and recipient had a sequence taken) the distance in units of time between their sequences in the true phylogeny, as well as the inferred phylogeny. We then calculated the mean absolute error of these distances across pairs. These distance measures provided an assessment of tree reconstruction accuracy in terms of local HIV-1 transmission chains and sampled transmission pairs.
Results
PANGEA-HIV next-generation sequences
Table 1 characterizes the first 3,985 PANGEA-HIV consensus sequences. Next-generation sequencing data are available through the European Nucleotide Archive (www.ebi.ac.uk/ena/data/view/PRJEB19239) and HIV-1 consensus sequences are available upon request to the PANGEA-HIV steering committee (Supplementary Data).
Table 1.
Characteristics of the First PANGEA-HIV Consensus Sequences
| Africa Health Research Institute Resistance Cohort | Mochudi Prevention Project | Rakai Community Cohort Study | MRC/UVRI Uganda | |
|---|---|---|---|---|
| Number of sequences | 92 | 359 | 2,833 | 701 |
| Number of individuals | 92 | 351 | 2,820 | 694 |
| Sex, % | ||||
| F | 73 | 73 | 56 | 51 |
| M | 27 | 24 | 44 | 32 |
| Missing | 0 | 3 | 0 | 16 |
| Age at time of sampling, % | ||||
| <25 | 10 | 17 | 25 | 11 |
| 25–29 | 20 | 21 | 28 | 13 |
| 30–34 | 16 | 18 | 22 | 20 |
| 35–39 | 24 | 14 | 15 | 19 |
| 40 or older | 30 | 24 | 10 | 15 |
| Missing | 0 | 6 | 0 | 23 |
| Serum/plasma HIV-1 RNA within 1 year of sampling date (copies/ml), % | ||||
| <10,000 | 7 | 30 | 19 | 1 |
| 10,000–49,999 | 36 | 22 | 8 | 0 |
| 50,000–99,999 | 13 | 16 | 3 | 1 |
| 100,000 or higher | 35 | 19 | 2 | 2 |
| Missing | 9 | 13 | 68 | 96 |
| Self-reported ART use before sampling, % | ||||
| Yes | 100 | 3 | 6 | 0 |
| No | 0 | 91 | 94 | 90 |
| Missing | 0 | 6 | 0 | 10 |
| Year of sampling, % | ||||
| 2009 | 0 | 0 | 0 | 35 |
| 2010 | 0 | 38 | 0 | 5 |
| 2011 | 55 | 36 | 25 | 0 |
| 2012 | 42 | 17 | 46 | 0 |
| 2013 | 2 | 7 | 19 | 40 |
| 2014 | 0 | 0 | 11 | 10 |
| Missing | 0 | 2 | 0 | 10 |
| HIV-1 subtype, % | ||||
| A1 | 0 | 0 | 19 | 23 |
| B | 0 | 0 | 0 | 2 |
| C | 94 | 93 | 3 | 1 |
| D | 0 | 0 | 30 | 21 |
| Other | 0 | 0 | 0 | 1 |
| potentially recombinanta | 6 | 3 | 33 | 38 |
| <500 nt to determine subtype | 0 | 4 | 15 | 14 |
As identified with the COMET HIV-1 subtyping tool52 on four partial amplicon sequences, see Materials and Methods. More refined approaches are underway to confirm recombinant sequences among potentially recombinant sequences.
Two thousand eight hundred thirty-three sequences are from 26 communities of the Rakai Community Cohort study, Uganda.53 Serum samples were obtained from household residents (aged 15–49 years) in three survey rounds between 2011 and 2014 in fisherfolk communities at the shores of Lake Victoria, and predominantly agrarian or trading communities inland. Participants were recruited at central community locations after a community mobilization event. Samples were sequenced regardless of viral load.
Two hundred thirty-one sequences were obtained from 25 neighboring communities in Kalungu district, Uganda, and from fisherfolk communities on the shores of Lake Victoria, Uganda, through MRC/UVRI. Plasma samples were obtained from household residents (aged 13+ years) through house-to-house census rounds in Kalungu district between 2013 and 2014, and from a subset of residents (aged 13+) in fisherfolk communities between 2009 and 2010.14–16 Fifty-two sequences were from a historic sample collection of the 1980s from MRC-UVRI. Four hundred eighteen sequences were obtained from female sex workers in Kampala, Uganda, as part of the Good Health for Women Project by MRC-UVRI.17 Women (aged 15+ years) involved in commercial sex or employed in entertainment facilities were enrolled through peers between 2009 and 2014. Samples were sequenced regardless of viral load.
Three hundred fifty-nine sequences are from the Mochudi Prevention Project, Botswana. Plasma samples were obtained from ART-naive individuals (aged 16–64 years) who tested positive during three rounds of an enhanced HIV testing and counseling campaign in households in northeastern Mochudi between 2010 and 2013.12,13 Samples were sequenced regardless of viral load.
Finally, 92 sequences are from the Africa Health Research Institute resistance cohort, South Africa. Plasma samples were obtained from primary health clinic attendees who failed ART in the Hlabisa sub-district of KwaZulu-Natal. Patients (>18 years) had been on ART for at least 1 year, had two successive plasma HIV-1 RNA measurements >1,000 copies/ml, at least 18 years old, and were seen between 2011 and 2013.
NGS success rates
Table 2 and Supplementary Figure S1 characterize sequencing success rates on the first PANGEA-HIV samples. More than 80% of the HIV-1 genome from the gag to the nef gene could be determined for all samples from the Africa Health Research Institute resistance cohort, 75% of samples from Mochudi, 60% of samples from MRC/UVRI Uganda, and 22% of samples from the Rakai Community Cohort. Sequencing success rates varied considerably across the genome. Figure 1 shows the alignment of PANGEA-HIV consensus sequences, directly obtained from paired consensus and assembly reference sequences (see the Materials and Methods section). As a result of alignment uncertainty, on average 0.11 additional missing nucleotide characters were introduced in the PANGEA-HIV alignment per missing character in unaligned consensus sequences (Supplementary Fig. S2).
Table 2.
Sequencing Success Rates Among the First PANGEA-HIV Consensus Sequences
| Average proportion of nonmissing nucleotide characters per sequence (relative to corresponding de novo reference sequence) | |||||
|---|---|---|---|---|---|
| Length in HXB2 (nt) | Africa Health Research Institute Resistance Cohort (%) | Mochudi Prevention Project (%) | Rakai Community Cohort Study (%) | MRC/UVRI Uganda (%) | |
| Partial genome | |||||
| gag | 1,503 | 99 | 91 | 82 | 85 |
| pol | 2,844 | 100 | 81 | 37 | 68 |
| env | 2,571 | 100 | 80 | 37 | 66 |
| gag (p17)-nef | 8,628 | 99 | 82 | 46 | 71 |
| Genomic region between primersa | |||||
| gag start-2F | 241 | 98 | 88 | 78 | 81 |
| 2F-1R | 879 | 100 | 93 | 84 | 89 |
| 1R-3F | 2,375 | 100 | 79 | 33 | 66 |
| 3F-2R | 231 | 100 | 91 | 48 | 76 |
| 2R-4F | 908 | 99 | 81 | 40 | 68 |
| 4F-3R | 1,844 | 100 | 84 | 42 | 71 |
| 3R-nef end | 1,048 | 99 | 71 | 30 | 59 |
See Figure 1 for location of the four forward and reverse primer sets.
Factors associated with partial sequencing failure
Serum and plasma samples were processed in batches from shipment to sequencing. Twenty-one of 110 batches were significantly associated with partial sequencing failure across the four amplicons of unaligned consensus sequences, after controlling for recent viral load, sampling location, amplicon, and ART use before sampling (Supplementary Fig. S3). The 21 batches were processed consecutively up to and inclusive of RNA extraction, and contained 770 sequences from Rakai.
Among the remaining sequences, serum or plasma viral load below 50,000 copies/ml within 1 year of sampling was significantly associated with partial sequencing failure across the four amplicons [adjusted odds ratio (OR) 2.47 (1.81–3.41) for viral loads within 10,000–49,999 copies/ml and 13.82 (10.34–18.79) for viral loads below 10,000 copies/ml; analysis 1 in Table 3]. After the 21 sequencing batches in Supplementary Fig S3 were excluded from analysis, we found sequencing success rates steadily decreased from amplicon 1 to amplicon 4. Sampling location remained significantly associated with partial sequencing failure after controlling for viral load, prior ART use, and differential amplicon success rates (Table 3).
Table 3.
Adjusted Odds Ratios of Partial Sequencing Failure Among the First PANGEA-HIV Sequences
| Sample characteristic | Adjusted odds ratio for sequencing failure (>80% missing characters) in partial amplicon sequences | |||
|---|---|---|---|---|
| Analysis 1 | Analysis 2 | |||
| Excluding sequences from 21 batches that were significantly associated with sequencing failure | Excluding sequences from 21 batches as in analysis 1, and short sequences of less than 500 nt whose subtype could not be determined | |||
| (n = 3,125 PANGEA-HIV sequences with 12,214 partial amplicon sequences) | (n = 2,725 PANGEA-HIV sequences with 10,635 partial amplicon sequences) | |||
| Odds ratio | 95% confidence interval | Odds ratio | 95% confidence interval | |
| Serum/plasma HIV-1 RNA within 1 year of sampling date (copies/ml) | ||||
| <10,000 | 13.82 | 10.34–18.79 | 12.81 | 9.03–18.69 |
| 10,000–49,999 | 2.47 | 1.81–3.41 | 3.6 | 2.5–5.32 |
| 50,000–99,999 | 1.02 | 0.69–1.5 | 1.36 | 0.87–2.14 |
| 100,000 or higher | 0.02 | 0.01–0.02 | 0.01 | 0.01–0.02 |
| Missing | 5.76 | 4.34–7.79 | 6.1 | 4.33–8.84 |
| Self-reported ART use before sampling | ||||
| No | 1.0 | 1.0 | ||
| Yes | 1.05 | 0.95–1.16 | 0.96 | 0.85–1.08 |
| Cohort site | ||||
| Mochudi | 1.0 | 1.0 | ||
| Africa Health Research Institute Resistance Cohort | 0a | singulara | 0a | singulara |
| Rakai | 4.01 | 3.38–4.76 | 5.95 | 4.24–8.38 |
| MRC/UVRI Historic | 2.65 | 1.88–3.72 | 3.72 | 2.3–6.01 |
| MRC/UVRI FSW cohort | 1.69 | 1.38–2.07 | 1.75 | 1.21–2.53 |
| MRC/UVRI population cohorts | 1.26 | 1–1.59 | 1.55 | 1.04–2.31 |
| Ampliconb | ||||
| Amplicon 1 | 1.0 | 1.0 | ||
| Amplicon 2 | 3.7 | 3.27–4.2 | 8.19 | 6.86–9.82 |
| Amplicon 3 | 5.06 | 4.48–5.73 | 12.42 | 10.42–14.87 |
| Amplicon 4 | 6.97 | 6.16–7.91 | 18.24 | 15.29–21.86 |
| HIV-1 subtypec | ||||
| A1 | — | — | 1.0 | |
| B | — | — | 0a | singulara |
| C | — | — | 1.18 | 0.87–1.59 |
| D | — | — | 1.22 | 1.07–1.38 |
| Other | — | — | 1.18 | 0.35–4.11 |
| Potential recombinant | — | — | 0.64 | 0.54–0.76 |
No partial sequencing failure observed
The following genomic regions (partial amplicon sequences) in each amplicon were considered: gag start-2F in amplicon 1, 1R-3F in amplicon 2, 2R-4F in amplicon 3, 3R-nef end in amplicon 4. See Figure 1 for location of the partial amplicon sequences.
HIV-1 subtype was determined with the COMET HIV-1 subtyping tool,52 version 2.1, for each of the genomic regions 1F-1R, 3F-4F, 4F-3R, if these were determined to at least 500 nt. If all region-specific assignments agreed, corresponding sequences were classified as “A1,” “B,” “C,” “D,” “other” (pure subtype). All other sequences were classified as “potential recombinant.”
The distribution of HIV-1 subtypes varied across sampling locations, with relatively homogeneous subtype C epidemics in Botswana and South Africa,12,29 and more diverse epidemics in Uganda where subtypes A and D circulate predominantly.30,54 This prompted us to investigate if HIV-1 subtypes or recombinant forms could be associated with partial sequencing failure.
We conducted a subanalysis on sufficiently long sequences whose subtype could be determined with the COMET HIV-1 subtyping tool version 2.1.55 The short sequences that were excluded all represent partial sequencing failures, which led to changes in the ORs relative to the central analysis. Relative to subtype A1, sequences of subtype D were significantly associated with more frequent partial sequencing failure, although not very strongly [(adjusted OR 1.22 (1.07–1.38), analysis 2 in Table 3]. By contrast, no subtype B sequence had more than 80% missing characters in the partial amplicon sequences. Sample sizes were small for sequences of subtypes other than A1, B, C, and D (Table 1). Depending on the exclusion criteria, potentially recombinant sequences were or were not significantly associated with partial sequencing failure. This indicates that more detailed analyses are required to identify recombinants among PANGEA-HIV sequences, and to evaluate their impact on sequencing success rates.
Large impact of missing characters in NGS on estimating HIV-1 phylogenies when sequences are sparsely sampled
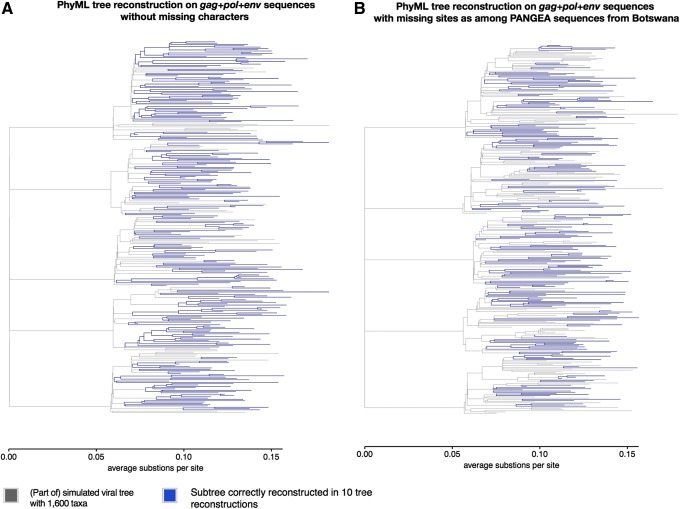
We generated 921 phylogenies with IQ-TREE,42,43 PhyML,44 RAxML,45 and FastTree46 from simulated sequence alignments that varied in size and missing data patterns (Supplementary Table S1). On the sequence alignments with 1,600 taxa (6% sequence sampling coverage of individuals living with HIV-1 by 2020 in the simulations), increased phylogenetic error was readily visible even when the gag+pol+env sequences contained relatively few missing characters, as among the PANGEA-HIV sequences from Botswana. Figure 2 illustrates this when using PhyML; similar results were obtained with IQ-TREE, RAxML, and FastTree.
FIG. 2.
Correctly reconstructed clades in simulated HIV-1 phylogenies from sequence alignments of 1,600 taxa with and without missing characters. Viral phylogenies of a generalized HIV-1 epidemic in a hypothetical sub-Sahara African setting were simulated, and HIV-1 gag, pol, and env sequences were generated along this phylogeny. The sampling coverage was 6% of individuals living with HIV-1 by 2020 in the simulation, corresponding to 1,600 taxa. PhyML was used to reconstruct the simulated viral tree. (A) Parts of the simulated viral phylogeny (blue) that were correctly reconstructed in 10 out of 10 replicate runs of PhyML from the sequence alignment of gag+pol+env sequences without missing characters (data set D1, see Supplementary Table S1). (B) Parts of the same simulated viral phylogeny that were correctly reconstructed in 10 out of 10 replicate runs of PhyML from a patchy sequence alignment, obtained by copying missing characters of randomly selected PANGEA-HIV sequences from Botswana into the sequence alignment D1 (data set D2). For visualization purposes, only the first five clades of the phylogeny are shown, each corresponding to a distinct transmission chain in the simulation. Results were similar with other tree reconstruction methods, and PhyML was chosen for illustration purposes.
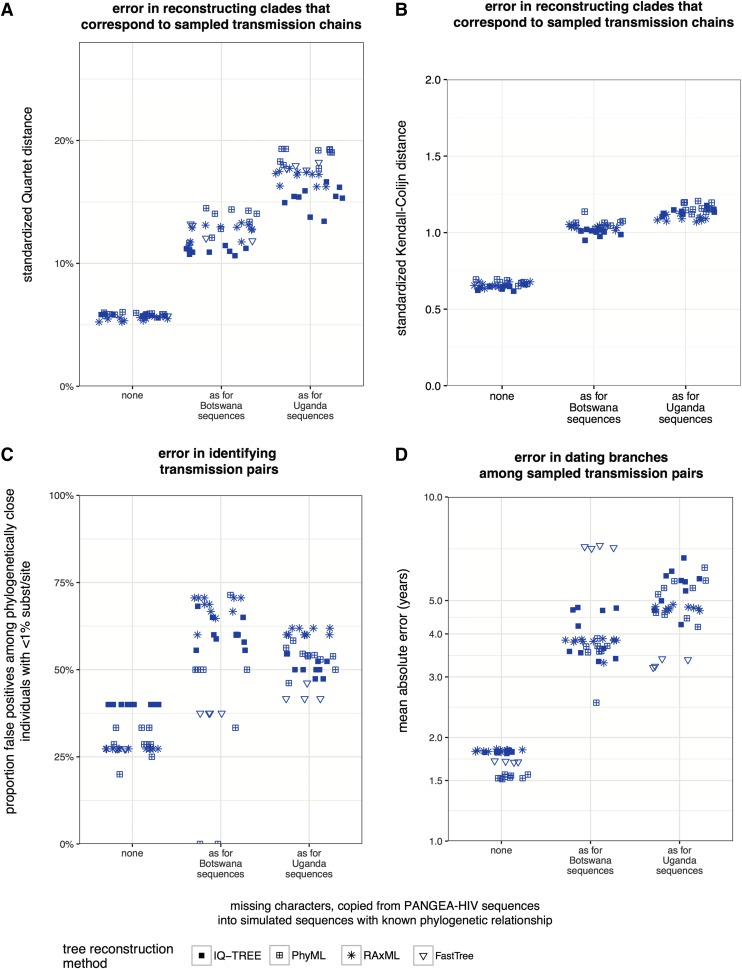
Figure 3 summarizes results from four error measures. We first assessed the accuracy with which clades that correspond to sampled HIV-1 transmission chains were reconstructed. When phylogenies were inferred from gag+pol+env sequences without missing characters using IQ-TREE, the average Quartet distance between true and reconstructed clades was 5.8% (meaning that 5.8% of all unrooted and unlabeled subtrees of four taxa of these clades were not correct). The average Quartet distance rose to 11% when phylogenies were reconstructed from sequences with missing characters as seen in PANGEA-HIV sequences from Botswana, and to 15.3% when simulated sequences had missing characters as seen in PANGEA-HIV sequences from Uganda (Fig. 3A).
FIG. 3.
Impact of missing characters in PANGEA-HIV sequences on phylogeny reconstruction when sequences are sparsely sampled. Three sequence data sets of 1,600 taxa of concatenated HIV-1 gag, pol, env genes were simulated. For each data set, missing characters in real PANGEA-HIV sequences from specific sampling locations (see x-axis) were copied into simulated sequences (data sets D1–D3, see Supplementary Table S1). Phylogenies were reconstructed in replicate with several tree reconstruction algorithms and compared to the true phylogeny. (A) Quartet distance between reconstructed and true subtrees that correspond to sampled transmission chains in the simulations. (B) Kendall-Colijn distance between reconstructed and true subtrees that correspond to sampled transmission chains in the simulations. (C) Proportion of false-positive transmission pairs among pairs of individuals that diverged less than 1% substitution/site in reconstructed phylogenies. (D) Mean absolute error (years) in estimated divergence times between sequences from sampled transmission pairs. Across all error measures, reconstructed phylogenies were considerably less accurate when sequences were sparsely sampled and contained missing characters as seen among PANGEA-HIV sequences from Botswana or Uganda, compared to gag+pol+env sequences without missing characters.
This trend was consistent regardless of tree distance measure (see Fig. 3B for results using the Kendall-Colijn distance), and results using other reconstruction methods were broadly comparable. Trees generated with FastTree did not have larger Quartet distances than other methods, despite significantly shorter run times of the method.
We also observed similar problems in accurately reconstructing basic topological relationships as the extent of missing characters increased in simulations. Specifically, we evaluated the proportion of incorrect transmission pairs among phylogenetically very close individuals. Despite tight selection criteria (<1% substitutions per site for classifying any pair as phylogenetically close), the false-positive rate was 28% on gag+pol+env sequences without missing characters using PhyML, and rose to 41% when gag+pol+env sequences contained missing characters as seen for PANGEA-HIV sequences from Botswana (Fig. 3C). Results were broadly comparable using other reconstruction methods.
The impact of missing data on falsely identified transmission pairs depended primarily on how well branch lengths for these topologically fundamental units were estimated, which was increasingly challenging and variable on patchy sequence alignments when sequence sampling was sparse. Specifically, the mean absolute error in divergence time estimates of sampled transmission pairs was 1.83 years with fully determined gag+pol+env sequences using IQ-TREE, and increased to 5.51 years with simulated gag+pol+env sequences that had missing characters as seen among PANGEA-HIV sequences from Uganda (Fig. 3D). Shorter branch lengths were typically inferred with increasing proportions of missing characters, suggesting that detection of multiple nucleotide substitutions was increasingly difficult (Supplementary Fig. S4). This led to more individuals estimated to be phylogenetically very closely related and increased false-positive rates (Supplementary Fig. S5).
Overall, trees reconstructed with FastTree had longer branch lengths compared to trees reconstructed with other methods, implying that the criteria for selecting phylogenetically close pairs were implicitly tighter for trees reconstructed with FastTree. This explains why phylogenetially close pairs identified with FastTree were overall more accurate with our error measure, compared to using IQ-TREE or RAxML.
Irregular distribution of missing characters in NGS exacerbates tree reconstruction errors, but only when sequences are sparsely sampled
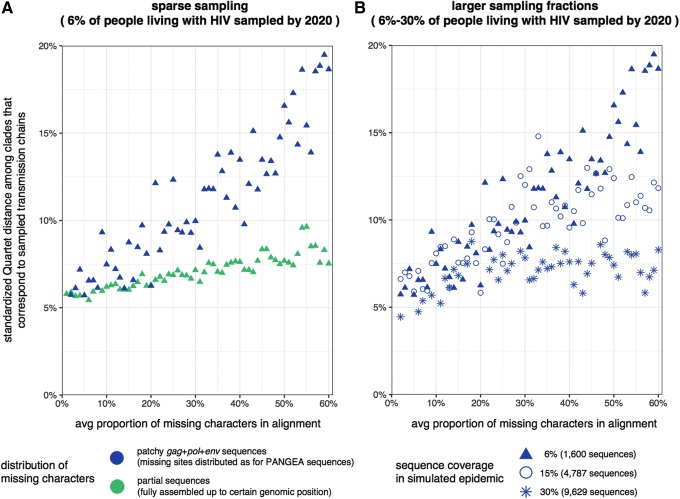
HIV-1 phylogenies have been more successfully reconstructed from partial gag or pol sequences even when taxon sampling is limited.29,30,56 We therefore suspected that the large increases in tree reconstruction error of Figure 3 were related to the irregular, nonrandom distribution of missing data patches seen in Figure 1. To test this hypothesis, we compared tree reconstructions from increasingly patchy gag+pol+env sequences to those from partial sequences that were fully determined up to a certain genome position. Thus, in simulated alignments of partial sequences, missing characters formed a contiguous block from a certain genome position to the end of the gag+pol+env sequence.
We then compared trees from patchy and partial sequences, while maintaining the overall proportion of missing nucleotides constant. For the same level of missing characters, viral trees were substantially less accurately reconstructed from gag+pol+env sequence alignments with irregularly distributed missing data than from alignments of partial sequences. (Fig. 4A). Thus, the poor performance of tree reconstruction methods is attributable to an excess negative impact of missing characters when these are irregularly distributed.
FIG. 4.
Excess negative impact of irregularly distributed missing characters on HIV-1 phylogeny reconstruction. Four times 60 sequence alignments of varying size (1,600 to 9,629 sequences, shape of points) and varying missing site patterns (either patchy or allocated in a single block after a certain genome position, color of points) were simulated (data sets D1-Mxx, D4-Mxx, D5-Mxx, D6-Mxx, D1-Pyy, see Supplementary Table S1). For each alignment, the average proportion of missing characters per sequence in alignments relative to the length of the gag+pol+env genome (6,807 nt) was calculated. One phylogeny per alignment was reconstructed with RAxML. (A) We first compared Quartet distances of trees reconstructed from patchy sequence alignments of 1,600 taxa to those of trees reconstructed from partial sequence alignments of 1,600 taxa. For the same average number/average proportion of missing characters, viral trees were less accurately reconstructed when missing characters were irregularly distributed. (B) We then compared Quartet distances of trees reconstructed from patchy sequence alignments of that increased in the number of viral sequences sampled. The excess error in Quartet distances associated with irregularly distributed missing characters vanished as sampling coverage approached 30% of individuals living with HIV-1 by 2020 in the simulations (∼10,000 taxa).
In addition, when a larger number of patchy sequences were available for tree reconstruction, accuracy increased and approached that of alignments of partial sequences with the same average proportion of missing characters (Fig. 4B). With a sequence sampling coverage above 30%, trees from patchy gag+pol+env sequence alignments were not significantly less accurately reconstructed as trees from partial sequence alignments with a comparable level of missing characters. This indicates that the excess negative impact of irregularly distributed missing characters in HIV-1 sequence alignments is sidestepped when sufficiently many patchy sequences are available for analysis. Below this sequence coverage threshold, error in phylogeny reconstructions from patchy sequence alignments was larger than expected from alignments of partial sequences with the same overall level of missing characters.
Alignment trimming to reduce tree reconstruction artifacts
The excess negative impact of irregularly distributed missing characters arises through a combination of direct and indirect effects, including disproportionally fewer informative sites that are shared between any two sequences, and accumulating tree reconstruction artifacts. Intriguingly, the indirect effects could potentially be mitigated by excluding alignment columns with disproportionally many missing characters (“trimming”),21,57 at the expense of fewer shared informative sites.
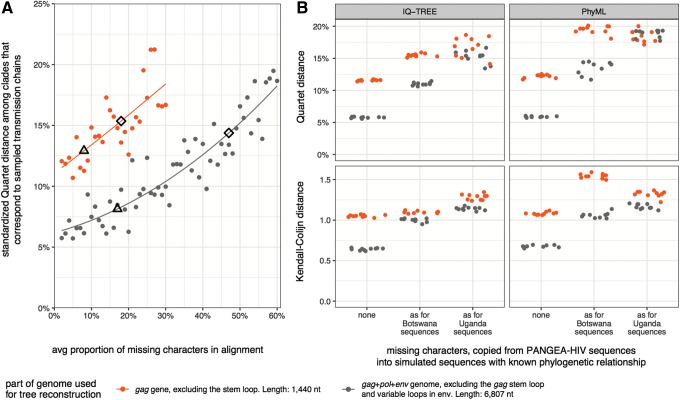
Sequencing success rates were highest for amplicon 1 among PANGEA-HIV sequences, prompting us to compare tree reconstructions from more complete gag genes (1,440 nt without stem loop) to tree reconstructions from more patchy gag+pol+env sequences (6,807 nt). Figure 5A shows that trees reconstructed from gag sequence alignments of 1,600 taxa with <10% missing characters were on average more accurate than trees reconstructed from gag+pol+env sequence alignments of 1,600 taxa with >40% missing characters. Thus, more accurate HIV-1 phylogenies are only expected from trimmed alignments when sequencing success rates are highly uneven. This explains why we did not reconstruct more accurate phylogenies from gag genes compared to gag+pol+env sequences when both alignments had missing characters as seen among PANGEA-HIV sequences from Botswana, nor when both alignments had missing characters as seen among PANGEA-HIV sequences from Uganda (Fig. 5B).
FIG. 5.
Alignment trimming to reduce tree reconstruction artifacts. (A) Sixty alignments of 1,600 gag+pol+env sequences (6,807 nt) with increasing proportions of missing characters were simulated. Missing site patterns were copied at random from PANGEA-HIV sequences (data sets D1-Mxx, see Supplementary Table S1). Thirty alignments were trimmed to the gag gene. One phylogeny per alignment was reconstructed with RAxML. We compared Quartet distances of trees reconstructed from patchy gag+pol+env sequences (gray) to those of patchy gag sequences (orange). It is possible to reconstruct more accurate phylogenies from shorter gag sequences, but only when the trimmed alignment harbors substantially fewer missing characters than the longer original alignment and sequence sampling coverage is low (6%). The proportion of missing characters in gag and gag+pol+env sequences among PANGEA-HIV sequences from Botswana and Uganda is indicated with triangles and diamonds. (B) The three sequence data sets of 1,600 gappy gag+pol+env sequences of Figure 2 were trimmed to the gag gene. Ten phylogenies were reconstructed with IQ-TREE, PhyML, and RAxML per alignment, and results are shown for IQ-TREE and PhyML. Tree reconstructions from gag genes that harbored missing characters as seen in PANGEA-HIV sequences from Botswana or Uganda were not more accurate than those from patchy gag+pol+env sequences, regardless of distance measure and tree reconstruction method. The differences in missing character patterns between the trimmed and original alignments were not large enough to result in more accurate tree reconstructions with the trimmed alignment.
Discussion
NGS data of HIV-1 viruses offer unprecedented opportunities for studying disease progression,58,59 evolution of resistance to antiretrovirals,60,61 as well as aspects of transmission dynamics.51,62,63 Obtaining NGS data from serum or plasma samples is fraught with difficulties, owing, in part, to the extreme genetic diversity of the virus, large variation in copy numbers in samples, as well as sample degradation.64 PANGEA-HIV adopted a sequencing protocol that combined automated RNA extraction with amplification-dependent next-generation sequencing under the Gall protocol.10 With this approach, consensus sequences of the HIV-1 genome could be generated from a diverse set of samples in high throughput. Sequencing success rates varied across the genome and were particularly low on samples from the Rakai Community Cohort Study, Uganda.
Our phylogenetic simulation study indicates that missing nucleotide characters in PANGEA-HIV sequences have limited impact on phylogeny reconstruction when a sufficiently high proportion of viral sequences from epidemics are sampled. Specifically, the particular missing data patterns in PANGEA-HIV sequences did not have a significant excess negative impact on reconstructing phylogenies of simulated HIV-1 transmission chains when sequence sampling coverage was at least 30% (of individuals living with HIV-1 by the end of the simulation, Fig. 4). Above this threshold, phylogenetic inference error from alignments with missing characters at differing positions did not increase faster than on alignments with missing characters at the same positions, and overall relatively slowly.
The Mochudi Prevention Project, the Rakai Community Cohort Study, and other sites have collected samples at higher coverage within the surveillance sites.30,56 At larger geographical areas (e.g., the regions that encompass individual surveillance sites), current sequence sampling coverage remains below 30% of all individuals living with HIV-1. Large data sets of partial HIV-1 sequences are now available for Southern and Central Africa, including historical samples from partnering cohort sites. These sequence data sets should be used to increase taxon sampling in future analyses, and further mitigate the impact that missing characters could have on phylogeny reconstruction.29,69
Phylogenetic analysis of real HIV-1 sequences is more challenging than that of the simulated sequences in this study. Simulations were generated under standard nucleotide evolution models and did not account for recombination between viral strains. Failure to appropriately account for recombination65 as well as differences in relative nucleotide substitution rates,66 evolutionary rates,67 and nucleotide composition bias across the genome68 can substantially increase systematic bias and lead to incorrect phylogenies that are highly supported in bootstrap analyses.25,57 It is not clear to what extent these factors could exacerbate the impact of irregularly distributed missing data on phylogeny reconstruction and subsequent molecular epidemiologic investigations.
Conversely, our accuracy measures were not restricted to phylogenetically credible clades due to computational limitations in generating large numbers of replicate trees. Phylogenetically credible clades (that occur in a high proportion of replicate trees) could be considerably more robust to the impact of missing sequence data than our error analyses across all clades suggest.
Several previous studies support our finding that irregularly distributed missing data patterns have a substantial excess negative impact on HIV-1 phylogeny reconstruction below a certain sampling level. Missing nucleotide characters can exacerbate long branch attraction artifacts,33,34 while increased taxon sampling reduces systematic errors in tree reconstruction.31,70 In settings with sparse sequence sampling, our findings also support previous arguments against ad hoc alignment trimming71: we found that missing characters must be highly unevenly distributed across the genome for this trimming to have a net positive impact on phylogeny reconstruction. We suggest evaluating the impact of alignment trimming in simulations before application to real data. Simulation routines for this purpose are available as part of the PANGEA-HIV simulation tool (https://github.com/olli0601/PANGEA.HIV.sim).
Even when using NGS without missing characters, a large proportion (25%–35%) of phylogenetically very close pairs of individuals (patristic distance <1% substitutions/site) were not transmission pairs in our simulations. The fact that complete NGS cannot confirm HIV-1 transmission events is primarily a consequence of sparse sampling in our simulations and also of viral evolution within hosts, which can lead to incongruencies between phylogenies and transmission trees regardless of sampling.72 Further work is needed to characterize false-positive rates when sequencing is targeted at subpopulations, for example, at young women and their sexual partners.
Considering the observed variation in sequencing success rates, there is clear scope for improving the PANGEA-HIV sequencing protocol. Our retrospective analyses indicate that in addition to low viral load, amplicon order and sampling locations were also strongly associated with partial sequencing failure.
This could be due to several factors. One study found that manual extraction of viral RNA led to improved recovery of near full-length viral sequences compared to automated RNA extraction.73 Considering differential amplicon sequencing success rates, modified protocols74 or amplification-independent sequencing techniques75–77 could also potentially improve NGS success rates. Data from the first PANGEA sequences were not sufficient to robustly evaluate the potential impact of nucleotide mutations at specific primer sites: at the 2R primer, we found larger proportions of sequences with a mutation toward the 3′-end, although the percent difference was not significant between sequences with >80% and <60% missing characters in 1R-3F, and mutations at the 2R primer were also not independently associated with partial sequencing failure in a subanalysis. A systematic comparison of NGS protocols on the same specimen is needed to identify more robust NGS approaches.
This study provides evidence that the missing data patterns in PANGEA-HIV sequences do not substantially impact on phylogeny reconstruction when sufficiently many viral sequences are sampled. Current sequence sampling levels of regional HIV-1 epidemics in sub-Saharan Africa remain considerably below the sampling coverage threshold of ∼30% that was identified on simulated data. Further efforts to develop more robust NGS protocols would be highly beneficial for using NGS data to characterize patterns of HIV-1 transmission and HIV-1 prevention opportunities.
Supplementary Material
Contributor Information
Collaborators: on behalf of the PANGEA-HIV Consortium
Acknowledgments
S.G. acknowledges the Centre for eResearch at the University of Auckland for general support. Computational results were partly achieved using the Vienna Scientific Cluster, the NeSI high-performance computing facilities (www.nesi.org.nz), and the Imperial College High Performance Computing Service (www3.imperial.ac.uk/ict/services/hpc). New Zealand's national facilities are provided by the NZ eScience Infrastructure and funded jointly by NeSIs collaborator institutions and through the Ministry of Business, Innovation and Employment's Research Infrastructure program. We thank the PANGEA-HIV steering committee (Supplementary Data) and the PANGEA-HIV/ICONIC sequence assembly working group (in particular Francois Blanquart, Dan Frampton, and Swee Hoe Ong) for their input, and the PANGEA-HIV steering committee for comments on a previous version of this article. O.R., A.L.B., P.K., Td.O., D.P., C.F. are supported through the PANGEA-HIV consortium by the Bill and Melinda Gates Foundation; C.W., C.F. by the European Research Council (Advanced Grant PBDR-339251); C.C. by the Engineering and Physical Sciences Research Council (EPSRC EP/K026003/1 and EP/I031626/1); S.F. by the Medical Research Council (MR/J013862/1); T.Q. by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH; A.S. and A.K. by the Klaus Tschira Foundation; J.T., B.Q.M., and Av.H. by the Austrian Science Fund (FWF I-2805-B29). The MRC/UVRI is funded jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement.
Data Availability
PANGEA-HIV sequencing output is available from the European Nucleotide Archive project page (www.ebi.ac.uk/ena/data/view/PRJEB19239). PANGEA-HIV consensus sequences and associated demographic data are available on request to the PANGEA-HIV steering committee (Supplementary Data). Simulated sequence data and viral trees are available as from https://doi.org/10.6084/m9.figshare.5056837.v1
Authors' Contributions
O.R., A.L.B., C.F., P.K., Td.O., D.P. conceived the study; D.S., P.K., K.G., T.Q., M.W., D.S., R.G., V.N., S.M., S.G., M.E., Td.O. selected and prepared samples; C.W., O.R., C.F., A.G., D.P. generated PANGEA-HIV consensus sequences; O.R., C.F. performed statistical analyses on sequencing failure; O.R. generated the simulated sequence data sets; S.G., D.G., Av.H., A.K., M.C.M., B.Q.M., A.S., J.T., O.R. reconstructed viral trees; O.R., C.C., S.F., M.K. evaluated submitted trees; O.R. wrote the first version of the manuscript; all authors reviewed and approved the statistical analysis and the final version of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. : High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis 2007;195:951–959 [DOI] [PubMed] [Google Scholar]
- 2.Oster AM, Dorell CG, Mena LA, Thomas PE, Toledo CA, Heffelfinger JD: HIV risk among young African American men who have sex with men: A case-control study in Mississippi. Am J Public Health 2011;101:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volz E, Ionides E, Romero-Severson E, Brandt MG, Mokotoff E, Koopman J: HIV-1 transmission during early infection in men who have sex with men: A phylodynamic analysis. PLoS Med 2013;10:e1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratmann O, van Sighem A, Bezemer D, Gavryushkina A, Juurrians S, Wensing AM, et al. : Sources of HIV infection among men having sex with men and implications for prevention. Sci Transl Med 2016;8:320ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon AF, Gustafson R, Daly P, Zerr L, Demlow SE, Wong J, et al. : Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: An implementation case study. Lancet HIV 2016;3:e231–e238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillay D, Herbeck J, Cohen MS, de Oliveira T, Fraser C, Ratmann O, et al. : PANGEA-HIV: Phylogenetics for generalised epidemics in Africa. Lancet Infect Dis 2015;15:259–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.HPTN 071 (PopART) Phylogenetics Protocol Team. HPTN 071-2 Phylogenetics in HPTN 071: An ancillary study to “Population Effects of Antiretroviral Therapy to Reduce HIV Transmission (PopART): A cluster-randomized trial of the impact of a combination prevention package on population-level HIV incidence in Zambia and South Africa” 2015. Available at https://www.hptn.org/sites/default/files/2016-05/HPTN?20071-2_Phylogenetics?20Ancillary?20Protocol_v?201.0_15Jan2015.pdf Accessed June17, 2017
- 8.Fraser C: Bridging the evolution and epidemiology of HIV in Europe 2014. Available at http://cordis.europa.eu/project/rcn/185401_en.html Accessed June17, 2017
- 9.Kozlakidis Z: ICONIC: Infection response through virus genomics 2014. Available at www.hra.nhs.uk/news/research-summaries/infection-response-through-virus-genomics-iconic/ Accessed June17, 2017
- 10.Gall A, Ferns B, Morris C, Watson S, Cotten M, Robinson M, et al. : Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J Clin Microbiol 2012;50:3838–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabowski MK, Lessler J, Bazaale JM, Chang LW, Nabukalu D, Wawer M, et al. : Migration, HIV-infection and access to combination HIV prevention in Rakai district, Uganda. CROI 20162016
- 12.Novitsky V, Bussmann H, Logan A, Moyo S, van Widenfelt E, Okui L, et al. : Phylogenetic relatedness of circulating HIV-1C variants in Mochudi, Botswana. PLoS One 2013;8:e80589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novitsky V, Bussmann H, Okui L, Logan A, Moyo S, van Widenfelt E, et al. : Estimated age and gender profile of individuals missed by a home-based HIV testing and counselling campaign in a Botswana community. J Int AIDS Soc 2015;18:19918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asiki G, Mpendo J, Abaasa A, Agaba C, Nanvubya A, Nielsen L, et al. : HIV and syphilis prevalence and associated risk factors among fishing communities of Lake Victoria, Uganda. Sex Trans Infect 2011;87:511–515 [DOI] [PubMed] [Google Scholar]
- 15.Seeley J, Nakiyingi-Miiro J, Kamali A, Mpendo J, Asiki G, Abaasa A, et al. : High HIV incidence and socio-behavioral risk patterns in fishing communities on the shores of Lake Victoria, Uganda. Sex Trans Dis 2012;39:433–439 [DOI] [PubMed] [Google Scholar]
- 16.Asiki G, Murphy G, Nakiyingi-Miiro J, Seeley J, Nsubuga RN, Karabarinde A, et al. : The general population cohort in rural south-western Uganda: A platform for communicable and non-communicable disease studies. Int J Epidemiol 2013;42:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandepitte J, Bukenya J, Weiss HA, Nakubulwa S, Francis SC, Hughes P, et al. : HIV and other sexually transmitted infections in a cohort of women involved in high-risk sexual behavior in Kampala, Uganda. Sex Trans Dis 2011;38:316–323 [PMC free article] [PubMed] [Google Scholar]
- 18.Danaviah S, Manasa J, Wilkinson E, Pillay S, Sibisi Z, Msweli S, et al. : Near Full Length HIV-1 Sequencing to Understand HIV Phylodynamics in Africa in Real Time. CROI, Seattle, Washington, 2015 [Google Scholar]
- 19.Rokas A, Williams BL, King N, Carroll SB: Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003;425:798–804 [DOI] [PubMed] [Google Scholar]
- 20.Wiens JJ: Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol 2003;52:528–538 [DOI] [PubMed] [Google Scholar]
- 21.Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D: Phylogenomics of eukaryotes: Impact of missing data on large alignments. Mol Biol Evol 2004;21:1740–1752 [DOI] [PubMed] [Google Scholar]
- 22.Delsuc F, Brinkmann H, Philippe H: Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 2005;6:361–375 [DOI] [PubMed] [Google Scholar]
- 23.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, et al. : Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 2010;463:1079–1083 [DOI] [PubMed] [Google Scholar]
- 24.Wiens JJ, Morrill MC: Missing data in phylogenetic analysis: Reconciling results from simulations and empirical data. Syst Biol 2011;60:719–731 [DOI] [PubMed] [Google Scholar]
- 25.Philippe H, Brinkmann H, Lavrov DV, Littlewood DT, Manuel M, Worheide G, et al. : Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol 2011;9:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M: Importance of viral sequence length and number of variable and informative sites in analysis of HIV clustering. AIDS Res Human Retroviruses 2015;31:531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yebra G, Hodcroft EB, Ragonnet-Cronin ML, Pillay D, Brown AJ; PANGEA HIV Consortium, et al. : Using nearly full-genome HIV sequence data improves phylogeny reconstruction in a simulated epidemic. Sci Rep 2016;6:39489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felsenstein J: Inferring Phylogenies. Sinauer Associates, Inc.; Sunderland MA: 2004. 580 p [Google Scholar]
- 29.Wilkinson E, Rasmussen DA, Ratmann O, Stadler T, Engelbrecht S, de Oliveira T: Origin, imports and exports of HIV-1 subtype C in South Africa: A historical perspective. Infect Genet Evol. 2016;46:200–208 [DOI] [PubMed] [Google Scholar]
- 30.Grabowski MK, Lessler J, Redd AD, Kagaayi J, Laeyendecker O, Ndyanabo A, et al. : The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: Evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med 2014;11:e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwickl DJ, Hillis DM: Increased taxon sampling greatly reduces phylogenetic error. Syst Biol 2002;51:588–598 [DOI] [PubMed] [Google Scholar]
- 32.Pybus OG, Rambaut A: Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet 2009;10:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiens JJ: Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst Biol 1998;47:625–640 [DOI] [PubMed] [Google Scholar]
- 34.Roure B, Baurain D, Philippe H: Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Mol Biol Evol 2013;30:197–214 [DOI] [PubMed] [Google Scholar]
- 35.Ratmann O, Hodcroft E, Pickles M, Cori A, Hall M, Lycett S, et al. : Phylogenetic tools for generalised HIV epidemics: Findings from the PANGEA-HIV methods comparison. Mol Biol Evol 2017;34:185–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gall A, Morris C, Kellam P, Berry N: Complete genome sequence of the WHO International Standard for HIV-1 RNA determined by deep sequencing. Genome Announc 2014;2:e01254–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wymant C, Blanquart F, Gall A, Bakker M, Bezemer D, Croucher NJ, et al.: Easy and accurate reconstruction of whole HIV genomes from short-read sequence data. bioRxiv 2016 [DOI] [PMC free article] [PubMed]
- 38.Hunt M, Gall A, Ong SH, Brener J, Ferns B, Goulder P, et al. : IVA: Accurate de novo assembly of RNA virus genomes. Bioinformatics 2015;31:2374–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuiken C, Foley B, Leitner T, Apetrei V, Hahn B, Mizrachi I, et al. : HIV Sequence Compendium 2012. In: Theoretical Biology and Biophysics Group (LANL, ed.) NM, LA-UR 12-246532012 [Google Scholar]
- 40.Stasinopoulos DM, Rigby RA: Generalized additive models for location scale and shape (GAMLSS) in R. J Stat Softw 2007;23 [Google Scholar]
- 41.Rambaut A, Grassly NC: Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci 1997;13:235–238 [DOI] [PubMed] [Google Scholar]
- 42.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ: IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chernomor O, von Haeseler A, Minh BQ: Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol 2016;65:997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O: New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 2010;59:307–321 [DOI] [PubMed] [Google Scholar]
- 45.Stamatakis A: RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price MN, Dehal PS, Arkin AP: FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.To TH, Jung M, Lycett S, Gascuel O: Fast dating using least-squares criteria and algorithms. Syst Biol 2015;65:82–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sand A, Holt MK, Johansen J, Fagerberg R, Brodal GS, Pedersen CN, et al. : Algorithms for computing the triplet and quartet distances for binary and general trees. Biology (Basel) 2013;2:1189–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kendall M, Colijn C: Mapping phylogenetic trees to reveal distinct patterns of evolution. Mol Biol Evol 2016;33:2735–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steel M, Penny D: Distributions of tree comparison metrics—Some new results. Syst Biol 1993;42:126–141 [Google Scholar]
- 51.Eshleman SH, Hudelson SE, Redd AD, Wang L, Debes R, Chen YQ, et al. : Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis 2011;204:1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell MS, Mullins JI, Hughes JP, Celum C, Wong KG, Raugi DN, et al. : Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One 2011;6:e16986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang LW, Grabowski MK, Ssekubugu R, Nalugoda F, Kigozi G, Nantume B, et al. : Heterogeneity of the HIV epidemic in agrarian, trading, and fishing communities in Rakai, Uganda: An observational epidemiological study. Lancet HIV 2016;3:e388–e396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yebra G, Ragonnet-Cronin M, Ssemwanga D, Parry CM, Logue CH, Cane PA, et al. : Analysis of the history and spread of HIV-1 in Uganda using phylodynamics. J Gen Virol 2015;96(Pt 7):1890–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP: COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014;42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novitsky V, Kühnert D, Moyo S, Widenfelt E, Okui L, Essex M: Phylodynamic analysis of HIV sub-epidemics in Mochudi, Botswana. Epidemics 2015;13:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeffroy O, Brinkmann H, Delsuc F, Philippe H: Phylogenomics: The beginning of incongruence? Trends Genet 2006;22:225–231 [DOI] [PubMed] [Google Scholar]
- 58.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, et al. : Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 2012;8:e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanini F, Brodin J, Thebo L, Lanz C, Bratt G, Albert J, et al. : Population genomics of intrapatient HIV-1 evolution. Elife 2015;4:e11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hedskog C, Mild M, Jernberg J, Sherwood E, Bratt G, Leitner T, et al. : Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS One 2010;5:e11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buzon MJ, Codoner FM, Frost SD, Pou C, Puertas MC, Massanella M, et al. : Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog 2011;7:e1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer W, Ganusov VV, Giorgi EE, Hraber PT, Keele BF, Leitner T, et al. : Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 2010;5:e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redd AD, Quinn TC, Tobian AA: Frequency and implications of HIV superinfection. Lancet Infect Dis 2013;13:622–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brumme CJ, Poon AF: Promises and pitfalls of Illumina sequencing for HIV resistance genotyping. Virus Res 2016 [DOI] [PubMed]
- 65.Hue S, Hassan AS, Nabwera H, Sanders EJ, Pillay D, Berkley JA, et al. : HIV type 1 in a rural coastal town in Kenya shows multiple introductions with many subtypes and much recombination. AIDS Res Human Retroviruses 2012;28:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wertheim JO, Fourment M, Kosakovsky Pond SL: Inconsistencies in estimating the age of HIV-1 subtypes due to heterotachy. Mol Biol Evol 2012;29:451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alizon S, Fraser C: Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirology 2013;10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Kuyl AC, Berkhout B: The biased nucleotide composition of the HIV genome: A constant factor in a highly variable virus. Retrovirology 2012;9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamers S, Barbier A, Ratmann O, Fraser C, Rose R, Laeyendecker O, et al. : HIV-1 sequence data coverage in Central East Africa from 1959–2013. AIDS Res Hum Retroviruses. 2016;32:904–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baurain D, Brinkmann H, Philippe H: Lack of resolution in the animal phylogeny: Closely spaced cladogeneses or undetected systematic errors? Mol Biol Evol 2007;24:6–9 [DOI] [PubMed] [Google Scholar]
- 71.Tan G, Muffato M, Ledergerber C, Herrero J, Goldman N, Gil M, et al. : Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst Biol 2015;64:778–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Didelot X, Gardy J, Colijn C: Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol 2014;31:1869–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cornelissen M, Gall A, Vink M, Zorgdrager F, Binter S, Edwards S, et al. : From clinical sample to complete genome: Comparing methods for the extraction of HIV-1 RNA for high-throughput deep sequencing. Virus Res. 2016 [DOI] [PubMed]
- 74.Novitsky V, Zahralban-Steele M, McLane MF, Moyo S, van Widenfelt E, Gaseitsiwe S, et al. : Long-range HIV genotyping using viral RNA and proviral DNA for analysis of HIV drug resistance and HIV clustering. J Clin Microbiol 2015;53:2581–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Batty EM, Wong TH, Trebes A, Argoud K, Attar M, Buck D, et al. : A modified RNA-Seq approach for whole genome sequencing of RNA viruses from faecal and blood samples. PLoS One 2013;8:e66129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, et al. : Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res 2013;41:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonsall D, Ansari MA, Ip C, Trebes A, Brown A, Klenerman P, et al. : ve-SEQ: Robust, unbiased enrichment for streamlined detection and whole-genome sequencing of HCV and other highly diverse pathogens. F1000Res 2015;4:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PANGEA-HIV sequencing output is available from the European Nucleotide Archive project page (www.ebi.ac.uk/ena/data/view/PRJEB19239). PANGEA-HIV consensus sequences and associated demographic data are available on request to the PANGEA-HIV steering committee (Supplementary Data). Simulated sequence data and viral trees are available as from https://doi.org/10.6084/m9.figshare.5056837.v1