Abstract
Tsetse flies (Glossina spp.) are the sole vectors of the protozoan parasites of the genus Trypanosoma, the causative agents of African Trypanosomiasis. Species of Glossina differ in vector competence and Glossina morsitans morsitans is associated with transmission of Trypanosoma brucei rhodesiense, which causes an acute and often fatal form of African Trypanosomiasis. Heat shock proteins are evolutionarily conserved proteins that play critical roles in proteostasis. The activity of heat shock protein 70 (Hsp70) is regulated by interactions with its J-protein (Hsp40) co-chaperones. Inhibition of these interactions are emerging as potential therapeutic targets. The assembly and annotation of the G. m. morsitans genome provided a platform to identify and characterize the Hsp70s and J-proteins, and carry out an evolutionary comparison to its well-studied eukaryotic counterparts, Drosophila melanogaster and Homo sapiens, as well as Stomoxys calcitrans, a comparator species. In our study, we identified 9 putative Hsp70 proteins and 37 putative J-proteins in G. m. morsitans. Phylogenetic analyses revealed three evolutionarily distinct groups of Hsp70s, with a closer relationship to orthologues from its blood-feeding dipteran relative Stomoxys calcitrans. G. m. morsitans also lacked the high number of heat inducible Hsp70s found in D. melanogaster. The potential localisations, functions, domain organisations and Hsp70/J-protein partnerships were also identified. A greater understanding of the heat shock 70 (Hsp70) and J-protein (Hsp40) families in G. m. morsitans could enhance our understanding of the cell biology of the tsetse fly.
Introduction
African trypanosomiasis is a parasitic disease giving rise to infection in both humans and animals. Human African trypanosomiasis (HAT) is a neglected tropical disease that burdens 37 countries in sub-Saharan Africa, with an estimated population of 70 million at risk of contracting this potentially lethal disease [1]. Animal African trypanosomiasis (AAT), also known as Nagana, afflicts both wild animals and domesticated livestock and has a detrimental impact on the economic development within sub-Saharan Africa as rearing livestock is nearly impossible in endemic areas [2]. The etiological agent of African trypanosomiasis belongs to the genus Trypanosoma, an extracellularly blood- and tissue-borne unicellular parasitic protozoan. The parasite is comprised of three subspecies: Trypanosoma brucei, Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense, with the latter two being human-infective [3] and all three-subspecies having the potential to be vectors for AAT. T. b. gambiense gives rise to a chronic infection with symptoms that may be dormant for months and even years and represents over 90% of reported cases [4]. T. b. rhodesiense is mainly a zoonotic disease responsible for less than 10% of reported cases and causes an acute infection, which is rapidly fatal if untreated [5].
The tsetse fly, which belongs to the Glossinidae family, which is comprised of only the Glossina genus [6], is the sole insect vector for all the Trypanosoma spp. residing in sub-Saharan Africa [7]. The trypanosomes are transmitted to its mammalian host when an infected tsetse fly vector takes a blood meal, which ensures the cyclical transmission of the parasite between hosts [1]. Thirty-three species and subspecies of tsetse flies have been identified [8], and classified into three subgenera: the Palpalis group, the Morsitans group and the Fusca group [9–10]. Host specificity of these groups differs, with the Palpalis group associating with humans and human activities, while the Morsitans and Fusca groups are associated with wild animals and cattle [11]. Flies of the Morsitans group prefer savannah and woodland habitats and are found mainly in East Africa and might be involved in the transmission of T. b. rhodesiense [12].
Tools for controlling the neglected tropical disease are limited, due to the inability to develop a vaccine, slow development of new and effective drugs, and the ever-increasing drug resistance in African trypanosomes to the current drug treatment regiments [13]. Strategies to control the vector have gained prominence in recent years and vector control could be improved by genome analysis [14–15]. An International Glossina Genome Initiative was established in 2004 to expand research capacity in sub-Saharan Africa, with the goal of sequencing a Glossina species [15]. This goal was realised in 2014 with the release of the Glossina morsitans morsitans genome which has enabled exploration of the cell biology of the insect vector, essentially aiding in the search for alternative strategies in controlling African trypanosomiasis [16]. Part of the original International Glossina Genome Initiative also included the sequencing of the non-vector obligate blood feeder Stomoxys calcitrans, also known as the stable fly [16–17]. Knowledge is lacking on growth and differentiation of trypanosomes in the tsetse fly, as well as vector-parasite interactions [18]. Heat shock proteins and the complexes that they form have gained significant interest as potential drug targets for a variety of diseases [19].
Heat shock proteins (Hsps) play a prominent role in protein biosynthesis, and maintaining homeostasis within the cell under both normal and stressful conditions [20]. Hsps are either constitutively expressed (heat shock cognates, Hsc), and maintain cellular homeostasis, or are up-regulated in response to external stimuli (heat shock proteins, Hsp) [21]. Hsps are traditionally classified according to their molecular weight (kDa), although an alternative nomenclature has been proposed for the major human Hsp families [22]. Members of the Hsp70 superfamily, comprising of the Hsp70/HSPA family and the Hsp110/HSPH family, are the most highly conserved heat shock protein family due to the indispensable role played in maintaining cellular homeostasis, as well as a host of other cellular processes [23]. The Hsp70 and J-proteins function together to bind polypeptides in a variety of essential cellular processes, including folding and unfolding of polypeptides, protein translocation and degradation [24]. Hsp70 proteins function in all major subcellular compartments of the cell, including the cytosol, nucleus, endoplasmic reticulum (ER), and the mitochondria. The number of J-proteins typically exceeds the number of Hsp70s in the cell, and as a result multiple J-proteins can interact with a single Hsp70, which enhances the functional diversity of Hsp70s [25]. The Hsp110s are divergent members of the Hsp70 superfamily and belong to one of the four classes of nucleotide exchange factors of the eukaryotic Hsp70 cycle that accelerate ADP-ATP exchange [26]. A few Hsp110s are able to bind substrate and prevent aggregation by functioning as “holdases” as the interaction cannot be modulated [27]. In addition, Hsp110s have been shown to play a prominent role in the protein disaggregation and reactivation machinery [28–29].
The Hsp70-based chaperone machineries are ATP-dependent processes that involve repetitive cycles of peptide binding and release that are facilitated by ATP binding and hydrolysis [30]. J-proteins play a crucial function of stimulating the basal ATPase activity of Hsp70 partners, while nucleotide exchange factors facilitate the exchange of ADP for ATP resulting in a conformational change in the substrate binding domain and bound substrates are released as the affinity of Hsp70 for its client protein is reduced [31]. The ~ 70 amino acid signature region known as the J-domain possesses an invariant His-Pro-Asp (HPD) motif that has been shown to play a vital role in stimulating the ATPase activity [32]. J-proteins are generally grouped into four classes based on their structural homology to the E. coli DnaJ [33–34]. While all J-proteins contain the canonical J-domain, most have additional domains that perform a variety of functions, including binding client proteins for subsequent transfer to Hsp70, targeting J-proteins to a particular cellular location or obtaining further factors necessary for their function [35].
The aim of this study was to analyse both the Hsp70 and J-protein complements found in the G. m. morsitans genome. Many Hsp70 proteins from Drosophila spp. have been characterised and D. melanogaster hsp70 genes are often used as a reference for comparative genome studies in other organisms [36–37]. Dipteran insects often display an evolutionary proliferation of their hsp70 genes and the Hsp70s from D. melanogaster include multiple constitutively expressed proteins (Hsc) and heat-inducible heat shock proteins (Hsp) [38]. This paper provides a comprehensive depiction of the Hsp70 and J-protein family from G. m. morsitans based on structural, functional and evolutionary analyses. In silico tools were used to evaluate the domain conservation, predicted subcellular localisation, syntenic and phylogenetic analysis of the Hsp70 and J-protein complements within G. m. morsitans. The Hsp70 and J-protein complements were also comparatively analysed in relation to those found in D. melanogaster, H. sapiens, and S. calcitrans, with the aim of identifying all Hsp70 and J-protein members, and potentially identifying Hsp70-J-protein partnerships. It is envisioned that the results of this study will provide a future context for studying the biology of the tsetse fly.
Methodology
Database mining and sequence analyses
In order to identify the Hsp70 complement of G. m. morsitans, the full set of Hsp70 genes from D. melanogaster were retrieved from FlyBase v6 (http://flybase.org/; [39]), and submitted as queries in a BLASTP search of the G. m. morsitans genome on the VectorBase (https://www.vectorbase.org; [17]) database. The e-value was set at an intermediately stringent level of e-10 for collecting as many potential hsp70-related sequences for further analysis. Keywords were also used to scan the genome of G. m. morsitans for hsp70 genes on the VectorBase database, and these included “Hsp70”, “Heat shock protein”, and “molecular chaperone”. The retrieved amino acid sequences were then screened for the Hsp70 domain using SMART 7 (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/; [40]), and Prosite (http://prosite.expasy.org/; [41]).
Retrieval of the protein sequences for the J-protein complement of G. m. morsitans was conducted in the same manner, except the J-domain (1-77aa) of Escherichia coli DnaJ (EcDnaJ) was used as the query, as the signature region for all J-proteins is the J-domain [33], and J-proteins are divided into the four type classes based on their structural homology to Escherichia coli DnaJ [33–34]. The keywords: “Hsp40”, “DnaJ”, “Heat shock protein”, and “molecular chaperone” were also used to scan the genome of G. m. morsitans for J-protein genes on the VectorBase database. The retrieved amino acid sequences from the various keyword searches were then screened for the J-domain using SMART 7 (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/; [40]), and Prosite (http://prosite.expasy.org/; [41]). The molecular weight (Da) of each gene were calculated using compute pI/Mw tool from ExPASy [42].
Phylogenetic and conserved syntenic analyses
Phylogenetic trees were constructed to analyse the phylogenetic relationship of the HSPA/Hsp70, HSPH/Hsp110, and J-protein complements of G. m. morsitans, Stomoxys calcitrans (S. calcitrans; stable fly), Homo sapiens (H. sapiens; humans), and Drosophila melanogaster (D. melanogaster; fruit fly). Separate phylogenetic trees for HSPA/Hsp70 and HSPH/Hsp110 were constructed, as the two Hsp70 subfamilies are very divergent. The Type III J-protein subfamily was omitted from the phylogenetic analysis, as the subfamily is diverse with regards to amino acid composition and protein length, with the only common feature being the J-domain. The full length amino acid sequences for the Hsp70 superfamily and selected J-protein subfamilies in the tsetse fly and stable fly were obtained from VectorBase [17], fruit fly protein sequences were obtained from FlyBase v6 [38], and human protein sequences were obtained from the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov). Multiple sequence alignments were performed using the in-built ClustalW program [43] with default parameters in MEGA 7.0 [44], and are provided in the supplementary data, S1–S3 Figs. Maximum-likelihood was utilized to find the best model of evolution, and based on the Bayesian Information Criteria (BIC) the substitution pattern that was best described for the protein families was the Le Gascuel (LG) model matrix [45] with a discrete Gamma (G) distribution to model evolutionary rates amongst sites (Hsp70/HSPA, gamma value = 1.1925; Hsp110/HSPH, gamma value = 1.3525; J-protein, gamma value = 2.3202). Maximum likelihood phylogenetic trees were constructed using MEGA 7.0 [45]. The accuracy of the reconstructed trees was assessed using a bootstrap test using a 1000 replicates with a pairwise gap deletion mode. The phylogenetic trees for Hsp70/HSPA and Hsp110/HSPH were rooted with the Escherichia coli HscC (EcHscC) sequence. The phylogenetic tree for the J-proteins was unrooted.
In order to provide additional evidence for orthology, conserved syntenic regions surrounding selected Hsp70 genes were searched by examining the conserved co-localization of neighbouring genes on a scaffold of G. m. morsitans and the selected organisms for this study using genome information from VectorBase, FlyBase, and NCBI database. The identities of unknown neighbour genes of the selected Hsp70 genes were conducted using a BLASTP search on the NCBI database.
Protein domain mapping, subcellular localisation and determination of fruit fly, stable fly and human orthologues
The protein domain mapping for the Hsp70 and J-protein complements from G. m. morsitans was conducted using a combination of online programs that included TPRpred (http://toolkit.tuebingen.mpg.de/tprpred; [46]), SMART 7 (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/; [40]), and Prosite (http://prosite.expasy.org/; [41]). The organelle distribution for the Hsp70 and J-protein complements were conducted, in the absence of experimental data, using a number of online programs that included NucPred (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi; [47]), MitoPROT (http://ihg.gsf.de/ihg/mitoprot.html; [48]), MultiLoc (http://abi.inf.uni-tuebingen.de/Services/MultiLoc; [49]), SignalP version 4.1 (http://www.cbs.dtu.dk/services/SignalP/; [50]), and WoLF PSORT (http://www.genscript.com/wolf-psort.html.; [51]).
Aside from phylogenetic and syntenic analysis, identification of orthologues for the G. m. morsitans Hsp70 and J-protein genes in stable fly (Stomoxys calcitrans), humans (Homo sapiens), and the fruit fly (Drosophila melanogaster) were also conducted using reciprocal BLASTP. In the first query, the putative amino acid sequences of the 9 Hsp70 and 37 J-proteins of G. m. morsitans were used as queries in a BLASTP search on the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov), using the default parameters. The amino acid sequences of the putative orthologues were then used as second queries in BLASTP searches using default parameters on the VectorBase database. If the most similar orthologue in G. m. morsitans was exactly the Hsp70 or J-protein sequence used as the first query, the sequence of the second query was selected as an orthologue.
Results and discussion
The Hsp70 complement of G. m. morsitans
As G. m. morsitans (referred to as Gmm in this study) and D. melanogaster (referred to as Dmel in this study) are both dipteran insects, the already well characterised Hsp70 proteins from D. melanogaster were used as queries and as a reference to explore the Hsp70 superfamily from G. m. morsitans, which has not been previously analysed. The nomenclature of the G. m. morsitans Hsp70s from VectorBase were derived from the nomenclature used for D. melanogaster, though, the nomenclature proposed in this study for the members of the Hsp110/HSPH family were based on their sequence similarity to their Drosophila and human orthologues. The nomenclature for the Hsp70 superfamily from the stable fly, Stomoxys calcitrans (referred to as Scal in this study) was derived in the same manner. A total of 9 putative Hsp70 genes, listed in Table 1, were identified in G. m. morsitans, with 3 of these belonging to the Hsp70 subfamily, Hsp110/HSPH. The domain architecture of the members of the GmmHsp70 and GmmHsp110 families are shown in S4 Fig.
Table 1. The predicted Hsp70 proteins from G. m. morsitans with their respective D. melanogaster, S. calcitrans, and H. sapiens orthologues.
| G. m. morsitans | D. melanogaster | S. calcitrans | H. sapiens | ||||
|---|---|---|---|---|---|---|---|
| Gene IDa | Nameb | Gene IDa | Name | Gene IDa | Name | Name | Localisationc |
| A: Hsp70s | |||||||
| GMOY009493 | GmmHsp70A | FBgn0013275 | Hsp70Aa | SCAU008520 | Hsp70 | - | CYT/NUC |
| GMOY009495 | GmmHsp68 | FBgn0001230 | Hsp68 | SCAU003728 | Hsp68 | - | CYT/NUC |
| GMOY004286 | GmmHsc70-1 | FBgn0001216 | Hsc70-1 | SCAU005225 | Hsc70-1 | - | CYT |
| GMOY003216 | GmmHsc70-3 | FBgn0001218 | Hsc70-3 | SCAU000678 | Hsc70-3 | HSPA5 | ER |
| GMOY012049 | GmmHsc70-4 | FBgn0266599 | Hsc70-4 | SCAU015347 | Hsc70-4 | HSPA8 | CYT |
| GMOY010851 | GmmHsc70-5 | FBgn0001220 | Hsc70-5 | SCAU003620 | Hsc70-5 | HSPA9 | MITO |
| B: NEFs | |||||||
| GMOY011246 | GmmHsp110-1 | FBgn0026418 | Hsp110 | SCAU005995 | Hsp110 | HSPH1 | CYT |
| GMOY013289 | GmmHsp110-2 | - | - | - | - | - | CYT |
| GMOY006943 | GmmGrp170 | FBgn0023529 | Grp170 | SCAU010922 | Grp170 | HSPH4 | ER |
a The Gene IDs for the G. m. morsitans, S. calcitrans and D. melanogaster Hsp70 proteins were acquired from the VectorBase database (https://www.vectorbase.org/; [17]), and FlyBase v6 database (http://flybase.org/; [39]) respectively.
b The nomenclature for the Hsp70 proteins from G. m. morsitans was derived from the VectorBase database (https://www.vectorbase.org/; 38]); the proposed names in this study for the G. m morsitans NEFs were derived from their orthologues in D. melanogaster.
c The subcellular localisations for the G. m. morsitans and S. calcitrans Hsp70 proteins were predicted using various online prediction servers, which are listed in the methods and D. melanogaster and H. sapiens have been experimentally determined (see text for details).
CYT-Cytosol; MITO- Mitochondria; ER- Endoplasmic reticulum; NUC-nucleus.
All retrieved amino acid sequences from VectorBase were full-length except for GmmHsp68 (GMOY009495). Analysis of the coding region of GmmHsp68 indicated that the protein was truncated to 619 amino acids due to a premature stop codon, and an additional 15 amino acids were found in the flanking 3’ downstream sequence that ended in a variant C-terminal EEID motif. Though the isoleucine substitution in the C-terminal EEVD motif could be a result of a sequencing error as the amino acids share similar side chains, but further sequence validation is needed to identify if this is indeed a miss-annotation. The amino acid sequence of GmmHsp110-2 (GMOY010029) was also re-annotated as it was found to possess all the functional domains of a typical Hsp110 protein but also possessed unusually an alkyl hydro peroxide reductase subunit C (AhpC) at the C-terminus of the protein. Insertion of a stop codon at the C-terminus of the protein prior to the AhpC yields a full-length protein but further sequence validation is needed to identify if this is indeed a miss-annotation.
The predicted subcellular localisations and the orthologous relationships of the Hsp70 and Hsp110 proteins from G. m. morsitans to the selected organisms in this study, as determined by pBLAST analysis (S1 Table) are presented in Table 1. Hsp70s are one of the most conserved groups of proteins [52–54], and thus, it was not surprising that the members of the Hsp70/HSPA family of G. m. morsitans showed a high degree of sequence identity to its orthologues in humans, and the selected dipteran species in this study (S1 Table). Though, there were no human orthologues of GmmHsp70A, GmmHsp68 or GmmHsc70-1 (Fig 1, Table 1). Notably absent from the Hsp70/HSPA family in G. m. morsitans is the six highly conserved copies of the inducible Hsp70 gene (Hsp70Aa, Hsp70Ab, Hsp70Ba, Hsp70Bb, Hsp70Bbb, and Hsp70Bc) that are found in D. melanogaster. The inducible Hsp70 gene in D. melanogaster has gone through extensive duplication during evolution [55], as this system has been specialized for intense expression during heat shock [56]. DmelHsp70 is virtually undetectable at normal growing temperatures of 25°C and is rapidly induced during heat shock where it plays an essential role in thermotolerance [57]. The absence of these inducible Hsp70 genes could imply that the duplication event did not occur in G. m. morsitans.
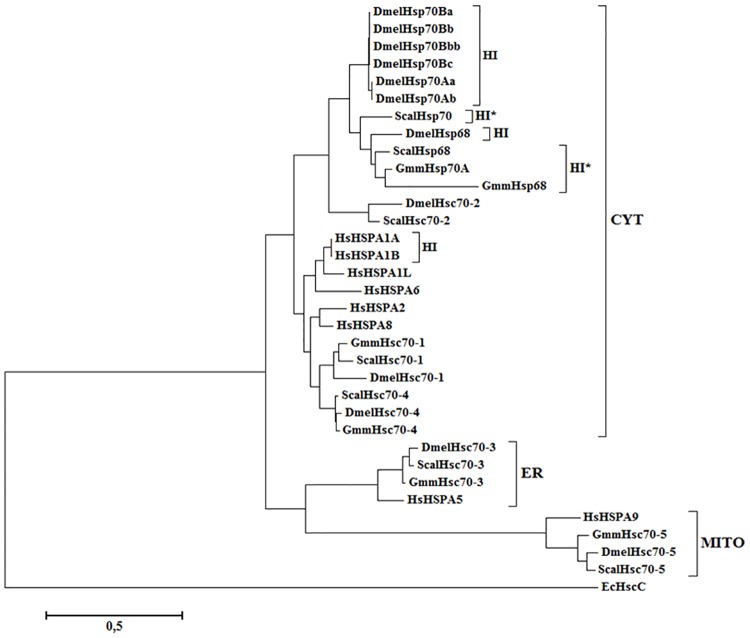
Fig 1. Phylogenetic analysis of the predicted Hsp70/HSPA family from G. m. morsitans in relation to D. melanogaster, H. sapiens and S. calcitrans.
Multiple sequence alignment of the full-length amino acid sequences of the Hsp70/HSPA gene families in humans, tsetse flies, fruit flies, and stable flies. The multiple sequence alignment (S1 Fig) was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Le Gascuel (LG) matrix-based model of amino acid substitution. A discrete Gamma distribution was used to model evolutionarily rate differences among sites (2 categories (+G, parameter = 1.1925). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 457. Bootstrap analysis was computed with 1000 replicates. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp70 (SCAU008520); Hsp68 (SCAU003728); Hsc70-1 (SCAU005225); Hsc70-2 (SCAU008036); Hsc70-3 (SCAU000678); Hsc70-4 (SCAU015347); Hsc70-5 (SCAU003620). D. melanogaster: Hsp68 (NP_524474.1); Hsp70Aa (NP_731651.1); Hsp70Ab (NP_524798.2); Hsp70Ba (NP_731716.1); Hsp70Bb (NP_524927.2); Hsp70Bbb (NP_788663.1); Hsp70Bc (NP_650209.1); Hsc70-1 (NP_524063.1); Hsc70-2 (NP_524339.1); Hsc70-3 (NP_727563.1); Hsc70-4 (NP_524356.1); Hsc70-5 (NP_523741.2). H. sapiens: HSPA1A (NP_005336.3); HSPA1B (NP_005337.2); HSPA1L (NP_005518.3); HSPA2 (NP_068814.2); HSPA5 (NP_005338.1); HSPA6 (NP_002146.2); HSPA8 (NP_006588.1); HSPA9 (NP_004125.3). Accession numbers for the G. m. morsitans Hsp70 sequences can be found in Table 1. The subcellular localisation for Hsp70s is indicated by a bracket on the right-hand side. CYT: cytosolic; ER: endoplasmic reticulum; MITO: mitochondrion. Hsp70 genes that are heat-inducible are depicted with HI. Hsp70 genes in G. m. morsitans that are predicted to be heat inducible are depicted with HI*.
Hsp68 proteins are very closely related to the Hsp70 proteins. Interestingly, the sequence similarity of GmmHsp68 was 70% identical to DmelHsp70Aa and 69% identical to DmelHsp68 (S1 Table). DmelHsp68 has been shown to partially compensate for the loss of Hsp70 in Hsp70-deficient flies, as the Hsp68 expression increased in the absence of Hsp70 [55]. Recently, DmelHsp68 has been shown to assist Hsp70-null larvae in cold acclimation when exposed to relatively mild doses of cold [58]. DmelHsp68 has also been implicated as a component in JNK-signalling where this gene regulatory network utilizes the chaperone in limiting oxidative damage, and thus extending the lifespan of the fly [59]. Both GmmHsp70A and GmmHsp68 were predicted to localise in the nucleus and cytosol (Table 1). Heat shock has shown to cause a concentration of DmelHsp70 in the nuclei, with some remaining in the cytosol, and during recovery the protein returns to the cytosol [60].
The GmmHsc70s were predicted to localise to the same subcellular compartments as their D. melanogaster and S. calcitrans orthologues, however no Hsc70-2 orthologue was found for G. m. morsitans (Table 1). GmmHsc70-3 was predicted to be localised to the ER as a hydrophobic leader sequence and C-terminal KDEL motif, characteristic of ER proteins, was reported, and a mitochondrial leader sequence detected for the larger Hsc70-5 protein (S1 and S4 Figs). The Hsc70 family from D. melanogaster carries out critical functions at normal temperatures as mutations in several of these proteins caused lethality [61]. Transcription of D. melanogaster Hsc70 genes are regulated during development, Hsc70-4 was present at high levels during embryonic, larval, and adult developmental stages, while Hsc70-1 and Hsc70-2 were detected in adults at low levels [62]. Proteomic profiling in D. melanogaster revealed that Hsc70-3, Hsc70-4 and Hsc70-5 as well as Hsp70Bb were significantly up-regulated during thermal acclimation [63]. The localisations of the human Hsp70 orthologues have been experimentally determined and corresponded to those of G. m. morsitans and the other selected dipteran species [64].
The phylogenetic tree in Fig 1 shows the classification of Hsp70 genes into three major monophyletic groups based on sub-cellular localisation (CYT, ER, MITO) among the different eukaryotes in this study, with heat inducible Hsp70s highlighted in the CYT subfamily. Functional differences of the three groups is reinforced by the phylogenetic analysis. The six inducible DmelHsp70 proteins phylogenetically clustered together, while DmelHsp68 appeared to be more closely related to the Hsp70 and Hsp68 proteins from G. m. morsitans and S. calcitrans (Fig 1). The Gmm cytosolic, mitochondrial and ER Hsp70s phylogenetically clustered with the dipteran Hsp70s, suggesting that they may be more functionally similar to the fruit fly and stable fly than to their human Hsp70 orthologues (Fig 1). Based on phylogenetic analysis both GmmHsp68 and GmmHsp70A are probably heat inducible, although the presence of heat shock elements needs to be confirmed. Not surprisingly the GmmHsc70 proteins clustered with their respective orthologues from D. melanogaster and S. calcitrans (Fig 1). DmelHsc70-2 and ScalHsc70-2 forms a unique clade and appears to be derived from the inducible Hsp70s, although an orthologue is absent from G. m. morsitans as well as H. sapiens (Fig 1). Based on phylogenetic analysis, the constitutive GmmHsc70 proteins appear to follow the same model of divergent evolution evident in D. melanogaster (Fig 1) [65]. Thus, it is possible that the Hsc70 proteins from G. m. morsitans and S. calcitrans could play a similar functional role to those from Drosophila.
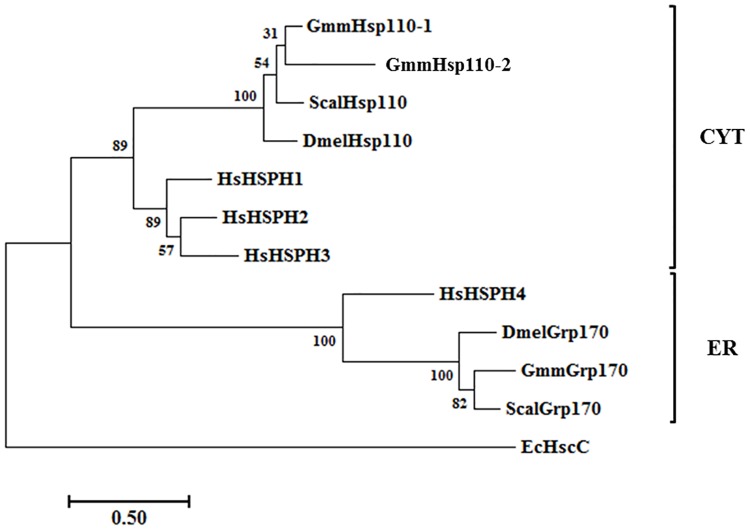
Two GmmNEFs were predicted to reside in the cytosol, whilst no Hsp110 orthologue was found in the mitochondria (Table 1, S4 Fig). This is not surprising as the mitochondrial GmmNEF is probably GmmRoe1 (GMOY010619), an orthologue of Mge1 which belongs to the GrpE class of NEFs for eukaryotic Hsp70s [66]. According to FlyBase, eight splice variants exist for Dmel Hsc70Cb/Hsp110, and further analysis of the NEFs revealed that GmmHsp110-1 (GMOY011246) exhibited the highest sequence identities to Hsc70Cb isoforms G and H (68.2%), while Hsp110-2 protein from G. m. morsitans (GMOY010029) exhibited the highest sequence identities to DmelHsc70Cb isoforms A, B, C, E, F and I (58.4%). The Hsp110s can be classified into the polyphyletic CYT group and the monophyletic ER group. Unlike D. melanogaster and S. calcitrans, G. m. morsitans has evolved three Hsp110 proteins, and the expression of additional isoforms cannot be ruled out (Fig 2). The human genome encodes three Hsp110 homologues that reside in the cytosol and one Grp170 homologue in the ER (Fig 2) [reviewed by 25]. A single Hsp110 homologue was found in the ER for all species (Fig 2).
Fig 2. Phylogenetic relationship of the Hsp110/HSPH protein family from G. m. morsitans (Gmm), D. melanogaster (Dmel), S. calcitrans (Scal), and H. sapiens (Hs).
Multiple sequence alignment of the full-length amino acid sequences of the Hsp110/HSPH gene families in humans, tsetse flies, fruit flies, and stable flies. The multiple sequence alignment (S2 Fig) was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Le Gascuel (LG) matrix-based model of amino acid substitution. A discrete Gamma distribution was used to model evolutionarily rate differences among sites (2 categories (+G, parameter = 1.3525). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 544. Bootstrap analysis was computed with 1000 replicates. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp110 (SCAU005995); Grp170 (SCAU010922). D. melanogaster: Hsp110 (NP_648687.1); Grp170 (NP_569995.1). H. sapiens: HSPH1 (NP_006635.2); HSPH2 (NP_002145.3); HSPH3 (NP_055093.2); HSPH4 (NP_006380.1). Accession numbers for the G. m. morsitans Hsp110 sequences can be found in Table 1. The subcellular localisation for Hsp110s is indicated by a bracket on the right. CYT: cytosolic; ER: endoplasmic reticulum.
GmmHsp110-1, GmmHsp110-2 and GmmGrp170 were found to be considerably longer in sequence length than the other identified Hsp70 members (S4 Fig). These are features typical of the Hsp110/HSPH family. Hsp110 and Grp170 have similar domains as canonical Hsp70s but have long insertions and C-terminal extensions [26]. DmelHsp110 was shown in a genome-wide RNAi screen to be a mitigating factor for aggregation of Huntington proteins [67]. The ATPase domain and C-terminal helical lid of Hsp110 have been shown to mediate the interaction with Hsp70 [27]. The putative peptide binding domain of Hsp110 is also unique with regards to the molecular basis on which the chaperone binds its client proteins, Hsp110 prefer to bind aromatic rings as opposed to canonical Hsp70s that prefer aliphatic side chains and proline residues [68–69]. Additionally, the putative peptide binding domain of the Plasmodium falciparum Hsp110c was shown to be modified to handle the asparagine repeat-rich proteome of the parasite particularly during febrile episode [70].
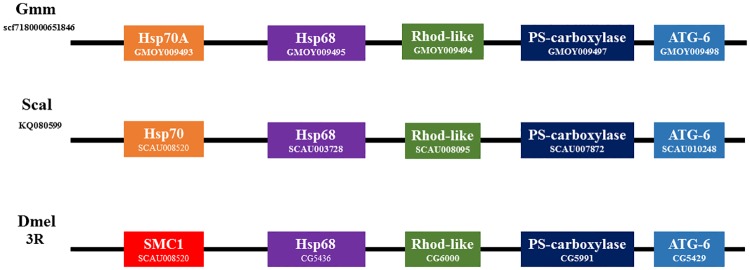
Syntenic analysis provided additional evidence for orthology of selected members of the Hsp70 complement from G. m. morsitans which included GmmHsp68, GmmHsp70A, and GmmHsp110-2. Even though the genome of G. m. morsitans has yet to be assembled into chromosomes, position of these genes and their neighbouring genes were identified from the genome scaffolds on VectorBase [17]. GmmHsp68 and GmmHsp70A formed a clade with the Hsp68 and Hsp70A proteins from D. melanogaster and S. calcitrans, but as observed in Fig 1 the proteins did not exclusively phylogenetically cluster with their respective orthologues, and pBLAST analysis illustrated that the sequence identity of GmmHsp68 and GmmHsp70A is relatively similar to both the Hsp68 and Hsp70A proteins from D. melanogaster and S. calcitrans (S1 Table). Syntenic analysis revealed that the GmmHsp70A and GmmHsp68 genes are located on the same chromosome in a head to head orientation, with the same genomic organisation being observed in S. calcitrans (Fig 3). Physical mapping of gene regions from Drosophila serrata illustrated that the Hsp70 and Hsp68 genes are located on the same chromosome [71], and the chromosomal gene position changes observed in D. melanogaster (Fig 3) may be a result of duplication/deletion events, and the movement of transposable elements [72–73]. Despite this, the neighbouring genes of Hsp68 are shown to be conserved among the three-dipteran species (Fig 3), supporting the orthologous relationship of GmmHsp68 to the Hsp68 proteins in D. melanogaster and S. calcitrans, and the orthologous relationship of GmmHsp70A to ScalHsp70.
Fig 3. Schematic representation of the conserved synteny blocks neighbouring GmmHsp70A and GmmHsp68.
Note that the gene order and orientation is relatively conserved. Abbreviations: SMC1, Structural maintenance of chromosomes 1; Rhod-like, Rhodanese-like domain protein; PS-carboxylate, Phosphatidylserine decarboxylase; ATG-6, autophagy-related protein 6. Accession numbers for each gene is shown in each synteny block.
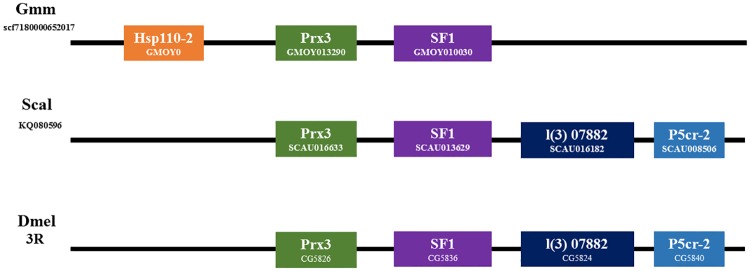
Syntenic analysis of GmmHsp110-2, as shown in Fig 4, was conducted in order to validate that the Hsp110 protein is specific to G. m. morsitans, as it was the only dipteran species in our study to possess two cytosolic Hsp110 protein members (Fig 2, Table 1). Syntenic analysis illustrated that GmmHsp110-2 is on the same region of the chromosome as Peroxiredoxin 3 (Prx3) and Splicing factor 1 (SF1) (Fig 3). The gene order and orientation of Peroxiredoxin 3 (Prx3) and Splicing factor 1 (SF1), as shown in Fig 4, is conserved in all three-dipteran species, but notably absent is a Hsp110 protein in S. calcitrans and D. melanogaster. Further neighbouring genes of Peroxiredoxin 3 (Prx3) and Splicing factor 1 (SF1) in S. calcitrans and D. melanogaster were shown to be conserved. However, these genes were also absent in G. m. morsitans (Fig 4). Overall, the genomic organisation of GmmHsp110-2 shows that it is a unique cytosolic Hsp110 protein to G. m. morsitans and may have arisen due to a duplication event.
Fig 4. Schematic representation of the conserved synteny blocks neighbouring GmmHsp110-2.
Note that the gene order and orientation is relatively conserved. Abbreviations: Prx3, Peroxiredoxin 3; SF1, Splicing factor 1; l (3) 07882, lethal (3) 07882; P5cr-2, Pyrroline-5-carboxylate reductase-like 2.
The J-protein complement
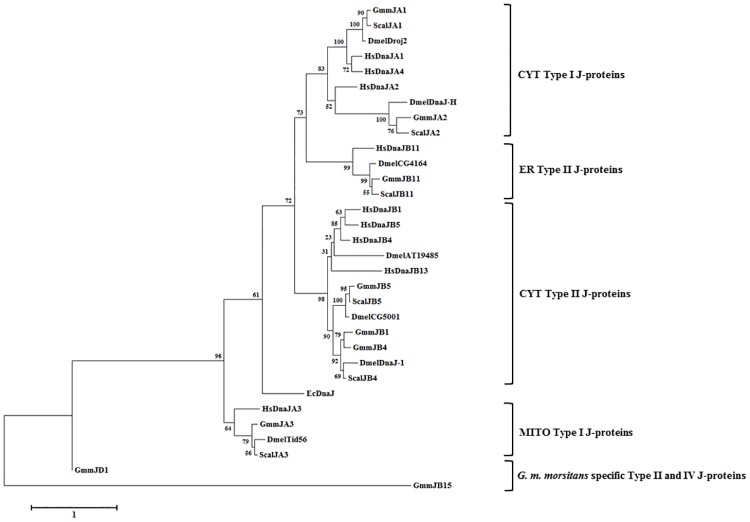
The J-protein complement for G. m. morsitans was identified through a genome-wide search using the J-domain from Escherichia coli DnaJ, as the J-domain is the signature region for all J-proteins [33]. A total of 37 J-proteins were identified in the G. m. morsitans genome. All retrieved amino acid sequences of the J-proteins were full-length sequences on VectorBase database except for GmmJC33 (GMOY003881) and GmmJC34 (GMOY004160/1), which are partial sequences. All J-proteins were further categorized into the 4 J-protein subfamilies, I-IV. Nomenclature proposed for the Gmm J-proteins was based on the guidelines in Kampinga et al. [22], except GmmJD was devised to incorporate Type IV J-proteins. Types I to IV in Gmm are designated as A-D respectively. Nomenclature for the Scal J-proteins were derived in the same manner. The predicted subcellular localisations, identification of orthologues and functional diversity of the Gmm J-proteins are summarized in Table 2. The results of the pBLAST analysis to determine the orthologous relationship of the Gmm J-proteins to the selected organisms in this study are presented in S1 Table. Phylogenetic analysis of the selected J-protein subfamilies as illustrated in Fig 5, shows that the J-proteins cluster based on their different classes and subcellular localisation. A comprehensive domain organisation of the predicted Gmm J-proteins is illustrated in Fig 6.
Table 2. The predicted J-proteins from G. m. morsitans with their respective D. melanogaster, H. sapiens and S. calcitrans orthologues.
| G. m. morsitansb | D. melanogasterc | S. calcitrans | H. sapiensc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | Namea | Gene ID | Namea | Gene ID | Name | Gene ID | Localisationd | Functione | References | |
| I | GmmJA1 | GMOY007075 | Droj2 | FBgn0038145 | ScalJA1 | SCAU009538 | DnaJA1 | CYT | Androgen receptor signalling | [76; 107] |
| NUC | Spermatogenesis | |||||||||
| hERG maturation and trafficking | ||||||||||
| Protein aggregation and refolding | ||||||||||
| Protein import-MITO | ||||||||||
| GmmJA2 | GMOY002478 | DnaJ-H | FBgn0032474 | ScalJA2 | SCAU013613 | DnaJA2 | CYT | hERG maturation and trafficking | [77; 107–108] | |
| JNK signalling | ||||||||||
| G-protein signalling | ||||||||||
| GmmJA3 | GMOY006702 | Tid56 | FBgn0002174 | ScalJA3 | SCAU003912 | DnaJA3 | MITO | Intracellular signalling pathways | [95] | |
| Tumour suppressor | ||||||||||
| Protein aggregation and folding | ||||||||||
| mtDNA maintenance | ||||||||||
| II | GmmJB1 | GMOY005834 | - | - | - | - | DnaJB1 | CYT | Protein (re)folding | [109] |
| GmmJB4 | GMOY003219 | DnaJ-1 | FBgn0263106 | ScalJB4 | SCAU013247 | DnaJB4 | CYT | ERAD | [110] | |
| NUC | ||||||||||
| GmmJB5 | GMOY007851 | CG5001 | FBgn0031322 | ScalJB5 | SCAU015003 | DnaJB5 | CYT | HDAC shuttling | [111] | |
| NUC | ||||||||||
| GmmJB11 | GMOY005166 | CG4164 | FBgn0031256 | ScalJB11 | SCAU015416 | DnaJB11 | ER | Protein folding | [112] | |
| mRNA editing | ||||||||||
| GmmJB15 | GMOY001519 | - | - | - | - | - | MITO? | ? | - | |
| III | GmmJC1 | GMOY000574 | CG7556 | FBgn0030990 | ScalJC1 | SCAU000575 | DnaJC1 | ER | ACT secretion | [113–114] |
| PLASMA | Protein folding | |||||||||
| MEM | ||||||||||
| GmmJC2 | GMOY000389 | CG10565 | FBgn0037051 | ScalJC2 | SCAU003775 | DnaJC2 | CYT | Translation | [115] | |
| NUC | ||||||||||
| GmmJC3 | GMOY009982 | P58IPK | FBgn0037718 | ScalJC3 | SCAU013977 | DnaJC3 | ER | Viral pathogenesis | [116–117] | |
| ER protein synthesis | ||||||||||
| UPR | ||||||||||
| GmmJC4 | GMOY002749 | DnaJ60 | FBgn0260775 | ScalJC4 | SCAU004329 | DnaJC4 | CYT | Spermatogenesis | [118] | |
| GmmJC5 | GMOY000122 | Cysteine string protein? | FBgn0004179 | ScalJC5 | SCAU013958 | DnaJC5? | CYT | Synaptic transmission | [119] | |
| Exocytosis | ||||||||||
| GmmJC6 | GMOY012999 | Auxilin | FBgn0037218 | ScalJC6 | SCAU011318 | DnaJC6 | CYT | Clathrin uncoating | [120] | |
| GmmJC7 | GMOY010330 | TPR2A | FBgn0032586 | ScalJC7 | SCAU011528 | DnaJC7 | CYT | Steroid hormone maturation | [121] | |
| NUC | Protein folding quality control | |||||||||
| GmmJC9 | GMOY010007 | CG6693 | FBgn0037878 | ScalJC9 | SCAU003671 | DnaJC9 | NUC | Anti-protein aggregation | [122] | |
| Nuclear exit upon stress | ||||||||||
| GmmJC11 | GMOY010852 | CG8531 | FBgn0033918 | ScalJC11 | SCAU005931 | DnaJC11 | MITO | Mitochondrial cristae morphology | [123] | |
| PLASM | ||||||||||
| GmmJC12 | GMOY000552 | Jdp | FBgn0027654 | ScalJC12 | SCAU015463 | DnaJC12 | CYT | Anti-protein aggregation | [124] | |
| GmmJC13 | GMOY009168 | Rme-8 | FBgn0015477 | ScalJC13 | SCAU006475 | DnaJC13 | CYT | EGFR trafficking | [125–126] | |
| MITO | Endosome trafficking | |||||||||
| GmmJC16 | GMOY002603 | CG40178 | FBgn0058178 | ScalJC16 | SCAU006018 | DnaJC16 | PLASM | ? | - | |
| GmmJC17 | GMOY011131 | CG17187 | FBgn0037882 | ScalJC17 | SCAU016288 | DnaJC17 | CYT | Pre-mRNA splicing | [127] | |
| NUC | ||||||||||
| GmmJC18 | GMOY004658 | - | - | ScalJC18 | SCAU005547 | - | PLASM | ? | - | |
| GmmJC19 | GMOY002685 | CG7394 | FBgn0036173 | ScalJC19 | SCAU007810 | DnaJC19 | MITO | Protein import-MITO | [127] | |
| GmmJC20 | GMOY000945 | l (3)72Dp | FBgn0263607 | ScalJC20 | SCAU000691 | DnaJC20 | MITO | FeS cluster biogenesis | [128–129] | |
| GmmJC21 | GMOY006819 | CG2790 | FBgn0027599 | ScalJC21 | SCAU016142 | DnaJC21 | CYT | Ribosome biogenesis | [127] | |
| NUC | ||||||||||
| GmmJC22 | GMOY009242 | Wurst | FBgn0030805 | ScalJC22 | SCAU013054 | DnaJC22 | PLASM | Clathrin-mediated endocytosis | [130] | |
| GmmJC23 | GMOY008009 | Sec63 | FBgn0035771 | ScalJC23 | SCAU014339 | DnaJC23 | ER | Protein import | [131] | |
| PLASM | ||||||||||
| GmmJC24 | GMOY009973 | CG2911 | FBgn0037350 | ScalJC24 | SCAU012042 | DnaJC24 | CYT | Dipthamide synthesis | [132] | |
| NUC | ||||||||||
| GmmJC25 | GMOY003438 | CG7872 | FBgn0030658 | ScalJC25 | SCAU002837 | DnaJC25 | SEC | ? | - | |
| GmmJC28 | GMOY003297 | CG43322 | FBgn0263027 | ScalJC28 | SCAU012595 | DnaJC28 | CYT | ? | - | |
| GmmJC30 | GMOY001329 | CG11035 | FBgn0037544 | ScalJC30 | SCAU007712 | DnaJC30 | MITO | ? | - | |
| GmmJC31 | GMOY007250 | Mrj | FBgn0034091 | ScalJC31 | SCAU012526 | DnaJB3 | CYT | Protein folding in sperm | [127] | |
| GmmJC32 | GMOY005661 | CG3061 | FBgn0038195 | ScalJC32 | SCAU005586 | DnaJB12 | CYT | ERAD | [133] | |
| GmmJC33 | GMOY003881 | CG17187? | FBgn0037882 | ScalJC17 | SCAU016288 | DnaJC17? | CYT | ? | - | |
| GmmJC34 | GMOY004160 | CG17187? | FBgn0037882 | ScalJC17 | SCAU016288 | DnaJC17? | CYT | ? | - | |
| GMOY004161 | NUC | |||||||||
| GmmJC35 | GMOY007968 | CG17187? | FBgn0037882 | ScalJC17 | SCAU016288 | DnaJC17? | CYT | ? | - | |
| IV | GmmJD1 | GMOY011949 | - | - | - | - | - | CYT | ? | - |
a The proposed nomenclature for the J-proteins of G. m. morsitans and S. calcitrans; these J-proteins were classified into Types I-IV (A-D).
b Gene IDs for G. m. morsitans and S. calcitrans were obtained from VectorBase (https://www.vectorbase.org/; [17]).
c Orthologues identified from Homo sapiens, Stomoxys calcitrans, and Drosophila melanogaster by NCBI database analysis.
d Subcellular localizations for the G. m. morsitans and S. calcitrans J-proteins were predicted using the online prediction servers listed in the methods. Subcellular localizations have been experimentally determined for certain J-proteins from Homo sapiens, and Drosophila melanogaster (see text for details).
CYT-Cytosol; MITO- Mitochondria; NUC- Nucleus; ER- Endoplasmic reticulum; PLASM- Plasma membrane; SEC- Secreted.
e The predicted cellular role and functions for each J-protein from G. m. morsitans were implied from either Gene Ontology [127], or published literature on the identified functions/cellular roles of their identified Drosophila and human orthologues.
Fig 5. Phylogenetic analysis of the predicted Type I, II, and IV J-protein subfamilies from G. m. morsitans in relation to D. melanogaster, H. sapiens and S. calcitrans.
A neighbour-joining tree was constructed using a multiple sequence alignment of the full-length amino acid sequences of the Type I, II, and IV J-protein subfamilies in humans, tsetse flies, fruit flies, and yeast. The multiple sequence alignment (S3 Fig) was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Le Gascuel (LG) matrix-based model of amino acid substitution. A discrete Gamma distribution was used to model evolutionarily rate differences among sites (2 categories (+G, parameter = 2.3202). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 159. Bootstrap analysis was computed with 1000 replicates. Accession numbers of the sequences used: E. coli: DnaJ (NP_308042.1). S. calcitrans: ScalJA1 (SCAU009538); ScalJA2 (SCAU013613); ScalJA3 (SCAU003912); ScalJB4 (SCAU013247); ScalJB5 (SCAU015003); ScalB11 (SCAU015416). D. melanogaster: DnaJ-1 (NP_523936.2); CG5001 (NP_608586.2); AT19485 (NP_572633.1); Droj2 (NP_650283.1); Tid56 (NP_524932.2); DnaJ-H (NP_609605.1); CG4164 (NP_608525.1). H. sapiens: DnaJA1 (NP_001530.1); DnaJA2 (NP_005871.1); DnaJA3 (NP_005138.3); DnaJA4 (NP_061072.3); DnaJB1 (NP_006136.1); DnaJB4 (NP_008965.2); DnaJB5 (NP_001128476.2); DnaJB11 (NP_057390.1); DnaJB13 (NP_705842.2). Accession numbers for the G. m. morsitans J-protein sequences can be found in Table 2. The subcellular localisation for the J-proteins are indicated by a bracket on the right-hand side. CYT: cytosolic; ER: endoplasmic reticulum; MITO: mitochondrion.
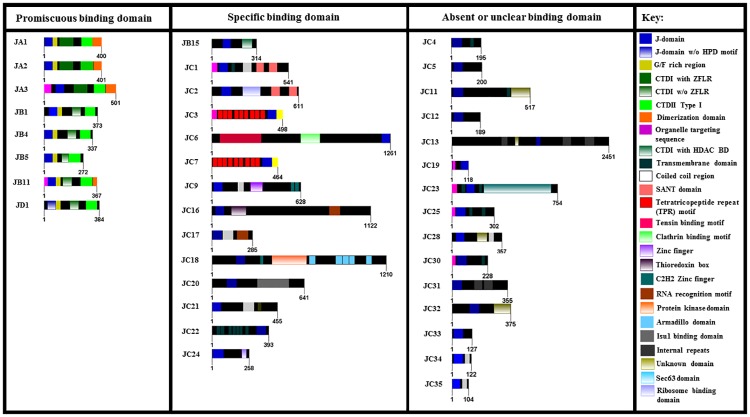
Fig 6. Schematic representation of the domain architecture of the different classes of J-proteins in G. m. morsitans.
Each protein sequence for the G. m. morsitans J-protein family is represented by an open bar with the number of amino acids indicated on either side of the protein bar. The name of the respective J-protein is indicated on the left-hand side. The various domains are highlighted by coloured blocks within the protein bar. A key is provided to give a short description of the various domains and features. The J-proteins were also categorized based on assumed client binding ability and mechanistic mode of functioning as proposed by Kampinga et al. [22].
The large and diverse family of J-proteins contain a number of domains, which have been used as the basis for classification of J-proteins into four different classes [33–34]. The basis for classification of a J-protein is their homology to the prokaryotic canonical J-protein, DnaJ [33]. The domain architecture of DnaJ is divided into an N-terminal J-domain, glycine-phenylalanine (G/F) rich region, zinc finger-like region (ZFLR), and a C-terminal peptide binding domain [33]. The C-terminal domain is comprised of two-barrel topology domains, CTDI and CTDII. CTDI has a hydrophobic pocket for peptide binding and a zinc-finger domain which may also bind peptides [25]. Type I J-proteins possess all these canonical domains, and thus, are highly conserved with respect to DnaJ [32]. Type II J-proteins lack the zinc finger-like region, which is substituted by a glycine-methionine (G/M) rich region [32]. Type III J-proteins contain only the signature J-domain which can occur anywhere along the protein sequence [33]. Type IIIs also possess specialized domains that assist in localizing the J-protein to certain locations within the cell, and specifying the clientele for substrate binding [34]. Type IV proteins possess a J-domain with a compromised or absent HPD motif and may also possess domain structures from other J-protein types [33].
Type I J-proteins
This study identified that the Type I J-protein subfamily in G. m. morsitans has three members: GmmJA1, GmmJA2, and GmmJA3 (Table 2). GmmJA1 and GmmJA2 are Type I J-proteins that are predicted to reside in the cytosol based on their orthology and phylogeny (Fig 5, Table 2), and thus are proposed to assist the predicted cytosolic Gmm Hsp70s in promoting the folding of nascent polypeptides. Though the main role of J-proteins is co-chaperone to their Hsp70 partner, a growing number of cellular roles independent of Hsp70 have been established [74]. The mammalian orthologue of GmmJA1, DnaJA1 has been shown to independently associate and prevent the aggregation of unfolded proteins [75], and is a regulator in the maturation of the androgen receptor (AR) [76]. DnaJA2, mammalian orthologue of GmmJA2, is an enhancer of G-protein-coupled signalling by the β2-adrenergic receptor [77], and assists in the ER-associated degradation of HERG potassium channels by the ubiquitin-proteasome system [78]. Despite their strong homology, the deletion of DnaJA1 in mammalian cells and mice could not be compensated by DnaJA2 and vice versa [76]. A study conducted by Baaklini and colleagues [79] showed that the substrate release mechanism and apparent conformations of DnaJA2 is biochemically different to DnaJA1, and it is inferred as one of the reasons for their functional divergence in their Hsp70 dependent and independent roles.
Loss of DnaJA1 in mice results in severe defects in the late stages of spermatogenesis due to aberrant AR signalling [76]. However, the biological and biochemically properties of GmmJA1 need to be first elucidated. Both GmmJA1 and GmmJA2 were found to possess CTSS and CQTG C-terminal CaaX motifs respectively (S1 Fig), which play a role in protein isoprenylation and farnesylation [80–82]. This post-translational modification has been observed to be integral to the proper functioning of Type I J-proteins, as alternation of this motif within Ydj1 (Type I J-protein from Saccharomyces cerevisiae) resulted in the development of a temperature-sensitive growth phenotype in S. cerevisiae as the motif redirects J-proteins to the plasma membrane or to multi-protein complexes that require its function under stressful conditions [80]. Farnesylation of the CaaX motif has been shown to influence Ydj1 co-operation with Hsp90 [82], and the transferring of substrates to Hsp70 [83].
GmmJA3 is predicted to localise in the mitochondria as it clusters with the known mitochondrial Type I J-proteins HsDnaJA3, and DmelTid56 (Fig 5), and has a N-terminal mitochondrial signal peptide (Fig 6). DmelTid56 is a J-protein that was first discovered as a tumour suppressor, as the deletion of the tid56 gene lead to malignant growth of imaginal disc cells and subsequent embryonic lethality [84]. DnaJA3, the mammalian counterpart of Tid56, has also been shown to be critical for early embryonic development [85], though its role in oncogenesis is controversial [86]. The dnaJA3 gene encodes for two alternatively spliced forms of the protein, which exhibit opposing biological functions in response to exogenous cytotoxic stimuli [87]. The human and Drosophila orthologues of GmmJA3, have been shown to co-operate with mitochondrial Hsp70 in the folding of mitochondrial synthesized and newly imported proteins within the mitochondrial matrix. Thus, GmmJA3 potentially interacts with GmmHsc70-5 in the mitochondria to promote protein folding and disaggregate toxic proteins.
Type II J-proteins
Our study revealed that 5 Type II J-proteins are present in the G. m. morsitans J-protein complement (Table 2). The domain architecture of Type II J-proteins is similar to the Type I J-proteins, except that the zinc finger region that is protruding from the client binding cleft (CTDI) in Type I J-proteins is absent [88]. Despite the difference in the CTDI, Type II J-proteins have been shown to bind non-native substrates, and promote folding in conjunction with Hsp70 [89]. Interestingly, the CTDI of Type II J-proteins have been shown to bind to their cytosolic Hsp70 partners via the C-terminal EEVD motif [90], and it would be interesting to investigate whether GmmJB1 displays the same stringent binding requirements in order to mediate the (re)folding of client proteins. DnaJ-1, Drosophila orthologue of GmmJB4, was shown to interact with Hsc70Cb/Hsp110 in suppressing polyglutamine-induced cell death in Drosophila, and thus, these proteins may function together to maintain protein homeostasis [91]; whilst DnaJ-1 suppressed the toxicity of aggregated proteins, Droj2 and CG5001 lacked this function. It could be proposed that GmmJB4 may also interact with either or both GmmHsp110-1 and GmmHsp110-2 in the same manner within the cell, and carry out the same role in maintaining protein homeostasis as its Drosophila orthologues.
GmmJB11 is a predicted ER Type II J-protein as it forms a clade with the ER luminal Type II J-protein, HsDnaJB11 (Fig 5), and the domain architecture of GmmJB11 shows it possesses an N-terminal signalling peptide (Fig 6). HsDnaJB11 has been experimentally shown to localize within the ER [92], where it binds directly to several nascent, unfolded and mutant secretory proteins, and presents them for HSPA5-dependent folding [93]. The expression of DnaJB11 has also been shown to be up-regulated in response to unfolded secretory protein stress, and is an integral part of the ER stress response [92–93]. The functionality of HSPA5 is highly dependent on its interaction with ER J-proteins during homeostasis and stress, as blocking the partnership will significantly impact HSPA5-dependent folding in vivo [94]. Thus, knockdown or inhibition of GmmJB11 and subsequently its partnership with GmmHsc70-3 could impede the secretion of nascent proteins from the ER. Though, elucidating the role of GmmJB11 and potential GmmJB11-GmmHsc70-3 partnership needs to be conducted.
Thioredoxin1 (Trx1) targets the dnaJB5 gene, the human orthologue of GmmJB5, resulting in an up-regulation of gene expression; DnaJB5 then recruits TBP-2, and orchestrates the formation of the Trx1-DnaJB5-TBP2 complex which mediates the reduction of class II histone deacetylases (HDAC4), essentially restoring its nuclear localisation [95–96]. The reduction of HDAC4 enables the transfer of NADPH-generated electrons to downstream targets, which in turn regulates cardiac hypertrophy [95–96]. The RNA-mediated knockdown of HDAC4 within Drosophila clock cells has been shown to impair the circadian rhythm [97], and long-term memory development within Drosophila [98]. Therefore, it will be interesting to explore the effect of knockout or inhibition of DnaJB5, and its subsequent effect on the function of HDAC4.
GmmJB15 is a Type II J-protein that is unique to the tsetse fly as it has no orthologues in the selected organisms in this study (Table 2). Phylogenetic analysis reinforces that this Type II J-protein is specific to G. m. morsitans as it forms a distinct clade on the tree (Fig 5). However, the domain architecture of GmmJB15 is similar to the human Type II J-proteins: DnaJB6, DnaJB7, and DnaJB8 due to the presence of a HDAC binding domain in the CTDI of GmmJB15 (Fig 6). DnaJB6 and DnaJB8 have been shown to be the two most potent suppressors of aggregation and related toxicity of expanded polyQ proteins [99]. Though, the functional role of DnaJB7 and the HDAC domain have not yet been determined, and therefore no infer of possible function can be made for GmmJB15. However, it raises interesting questions on the biological role of GmmJB15 within the tsetse fly, and it should be prioritized for future studies.
Type III J-proteins
The majority of J-proteins are often comprised of the Type III J-protein subfamily, and G. m. morsitans is no exception as 76% of the J-proteins are Type III J-proteins (Table 2). The functional diversity of the J-protein complement is predominately due to the Type III J-proteins as these members possess a variety of protein domains and motifs, as illustrated in Fig 6, that enable these members to carry out diverse functions within the cell [35]. Eleven of the identified Type III J-proteins (GmmJC2, GmmJC4-7, GmmJC12-13, GmmJC17, GmmJC21, GmmJC24, GmmJC28, and GmmJC31-35) were predicated to localize within the cytosol, with six of these also predicted to be exported to the nucleus, and one exported to the mitochondria (Table 2). Three Type III J-proteins were predicted to localize in the mitochondria (GmmJC19, GmmJC20, and GmmJC30) and one in the ER (GmmJC3) (Table 2). Many of the J-proteins were predicted to associate with the plasma membrane of the cell or subcellular compartments (Table 2), as several of the J-proteins were shown to possess transmembrane domains (Fig 6).
Despite the fact that the human J-proteins DnaJB3 and DnaJB12 are categorised as Type II J-proteins [25], their Gmm orthologues, GmmJC31 and GmmJC32, have been categorised as Type III due to the identification of only a J-domain (Fig 6, Table 2). All of the predicted Type III J-proteins were found to possess human and Drosophila orthologues, and thus could possess similar functions/roles to their identified orthologues (Table 2). GmmJC5, despite its orthology to HsDnaJC5 and the cysteine string protein in D. melanogaster, does not contain the characteristic cysteine-rich region for palmitoylation, and subsequent export to the post-Golgi membranes (Fig 5) [100]. Studies conducted on Drosophila demonstrated that the loss of cysteine string protein expression has been reported to result in very rapid death of adult flies [101]. However, the absence of the cysteine-rich region may possibly be the result of a miss-annotation of the coding region, or sequencing error of GmmJC5.
Additional investigations of miss-annotations/sequencing errors of the G. m. morsitans genome include GmmJC33 and GmmJ34 as these appear to be partial amino acid sequences. The domain architecture for these J-proteins is entirely comprised of the J-domain (Fig 6). Both these J-proteins are putative orthologues of HsDnaJC17, and indicated to be involved in pre-mRNA splicing [102]. Though, this is inconclusive due to the absence of the RNA recognition motif and spliceosome interaction domain that are present in its human counterpart [25].
Type IV J-proteins
Type IV J-proteins are characterized by a J-domain with an abrogated or absent HPD motif [34]. GmmJD1 was the only Type IV J-protein identified in the G. m. morsitans genome and phylogenetic analysis revealed that this J-protein is a unique Type IV J-protein to G. m. morsitans (Fig 4). GmmJD1 was shown to possesses a HNY motif, but also the canonical domains of a typical Type I J-protein (Fig 6). DnaJB13 is the only mammalian J-protein that has a J-domain with an imperfect HPD motif, as it has a HPL motif instead [22]. Due to the abrogated HPD motif, it was questioned whether DnaJB13 could serve as a typical J-protein as the HPD residues are critical to the function of the J-domain. However, it has been shown that DnaJB13 is a cytosolic J-protein involved in the process of spermiogenesis, and sperm movement [103–104]. It would be interesting to investigate the cellular role of GmmJD1, and whether it forms a potential partnership with the Gmm Hsp70s. It marks another J-protein that should be prioritized for future studies.
Expression of Hsp70 and J-protein genes in G. m. morsitans after trypanosome infection
Transcriptomic analysis of trypanosome-infected tsetse flies revealed an increase in the expression of GmmHsp70s, in particular Hsc70-3 (GMOY003216), 4 (GMOY012049), 5 (GMOY010851) and Hsp110-1 (GMOY011246), linked to structural damage of the salivary glands in comparison to uninfected flies, and induction of the stress response could be used as a tool to aid cell renewal [18]. An additional detailed transcriptomic study to determine the effect of trypanosome infection on the salivary gland functions of tsetse flies revealed that a number of genes encoding heat shock proteins were differentially expressed [105]. In a comparison of flies with a mature parasite infection in the salivary glands versus non-infected flies, Hsp70/Hsp90 organising protein (HOP; GMOY003596), Hsc70-5, Hsp110-2 (GMOY013289), JA1 (GMOY007075) were moderately upregulated. Whilst Hsc70-3, Grp170 (GMOY006943), JC16 (GMOY002603) and JC31 (GMOY007250) were moderately down regulated [105]. A further comparison of flies with a mature parasite infection in the salivary glands versus flies with only an established midgut infection revealed that Hsp70A (GMOY009493), HOP and JB4 (GMOY003219) were moderately upregulated [105]. Interestingly, GmmHsp110-2, the Hsp110 unique to G. m. morsitans identified also in this study, was the only heat shock protein that showed increased expression in the salivary glands of flies with an existing trypanosome infection in the midgut in comparison to uninfected flies, which suggests a preliminary response in the salivary glands ahead of parasite infection [105]. GmmHsp110-2 may also be essential for viability and prevention of protein aggregation during stress conditions in the tsetse fly.
Conclusion
The Hsp70 and J-protein complements were comparatively analysed in relation to those found in D. melanogaster, H. sapiens, and S. calcitrans. This study resulted in the identification of 9 putative Hsp70 proteins. The arrangement of the 6 inducible Hsp70 proteins in Drosophila was absent in G. m. morsitans and S. calcitrans. The Hsc70 proteins in Drosophila are regulated during development and exhibit cell and tissue specificity, the same will probably be true of G. m. morsitans.
In this study 37 J-proteins were identified, with two of these being partial sequences. Based on the available data from the eukaryotic orthologues, it was possible to infer functions of many of the Hsp70 and J-proteins from G. m. morsitans. Obviously, many of our inferences will need be to be confirmed experimentally. The diversity of the J-protein complement has evolved to fulfil specific functions. Some heat shock proteins from the trypanosomes have been studied and those essential for differentiation and survival have been identified [106]. A comparative analysis of the Hsp70-J-protein complex from the human and animal hosts, as well as the Trypanosoma brucei parasites and the insect vectors will enhance our understanding of the differences in host specificities, in addition it will be possible to gain a better understanding of vector-parasite and host-parasite interactions.
Supporting information
Multiple sequence alignment of the full-length amino acid sequences of the Hsp70/HSPA gene families in humans, tsetse flies, fruit flies, and stable flies. The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp70 (SCAU008520); Hsp68 (SCAU003728); Hsc70-1 (SCAU005225); Hsc70-2 (SCAU008036); Hsc70-3 (SCAU000678); Hsc70-4 (SCAU015347); Hsc70-5 (SCAU003620). D. melanogaster: Hsp68 (NP_524474.1); Hsp70Aa (NP_731651.1); Hsp70Ab (NP_524798.2); Hsp70Ba (NP_731716.1); Hsp70Bb (NP_524927.2); Hsp70Bbb (NP_788663.1); Hsp70Bc (NP_650209.1); Hsc70-1 (NP_524063.1); Hsc70-2 (NP_524339.1); Hsc70-3 (NP_727563.1); Hsc70-4 (NP_524356.1); Hsc70-5 (NP_523741.2). H. sapiens: HSPA1A (NP_005336.3); HSPA1B (NP_005337.2); HSPA1L (NP_005518.3); HSPA2 (NP_068814.2); HSPA5 (NP_005338.1); HSPA6 (NP_002146.2); HSPA8 (NP_006588.1); HSPA9 (NP_004125.3). Accession numbers for the G. m. morsitans Hsp70 sequences can be found in Table 1.
(PDF)
Multiple sequence alignment of the full-length amino acid sequences of the Hsp110/HSPH gene families in humans, tsetse flies, fruit flies, and yeast. The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters in the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp110 (SCAU005995); Grp170 (SCAU010922). D. melanogaster: Hsp110 (NP_648687.1); Grp170 (NP_569995.1). H. sapiens: HSPH1 (NP_006635.2); HSPH2 (NP_002145.3); HSPH3 (NP_055093.2); HSPH4 (NP_006380.1). Accession numbers for the G. m. morsitans Hsp110 sequences can be found in Table 1.
(PDF)
The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: DnaJ (NP_308042.1). S. calcitrans: ScalJA1 (SCAU009538); ScalJA2 (SCAU013613); ScalJA3 (SCAU003912); ScalJB4 (SCAU013247); ScalJB5 (SCAU015003); ScalB11 (SCAU015416). D. melanogaster: DnaJ-1 (NP_523936.2); CG5001 (NP_608586.2); AT19485 (NP_572633.1); Droj2 (NP_650283.1); Tid56 (NP_524932.2); DnaJ-H (NP_609605.1); CG4164 (NP_608525.1). H. sapiens: DnaJA1 (NP_001530.1); DnaJA2 (NP_005871.1); DnaJA3 (NP_005138.3); DnaJA4 (NP_061072.3); DnaJB1 (NP_006136.1); DnaJB4 (NP_008965.2); DnaJB5 (NP_001128476.2); DnaJB11 (NP_057390.1); DnaJB13 (NP_705842.2). Accession numbers for the G. m. morsitans J-protein sequences can be found in Table 2.
(PDF)
Each protein sequence for the G. m. morsitans Hsp70 superfamily is represented by an open bar with the various protein domains and other associated features that were identified using Prosite [44] and SMART 7 [43] are displayed as colour blocks within the open bar. These domains and associated features include the N-terminal ATPase domain (red), substrate binding domain (SBD; green), putative substrate binding domain for NEFs (SBD; dark green), C-terminal region (C-terminal; purple) and targeting signal peptides (S; dark blue).
(TIF)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by a grant from the National Research Foundation (NRF); grant number 87663. S.J.B. is the recipient of an NRF Doctoral Innovation Scholarship.
References
- 1.Malvy D, Chappuis F. Sleeping sickness. Clin Microbiol Infect. 2011; 17(7):986–995, doi: 10.1111/j.1469-0691.2011.03536.x [DOI] [PubMed] [Google Scholar]
- 2.Alsan M. The effect of the tsetse fly on african development. Am Econ Rev. 2015; 105: 382–410, doi: 10.1257/aer.20130604 [Google Scholar]
- 3.Odiit M, Kansiime F, Enyaru JCK. Duration of symptoms and case fatality of sleeping sickness caused by Trypanosoma brucei rhodesiense sleeping sickness in Tororo, Uganda. East Afr Med J. 1997; 74: 792–795. [PubMed] [Google Scholar]
- 4.Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: Where do we stand and what comes next? PLoS Med. 2008; 5:174–180, doi: 10.1371/journal.pmed.0050055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010; 375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- 6.Leak SGA. Tsetse biology and ecology: their role in the epidemiology and control of trypanosomosis. 1st ed CABI publishing in association with the ILRI; 1999. [Google Scholar]
- 7.Aksoy S. Control of tsetse flies and trypanosomes using molecular genetics. Vet Parasitol. 2003; 115(2):125–145, doi: 10.1016/s0304-4017(03)00203-6 [DOI] [PubMed] [Google Scholar]
- 8.Gooding RH, Krafsur ES. Tsetse genetics: contributions to biology, systematics, and control of tsetse flies. Annu Rev Entomol. 2005; 50:101–123. doi: 10.1146/annurev.ento.50.071803.130443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAlpine JF. Phylogeny and classification of the Muscomorpha In: McAlpine JF. Manual of Nearctic Diptera. Ottawa: Res. Branch Agric. Can; 1989. pp. 1397–1518. [Google Scholar]
- 10.Krafsur ES. Tsetse flies: genetics, evolution, and role as vectors. Infect Genet Evol. 2009; 9:124–141. doi: 10.1016/j.meegid.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benoit JB, Attardo GM, Baumann AA, Michalkova V, Aksoy S. Adenotrophic viviparity in tsetse flies: potential for population control and as an insect model for lactation. Annu Rev Entomol. 2015; 60:351–371, doi: 10.1146/annurev-ento-010814-020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers D, Robinson T. Tsetse distribution In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. Oxford, UK: CABI; 2004. pp. 139–179. [Google Scholar]
- 13.Barrett MP, Croft SL. Management of trypanosomiasis and leishmaniasis. Br Med Bull. 2012; 104:175–196, doi: 10.1093/bmb/lds031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welburn SC, Maudlin I, Simarro PP. Controlling sleeping sickness-a review. Parasitology. 2009: 136: 1943–1949. doi: 10.1017/S0031182009006416 [DOI] [PubMed] [Google Scholar]
- 15.Christoffels A, Masiga D, Berriman M, Lehane M, Touré Y, Aksoy S. International glossina genome initiative 2004–2014: a driver for post-genomic era research on the African continent. PLoS Negl Trop Dis. 2014; 8: e3024, doi: 10.1371/journal.pntd.0003024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe J, Hattori M, Berriman M, Lehane M, Hall N, Solano P. et al. Genome sequence of the tsetse fly (glossina morsitans): vector of african trypanosomiasis. Science 2014; 344:380–386, doi: 10.1126/science.1249656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–713, doi: 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telleria EL, Benoit JB, Zhao X, Savage AF, Regmi S, Alves e Silva TL et al. Insights into the trypanosome-host interactions revealed through transcriptomic analysis of parasitized tsetse fly salivary glands. PLoS Negl Trop Dis. 2014; 8:e2649, doi: 10.1371/journal.pntd.0002649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assimon VA, Gillies AT, Rauch JN, Gestwicki JE. Hsp70 protein complexes as drug targets. Curr Pharm Des. 2013; 19:404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009; 583:2647–2653, doi: 10.1016/j.febslet.2009.04.029 [DOI] [PubMed] [Google Scholar]
- 21.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002; 295:1852–1858. doi: 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 22.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones, 2009; 14:105–111, doi: 10.1007/s12192-008-0068-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994; 38:1–17. [DOI] [PubMed] [Google Scholar]
- 24.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005; 62:670–684. doi: 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010; 11:579–592, doi: 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 2000; 5:276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008; 133:1068–1079, doi: 10.1016/j.cell.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 28.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 2011; 6:e26319, doi: 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattoo RUH, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013; 288:21399–21411, doi: 10.1074/jbc.M113.479253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer MP. Gymnastics of molecular chaperones. Mol Cell 2010; 39:321–331, doi: 10.1016/j.molcel.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 31.Rampelt H, Mayer MP, Bukau B. Nucleotide exchange factors for Hsp70 chaperones. Methods Mol Biol 2011; 787:83–91, doi: 10.1007/978-1-61779-295-3_7 [DOI] [PubMed] [Google Scholar]
- 32.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005; 14:1697–1709. doi: 10.1110/ps.051406805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998; 3(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botha M, Pesce ER, Blatch GL. The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: Regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol. 2007; 39:1781–1803. doi: 10.1016/j.biocel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- 35.Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta 2015; 1853:1035–1045, doi: 10.1016/j.bbamcr.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severson DW, Knudson DL, Soares MB, Loftus BJ. Aedes aegypti genomics. Insect Biochem Mol Biol. 2004; 34:715–721. doi: 10.1016/j.ibmb.2004.03.024 [DOI] [PubMed] [Google Scholar]
- 37.Waterhouse RM, Wyder S, Zdobnov EM. The Aedes aegypti genome: A comparative perspective. Insect Mol Biol. 2008; 17:1–8, doi: 10.1111/j.1365-2583.2008.00772.x [DOI] [PubMed] [Google Scholar]
- 38.Lakhotia SC, Prasanth KV. Tissue- and development-specific induction and turnover of hsp70 transcripts from loci 87A and 87C after heat shock and during recovery in Drosophila melanogaster. J Exp Biol. 2002; 205:345–358. [DOI] [PubMed] [Google Scholar]
- 39.Dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J et al. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015; 43:D690–697, doi: 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012; 40:D302–305, doi: 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigrist CJ, Cerutti L, De Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2009; 38:161–166, doi: 10.1093/nar/gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al. Protein Identification and Analysis Tools on the ExPASy Server; In: Walker JM, editors: The Proteomics Protocols Handbook, Humana Press; 2005. [Google Scholar]
- 43.Larkin MA, Blackshield G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23(21): 2947–2948, doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1879. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008; 25(7): 1307–1320. doi: 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 46.Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007; 8, 2 doi: 10.1186/1471-2105-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brameier M, Krings A, MacCallum RM. NucPred—Predicting nuclear localization of proteins. Bioinformatics. 2007; 23:1159–1160. doi: 10.1093/bioinformatics/btm066 [DOI] [PubMed] [Google Scholar]
- 48.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996; 241:779–786. [DOI] [PubMed] [Google Scholar]
- 49.Höglund A, Dönnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: Prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006; 22:1158–1165. doi: 10.1093/bioinformatics/btl002 [DOI] [PubMed] [Google Scholar]
- 50.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011; 8:785–786. doi: 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 51.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007; 35:W585–587. doi: 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindquist S, Craig EA. The heat shock proteins. Ann. Rev. Gen. 1988; 22: 631–677. [DOI] [PubMed] [Google Scholar]
- 53.Gupta RS, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggest a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 1994; 4: 1104–1114. [DOI] [PubMed] [Google Scholar]
- 54.Hunt C, Morimoto RI. Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70. Proc. Natl. Acad. Sci. USA. 1985; 82: 6455–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feder ME, Krebs RA. Natural and genetic engineering of the heat-shock protein Hsp70 in Drosophila melanogaster: Consequences for thermotolerance. Am Zool. 1998; 38(3): 503–517. [Google Scholar]
- 56.Lindquist S. Autoregulation of the heat-shock response In Ilan J., editors: Translational regulation of gene expression, Plenum Press, New York; 1993. [Google Scholar]
- 57.Ingolia TD, Craig EA. Drosophila gene related to the major heat shock-induced gene is transcribed at normal temperatures and not induced by heat shock. Proc Natl Acad Sci U S A. 1982; 79:525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Štětina T, Koštál V, Korbelová J. The role of inducible Hsp70, and other heat shock proteins, in adaptive complex of cold tolerance of the fruit fly (Drosophila melanogaster). PLoS ONE. 2015; 10(6): e0128976 https://doi.org/10.1371/journal.pone.0128976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang MC, Bohmann D, Jasper H. JNK signalling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell. 2003; 5(5): 811–816. [DOI] [PubMed] [Google Scholar]
- 60.Gong WJ, Golic KG. Loss of Hsp70 in drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics. 2006; 172:275–286, doi: 10.1534/genetics.105.048793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elefant F, Palter KB. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol. Biol. Cell. 1999; 10:2101–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Craig EA, Ingolia TD, Manseau LJ. Expression of Drosophila heat-shock cognate genes during heat shock and development. Dev Biol. 1983; 99:418–426. [DOI] [PubMed] [Google Scholar]
- 63.Colinet H, Overgaard J, Com E, Sørensen JG. Proteomic profiling of thermal acclimation in Drosophila melanogaster. Insect Biochem Mol Biol. 2013; 43:352–365. doi: 10.1016/j.ibmb.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 64.Daugaard M, Rohde M. Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007; 581:3702–3710. doi: 10.1016/j.febslet.2007.05.039 [DOI] [PubMed] [Google Scholar]
- 65.Nikolaidis N, Nei M. Concerted and nonconcerted evolution of the Hsp70 gene superfamily in two sibling species of nematodes. Mol Biol Evol. 2004; 21(3): 498–505, https://doi.org/10.1093/molbev/msh041. [DOI] [PubMed] [Google Scholar]
- 66.Hu C, Lin S, Chi W, Charng Y. Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiology. 2012; 158(2):747–58. doi: 10.1104/pp.111.187674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doumanis J, Wada K, Kino Y, Moore AW, Nukina N. RNAi screening in Drosophila cells identifies new modifiers of mutant huntingtin aggregation. PLoS One. 2009; 4:e7275, doi: 10.1371/journal.pone.0007275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rüdiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol. 1997; 4: 342–349. [DOI] [PubMed] [Google Scholar]
- 69.Zahn M, Berthold N, Kieslich B, Knappe D, Hoffmann R, Sträter N. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol. 2013; 425:2463–79, doi: 10.1016/j.jmb.2013.03.041 [DOI] [PubMed] [Google Scholar]
- 70.Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun. 2012; 3:1310, doi: 10.1038/ncomms2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stocker AJ, Rusuwa BB, Blacket MJ, Frentiu FD, Sullivan M, Foley BR et al. Physical and linkage maps for Drosophila serrata, a model species for studies of clinal adaptation and sexual selection. G3 genes genom genet, 2012; 2(2): 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994; 15: 269–275. [DOI] [PubMed] [Google Scholar]
- 73.Gray HM. It takes two transposons to tango. Trends Genet. 2000; 16: 461–468. [DOI] [PubMed] [Google Scholar]
- 74.Sterrenberg JN, Blatch GL, Edkins AL. Human DNAJ in cancer and stem cells. Cancer Lett. 2011; 312(3): 129–142. [DOI] [PubMed] [Google Scholar]
- 75.Terada K, Oike Y. Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. J Biol Chem. 2010; 285(22): 16789–16797. doi: 10.1074/jbc.M110.101501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terada K, Yomogida K, Imai T, Kiyonari H, Takeda N, Kadomatsu T et al. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 2005; 24:611–622. doi: 10.1038/sj.emboj.7600549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosales-Hernandez A, Beck KE, Zhao X, Braun AP, Braun JEA. RDJ2 (DNAJA2) chaperones neural G protein signaling pathways. Cell Stress Chaperones 2009; 14:71–82, doi: 10.1007/s12192-008-0056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walker VE, Wong MJ, Atanasiu R, Hantouche C, Young JC, Shrier A. Hsp40 chaperones promote degradation of the HERG potassium channel. J Biol Chem. 2010; 285(5): 3319–3329. doi: 10.1074/jbc.M109.024000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baaklini I, Wong MJ, Hantouche C, Patel Y, Shrier A, Young JC. The DNAJA2 substrate release mechanism is essential for chaperone-mediated folding. J Biol Chem. 2012; 287(50): 41939–41954. doi: 10.1074/jbc.M112.413278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kitten GT, Nigg EA. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J Cell Biol. 1991; 113:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu JK, Bressan RA, Hasegawa PM. Isoprenylation of the plant molecular chaperone ANJ1 facilitates membrane association and function at high temperature. Proc Natl Acad Sci U S A. 1993; 90:8557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol Biol Cell. 2008; 19: 5249–5258. doi: 10.1091/mbc.E08-04-0435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem. 2005; 280(1): 695–702. doi: 10.1074/jbc.M410645200 [DOI] [PubMed] [Google Scholar]
- 84.Kurzik-Dumke U, Gandacker D, Renthrop M, Gateff E. Tumor suppression in Drosophila is causally related to the function of the lethal (2) tumorous imaginal discs gene, a dnaJ homolog. Dev Genet. 1995; 16(1): 64–76. doi: 10.1002/dvg.1020160110 [DOI] [PubMed] [Google Scholar]
- 85.Lo JF, Hayashi M, Woo-Kim S, Tian B, Huang JF, Fearns C et al. Tid1, a cochaperone of the heat shock 70 protein and the mammalian counterpart of the Drosophila tumor suppressor l(2)tid, is critical for early embryonic development and cell survival. Mol Cell Biol. 2004; 24(6): 2226–2236. doi: 10.1128/MCB.24.6.2226-2236.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Niu G, Zhang H, Liu D, Chen L, Belani C, Wang HG, Cheng H. Tid1, the mammalian homologue of drosophila tumor suppressor Tid56, mediates macroautophagy by interacting with Beclin1-containing autophagy protein complex. J Biol Chem. 2015; 290: 18102–18110. doi: 10.1074/jbc.M115.665950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edwards KM, Munger K. Depletion of physiological levels of the human TID1 protein renders cancer cell lines resistant to apoptosis mediated by multiple exogenous stimuli. Oncogene. 2004; 23:8419–8431. doi: 10.1038/sj.onc.1207732 [DOI] [PubMed] [Google Scholar]
- 88.Li J, Qian X, Sha B. The Crystal Structure of the Yeast Hsp40 Ydj1 Complexed with Its Peptide Substrate. Structure. 2003; 11:1475–1483. [DOI] [PubMed] [Google Scholar]
- 89.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998; 23(8): 222–227. [DOI] [PubMed] [Google Scholar]
- 90.Yu HY, Ziegelhoffer T, Craig EA. Functionality of Class A and Class B J-protein co-chaperones with Hsp70. FEBS Lett. 2015a; 589: 2825–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuo Y, Ren S, Lao U, Edgar BA, Wang T. Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis. 2013; 4: e833 doi: 10.1038/cddis.2013.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu M, Haslam RHA, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000; 275: 24984–92. doi: 10.1074/jbc.M000739200 [DOI] [PubMed] [Google Scholar]
- 93.Shen Y, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002; 277: 15947–15956. doi: 10.1074/jbc.M112214200 [DOI] [PubMed] [Google Scholar]
- 94.Guo F, Snapp EL. ERdj3 regulates BiP occupancy in living cells. J Cell Sci. 2013; 126(6): 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iosefson O, Sharon S, Goloubinoff P, Azem A. Reactivation of protein aggregates by mortalin and Tid1-the human mitochondrial Hsp70 chaperone system. Cell Stress Chaperones 2012; 17:57–66 doi: 10.1007/s12192-011-0285-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oka SI, Ago T, Kitazono T, Zablocki D, Sadoshima J. The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy. J Mol Med. 2009; 87:785–791. doi: 10.1007/s00109-009-0471-2 [DOI] [PubMed] [Google Scholar]
- 97.Fogg PCM, O’Neill JS, Dobrzycki T, Calvert S, Lord EC, McIntosh RLL et al. Class IIa histone deacetylases are conserved regulators of circadian function. J Biol Chem. 2014; 289:34341–34348. doi: 10.1074/jbc.M114.606392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fitzsimons HL, Schwartz S, Given FM, Scott MJ. The histone deacetylase HDAC4 regulates long-term memory in Drosophila. PLoS One. 2013; 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van den Nieuwendijk R et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013; 288(24): 17225–17237. doi: 10.1074/jbc.M112.421685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burgoyne RD, Morgan A. Cysteine string protein (CSP) and its role in preventing neurodegeneration. Semin Cell Dev Biol. 2015; 40:153–159. doi: 10.1016/j.semcdb.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Umbach JA, Zinsmaier KE, Eberle KK, Buchner E, Benzer S et al. Presynaptic dysfunction in Drosophila csp mutants. Neuron. 1994; 13:899–907. [DOI] [PubMed] [Google Scholar]
- 102.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006; 63(22): 2560–2570. doi: 10.1007/s00018-006-6192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guan J, Yuan L. A heat-shock protein 40, DNAJB13, is an axoneme-associated component in mouse spermatozoa. Mol Reprod Dev. 2008; 75(9): 1379–1386. doi: 10.1002/mrd.20874 [DOI] [PubMed] [Google Scholar]
- 104.Li W, Liu G. DNAJB13, a type II HSP40 family member, localizes to the spermatids and spermatozoa during mouse spermatogenesis. BMC Dev Biol. 2014; 14(38), doi: 10.1186/s12861-014-0038-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matetovic I, Caljon G, Van Den Abbeele J. Tsetse fly tolerance to T. brucei infection: transcriptome analysis of trypanosome-associated changes in the tsetse fly salivary gland. BMC Genomics. 2016; 17(1): 971 doi: 10.1186/s12864-016-3283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011; 21(6): 915–924. doi: 10.1101/gr.115089.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ahrendt E, Braun JE. Channel triage: Emerging insights into the processing and quality control of hERG potassium channels by DnaJA proteins 1, 2 and 4. Channels (Austin). 2010; 4(5):335–336. doi: 10.4161/chan.4.5.13090 [DOI] [PubMed] [Google Scholar]
- 108.Whitaker R, Faulkner S, Miyokawa R, Burhenn L, Henriksen M, Wood JG et al. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging. 2013; 5: 682–691. doi: 10.18632/aging.100599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996; 15(12):2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 110.Tsai MF, Wang CC, Chang GC, Chen YC, Chen HY, Cheng CL et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst. 2006; 98(12):825–838. doi: 10.1093/jnci/djj229 [DOI] [PubMed] [Google Scholar]
- 111.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD. A Redox-Dependent Pathway for Regulating Class II HDACs and Cardiac Hypertrophy. Cell. 2008; 133: 978–993. doi: 10.1016/j.cell.2008.04.041 [DOI] [PubMed] [Google Scholar]
- 112.Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009; 48(1):41–49. doi: 10.1021/bi8015923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chevalier M, Rhee H, Elguindi EC, Blond SY. Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J Biol Chem. 2000; 275:19620–19627. doi: 10.1074/jbc.M001333200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kroczynska B, Evangelista CM, Samant SS, Elguindi EC, Blond SY. The SANT2 domain of the murine tumor cell DnaJ-like protein 1 human homologue interacts with alpha1-antichymotrypsin and kinetically interferes with its serpin inhibitory activity. J Biol Chem. 2004; 279(12):11432–11443. doi: 10.1074/jbc.M310903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Otto H, Conz C, Maier P, Wölfle T, Suzuki CK, Jenö P et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A. 2005; 102(29):10064–10069. doi: 10.1073/pnas.0504400102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002; 99(25):15920–15925. doi: 10.1073/pnas.252341799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goodman AG, Tanner BC, Chang ST, Esteban M, Katze MG. Virus infection rapidly activates the P58(IPK) pathway, delaying peak kinase activation to enhance viral replication. Virology. 2011; 417(1):27–36. doi: 10.1016/j.virol.2011.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iliopoulos I, Torok I, Mechler BM. The DnaJ60 gene of Drosophila melanogaster encodes a new member of the DnaJ family of proteins. Biol Chem. 1997; 378(10): 1177–1181. [PubMed] [Google Scholar]
- 119.Bronk P, Nie Z, Klose MK, Dawson-Scully K, Zhang J, Robertson R M, et al. (2005). The multiple functions of cysteine-string protein analyzed at Drosophila nerve terminals. J Neurosci. 2005; 25:2204–2214. doi: 10.1523/JNEUROSCI.3610-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995; 378: 632–635. doi: 10.1038/378632a0 [DOI] [PubMed] [Google Scholar]
- 121.Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WM. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003; 22(14):3613–3623. doi: 10.1093/emboj/cdg362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Han C, Chen T, Li N, Yang M, Wan T, Cao X. HDJC9, a novel human type C DnaJ/HSP40 member interacts with and cochaperones HSP70 through the J domain. Biochem Biophys Res Commun. 2007; 353(2):280–285. doi: 10.1016/j.bbrc.2006.12.013 [DOI] [PubMed] [Google Scholar]
- 123.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007; 581(18):3545–3549. doi: 10.1016/j.febslet.2007.06.052 [DOI] [PubMed] [Google Scholar]
- 124.Choi J, Djebbar S, Fournier A, Labrie C. The co-chaperone DNAJC12 binds to Hsc70 and is upregulated by endoplasmic reticulum stress. Cell Stress Chaperones. 2014; 19(3): 439–446, doi: 10.1007/s12192-013-0471-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Girard M, Poupon V, Blondeau F, McPherson PS. The DnaJ-domain protein RME-8 functions in endosomal trafficking. J Biol Chem. 2005; 280: 40135–40143. doi: 10.1074/jbc.M505036200 [DOI] [PubMed] [Google Scholar]
- 126.Girard M, McPherson PS. RME-8 regulates trafficking of the epidermal growth factor receptor. FEBS Lett. 2008; 582(6): 961–966. doi: 10.1016/j.febslet.2008.02.042 [DOI] [PubMed] [Google Scholar]
- 127.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25(1):25–9. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE et al. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J.2003a; 22: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010; 19(19): 3816–3834. doi: 10.1093/hmg/ddq301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581(15):2783–2793. doi: 10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]
- 131.Skowronek MH, Rotter M, Haas IG. Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol Chem. 1999; 380(9): 1133–1138. doi: 10.1515/BC.1999.142 [DOI] [PubMed] [Google Scholar]
- 132.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004; 24(21): 9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sopha P, Kadokura H, Yamamoto YH, Takeuchi M, Saito M, Tsuru A. A novel mammalian ER-located J-protein, DNAJB14, can accelerate ERAD of misfolded membrane proteins. Cell Struct Funct. 2012; 37(2):177–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of the full-length amino acid sequences of the Hsp70/HSPA gene families in humans, tsetse flies, fruit flies, and stable flies. The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp70 (SCAU008520); Hsp68 (SCAU003728); Hsc70-1 (SCAU005225); Hsc70-2 (SCAU008036); Hsc70-3 (SCAU000678); Hsc70-4 (SCAU015347); Hsc70-5 (SCAU003620). D. melanogaster: Hsp68 (NP_524474.1); Hsp70Aa (NP_731651.1); Hsp70Ab (NP_524798.2); Hsp70Ba (NP_731716.1); Hsp70Bb (NP_524927.2); Hsp70Bbb (NP_788663.1); Hsp70Bc (NP_650209.1); Hsc70-1 (NP_524063.1); Hsc70-2 (NP_524339.1); Hsc70-3 (NP_727563.1); Hsc70-4 (NP_524356.1); Hsc70-5 (NP_523741.2). H. sapiens: HSPA1A (NP_005336.3); HSPA1B (NP_005337.2); HSPA1L (NP_005518.3); HSPA2 (NP_068814.2); HSPA5 (NP_005338.1); HSPA6 (NP_002146.2); HSPA8 (NP_006588.1); HSPA9 (NP_004125.3). Accession numbers for the G. m. morsitans Hsp70 sequences can be found in Table 1.
(PDF)
Multiple sequence alignment of the full-length amino acid sequences of the Hsp110/HSPH gene families in humans, tsetse flies, fruit flies, and yeast. The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters in the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: HscC (NP_415183.1). S. calcitrans: Hsp110 (SCAU005995); Grp170 (SCAU010922). D. melanogaster: Hsp110 (NP_648687.1); Grp170 (NP_569995.1). H. sapiens: HSPH1 (NP_006635.2); HSPH2 (NP_002145.3); HSPH3 (NP_055093.2); HSPH4 (NP_006380.1). Accession numbers for the G. m. morsitans Hsp110 sequences can be found in Table 1.
(PDF)
The multiple sequence alignment was performed using the in-built ClustalW program [43] with default parameters on the MEGA7 software [44]. Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers of the sequences used: E. coli: DnaJ (NP_308042.1). S. calcitrans: ScalJA1 (SCAU009538); ScalJA2 (SCAU013613); ScalJA3 (SCAU003912); ScalJB4 (SCAU013247); ScalJB5 (SCAU015003); ScalB11 (SCAU015416). D. melanogaster: DnaJ-1 (NP_523936.2); CG5001 (NP_608586.2); AT19485 (NP_572633.1); Droj2 (NP_650283.1); Tid56 (NP_524932.2); DnaJ-H (NP_609605.1); CG4164 (NP_608525.1). H. sapiens: DnaJA1 (NP_001530.1); DnaJA2 (NP_005871.1); DnaJA3 (NP_005138.3); DnaJA4 (NP_061072.3); DnaJB1 (NP_006136.1); DnaJB4 (NP_008965.2); DnaJB5 (NP_001128476.2); DnaJB11 (NP_057390.1); DnaJB13 (NP_705842.2). Accession numbers for the G. m. morsitans J-protein sequences can be found in Table 2.
(PDF)
Each protein sequence for the G. m. morsitans Hsp70 superfamily is represented by an open bar with the various protein domains and other associated features that were identified using Prosite [44] and SMART 7 [43] are displayed as colour blocks within the open bar. These domains and associated features include the N-terminal ATPase domain (red), substrate binding domain (SBD; green), putative substrate binding domain for NEFs (SBD; dark green), C-terminal region (C-terminal; purple) and targeting signal peptides (S; dark blue).
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.