Abstract
Hemigrapsus sanguineus, the Asian shore crab, has rapidly replaced Carcinus maenas, the green crab, as the most abundant crab on rocky shores in the northwest Atlantic since its introduction to the United States (USA) in 1988. The northern edge of this progressing invasion is the Gulf of Maine, where Asian shore crabs are only abundant in the south. We compared H. sanguineus population densities to those from published 2005 surveys and quantified genetic variation using the cytochrome c oxidase subunit I gene. We found that the range of H. sanguineus had extended northward since 2005, that population density had increased substantially (at least 10-fold at all sites), and that Asian shore crabs had become the dominant intertidal crab species in New Hampshire and southern Maine. Despite the significant increase in population density of H. sanguineus, populations only increased by a factor of 14 in Maine compared to 70 in southern New England, possibly due to cooler temperatures in the Gulf of Maine. Genetically, populations were predominantly composed of a single haplotype of Japanese, Korean, or Taiwanese origin, although an additional seven haplotypes were found. Six of these haplotypes were of Asian origin, while two are newly described. Large increases in population sizes of genetically diverse individuals in Maine will likely have a large ecological impact, causing a reduction in populations of mussels, barnacles, snails, and other crabs, similar to what has occurred at southern sites with large populations of this invasive crab species.
Keywords: Invasive species, Crab, Population genetics, Range expansion, Asian shore crab, Green crab, Mitochondrial DNA, Gulf of Maine
Introduction
Invasive species can have significant effects on biodiversity by reducing native species richness, altering evolutionary pathways, or even causing extinctions (Ruiz and Carlton 1997; Mooney and Cleland 2001; Clavero and García-Berthou 2005). In marine environments, introductions of lionfish in the northwest Atlantic (Albins 2013) and the ctenophore Mnemiopsis leidyi (Shiganova 1998) in the Black Sea have led to drops in the abundance and diversity of native fish species. One of the biggest challenges in dealing with biological invasions is preventing population growth and further spread after the initial introduction stage. In the northwest Atlantic, there are two ecologically dominant invasive crabs in markedly different stages of their invasion of rocky shore habitats: the green crab Carcinus maenas (Linnaeus, 1758) and the Asian shore crab Hemigrapsus sanguineus (De Haan, 1835).
Green crabs have established populations all over the world, mostly in the nineteenth and early twentieth centuries, and are among the most widespread marine invasive species (Grosholz and Ruiz 1996). In the northwest Atlantic, C. maenas was introduced to New Jersey, USA, by 1817 (Say 1817), and had spread northward to Maine, USA, by 1905 and the east coast of Canada by 1950 (Audet et al. 2003). Green crabs have had a substantial ecological impact, as they have negatively affected clam, mussel, barnacle, and snail populations and play an important role in shaping rocky intertidal communities (Lubchenco 1978; Floyd and Williams 2004; Tyrrell et al. 2006). However, green crabs have largely been replaced in rocky intertidal habitats along the mid-Atlantic and southern New England coast of the USA by Asian shore crabs over the last two decades.
Asian shore crabs were introduced near Cape May, NJ, USA, prior to 1988 via ship ballast water, a common vector for marine invasive species with a pelagic larval stage (McDermott 1991; Carlton and Geller 1993; Epifanio 2013). This species spread rapidly to North Carolina in the south and by 2002 had reached northward to southern Maine (Stephenson et al. 2009; Epifanio 2013). The northward spread of H. sanguineus is likely limited by thermal tolerance of the megalopa larval stage, though this has not been tested in the northern extent of its range (Epifanio 2013). Asian shore crabs have a combined larval (zooea + megalopa) duration of approximately 25 days, long enough for long-distance dispersal via coastal currents (Epifanio 2013; Epifanio et al. 2013). In addition to the east coast of the USA, H. sanguineus spread to France and the Netherlands by 1999, the Black Sea by 2008, and the UK by 2014, so this crab species is a burgeoning problem in several parts of the world (Breton et al. 2002; Micu et al. 2010).
Asian shore crabs are successful invasive species in part because of their life history. Females can produce over 40,000 eggs per brood, only brood for 22 days, and have multiple broods in one summer, unlike green crabs (McDermott 1998). They are omnivorous, as they consume mussels, snails, clams, oysters, barnacles, amphipods, and worms, but also scavenge and can consume a variety of algae (Brousseau et al. 2001; Ledesma and O’Connor 2001; Bourdeau and O’Connor 2003; Tyrrell et al. 2006; Blasi and O’Connor 2016). Asian shore crabs compete with green crabs for both food and space under rocks and are able to outcompete them because H. sanguineus is more aggressive than C. maenas (Jensen et al. 2002), has higher feeding rates (DeGraaf and Tyrrell 2004), has stronger claws for its size (Payne and Kraemer 2013), different feeding activity rhythms (Spilmont et al. 2015) and consume juvenile C. maenas (Lohrer and Whitlatch 2002a; Payne and Kraemer 2013). Competition between these crab species has led to massive declines in rocky shore populations of C. maenas, as well as several other intertidal species in the invaded range of H. sanguineus (Epifanio 2013; O’Connor 2014). The northwest Atlantic coast is the only location with substantial populations of both green and Asian shore crabs, as Asian shore crabs have only recently become established in the native European range of green crabs.
In western Long Island Sound, 25% of mussel (Mytilus edulis) mortality has been attributed to H. sanguineus predation, as these crabs can consume up to 150 juvenile mussels per day (Lohrer and Whitlatch 2002b; Brousseau et al. 2014). Asian shore crabs have also caused declines in populations of barnacles, spirorbid worms, littorine snails, mud crabs, and fiddler crabs, which they have recently begun to outcompete for burrows in salt marshes as they move into this new habitat (Tyrrell et al. 2006; Kraemer et al. 2007; Peterson et al. 2014). Asian shore crabs can also consume juvenile lobsters, which they have the additional potential to compete with in rocky cobble habitats (Demeo and Riley 2006; Lord and Dalvano 2015). The spread of H. sanguineus has been well-chronicled in the mid-Atlantic and southern New England, as surveys of H. sanguineus populations have documented the population explosion of this invasive crab (Stephenson et al. 2009; O’Connor 2014). Population densities at several sites in Massachusetts and Rhode Island increased from only a few crabs per square meter to over 100-m−2 in just a few years per site in the early 2000s (O’Connor 2014). While invasive H. sanguineus population densities have been tracked over the past few decades, there has been no documentation as to the genetic diversity of these individuals.
Population genetics is frequently used as a powerful tool to examine differences between individuals and populations and can be used to infer parameters related to dispersal and migration. Analyses using the maternally-derived mitochondrial cytochrome c oxidase subunit I gene have been widely utilized in crab population genetic studies, most notably to study the invasion history of the green crab along the northwest Atlantic coast (Roman 2006; Darling et al. 2008, 2014; Blakeslee et al. 2010; Darling 2011; Williams et al. 2015). Only one study, however, has documented H. sanguineus COI genetic diversity, and this was conducted in its native range (Yoon et al. 2011). Four sites in Korea and one in Japan were diverse, with 6–10 haplotypes identified at each site totaling 28 unique haplotypes (Yoon et al. 2011). Yoon et al. (2011) suggested that the high haplotype diversity and low nucleotide diversity was due to rapid population growth from a small ancestral population (Yoon et al. 2011). Using Korean populations and a different mitochondrial marker, the cytochrome b (Cytb) gene, assumptions about populations dynamics proposed in their 2011 study were supported (Hong et al. 2012). While H. sanguineus invaded the northwest Atlantic 28 years ago (McDermott 1991), no genetic analyses have been leveraged to learn about the origin of these initial invaders and the subsequent population dynamics along this range.
Genetic analysis is particularly important for invasive species, as it can reveal evidence of multiple invasions, even from source populations with different temperature tolerances. For example, green crabs introduced in the early nineteenth century were of a singular “southern” haplotype (Carlton and Cohen 2003). These crabs were successful at invading much of the northern east coast of the USA, but didn’ t extend much into colder waters of Canada until late in the twentieth century. Starting in the 1980s, there were subsequent introductions of genetically diverse, cold tolerant, “northern” haplotypes from Europe along the Nova Scotian coast (Roman 2006). These introductions assisted in the expansion of the range into Prince Edward Island and Newfoundland and the development of an introgression zone between northern and southern haplotypes as far north as Newfoundland (Roman 2006; Blakeslee et al. 2010; Darling et al. 2014; Williams et al. 2015). The origin of invasive species provides clues about their thermal tolerance and potential spread in their invaded range, and temperature is a factor that may play a large role in limiting spread of H. sanguineus (Stephenson et al. 2009; Epifanio 2013).
The aim of the present study was to compare invasive Asian shore crab population growth rates near its northern range limit with those in the central part of its geographic range and determine adult genetic variation along that same range. This is important from both an ecological and management perspective, as efforts to mitigate impacts of invasive species focus on preventing both spread to new sites and expansion of established populations. We examined the progression of the H. sanguineus invasion in northern New England and compared population densities to those of other intertidal crab species. The green crab invasion was not well-documented in the 1800s and early 1900s, and this is an important opportunity to determine the spreading rates of the H. sanguineus invasion in its early to mid-stages. Comparing H. sanguineus densities at 19 sites in 2015 to those from 2005 surveys by Stephenson et al. (2009) allowed us to calculate 10-year population expansion rates and quantify northward expansion of this species. We were then able to compare these expansion rates to 10-year expansion rates from 2001 to 2011 in southern New England where water temperatures are warmer (O’Connor 2014). Furthermore, this is the first published study to determine the extent of genetic variation of this crab outside its native range. We hypothesized that H. sanguineus would be found further north in 2015 and that populations in southern Maine would be substantially larger in 2015 than 2005, paralleling the expansion rates documented by O’Connor (2014) in Massachusetts and Rhode Island. In larger populations that were closer to the original site of invasion, it was expected that genetic variation would be greater as compared to more northern sites where populations were expected to be newer and smaller.
Methods
Sites
Surveys were conducted at 19 total rocky intertidal sites along the northwest Atlantic coast in June and early July 2015 in order to assess population densities of Hemigrapsus sanguineus. Surveys were conducted during the summer because Asian shore crabs bury in sediment or move to the subtidal zone during the winter (Stephenson et al. 2009). Seventeen of the 19 total survey sites were chosen because they were surveyed in 2005 by Stephenson et al. (2009) and were spaced approximately 30-km apart (Table 1; Fig. 1). This allowed for a 10-year comparison of population densities at these 17 sites. An additional survey site (Groton CT) from southern New England with a long-established H. sanguineus population was surveyed for the sake of comparison with the other 18 sites, which were in New Hampshire (1 site) or Maine (17 sites) (Table 1; Fig. 1). Initial surveys were conducted from south to north to minimize seasonal variability, with 3 additional sites in central Maine surveyed at later dates (still in June) to gather higher resolution data within a central Maine transition zone that displayed a rapid drop in H. sanguineus population densities (Sites 17–19 in Table 1). Site temperatures, locations, and sampling dates are shown in Table 1. Adult crabs were collected at an additional site in New Jersey (Table 1) solely for the purpose of genetic analysis and no survey was conducted. Temperatures for each site were obtained from NOAA’s AVHRR (Advanced Very High Resolution Radiometer) SST maps (resolution 1.1 km) for the nearest quadrant to each site during the 3-day window around the sampling date (Table 1) in order to show broad thermal patterns, though this was solely for general comparisons, as no analyses were based on temperature.
Table 1.
Dates and characteristics of all sites surveyed in summer 2015
| # | Town | Site location | Site label | Date | Latitude | Longitude | Percent cobble | SST (°C) |
|---|---|---|---|---|---|---|---|---|
| 20* | Avalon | Townsend Inslet | NJ | 8/15 | 39.1174 | −74.7158 | N/A | 22.0 |
| 1* | Groton CT | Avery Point | CT | 6/10 | 41.3170 | −72.0616 | 100 | 12.2 |
| 2 | Rye NH | Odiorne Point State Park | OD | 6/13 | 43.0527 | −70.7184 | 100 | 10.6 |
| 19 | Kittery | Seapoint Beach | KT | 6/25 | 43.0863 | −70.6593 | 95 | 10.8 |
| 3 | York | Long Sands Beach | YK | 6/13 | 43.1588 | −70.6205 | 60 | 10.5 |
| 4 | Biddeford Pool | East Point Sanctuary | BP | 6/14 | 43.4481 | −70.3348 | 30 | 10.2 |
| 5 | Cape Elizabeth | Two Lights Park | CE | 6/15 | 43.5584 | −70.2060 | 25 | 10.3 |
| 6 | Baileys Island | Lands End | LE | 6/16 | 43.7175 | −70.0031 | 80 | 10.3 |
| 7 | Bristol | Pemaquid Point Light | PP | 6/17 | 43.8364 | −69.5075 | 30 | 10.2 |
| 17 | New Harbor | Rachel Carson Salt Pond | NH | 6/24 | 43.8801 | −69.4826 | 70 | 10.1 |
| 18* | Chamberlain | Chamberlain Point | CH | 6/24 | 43.8855 | −69.4751 | 30 | 10.1 |
| 8 | St George | Marshall Point Light | MP | 6/17 | 43.9173 | −69.2670 | 40 | 9.4 |
| 9 | Owls Head | Owls Head Light | OH | 6/18 | 44.0900 | −69.0456 | 40 | 8.9 |
| 10 | Searsport | Moose Point State Park | MO | 6/19 | 44.4308 | −68.9422 | 90 | 8.8 |
| 11 | Stonington | Town Pier | ST | 6/20 | 44.1543 | −68.6655 | 15 | 8.3 |
| 12 | Bass Harbor | Town Pier | BH | 6/21 | 44.2403 | −68.3521 | 40 | 6.7 |
| 13 | Lamoine | Lamoine State Park | LM | 6/21 | 44.4510 | −68.2998 | 70 | 7.8 |
| 14 | Steuben | Petit Manan NWR | PM | 7/03 | 44.4236 | −67.8874 | 80 | 6.7 |
| 15 | Jonesport | Jonesport Campground | JP | 7/02 | 44.5313 | −67.5895 | 70 | 5.8 |
| 16 | Dennysville | Cobscook Bay State Park | CB | 7/03 | 44.8527 | −67.1470 | 40 | 5.5 |
Percent cobble is the approximate overall percent of that site that was composed of cobble habitat. Site labels correspond to those on the maps in Figs. 1 and 2. Site numbers 1–16 were initial sampling sites, while 17–19 were additional sites added to provide further detail in the transition zone. Site 20 was included for genetic analysis. Sites are displayed in order from south to north. The three site numbers with asterisks (*) were the only ones not included in the Stephenson et al. (2009) surveys from 2005
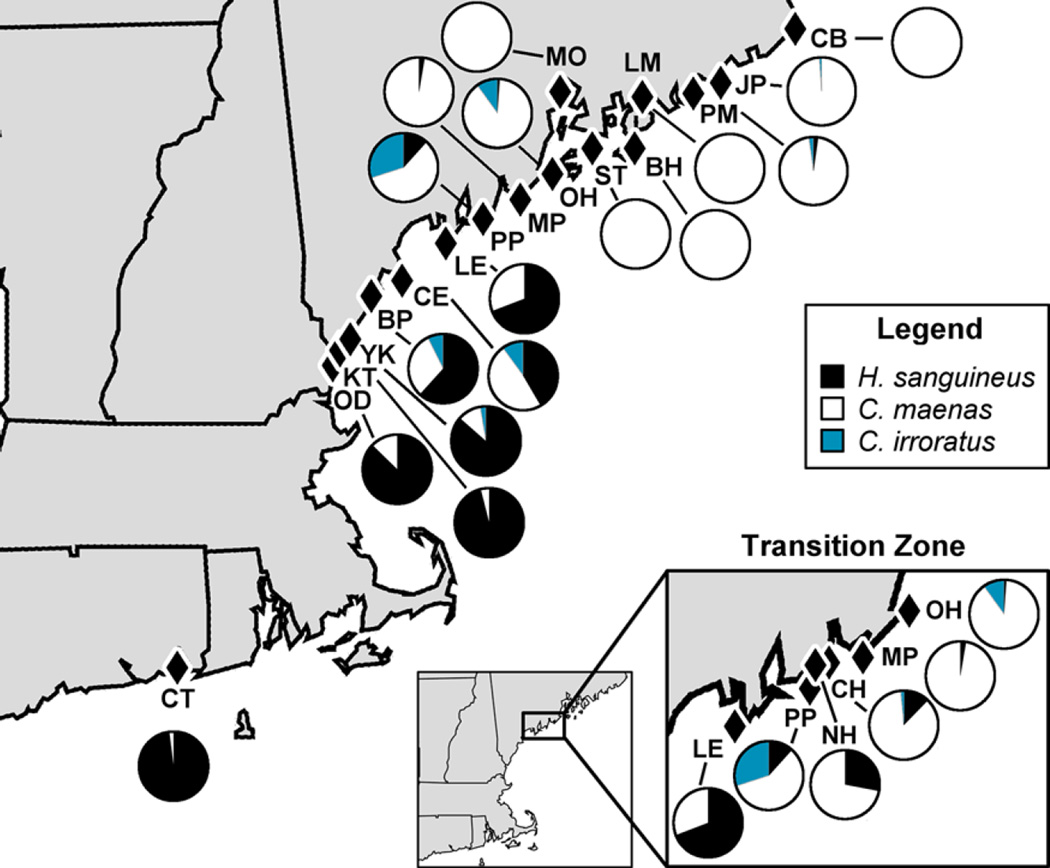
Fig. 1.
Proportion of all three crab species for all 19 sites surveyed in summer 2015. The highlighted transition zone in central Maine is the region in which numerical dominance in rocky cobble sites switches from Asian shore crabs (H. sanguineus) to green crabs (C. maenas). Rock crabs (C irroratus) were relatively sparse in the intertidal zone throughout the survey region. Site labels correspond to site descriptions and crab densities in Tables 1 and 2
Quadrat surveys
In order to directly compare crab densities with those found by previous researchers, we used methods modified from Stephenson et al. (2009). We did not use the vertical timed searches included in the Stephenson et al. study, but used the same quadrat sampling methods that those researchers used to estimate H. sanguineus density. As in Stephenson et al. (2009), 1 m2 quadrats were used to estimate crab density and were placed in locations with maximum suitable cobble habitat (non-random). At each site, we placed the quadrats in areas that were near 100% cobble, where we could flip over all of the rocks in the quadrat. We conducted quadrat surveys in the high (4 quadrats), mid (4 quadrats), and low (4 quadrats) intertidal zones at all 16 initial sites, then only in the high and mid zones at the three additional sites, due to tidal constraints. Tidal zones were determined by algal zonation, as in Stephenson et al. (2009), with high quadrats at the upper limit of macroalgae, mid quadrats in the center of the fucoid algae zone (mostly Ascophyllum nodosum), and low quadrats in the lowest algal zone (primarily Chondrus crispus and coralline algae) near 0.0 MLLW. We flipped over all rocks, moved seaweed, and scraped through sediment to find all crabs in each quadrat, including Asian shore crabs (H. sanguineus), green crabs (C. maenas), and rock crabs [Cancer irroratus (Say 1817)]. Carapace width (CW), sex, and the presence of eggs were recorded for each of the three crab species for all individuals over 10 mm CW. Stephenson et al. (2009) did not mention a minimum crab size in their study, but we excluded crabs under 10 mm CW because they could not be captured as reliably; as such, our population density estimates were conservative. With this exception, our quadrat sampling methods were as similar as possible to Stephenson et al. (2009) in order to make accurate comparisons of population density. The present study also included green crabs and rock crabs in the survey, which were not described by Stephenson et al. (2009).
DNA extraction and sequencing
For DNA analysis, 25 H. sanguineus individuals per sampling site were sequenced except for Steuben (PM), where only five crabs were found. Three locations (Kittery, KT; New Harbor, NH; Chamberlain, CH) were follow-up sites that were surveyed solely to gain higher resolution density data; at these sites, H. sanguineus was observed but was not included in DNA analysis. Owls Head (OH) was also excluded from genetic analysis as only a single Asian shore crab was found. A single site from New Jersey at the location of the initial introduction to the USA was included in the genetic diversity data set (Table 1). Samples from different tidal levels were pooled together (within each site) for genetic analysis. When more than 25 H. sanguineus were found at a site, crabs to be used for genetic analysis were haphazardly selected from a bucket containing all crabs collected at that site.
DNA was extracted from pereopod muscle tissue using the Qiagen DNeasy Tissue kit and purified by ethanol precipitation. The DNA pellet was rinsed with EtOH (95%) before it was air-dried and re-suspended in molecular-grade water. A 512 bp region of COI was amplified by PCR (2 min at 94 °C, then 30 cycles of 15 s at 94 °C, 30 s at 53 °C, 1 min at 72 °C, then 72 °C for 7 min) using REDTaq® ReadyMix™ PCR Reaction Mix (Sigma-Aldrich) and COI universal primers LCO1490f (5′-GGTCCAACAAATCATAAAGATATTGG-3′) and HC02198r (5′-TAAACTTCAGGGTGACCAAAAAATCA) (Folmer et al. 1994). The amplified product was purified using the QIAquick® PCR Purification Kit (Qiagen). The PCR products were sequenced directly using amplification primers in both directions.
Density and distribution analysis
We calculated the proportion and overall density of each of the three crab species at all 19 sites in order to show relative and absolute abundance, respectively. Rock crabs were not found at most sites, so were excluded from most of the analyses. We used a 3-way ANOVA to test the effect of site, tidal level, and species on crab densities for H. sanguineus and C. maenas. We analyzed the sex ratios and percent of ovigerous females for H. sanguineus and C. maenas separately with 2-way ANOVAs because there were few sites at which enough of both species were found to use a 3-way ANOVA setup. We arcsine-trans-formed all sex ratio and ovigerous female data prior to the 2-way ANOVA to statistically analyze non-normal proportion data.
Carapace width was also compared between sites and tidal levels with a 2-way ANOVA in separate tests for H. sanguineus and C. maenas. The surveys in this study were designed to estimate density and repeat a previous survey (Stephenson et al. 2009), not to conduct complex population structure analysis which would require larger and more uniform numbers of collected species per site. However, because H. sanguineus populations expanded and were the focus of this study, we used our collected size-frequency data to create histograms and visually compare H. sanguineus population structure between sites. To develop more robust histograms, we combined H. sanguineus across all tidal levels within each site for this analysis (H. sanguineus size did not vary significantly by tidal level).
After we calculated population densities of H. sanguineus we were able to compare to the 4 sites which had established H. sanguineus populations in 2005 (Stephenson et al. 2009) and determine the 10-year factor of increase (FOI) in population density. We then compared this with the 10-year FOI at 4 sites monitored by O’Connor (2014) in southern New England, using a two-sample T test to test for differences in FOI between regions.
Genetic analysis
Using forward and reverse sequences, consensus sequences were produced for each individual using ClustalW (Larkin et al. 2007) and subsequently checked by hand. All individuals were aligned in ClustalX 2.1 (Larkin et al. 2007). Using DnaSP version 5.10.1 (Librado and Rozas 2009), within and between population haplotype diversity (h) (Nei 1987) and nucleotide diversity (π) (Tajima 1983) were estimated. Pairwise population FST (a measure of pairwise population differentiation) and Nem (a measure of temporal gene flow between populations) values were calculated with the Arlequin program version 3.5 using the methods of Tajima and Nei (1984), Slatkin and Hudson (1991), and Slatkin (1993). A parsimony network connecting the haplotypes was plotted with TCS version 1.21 (Clement et al. 2000).
Results
At sites in Connecticut, New Hampshire, and southern Maine, Hemigrapsus sanguineus was the most abundant crab species in cobble habitats in 2015 surveys, with relatively small populations of Carcinus maenas and Cancer irroratus (Fig. 1). This shifted in an apparent transition zone in central Maine, as 69% of crabs at Baileys Island were H. sanguineus, compared to only 1% at Owls Head, 88 km to the northeast (Fig. 1). Rock crabs (C. irroratus) were relatively rare at most sites, so after the transition zone all sites were numerically dominated by green crabs (Fig. 1).
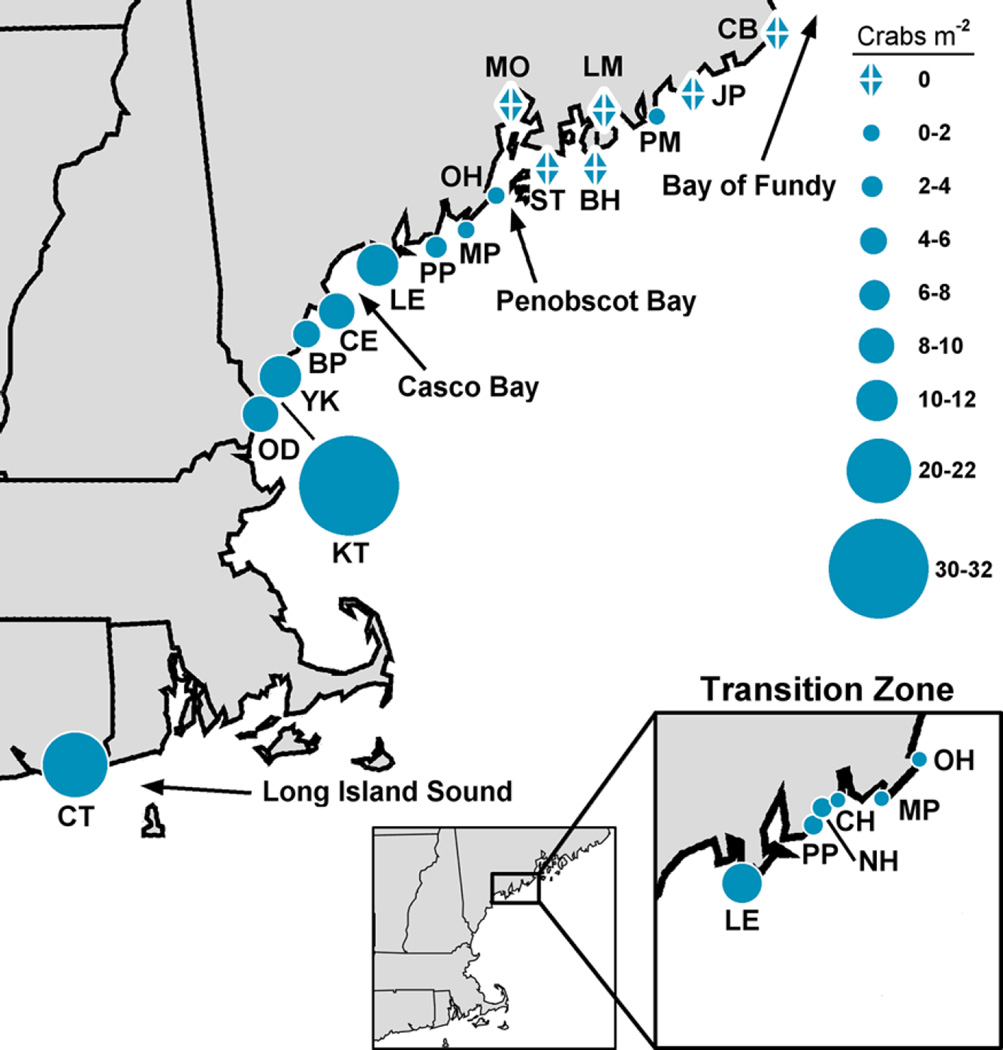
Crab population densities in 2015 varied significantly by site and species but not by tidal level (three-way ANOVA; site F15,288 = 9.26, P < 0.001; species F1,288 = 7.15, P = 0.008; tidal level F2,288 = 2.34, P = 0.10). Because of the differential south-north density patterns of C. maenas and H. sanguineus, there was a significant 3-way ANOVA interaction term for site × species and for site × tidal level × species. Densities of green crabs were substantially higher north of Baileys Island in central Maine (x̄ ± SE, 6.81 ± 1.00 crabs-m−2) than they were in southern sites where H. sanguineus was more abundant (2.96 ± 1.22 crabs-m−2). In contrast, Asian shore crabs were found in distinctly higher densities in the southern (13.40 ± 3.37 crabs-m−2) than northern (0.59 ± 0.31 crabs-m−2) Gulf of Maine, peaking at 30.13 crabs-m−2 in Kittery, ME (Table 2; Fig. 2). Densities of H. sanguineus varied widely by site in 2015 but were over 4.1 crabs-m−2 at all sites south of Casco Bay, ME, including Baileys Island on the north side of Casco Bay. This was a large increase from 2005 surveys by Stephenson et al. (2009) which found H. sanguineus at 1.75 crabs-m−2 in Kittery but no more than 1 crab-m−2 at any other site. Those 2005 surveys only found trace (1–3 total crabs per site) levels of H. sanguineus at sites north of Casco Bay, compared to considerable densities (1–3 crabs-m−2) in the present study in central Maine (Table 2; Fig. 2). Rock crabs were primarily found in the low intertidal zone but were not abundant enough across all sites to make any statistical comparisons including this species.
Table 2.
Density of crabs at all 19 sites sampled
| Town | Site label | Factor of increase | Mean density (crabs-m−2) |
Peak density (crabs-m−2) |
||||
|---|---|---|---|---|---|---|---|---|
| HS | CM | CI | HS | CM | CI | |||
| Groton CT | CT | 20.75 | 0.42 | 0 | 40 | 2 | 0 | |
| Rye NH | OD | 9.17 | 1.33 | 0 | 21 | 7 | 0 | |
| Kittery | KT | 17.21 | 30.13 | 1.25 | 0 | 51 | 6 | 0 |
| York | YK | 10.67 | 1.33 | 0.33 | 32 | 5 | 2 | |
| Biddeford Pool | BP | 10.43 | 4.17 | 2.08 | 0.50 | 8 | 7 | 4 |
| Cape Elizabeth | CE | 22.30 | 8.25 | 9.58 | 2.00 | 21 | 47 | 12 |
| Baileys Island | LE | 17.78 | 10.67 | 4.75 | 0 | 25 | 9 | 0 |
| Bristol | PP | 2.92 | 14.58 | 7.50 | 11 | 44 | 30 | |
| New Harbor | NH | 2.63 | 6.75 | 0 | 6 | 13 | 0 | |
| Chamberlain | CH | 1.00 | 7.00 | 0.13 | 5 | 11 | 1 | |
| St George | MP | 0.33 | 12.08 | 0 | 1 | 21 | 0 | |
| Owls Head | OH | 0.08 | 6.67 | 0.75 | 1 | 13 | 4 | |
| Searsport | MO | 0 | 6.50 | 0 | 0 | 14 | 0 | |
| Stonington | ST | 0 | 2.83 | 0 | 0 | 11 | 0 | |
| Bass Harbor | BH | 0 | 6.17 | 0 | 0 | 15 | 0 | |
| Lamoine | LM | 0 | 2.83 | 0 | 0 | 8 | 0 | |
| Steuben | PM | 0.08 | 3.50 | 0.08 | 1 | 13 | 1 | |
| Jonesport | JP | 0 | 6.92 | 0.08 | 0 | 12 | 1 | |
| Dennysville | CB | 0 | 5.83 | 0 | 0 | 13 | 0 | |
Mean density is the density averaged across all three tidal levels (N = 12), and peak density is the highest density found at that site for that species. Species are abbreviated as HS (H. sanguineus), CM (C. maenas), and CI (C. irroratus). Sites are shown in order from south to north. Density data are displayed in Fig. 2
Fig. 2.
Densities of Asian shore crabs (H. sanguineus) at all surveyed sites. Highest densities are in Connecticut, New Hampshire and southern Maine, decreasing in the transition zone highlighted in the lower right hand corner. Site labels correspond to site descriptions and crab densities in Tables 1 and 2
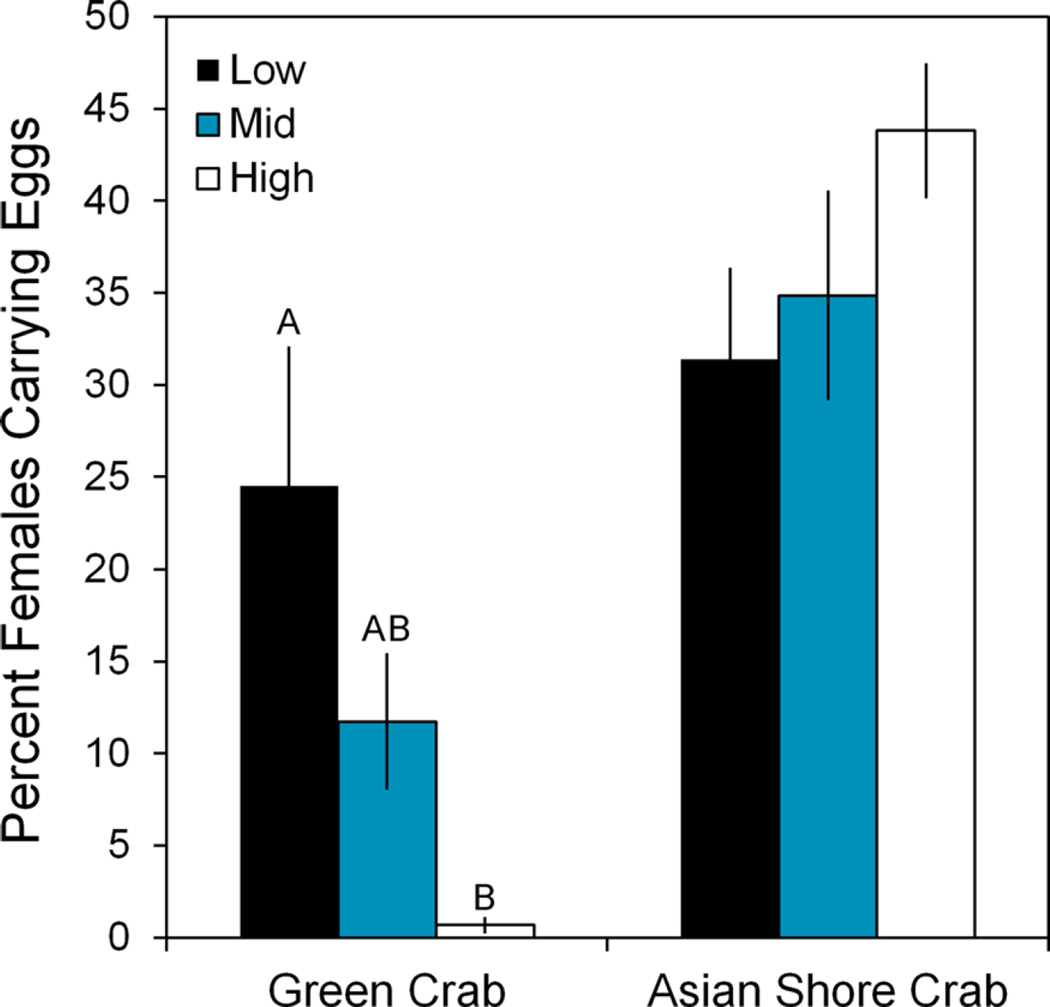
There was no significant difference in 2015 crab sex ratio with site or tidal level for either H. sanguineus (Two-way ANOVA, site P = 0.36; tidal level P = 0.39) or C. maenas (two-way ANOVA, site P = 0.17; tidal level P = 0.54). There was also no difference in the percent of ovigerous H. sanguineus females with site or tidal level (two-way ANOVA, site P = 0.37; tidal level P = 0.43). However, the percent of ovigerous female green crabs was significantly higher in the low intertidal zone (x̄ ± SE, 24.5 ± 7.58%) than in the mid (11.7 ± 3.97%) or high zone (0.7 ± 0.50%) across all sites (Two-way ANOVA, site F9,90 = 2.86, P = 0.005; tidal level F2,90 = 8.44, P < 0.001; interaction F18,90 = 1.93, P = 0.023) (Fishers LSD post hoc test, low v. high P < 0.001; low v. mid P = 0.057; mid v. high P = 0.032) (Fig. 3). The significant interaction term indicates that the percent of ovigerous female green crabs did not vary by tidal level in a consistent manner across all sites, as some sites had few ovigerous females even in the low intertidal zone.
Fig. 3.
Percent of females carrying eggs at each tidal level (±SE). Green crabs (C. maenas) had the highest egg carrying percentage in the low zone, followed by the mid zone, before dropping to less than 1% in the high zone. Asian shore crabs (H. sanguineus) did not show any significant difference with regard to tidal level. Letters indicate significant differences at P = 0.05. There were no significant differences (α = 0.05) between tidal levels for H. sanguineus
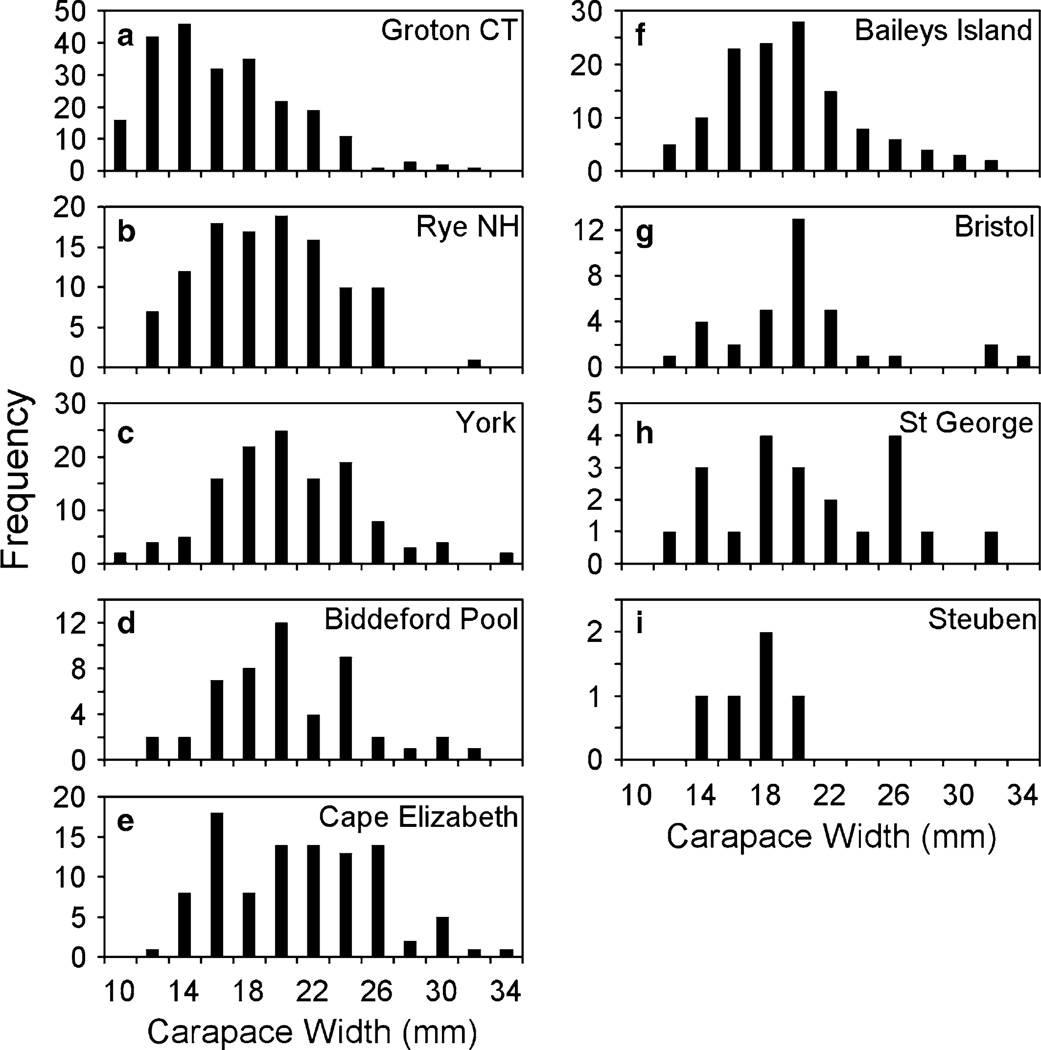
Green crab carapace width varied significantly with tidal level and site, though there was a significant interaction suggesting that the tidal pattern was not consistent across sites in 2015 (two-way ANOVA, tidal level F2,101 = 5.78, P = 0.004; site F11,101 = 7.64, P < 0.001; interaction F22,101 = 2.57, P < 0.001). Carapace widths of C. maenas in the low intertidal (29.45 ± 0.84 cm) were significantly higher than those in the high intertidal (x̄ ± SE, 25.51 ± 0.80 cm) (Fishers LSD post hoc test, P < 0.01). Asian shore crab carapace widths also varied significantly by site, though there was no difference with tidal level and the only site differences were between the Groton, CT, site and all northern sites (Fishers LSD post hoc tests, P < 0.05) (two-way ANOVA, tidal level F2,54 = 0.16, P = 0.85; site F6,54 = 4.23, P = 0.001; interaction F12,54 = 1.14, P = 0.35). While surveys were designed to estimate density, not conduct population structure analysis, visual comparison of size-frequency histograms for H. sanguineus at all sites showed that the Connecticut population had a higher relative abundance of small individuals than any of the sites in northern New England (Fig. 4). Crab size-frequency distributions were fairly similar for sites in New Hampshire and southern Maine (Fig. 4b–f) but were more uneven at the central Maine sites (Fig. 4g–i).
Fig. 4.
Histograms of Asian shore crab sizes at survey sites in New England. Sites are displayed from south to north, with highest H. sanguineus densities at more southern sites (a–f). Graphs that are not labeled with state abbreviations (c–i) were all sites in Maine. Southern sites show a wide range of sizes with no obvious cohorts, while sites in the transition zone (shown in Figs. 1, 2) have multiple peaks that likely correspond to successful cohorts (g–i)
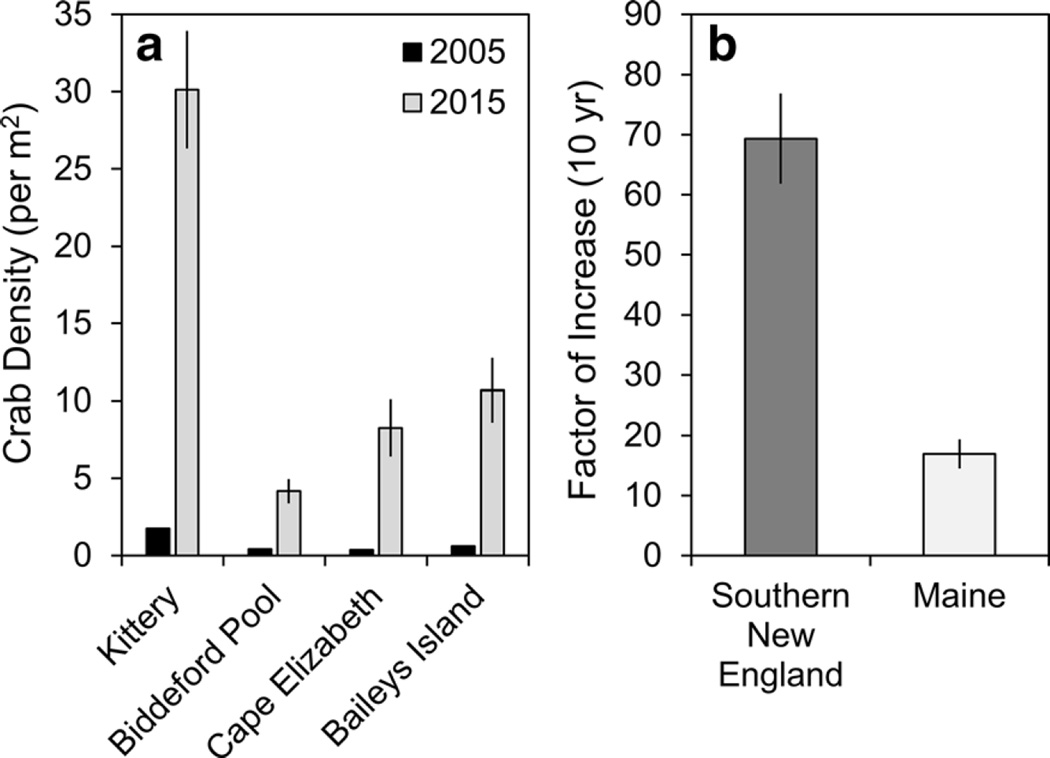
Increases in H. sanguineus population densities from 2005 (Stephenson et al. 2009) to 2015 were substantial, as densities at all sites increased by at least a factor of 10 (Fig. 5a). This 10-year factor of increase (FOI) was compared between Maine (2005–2015) and southern New England (2001–2011) (O’Connor 2014), showing a significantly more rapid increase in southern New England (two-sample T-test, df = 4, T = −6.60, P = 0.0027) (Fig. 5b). Asian shore crab densities at all sites in Rhode Island and Massachusetts increased by at least a factor of 47, for a mean FOI of 69.4, compared to 16.9 for Maine (Table 2; Fig. 5).
Fig. 5.
Changes in population density of H. sanguineus over 10 years (±SE). The 10-year factor of increase in H. sanguineus density in Maine (a) was over 17-fold at 4 sites from 2005 to 2015. This Maine factor of increase was ×4.1 lower than the 10-year increase in southern New England described by O’Connor (2014) for sites in Rhode Island and Massachusetts (b). Southern New England data were taken from O’Connor (2014) and northern New England 2005 survey data were taken from Stephenson et al. (2009)
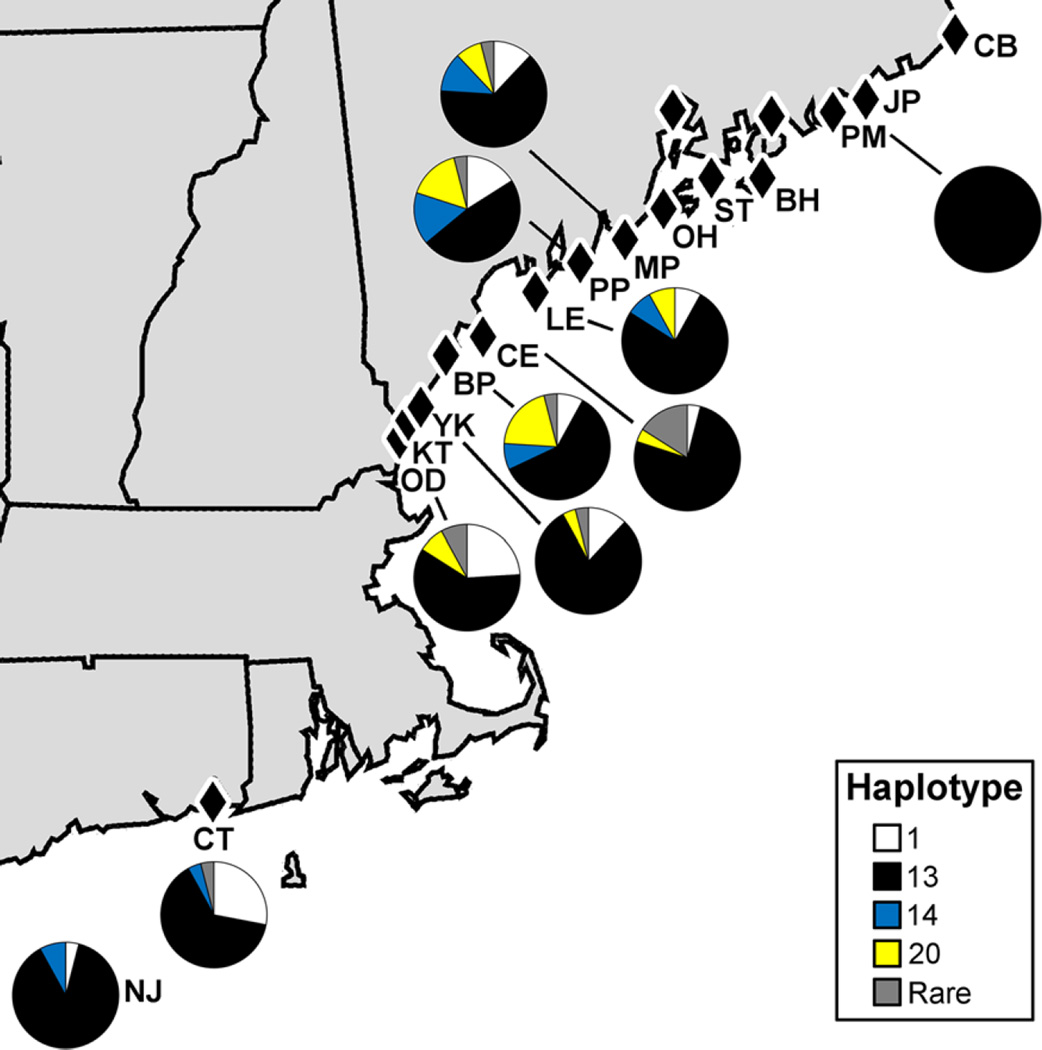
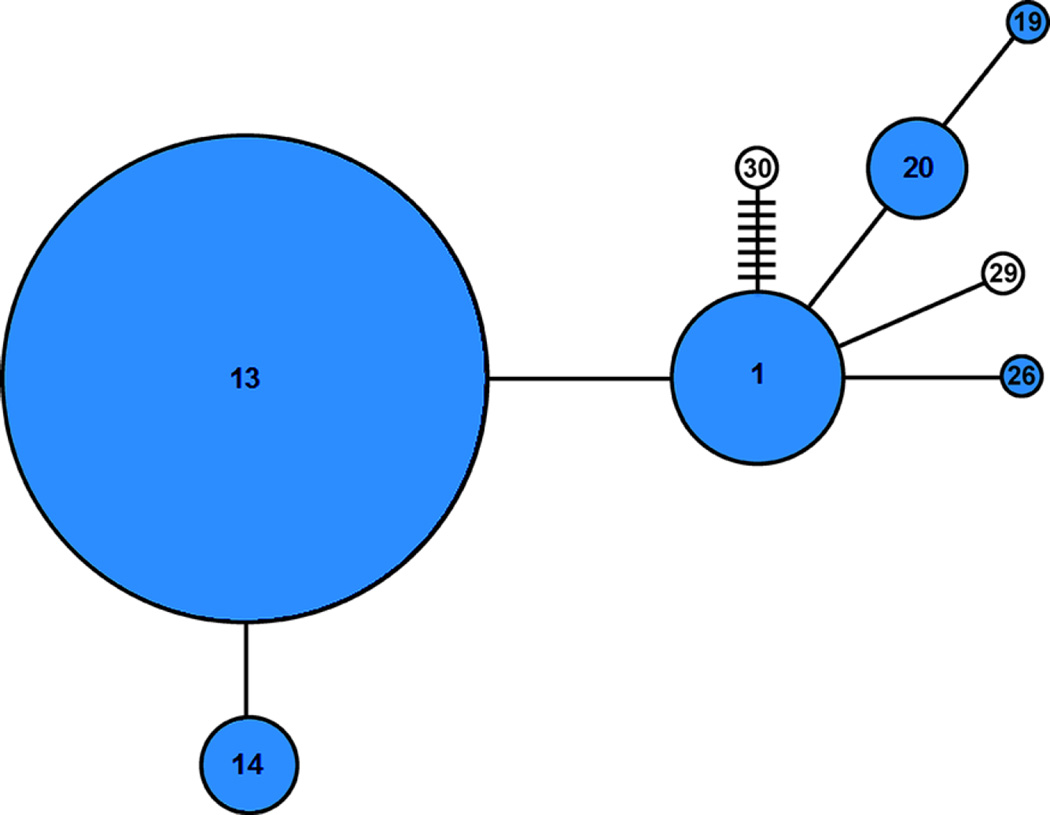
Among 230 individuals from 10 sampling sites that were sequenced, a 512 base pair region of the COI gene revealed 8 haplotypes (Table 3; Suppl Table 1) comprised of 13 biallelic variable sites (Suppl Table 1). Of these haplotypes, six were previously identified in Japan and/or Korea (Yoon et al. 2011: GenBank HQ702865, HQ70286577, HQ70286578, HQ70286583, HQ70286584, and HQ70286590). The most prominent and focal haplotype, haplotype 13 (HQ70286577), was found in all sampling sites (Figs. 6, 7; Suppl Table 2). Haplotype 1 (HQ702865) was found in all sites except for the most northern (Fig. 6; Suppl Table 2). Haplotypes 14 and 20 were also found consistently along the sampling range, with haplotype 14 ranging from NJ to MP and haplotype 20 ranging from OD to MP (Fig. 6; Suppl Table 2). Haplotype 19 was found in two individuals at 1 site in Maine (CE) and haplotype 26 was found in 1 individual from Connecticut (CT) and 1 from New Hampshire (OD) (Fig. 6; Suppl Table 2). Two newly identified haplotypes, named haplotype 29 and haplotype 30, respectively (GenBank KX579065, KX579066), were also found. Haplotype 29 was found in a single individual in each of four sites in New Hampshire and Maine (OD, BP, CE, MP) and haplotype 30 found in a single individual in each of 3 sites in southern Maine (YK, CE, PP) (Fig. 6; Suppl Table 2). As compared to the focal haplotype, haplotype 13, substitutions in all haplotypes were synonymous (Fig. 7; Suppl Table 1). In all sites except PM, the most northern site, haplotype diversity was high and ranged from 0.3072 to 0.7662 (Table 3). Nucleotide diversity, on the other hand, was very low and ranged from 0 to 0.0035 (Table 3).
Table 3.
Measures of COI genetic diversity in H. sanguineus across 10 sampling sites
| Sampling site | Number of haplotypes | Haplotype diversity (h ± SD) | Nucleotide diversity (π) |
|---|---|---|---|
| NJ | 3 | 0.3072 ± 0.025 | 0.0006 |
| CT | 4 | 0.6140 ±0.031 | 0.0014 |
| OD | 5 | 0.7417 ± 0.044 | 0.0019 |
| YK | 4 | 0.3834 ± 0.066 | 0.0021 |
| BP | 5 | 0.6623 ± 0.022 | 0.0022 |
| CE | 6 | 0.5380 ±0.114 | 0.0035 |
| LE | 4 | 0.4506 ± 0.019 | 0.0012 |
| PP | 5 | 0.7662 ± 0.012 | 0.0035 |
| MP | 5 | 0.6737 ± 0.022 | 0.0020 |
| PM | 1 | 0 | 0 |
Sites are shown in order from south to north. See Table 1 for location designations
Fig. 6.
Cytochrome c oxidase subunit I haplotype frequencies of H. sanguineus (n = 25 except for PM where n = 5) represented in more than one individual and more than one site. Pie charts indicate proportion of those named haplotypes found in more than three individuals and more than one site. Haplotypes ≤ 28 are based on Yoon et al. 2011. See Table 1 for site abbreviations and supplemental Tables 1 and 2 for additional details about the haplotypes
Fig. 7.
Minimum spanning tree of the eight mitochondrial COI haplotypes of H. sanguineus. Circle sizes reflect the abundance of each haplotype and the number within each circle indicates the haplotype designation. Each line indicates a single nucleotide difference and additional differences are indicated by hash marks. Haplotypes of known Asian origin are shown in blue, whereas newly identified haplotypes are shown in white. Haplotypes ≤ 28 are based on Yoon et al. (2011). See supplemental Tables 1 and 2 for additional details about the haplotypes
Pairwise population differentiation (FST) and gene flow (Nem) estimates indicate that there is little genetic difference between populations from Connecticut to Maine due to high gene flow (Table 4). Significant differentiation occurred between the most southern site, NJ, and three southern sites (CT, OD, and BP) (Table 4). The most northern site, PM, was also significantly differentiated from most sites due to the presence of a single haplotype in that site as compared to multiple haplotypes that were found in sites to the south (Table 4). At the genetic level, populations within southern and central Maine should be considered to be panmictic due to low FST and very high gene flow (Table 4); these sites are also the most genetically diverse (Fig. 6; Table 3; Suppl Table 2).
Table 4.
Pairwise FST (shown below the diagonal) and Nem (shown above the diagonal) values calculated from H. sanguineus haplotype frequencies
| NJ | CT | OD | YK | BP | CE | LE | PP | MP | PM | |
|---|---|---|---|---|---|---|---|---|---|---|
| NJ | - | 1.045 | 0.510 | 8.083 | 1.562 | 3.175 | ∞ | 2.044 | 4.136 | 6.160 |
| CT | 0.193* | - | 62.62 | 7.815 | 12.908 | ∞ | 3.426 | 49.75 | ∞ | 0.510 |
| OD | 0.329* | 0.004 | - | 1.816 | 7.563 | 6.507 | 1.154 | 10.167 | ∞ | 0.321 |
| YK | 0.030 | 0.031 | 0.121 | - | 8.908 | ∞ | ∞ | 13.639 | ∞ | 3.373 |
| BP | 0.138* | 0.019 | 0.032 | 0.027 | - | ∞ | 12.250 | ∞ | ∞ | 0.827 |
| CE | 0.073 | 0 | 0.037 | 0 | 0 | - | ∞ | ∞ | ∞ | 2.003 |
| LE | 0 | 0.068 | 0.178 | 0 | 0.020 | 0 | - | 9.009 | ∞ | 1.032 |
| PP | 0.109 | 0.005 | 0.024 | 0.018 | 0 | 0 | 0.027 | - | ∞ | ∞ |
| MP | 0.057 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | 1.247 |
| PM | 0.039 | 0.329* | 0.438* | 0.069 | 0.232* | 0.111 | 0.090 | 0.195* | 0.167* | - |
Significant differentiation of pairwise FST values (P < 0.05) on the exact test (Raymond and Rousset 1995). See Table 1 for location designations
Discussion
We found that Asian shore crab (Hemigrapsus sanguineus) populations, predominantly composed of a single haplotype (Fig. 6) originating from Japan, Korea, or Taiwan, have expanded over the last 10 years in southern Maine, mirroring the ecological takeover by this species in southern New England over the last 20 years. Much like in southern New England, H. sanguineus is now the dominant crab species on rocky shores in southern Maine (Fig. 1) (Lohrer and Whitlatch 2002a; O’Connor 2014). As the range and populations of H. sanguineus have expanded since its introduction to the USA in 1988, it has led to declines in populations of the invasive green crab (Carcinus maenas), and this appears to be happening in Maine as well (Epifanio 2013). At sites in New Hampshire and southern Maine where H. sanguineus was abundant, C. maenas was found at less than half the densities at which it was found in northern Maine where H. sanguineus was rare. Green crab densities in southern Maine were similar to those measured by Griffen et al. (2008) in 2006, but that previous study used a random sampling method as opposed to targeting the preferred cobble habitat like in the present study. As such, it is possible that C. maenas densities in these cobble areas have declined in conjunction with the substantial increase in H. sanguineus populations. It is also possible that green crabs in this region have moved into the subtidal zone or into alternative habitats like mud flats and salt marshes where H. sanguineus is not prevalent.
The difference in C. maenas density between northern and southern Maine does not show causation related to the effects of the expanding H. sanguineus population, but it would be in line with the ecological transition in southern New England as H. sanguineus spread and C. maenas populations declined (Epifanio 2013). However, it is important to note that C. maenas is more abundant in soft-sediment habitats than H. sanguineus, so those areas may be less affected by increasing H. sanguineus densities in the Gulf of Maine. In addition, a secondary green crab invasion in the 1980s introduced northern (cold-water) haplotype C. maenas to the Gulf of Maine, and these haplotypes are presently found solely in northern Maine and sites north in adults (Williams et al. 2015). The green crabs with the northern haplotypes may interact differently with Asian shore crabs or may be able to better compete in the colder waters in the northern Gulf of Maine, leading to higher green crab densities in the north. Ecological significance of the northern green crab haplotypes has yet to be tested, so any competitive differences or potential impacts on population density are purely speculative at this point.
Population densities of H. sanguineus increased a minimum of 10-fold by 2015 at all Maine sites which had established populations in 2005 (Stephenson et al. 2009), highlighting the success of these invasive crabs in the northern extent of their range (Fig. 2). Importantly, while populations have expanded rapidly in southern Maine, few northern populations have been established since 2005. While some sites including Owls Head and Jonesport did not have H. sanguineus in 2005 but did in 2015, population densities in these locations were extremely low. Regardless of time since invasion, populations along this range are relatively homogenous, with 69% of all individuals sharing the same haplotype (Fig. 6; Suppl Table 2). However, the Asian shore crab has more COI diversity (Fig. 6) in this initial time since invasion (~27 years) as compared to the green crab whose populations were of a single southern haplotype for over 150 years (Roman 2006).
Unlike green crabs, which primarily carry eggs in the low intertidal zone (Fig. 3), Asian shore crabs appear to carry eggs at similar rates across tidal levels (Fig. 3), which may allow them to gain a foothold in areas where low intertidal zones are dominated by competing species such as green crabs, rock crabs, juvenile Jonah crabs, and juvenile lobsters (all of which were observed in this study). This was supported by an absence of any significant difference in density, size, or sex ratio of H. sanguineus between different tidal levels at the sites included in this survey, as well as by Lohrer et al. (2000), who found no tidal preference for H. sanguineus. Along with the recent spread of H. sanguineus into salt marshes (Peterson et al. 2014), the lack of tidal level preference suggests a generality of habitats that could contribute to the continued success of this invasive crab.
The rapid expansion of southern Maine H. sanguineus populations will likely have substantial ecological impacts, as this species has contributed to major declines in crab and mussel populations throughout southern New England (Lohrer and Whit-latch 2002b; Brousseau et al. 2014; O’Connor 2014). Asian shore crabs have also reduced populations of barnacles, snails, worms, and algae in various parts of their range (Tyrrell et al. 2006; Kraemer et al. 2007), and may compete with juvenile lobsters as well (Demeo and Riley 2006; Lord and Dalvano 2015). The primary crab species that H. sanguineus is replacing in the northwest Atlantic is the green crab, which is ecologically relevant because while these crabs have similar diets, H. sanguineus has shorter egg brooding and larval duration (McDermott 1998), is more aggressive than C. maenas (Jensen et al. 2002), and can reach much higher population densities (O’Connor 2014). There are contradicting reports of whether C. maenas or H. sanguineus have higher per capita feeding rates, which is important because H. sanguineus are generally smaller and thus would have much less biomass at similar population densities (Lohrer and Whitlatch 2002a; DeGraaf and Tyrrell 2004). The feeding activity rhythm of H. sanguineus may also provide it with a competitive advantage, as starved individuals do not display the photophobic behavior displayed by other crab species (Spilmont et al. 2015). Similarity in feeding rates between species would suggest that they have similar ecological impacts despite their difference in body size and any resultant differences in population biomass.
While expanding H. sanguineus populations will likely have a large ecological impact, the 10-year factor of increase (FOI) in Maine populations was 3-fold lower than the 10-year FOI for populations in southern New England (O’Connor 2014) (Fig. 5). Small populations in Rhode Island increased to nearly 200 crabs-m−2 over ten years from 2001 to 2011, while the highest population size in the present study was just over 30 crabs-m−2 (Fig. 2). While crab populations may vary from year-to-year, the drastic differences in H. sanguineus population density between 2005 and 2015 (17-fold) and the 3-fold greater FOI in southern New England are unlikely to be strongly affected by annual variability. There are a couple possible reasons for the lower FOI in Maine, one of which is temperature limitation of the megalopa larval stage (Stephenson et al. 2009; Epifanio 2013). There is a strong temperature gradient in the Gulf of Maine (Table 1), with June seawater temperatures ranging from approximately 6 °C in the north to over 14 °C in the south (Kolber et al. 1990). This varies seasonally but would also explain the limited northern spread of H. sanguineus over the last decade, making it the most likely hypothesis. If temperature is the factor limiting northward spread and population growth due to limited thermal tolerance of megalopae, then H. sanguineus populations could expand within a single warm summer due to the short duration (approximately 2 weeks) of the megalopa stage (McDermott 1998; Epifanio 2013).
It is also possible that ocean currents and associated larval transport are limiting population expansion in Maine, as the Eastern and Western Maine Coastal Currents move water to the southwest, unlike major currents in southern New England and the mid-Atlantic Bight, which generally move northeast. As a result, Maine lobster larvae are typically transported southwest in surface currents (Incze and Naimie 2000), a pattern that may be similar for crab larvae. However, the C. maenas invasion spread northward through the Gulf of Maine and H. sanguineus spread southward from New Jersey to North Carolina shortly after introduction, both primarily against prevailing currents, so current-limited spread is less likely. Since the populations in Maine are genetically diverse (Fig. 6), it is possible that there is standing variation in either the nuclear or mitochondrial genomes that could be selected upon over time and allow for cold-water temperature tolerance allowing for an expanded range in the future.
Gaps in the size-frequency histograms of H. sanguineus populations may suggest episodic recruitment in the northern part of the range of this species or just that the populations in central and northern Maine have only recently become established (Fig. 4). FST and Nem estimates indicate that the most northern population is isolated compared to sites to the south that are relatively panmicitic (Table 4). If megalopae thermal tolerance is the limiting factor in the northward spread of this species, rapidly warming waters in the Gulf of Maine associated with climate change (Pershing et al. 2015) could facilitate further expansion of H. sanguineus populations in this region. Regardless of the factors preventing maximum rates of population expansion, it is clear that H. sanguineus populations have increased substantially in New Hampshire and south-central Maine over the last decade and could extend this upward trajectory, given that H. sanguineus can reach densities of hundreds per square meter in cobble habitats. A continued increase in the prevalence of H. sanguineus will likely lead to decreased abundance of various prey species and to declines in green crab populations as has occurred in southern New England. We have the opportunity to document the spread and mitigate the impact of H. sanguineus during the midst of its invasion of the Gulf of Maine, an opportunity that would have been valuable during the green crab invasion 100 years earlier.
Haplotype data from our study indicates that haplotype 13 animal(s) from either Korea, Japan, or Taiwan originally invaded North America (Figs. 6, 7; Suppl Table 1), a haplotype that is the second most predominant within its native range (Yoon et al. 2011) and has previously been identified in samples from Taiwan (GenBank: EU169911.1), Delaware (Gen-Bank: EU169902.1, EU169905.1, EU169923.1), New Hampshire (GenBank: EU169914-17.1), and Maine (GenBankEU169908.1 and EU169910.1). This haplotype was likely the first to invade due to its frequency within every sampled population and the extent of its range from New Jersey, the initial site of invasion, to northern Maine, the northern extent of its current range. In addition to haplotype 13, five others from Japan and/or Korea were identified in our sites (Suppl Tables 1,2), including haplotype 1 which is the most prominent haplotype in the native range (Yoon et al. 2011). The presence of these haplotypes along much of the northwest Atlantic range indicate that there were either multiple individuals during the initial introduction in the 1980s (McDermott 1991), or there have been subsequent introductions since that time. High haplotype and low nucleotide diversity are also very similar to crabs in their home range (Yoon et al. 2011). Haplotypes 1 and 14 have also been found in the shallow southern parts of the North Sea (Raupach et al. 2015). Given the presence of these Asian haplotypes in both the northwest Atlantic and Asia, it is unclear if European invasions (Breton et al. 2002; Micu et al. 2010) were caused by ballast water originating in North American and/or Asia. Further studies comparing haplotypes throughout the native range, the northwest Atlantic, and Europe may elucidate the origin. Interestingly, while Japanese and/or Korean haplotypes were predominant in the northwest Atlantic, haplotype 30 matched six previously identified Hong Kong (HK) individuals (Steinberg 2008) in 7 of 8 nucleotide substitutions (Fig. 7; Suppl Table 1), with only the substitution at nucleotide 252 not matching those HK samples. It is likely that this substitution does exist in HK, but was under-sampled in Steinberg (2008). The presence of haplotype 30 in our study provides further evidence for the hypothesis of multiple individuals in the first introduction or multiple introductions since that time. Regardless of the initial invasion history, it is clear that the populations in the northwest Atlantic, especially those in Maine, are genetically diverse and well connected which may lead to further invasion success in terms of distribution and total biomass in Maine and points north.
It is important for future research on Asian shore crabs to concentrate on the ecological impact and the genetics of this invasive crab species that is rapidly becoming the dominant predator on southern Maine rocky shores. It is of particular importance to determine the thermal tolerance of H. sanguineus megalopae or any other mechanism limiting the northward spread of this crab species. This could help explain the major findings of this study, in which we determined that Asian shore crab populations in the Gulf of Maine are diverse and are rapidly increasing in numbers. The northward spread appears to be slow and sporadic, in contrast to the first decade after the introduction of H. sanguineus to the USA, when northward and southward spread occurred rapidly and largely unimpeded. The 17-fold increase in Gulf of Maine H. sanguineus density of a relatively panmictic population that was revealed by this study suggests that while population expansion is happening at a slower rate than in the warmer waters of southern New England, Asian shore crabs are quickly becoming important ecological players in the Gulf of Maine.
Supplementary Material
Acknowledgments
Thanks to Brielle Dalvano and Dr. Zair Burris for assistance in the field and with measuring crabs in the laboratory, to Drs. Mia Steinberg, Charles Epifanio, and Shawn McCafferty for sharing their Hemigrapsus sanguineus COI sequences, and Alexandra Ulin for assistance with PCR. Additional thanks to the Bates College Biology Department for providing space and facilities for analysis. Funding provided by a Bates College startup fund 3457-2025 (to JPL), and a Bates College research reserve fund 3829–9155 (to LMW). Research reported in this project was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant Number P20GM103423.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10530-016-1304-1) contains supplementary material, which is available to authorized users.
Contributor Information
Joshua P. Lord, Monterey Bay Aquarium Research Institute, Moss Landing, CA 95039, USA, joshua.p.lord@gmail.com
Larissa M. Williams, Bates College, Lewiston, ME 04240, USA
References
- Albins MA. Effects of invasive Pacific red lionfish Pterois volitans versus a native predator on Bahamian coral-reef fish communities. Biol Invasions. 2013;15:29–43. [Google Scholar]
- Audet D, Davis D, Miron G, Moriyasu M. Geographical expansion of a nonindigenous crab, Carcinus maenas (L.), along the Nova Scotian shore into the southeastern Gulf of St. Lawrence, Canada. J Shellfish Res. 2003;22:255–262. [Google Scholar]
- Blakeslee A, McKenzie C, Darling J, Byers JE, Darling J, Pringle J, Roman J. A hitchhiker’s guide to the Maritimes: anthropogenic transport facilitates long-distance dispersal of an invasive marine crab to Newfoundland. Divers Distrib. 2010;16:879–891. [Google Scholar]
- Blasi JC, O’Connor NJ. Amphipods as potential prey of the Asian shore crab Hemigrapsus sanguineus: laboratory and field experiments. J Exp Mar Biol Ecol. 2016;474:18–22. [Google Scholar]
- Bourdeau P, O’Connor N. Predation by the nonindigenous Asian shore crab Hemigrapsus sanguineus on macroalgae and molluscs. Northeast Nat. 2003;10:319–334. [Google Scholar]
- Breton G, Faasse M, Noel P, Vincent T. A new alien crab in Europe: Hemigrapsus sanguineus (Decapoda: Brachyura: Grapsidae) J Crustac Biol. 2002;22:184–189. [Google Scholar]
- Brousseau DJ, Filipowicz A, Baglivo JA. Laboratory investigations of the effects of predator sex and size on prey selection by the Asian crab, Hemigrapsus sanguineus . J Exp Mar Biol Ecol. 2001;262:199–210. doi: 10.1016/s0022-0981(01)00290-8. [DOI] [PubMed] [Google Scholar]
- Brousseau DJ, Goldberg R, Garza C. Impact of predation by the invasive crab Hemigrapsus sanguineus on survival of juvenile blue mussels in western Long Island Sound. Northeast Nat. 2014;21:119–133. [Google Scholar]
- Carlton JT, Cohen AN. Episodic global dispersal in shallow water marine organisms: the case history of the European shore crabs Carcinus maenas and C. aestuarii . J Biogeogr. 2003;30:1809–1820. [Google Scholar]
- Carlton J, Geller J. Ecological roulette: the global transport of nonindigenous marine organisms. Science. 1993;261:78–82. doi: 10.1126/science.261.5117.78. [DOI] [PubMed] [Google Scholar]
- Clavero M, García-Berthou E. Invasive species are a leading cause of animal extinctions. Trends Ecol Evol. 2005;20:110. doi: 10.1016/j.tree.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Krandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Darling J. Interspecific hybridization and mitochondrial introgression in invasive Carcinus shore crabs. PLoS ONE. 2011;6(3):e17828. doi: 10.1371/journal.pone.0017828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling JA, Bagley MJ, Roman J, Tepolt CK, Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus . Mol Ecol. 2008;17:4992–5007. doi: 10.1111/j.1365-294X.2008.03978.x. [DOI] [PubMed] [Google Scholar]
- Darling J, Tsai Y-H, Blakeslee A, Roman J. Are genes faster than crabs? Mitochondrial introgression exceeds larval dispersal during population expansion of the invasive crab Carcinus maenas . R Soc Open Sci. 2014;1(2):140202. doi: 10.1098/rsos.140202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGraaf J, Tyrrell M. Comparison of the feeding rates of two introduced crab species, Carcinus maenas and Hemigrapsus sanguineus, on the blue mussel, Mytilus edulis . Northeast Nat. 2004;11:163–166. [Google Scholar]
- Demeo A, Riley JG. Hemigrapsus sanguineus (Asian Shore Crab) as predator of juvenile Homarus americanus (American Lobster) Tech Bull Maine Agric For Exp Stn. 2006;194:1–7. [Google Scholar]
- Epifanio CE. Invasion biology of the Asian shore crab Hemigrapsus sanguineus: a review. J Exp Mar Biol Ecol. 2013;441:33–39. [Google Scholar]
- Epifanio CE, Tilburg CE, Dittel AI. Abundance of invasive and native crab larvae in the mouth of Delaware Bay: Hemigrapsus sanguineus and Uca Pugnax . J Shellfish Res. 2013;32:543–550. [Google Scholar]
- Floyd T, Williams J. Impact of green crab (Carcinus maenas L.) predation on a population of softshell clams (Mya arenaria L.) in the southern Gulf of St. Lawrence. J Shellfish Res. 2004;23:457–162. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Griffen BD, Guy T, Buck JC. Inhibition between invasives: a newly introduced predator moderates the impacts of a previously established invasive predator. J Anim Ecol. 2008;77:32–10. doi: 10.1111/j.1365-2656.2007.01304.x. [DOI] [PubMed] [Google Scholar]
- Grosholz E, Ruiz G. Predicting the impact of introduced marine species: lessons from the multiple invasions of the European green crab Carcinus maenas . Biol Conserv. 1996;3207:59–66. [Google Scholar]
- Hong S-E, Kim J-K, Yu J-N, Yoon M. Genetic variation in the Asian shore crab Hemigrapsis sanguineus in Korean coastal waters as inferred from mitochondrial DNA sequences. Fish Aquat Sci. 2012;15(1):49–56. [Google Scholar]
- Incze LS, Naimie CE. Modelling the transport of lobster (Homarus americanus) larvae and postlarvae in the Gulf of Maine. Fish Oceanogr. 2000;9:99–113. [Google Scholar]
- Jensen G, McDonald P, Armstrong D. East meets west: competitive interactions between green crab Carcinus maenas, and native and introduced shore crab Hemigrapsus spp. Mar Ecol Prog Ser. 2002;225:251–262. [Google Scholar]
- Kolber Z, Wyman KV, Falkowski PG. Natural variability in photosynthetic energy conversion efficiency: a field study in the Gulf of Maine. Limnol Oceanogr. 1990;35:72–79. [Google Scholar]
- Kraemer G, Sellberg M, Gordon A, Main J. Eight-year record of Hemigrapsus sanguineus (Asian shore crab) invasion in western Long Island sound estuary. Northeast Nat. 2007;14:207–224. [Google Scholar]
- Larkin M, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ledesma M, O’Connor N. Habitat and diet of the non-native crab Hemigrapsus sanguineus in southeastern New England. Northeast Nat. 2001;8:63–78. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lohrer A, Whitlatch R. Interactions among aliens: apparent replacement of one exotic species by another. Ecology. 2002a;83:719–732. [Google Scholar]
- Lohrer A, Whitlatch R. Relative impacts of two exotic brachyuran species on blue mussel populations in Long Island Sound. Mar Ecol Prog Ser. 2002b;227:135–144. [Google Scholar]
- Lohrer A, Fuikui Y, Wada K, Whitlatch R. Structural complexity and vertical zonation of intertidal crabs, with focus on habitat requirements of the invasive Asian shore crab, Hemigrapsus sanguineus (de Haan) J Exp Mar Biol Ecol. 2000;244:203–217. [Google Scholar]
- Lord J, Dalvano B. Differential response of the American lobster Homarus americanus to the invasive Asian shore crab Hemigrapsus sanguineus and green crab Carcinus maenas . J Shell Res. 2015;34:1091–1096. [Google Scholar]
- Lubchenco J. Plant species diversity in a marine intertidal community: importance of herbivore food preference and algal competitive abilities. Am Nat. 1978;112:23–39. [Google Scholar]
- McDermott J. A breeding population of the western Pacific crab Hemigrapsus sanguineus (Crustacea: Decapoda: Grapsidae) established on the Atlantic coast of North America. Biol Bull. 1991;181:195–198. doi: 10.2307/1542503. [DOI] [PubMed] [Google Scholar]
- McDermott J. The western Pacific brachyuran Hemigrapsus sanguineus (Grapsidae) in its new habitat along the Atlantic coast of the United States: reproduction. J Crustac Biol. 1998;18:308–316. [Google Scholar]
- Micu D, Niţă V, Todorova V. First record of the Japanese shore crab Hemigrapsus sanguineus (de Haan, 1835) (Brachyura: Grapsoidea: Varunidae) from the Black Sea. Aquat Invasions. 2010;5:S1–S4. [Google Scholar]
- Mooney HA, Cleland EE. The evolutionary impact of invasive species. Proc Natl Acad Sci. 2001;98(10):5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- O’Connor NJ. Invasion dynamics on a temperate rocky shore: from early invasion to establishment of a marine invader. Biol Invasions. 2014;16:73–87. [Google Scholar]
- Payne A, Kraemer G. Morphometry and claw strength of the non-native Asian shore crab, Hemigrapsus sanguineus . Northeast Nat. 2013;20:478–492. [Google Scholar]
- Pershing A, Alexander MA, Hernandez CM, Kerr LA, Le Bris A, Mills KE, Nye JA, Record NR, Scannell HA, Scott JD, Sherwood GD, Thomas AC. Slow adaptation in the face of rapid warming leads to collapse of the Gulf of Maine cod fishery. Science. 2015;350:809–812. doi: 10.1126/science.aac9819. [DOI] [PubMed] [Google Scholar]
- Peterson BJ, Fournier AM, Furman BT, Carroll JM. Hemigrapsus sanguineus in Long Island salt marshes: experimental evaluation of the interactions between an invasive crab and resident ecosystem engineers. PeerJ. 2014;2:e472. doi: 10.7717/peerj.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach MJ, Barco A, Steinke D, Beermann J, Laakmann S, Mohrbeck I, Neumann H, Kihara TC, Pointner A, Segelken-Voigt A, Wesse C, Knebelsberger T. The application of DNA barcodes for the identification of crustaceans from the North Sea and Adjacent Regions. PLoS ONE. 2015;10(9):e0139421. doi: 10.1371/journal.pone.0139421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49(6):1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Roman J. Diluting the founder effect: cryptic invasions expand a marine invader’s range. Proc R Soc B. 2006;273:2453–2459. doi: 10.1098/rspb.2006.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz G, Carlton J. Global invasions of marine and estuarine habitats by non-indigenous species: mechanisms, extent, and consequences. Am Zool. 1997;37:621–632. [Google Scholar]
- Say T. An account of the Crustacea of the United States. J Acad Nat Sci Phila. 1817;1:57–63. [Google Scholar]
- Shiganova T. Invasion of the Black Sea by the ctenophore Mnemiopsis leidyi and recent changes in pelagic community structure. Fish Oceanogr. 1998;7:305–310. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47(1):264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129(2):555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilmont N, Gothland M, Seuront L. Exogenous control of the feeding activity in the invasive Asian shore crab Hemigrapsus sanguineus (De Haan, 1835) Aquat Invasions. 2015;10:327–332. [Google Scholar]
- Steinberg MK. Dissertation. University of Delaware; 2008. To be or not to be: the presence and absence of the Asian shore crab Hemigrapsus sanguineus in North America. [Google Scholar]
- Stephenson EH, Steneck RS, Seeley RH. Possible temperature limits to range expansion of non-native Asian shore crabs in Maine. J Exp Mar Biol Ecol. 2009;375:21–31. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105(2):437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1(3):269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Tyrrell M, Guarino P, Harris L. Predatory impacts of two introduced crab species: inferences from microcosms. Northeast Nat. 2006;13:375–390. [Google Scholar]
- Williams LM, Nivison CL, Ambrose WG, Dobbin R, Locke WL. Lack of adult novel northern lineages of invasive green crab Carcinus maenas along much of the northern US Atlantic coast. Mar Ecol Prog Ser. 2015;532:153–159. doi: 10.3354/meps11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Hong S-E, Nam Y, Kim D. Genetic diversity of the Asian shore crab, Hemigrapsus sanguineus, in Korea and Japan inferred from mitochondrial cytochrome c oxidase subunit I gene. Anim Cells Syst. 2011;15(3):243–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.