ABSTRACT
Selective nuclear import in eukaryotic cells involves sequential interactions between nuclear import receptors and phenylalanine-glycine (FG)-repeat nucleoporins. Traditionally, binding of cargoes to import receptors is perceived as a nuclear pore complex independent event, while interactions between import complexes and nucleoporins are thought to take place at the nuclear pores. However, studies have shown that nucleoporins are mobile and not static within the nuclear pores, suggesting that they may become engaged in nuclear import before nuclear pore entry. Here we have studied post-mitotic nuclear import of the tumor suppressor protein PML. Since this protein forms nuclear compartments called PML bodies that persist during mitosis, the assembly of putative PML import complexes can be visualized on the surface of these protein aggregates as the cell progress from an import inactive state in mitosis to an import active state in G1. We show that these post-mitotic cytoplasmic PML bodies incorporate a multitude of peripheral nucleoporins, but not scaffold or nuclear basket nucleoporins, in a manner that depends on FG-repeats, the KPNB1 import receptor, and the PML nuclear localization signal. The study suggests that nucleoporins have the ability to target certain nuclear cargo proteins in a nuclear pore-uncoupled state, before nuclear pore entry.
KEYWORDS: FG-repeats, KPNB1, nuclear import, nuclear pore complex, nucleoporins, PML, PML bodies
Introduction
In eukaryotic cells, transport of components across the nuclear envelope is mediated by nuclear pore complexes (NPCs). These structures are arranged in an eightfold radial symmetry surrounding a central channel and contain multiple copies of approximately 30 different proteins termed nucleoporins (NUPs).1-6
For most proteins that are destined to cross the nuclear membrane, the process of nuclear envelope translocation is facilitated by a family of transport receptors referred to as karyopherins. These proteins typically bind a nuclear import or export sequence present on the cargo protein and subsequently catalyze translocation of the cargo complex across the nuclear membrane by a mechanism that is facilitated by the Ran GTPase and energy.7-11 The best characterized nuclear import pathway, which often is referred to as the classical nuclear import pathway, is that mediated by the karyopherin KPNB1 (also known as importin β). KPNB1 binds classical nuclear localization signals (NLSs) through the adaptor protein importin α, but can also mediate transport through direct interactions with nuclear cargoes.12,13
Broadly, NUPs can be divided into 2 subclasses referred to as scaffold NUPs and peripheral NUPs. While scaffold NUPs are thought to mediate nuclear membrane anchoring and to provide NPC structural integrity, peripheral NUPs occupy the nuclear, cytoplasmic and central region of the pore, where they participate in regulation of nuclear import and export.14-16
Most peripheral NUPs comprise characteristic sequences termed phenylalanine-glycine (FG)-repeats, which represent flexible and dynamic regions consisting of multiple FXFG, FG or GLFG motifs interspersed by linker regions.17 The FG-repeats function as contact sites for incoming karyopherin-cargo complexes.18-20 In vitro experiments have suggested that FG-repeats create a hydrogel-like web throughout the pore channel that allows for passive diffusion of small, neutral molecules, while proteins larger than 30–40 kDa require receptor-mediated import or export.21-23 Notably, the ability to form a FG-hydrogel was found to be essential for viability in yeast.22 The passage of cargo through the NPC has also been explained by a brush-like conformation of FG-repeats that collapse upon binding to transport receptors24 and by the virtual gating model, which suggests that interactions between import complexes and FG-repeats lead to lowering of the energy required for nucleo-cytoplasmic translocation.25
For many years NPCs were perceived as being static channels in interphase cells. This view has later been challenged by several studies aimed at analyzing NUP dynamics. In one study, using inverse fluorescence recovery after photobleaching (FRAP), several of the peripheral NUPs were found to have a significantly shorter residence time at NPCs compared with scaffold NUPs.14 This result suggested that these proteins may have functions at NPC distal sites, in addition to their function at nuclear pores.26 Notably, several recent studies have suggested NPC-independent functions of NUPs in gene expression regulation during interphase.27-32 However, a role of nuclear pore-uncoupled NUPs in nucleo-cytoplasmic transport has not previously been reported.
The tumor suppressor protein promyelocytic leukemia (PML) is mostly confined to the nucleus during interphase, where it plays an essential role in forming distinct nuclear compartments called PML nuclear bodies (PML NBs).33,34 At the structural level the PML protein is organized into an N-terminal TRIM motif, common to all isoforms, and a variable C-terminal domain.35 While the TRIM motif seems to be important for PML body assembly,36-38 the C-terminal variable domain may confer isoform-specific functions. For nuclear import, most PML isoforms seem to rely on a lysine- and arginine-rich NLS present in the central region of the protein.35 However, nuclear import activity has also been reported to be present in the variable isoform-specific region of the splice variant PML II.39
The composition and morphology of PML bodies change as cells go through the cell cycle. Firstly, the mitotic PML bodies, which are referred to as mitotic accumulations of PML proteins (MAPPs), are larger than PML NBs, possibly resulting from PML body aggregation during mitotic entry.40 Secondly, several of the PML NB residence proteins, including SUMO, DAXX and SP100 are released from these structures as MAPPs are formed.40,41 Following cell division, MAPPs persist in the cytoplasm for a short period and they gradually become disassembled and recycled to the nucleus.40,42 During this period of early G1 phase they have been observed to recruit the 2 NUPs NUP98 and NUP214.42 These post-mitotic cytoplasmic PML bodies, which we refer to as cytoplasmic assemblies of PML and nucleoporins (CyPNs),42 may represent aggregates of PML proteins that are being prepared for nuclear import.
In the present study we investigated a total of 20 NUPs representing different nuclear pore subcomplexes and found that CyPNs exclusively recruit peripheral NUPs, and not scaffold NUPs. In addition, we show that the assembly of NUPs on the surface of CyPNs requires the KPNB1 import receptor and depends on the presence of FG-repeat regions. Together, the data provide insight into post-mitotic nuclear import of PML and emphasize that FG-repeat-containing NUPs (FG-NUPs) target PML nuclear import complexes in the cytoplasm.
Results
KPNB1 localizes to cytoplasmic assemblies of PML and NUPs
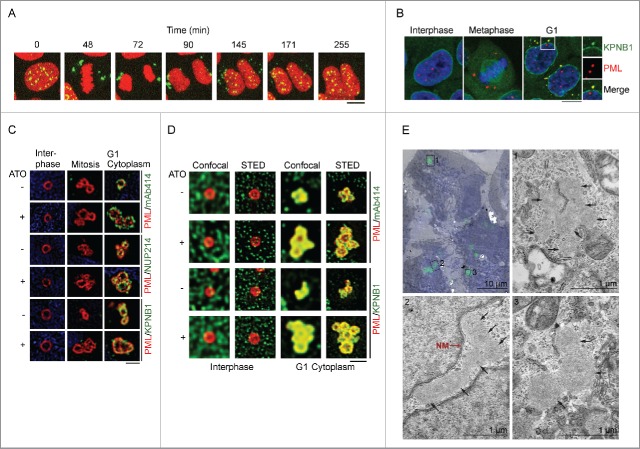
As demonstrated in previous studies,40,43 and as shown by time lapse microscopy in Fig. 1A and Video S1, nuclear PML bodies become released into the cytoplasm during mitosis and subsequently re-imported into progeny nuclei following cell division. To further investigate the transition from CyPNs to PML NBs, we first assessed if CyPNs contain KPNB1, which represents one of the most abundant and most studied of the nuclear import receptors. Immunofluorescence (IF) analysis of HaCaT cells revealed colocalization between endogenous KPNB1 and PML in the cytoplasm of newly divided cells. In contrast, we did not detect colocalization between these 2 proteins in the nuclear membrane, PML NBs or MAPPs (Fig. 1B). Thus, KPNB1 and NUPs appear to be co-recruited to CyPNs concomitant with post-mitotic nuclear import activation. We also investigated if CyPNs contain other karyopherins such as karyopherin β2 and CRM1 (also known as exportin 1). As shown in Fig. S1, CRM1, but not karyopherin β2, accumulates in cytoplasmic PML foci.
Figure 1.
PML and KPNB1 colocalize in the cytoplasm of post-mitotic cells. (A) Time lapse of a dividing HaCaT cell expressing EYFP-PML I (green) and mCherry-Histone H2B (red). PML bodies are released into the cytoplasm during mitosis and recycled back into the nucleus during G1. The complete time lapse series can be viewed in Video S1. Scale bar, 10 µm. (B) IF showing HaCaT cells at different stages of the cell cycle. Antibodies against PML (red) and KPNB1 (green) were used. DAPI is shown in blue. Selected CyPN is highlighted. Each image is a projection of multiple z-sections. Scale bar, 10 µm. (C and D) Super-resolution microscopy images of untreated or ATO-treated HaCaT cells using SIM (C) and gSTED (D). Cells were labeled using antibodies specific for PML (red), and NUPs (green) or KPNB1 (green). DAPI is shown in blue. One-plane representations of reconstructed (SIM) or deconvolved (gSTED) captures are shown. Scale bar, 1 µm. (E) HaCaT cells stably expressing EYFP-PML I were grown in the presence of ATO for 24 h before CLEM analysis. For recognition of cells with CyPNs, electron micrographs (EM) were overlaid with the respective fluorescence image of the same cell (shown in the upper left image). The nuclei are labeled with Hoechst (blue) and aggregations of EYFP-PML I are shown in green. Each micrograph, labeled 1–3, is a magnification of the respective area denoted by black squares in the overlay. CyPNs detected in the respective EM are marked by black arrows. The nuclear membrane (NM) can be seen in micrograph nr. 2 and is marked by a red arrow. Scale bars; 10 µm (overlay), 1 µm (EM).
To obtain insight into PML body structural dynamics during entry into and exit from mitosis, we imaged PML NBs, MAPPs and CyPNs at nanoscale resolution using structured illumination microscopy (SIM) and gated stimulated emission depletion (gSTED) microscopy. We confirmed previous results showing that PML nuclear bodies are organized as monomeric spheres.44 In addition, we found that MAPPs and CyPNs are composed of aggregates of multiple spheres (Fig. 1C and D). The multispherical composition of CyPNs was particularly evident in cells treated with a low concentration (0.25 µM) of arsenic trioxide (ATO), which is known to stabilize CyPNs and inhibit post-mitotic PML recycling (Fig. 1C and D).43 This result is in agreement with the notion that CyPNs represent aggregates of previous generation PML NBs.
NUPs and other nuclear import factors have previously been detected in annulate lamellae, cytoplasmic structures with unknown functions that consist of stacks of membrane embedded NPCs.45-47 To rule out the possibility that CyPNs are related to annulate lamellae, we investigated EYFP-tagged PML I in ATO-treated HaCaT cells using correlative light electron microscopy (CLEM). This experiment revealed the presence of cytoplasmic EYFP-PML I in globular, proteinaceous compartments that do not contain lipid membranes (Fig. 1E). Thus, CyPNs are distinct from annulate lamella and do not appear to contain NPCs.
Recruitment of NUPs to CyPNs is regulated by KPNB1 and the central PML NLS
KPNB1 is known to mediate contact between import complexes and nuclear pores through interactions with FG-NUPs.7,10,19,20 To investigate if KPNB1 is responsible for recruitment of NUPs to CyPNs, we performed targeted depletion of this protein using siRNAs. Since HaCaT cells exhibit relatively poor transfection efficiency, we decided to use U2OS cells for these experiments. Cells receiving KPNB1-targeted siRNAs exhibited reduced KPNB1 expression compared with control transfected cells at 1 day following transfection, and the decreased expression level persisted at day 2 and 3 (Fig. 2A). Notably, the KPNB1-depleted cells did not display altered cell cycle progression, as determined by flow cytometry, and did not exhibit increased cell death by apoptosis compared with control-depleted cells (Fig. S2A and B). By using IF we readily detected CyPN-like PML bodies in KPNB1-depleted cells. However, the recruitment of FG-NUPs to these bodies was found to be significantly impaired when compared with control-depleted cells (Fig. 2B and C). This result suggests that PML-mediated recruitment of KPNB1 is a prerequisite for NUP accumulation at CyPNs. Furthermore, the ratio of cytoplasmic PML foci per cell was increased on day 2 and 3 following KPNB1-depletion (Fig. 2D). This observation is consistent with the hypothesis that KPNB1 facilitates disassembly of CyPNs and nuclear import of its components in post-mitotic cells. Finally, we assessed if KPNB1 has the capacity to mediate nuclear import of Glutathione S-transferase (GST) fused to an 86 amino acid long peptide sequence corresponding to PML exon 6 (the exon that encodes the central NLS) in an in vitro import assay. We found that recombinant KPNB1 mediates nuclear import of GST-tagged PML exon 6 in semi-permeabilized HaCaT cells (Fig. S3).
Figure 2.
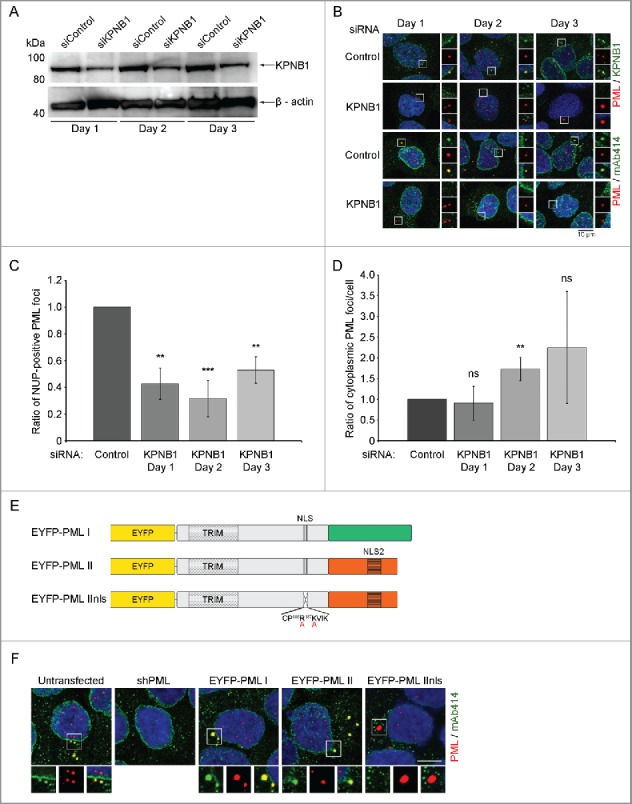
KPNB1 controls recruitment of NUPs to CyPNs. (A) Western blot showing expression of KPNB1 in KPNB1-depleted and control-depleted U2OS cells. β-actin was used as a loading control. The blot is representative of 3 independent experiments. (B) IF labeling of siRNA transfected cells using antibodies against PML (red) in combination with antibodies against FG-NUPs (mAb414; green) or KPNB1 (green). Each image represents projections of multiple z-sections. Selected cytoplasmic foci are highlighted. DAPI is shown in blue. (C) Quantification of the data shown in (B). The ratio of NUP-positive PML foci was determined by manual inspection of more than 300 cells in each sample and the data was normalized to the control. The bar graph shows mean ± standard deviation (SD) calculated from 3 independent experiments (**p < 0.01, ***p < 0.001). (D) Quantification of the data shown in (B). The ratio of cytoplasmic PML foci per cell was determined by manual inspection of more than 300 cells in each sample and the data was normalized to the control. The bar graph shows mean ± standard deviation (SD) calculated from 3 independent experiments (ns = non-significant; **p = 0.01). (E) Schematic of lentivirus-based constructs. The isoform-specific C-terminus is shown in green and orange for PML I and II respectively. The TRIM motif, the central NLS (in exon 6) and the PML II specific NLS (NLS2) are also depicted. Furthermore, the alanine substitutions in the central NLS of PML IInls are indicated with red letters. (F) The first 2 panels show IF labeling of endogenous FG-NUPs (mAb414; green) and endogenous PML (red) in untransfected and shPML-transfected HaCaT cells, respectively. The remaining panels show autofluorescent EYFP-tagged PML (red) and endogenous FG-NUPs (mAb414; green). DAPI is shown in blue. Each image represents projections of multiple z-sections. Selected cytoplasmic PML bodies are highlighted to demonstrate the absence or presence of FG-NUPs. Scale bar 10 µm.
It has previously been shown that mutations in the NLS of PML III disrupt interaction between PML and NUPs in the cytoplasm.42 However, since these mutations also result in redirection of PML to endosomes and prevent formation of PML NBs39 it was not clear from these data if the NLS had a direct role in NUP recruitment. To investigate if the NLS plays a direct role, we exploited the fact that the PML II isoform contains an extra noncanonical NLS in its C-terminal, isoform-specific region.39 We generated stable HaCaT cells using a previously described PML body regeneration system, which involves depletion of endogenous PML through lentiviral expression of PML-targeted shRNAs and subsequent introduction of EYFP-tagged PML I, PML II or a mutated version of PML II (PML IInls) containing point mutations within the central NLS (Fig. 2E). We observed PML NBs, MAPPs and CyPN-like PML bodies in all 3 cell lines. While the CyPNs generated by PML I and II recruited KPNB1 and FG-NUPs, the CyPN-like PML bodies that formed in PML IInls-expressing cells did not accumulate detectable levels of these import factors. Furthermore, ATO-treatment did not appear to affect the localization of KPNB1 and FG-NUPs in these experiments (Figs. 2F and S4). Our results show that interaction between KPNB1 and the central PML NLS is required for recruitment of NUPs to CyPNs after mitosis.
CyPNs preferentially recruit peripheral NUPs
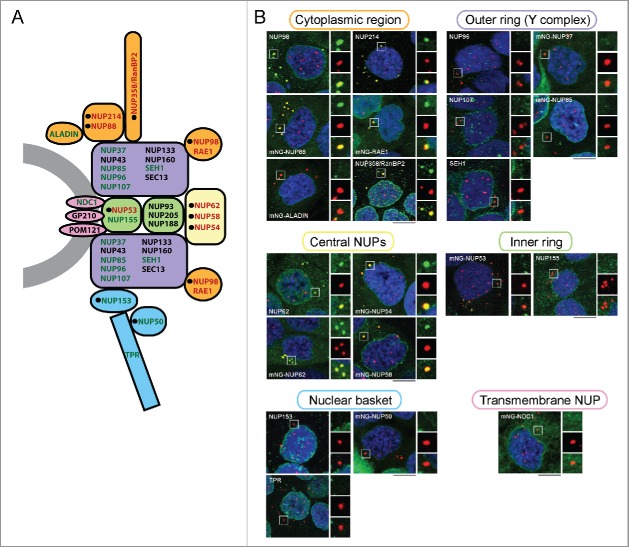
To obtain an overview of NUPs that are recruited to CyPNs, we used a panel of NUP-specific antibodies and a series of HaCaT-derived cell lines that stably express mNeonGreen (mNG)-tagged NUPs.48 The selection of NUPs included in this analysis was based on the availability of primary antibodies and the ability of tagged proteins to become stably expressed in HaCaT cells. We only included NUPs that were readily detected within the nuclear membrane of the cells. For each of the NUP-specific antibodies and NUP-expressing cell lines, we performed IF to determine the extent of CyPN localization in newly divided cells. The results from this experiment showed that peripheral NUPs containing FG-repeat motifs generally become recruited to CyPNs after mitosis, whereas scaffold NUPs are not (Fig. 3A and B; Table 1). However, 3 exceptions were noted: 1) The nuclear basket NUPs NUP153 and mNG-NUP50 could not be detected in CyPNs despite the presence of FG-repeats in these proteins. 2) mNG-RAE1, a peripheral NUP that does not contain FG-repeat motifs, was found to be recruited effectively by the CyPNs after mitosis. 3) The inner ring FG-NUP mNG-NUP53 displayed a weak accumulation at CyPNs (Fig. 3A and B; Table 1). Together, these results show that CyPNs primarily recruit peripheral NUPs, and that scaffold NUPs and NUPs in the nuclear basket do not participate in protein complexes formed at these sites.
Figure 3.
CyPNs preferentially recruit peripheral NUPs. (A) Cross section of a nuclear pore complex showing localization of NUPs within NPC subcomplexes. NUPs that we detect in CyPNs are shown in red and NUPs that we do not detect in CyPNs are shown in green. Black circles indicate NUPs with FG-motifs. NUPs that have not been analyzed in the present study are shown in black. The schematic representation of the NPC used here is based on previous publications.1,26,64 (B) IF labeling showing the localization of PML (red) and indicated NUPs (green) in G1 HaCaT cells. NUPs were detected either as stably expressed mNG fusions (autofluorescence) or as endogenously expressed proteins using NUP-specific antibodies. Each image represents projections of multiple z-sections. Selected CyPNs are highlighted to demonstrate the absence or presence of the indicated NUPs. Scale bars 10 µm.
Table 1.
Summary of NUP localization data.
| FG-containing NUP | CyPN-localization | |
|---|---|---|
| NUP50b | + | − |
| NUP53b,* | + | +/− |
| NUP54b | + | + |
| NUP58b | + | + |
| NUP62a,b | + | + |
| NUP88b | + | + |
| NUP98a | + | + |
| NUP153a | + | − |
| NUP214a | + | + |
| RAE1b | − | + |
| NUP358/RanBP2a | + | + |
| NUP37b | − | − |
| NUP85b | − | − |
| NUP96a | − | − |
| NUP107a | − | − |
| NUP155a | − | − |
| NDC1b | − | − |
| SEH1a | − | − |
| ALADINb | − | − |
| TPRa | − | − |
NUPs were detected by the use of NUP-specific antibodies.
NUPs were visualized by the use of mNeonGreen-tags.
Weak accumulation in some CyPNs was detected.
FG-repeat motifs are required and sufficient for recruitment of NUPs to CyPNs in post-mitotic cells
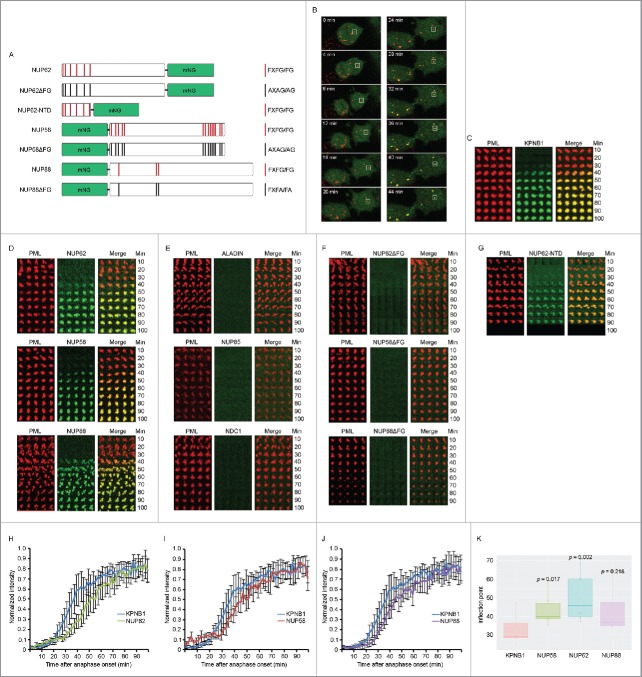
To study assembly of CyPNs in living cells we established HaCaT cells that stably express mRFP-tagged PML I together with mNG-tagged KPNB1 or mNG-tagged NUPs. The NUPs selected for this experiment were the peripheral NUPs NUP62, NUP58 and NUP88, and the scaffold NUPs ALADIN, NUP85 and NDC1. We also established stable cell lines that express FG-repeat mutants of NUP62, NUP58 and NUP88 (Fig. 4A). Furthermore, we produced a cell line that expresses a mNG-tagged truncated version of NUP62 consisting solely of the N-terminal FG-repeat region of this protein (Fig. 4A). Relative expression levels of wild type and mutated proteins were estimated by flow cytometry (Fig. S5A and B). To produce time lapse series, confocal stacks were acquired simultaneously in the red and green channel at 2 minute intervals between frames. Anaphase onset was used as a reference starting point in all data series. We designed an Image J plugin that tracked MAPPs/CyPNs in the red mRFP-PML I channel and subsequently plotted both channels (red and green) in diagrams as shown in Fig. 4C-G.
Figure 4.
Post-mitotic recruitment of KPNB1 and FG-NUPs to CyPNs in living cells. (A) Schematic overview of the mNG-tagged FG-repeat NUPs NUP62, NUP58, NUP88 and their mutated versions. (B) Time lapse of a dividing HaCaT cell expressing mRFP-PML I (red) and mNG-KPNB1 (green). Time point zero is defined by anaphase onset. The complete mitosis is shown in Video S2. (C) Tracking of a single PML body in HaCaT cells expressing mRFP-PML I (red) and mNG-KPNB1 (green). Confocal z-stacks were acquired at 2 min intervals between frames. Time point zero is defined by anaphase onset. (D) Tracking of a single PML body in HaCaT cells stably expressing mRFP-PML I (red) and one of the indicated mNG-tagged FG-NUPs (green). Data were acquired as aforementioned in (C). (E) Tracking of a single PML body in HaCaT cells stably expressing mRFP-PML I (red) and one of the indicated mNG-tagged scaffold NUPs (green). Data were acquired as aforementioned in (C). (F) Tracking of a single PML body in HaCaT cells stably expressing mRFP-PML I (red) and mNG-tagged mutated versions of the indicated FG-NUPs (green). Data were collected as aforementioned in (C). (G) Tracking of a single PML body in HaCaT cells stably expressing mRFP-PML I (red) and mNG-tagged N-terminal FG-repeat region of NUP62 (green). Data were collected as aforementioned in (C). (H-J) Quantification of PML body tracking data showing normalized fluorescence intensity measured through the center of a MAPP/CyPN in single z-sections. Images were acquired at 2 min intervals between frames. Time point zero is defined by anaphase onset. Graphs show mean ± SD of intensity measurements from 6–10 CyPNs assembled in different cell divisions. Lines represent detection of mNG-tagged KPNB1 and NUP62 (H), KPNB1 and NUP58 (I), as well as KPNB1 and NUP88 (J) respectively. (K) Statistical analysis of KPNB1 versus NUP recruitment to cytoplasmic PML foci. Boxplot represents the inflection points calculated from fitted sigmoid curves shown in Fig. S7B. Indicated p-values were obtained by a Wilcoxon Rank sum/Mann-Whitney test, with Bonferroni multiple testing correction.
The results from these experiments showed that KPNB1 and the peripheral NUPs NUP62, NUP58 and NUP88 are effectively recruited to CyPNs starting between 20 and 30 minutes after anaphase onset (Fig. 4B-D; Video S2). Conversely, we did not detect recruitment of the scaffold NUPs ALADIN, NUP85 and NDC1 to CyPNs during the first 100 minutes after anaphase onset (Fig. 4E). Furthermore, NUP62, NUP58 and NUP88 containing mutated FG-repeat motifs failed to accumulate within CyPNs after mitosis (Fig. 4F). Notably, while mutated NUP62 and NUP58 showed normal localization to the nuclear membrane, NUP88 containing FG-repeat mutations was not detected at the nuclear membrane suggesting that these mutations also affected localization at nuclear pore complexes (Fig. S5C). It should be noted that even though NUP88 contains 3 FG-motifs, this protein has not, to our knowledge, been reported to bind karyopherins directly. Notably, NUP88 may also be recruited to CyPNs through its ability to interact with NUP214.49 Finally, the N-terminal FG-repeat region of NUP62 alone exhibited recruitment kinetics similar to that observed for full-length NUP62 (Fig. 4G). The results show that assembly of NUPs in CyPNs is dictated largely by FG-repeats.
To verify that endogenous FG-NUPs have similar recruitment kinetics as the exogenously expressed mNG-tagged proteins, we correlated the appearance of NUP-positive CyPNs in fixed HaCaT cells against a nuclear growth curve. Consistent with the live cell imaging data, we detected endogenous FG-NUPs within CyPNs at 20 to 30 minutes after anaphase onset (Fig. S6A and B).
We also analyzed the appearance of mRFP-PML I positive PML bodies that form within the daughter nuclei after cell division. The first PML NBs were detected between 25 and 30 minutes after anaphase onset and they subsequently continued to increase in size and numbers (Fig. S7A). Thus, formation of NUP-positive CyPNs seems to correlate well with the formation of post-mitotic PML NBs.
Subsequently, we performed a more accurate kinetic analysis of mNG-NUP recruitment by combining data from multiple cell divisions. The result of this analysis showed that recruitment of NUP62, NUP58 and NUP88 to CyPNs consistently lag behind recruitment of KPNB1 (Fig. 4H-J). We confirmed this result statistically by determining the inflection point (the point where the curve has risen to half of its maximum value) of fitted sigmoid curves derived from individual CyPNs (Figs. 4K and S7B). This result is in agreement with a stepwise assembly process, where KPNB1 first form a complex with PML through the NLS and subsequently recruit FG-NUPs.
KPNB1 and FG-repeat NUPs are enriched in mobile cytoplasmic foci in interphase cells
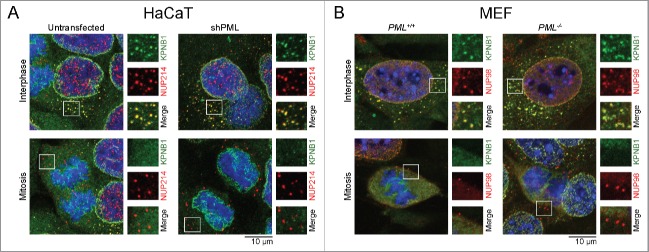
In addition to the CyPNs, we also observed an underlying pattern of smaller cytoplasmic foci that were enriched in FG-NUPs and KPNB1. Unlike CyPNs, these small structures were found to persist in PML-depleted HaCaT/shPML cells and Pml−/− MEF cells, showing that they form independently of PML. In addition, foci positive for KPNB1 and FG-NUPs were detected in interphase cells and not during mitosis, which could indicate that they depend on an active nuclear import machinery (Fig. 5A and B). Confocal live cell imaging at high temporal resolution (2 second interval between frames), using HaCaT cells stably expressing mNG-NUP88 or mNG-NUP54 revealed the presence of numerous small structures with high mobility in the cytoplasm, showing that the observed cytoplasmic foci do not represent fixation artifacts (Video S3 and S4). Thus, a fraction of FG-NUPs and KPNB1 appears to be enriched in mobile compartments within the cytoplasm of interphase cells.
Figure 5.
KPNB1 and FG-NUPs in mobile cytoplasmic foci. (A) Untransfected and shPML-transfected HaCaT cells were subjected to IF analysis using antibodies against KPNB1 (green) and NUP214 (red). (B) Pml+/+ and Pml−/− MEFs were labeled with antibodies against KPNB1 (green) and NUP98 (red). Numerous cytoplasmic foci enriched in KPNB1 and FG-NUPs are observed in interphase cells (upper panels), but not in mitotic cells (lower panels). Selected foci are highlighted. Each image represents projections of multiple z-sections.
Discussion
During mitosis, several proteins are released into the cytoplasmic environment due to breakdown of the nuclear envelope. As a consequence, a portion of the nuclear proteins present in the mother cell are subjected to re-import into progeny daughter nuclei shortly after completion of mitosis. Such post-mitotic recycling of protein components to the nucleus may play an important role for timely regeneration of the nuclear environment following cell division.
The present study demonstrates that the PML protein provides us with a unique opportunity to study spatio-temporal aspects of post-mitotic nuclear import. First, the PML proteins aggregate into nuclear structures that persist during mitosis and that are large enough for detection by conventional fluorescence microscopy. Second, the process of nuclear import activation and import complex assembly can be monitored at a defined time point of the cell cycle as the PML bodies traverse from late mitosis to early G1.
Although formation of CyPNs is probably not a prerequisite for nuclear PML body formation, these structures have been shown to contribute to efficient PML body regeneration after mitosis.40,43 Other sources of PML proteins that may participate in assembly of progeny PML NBs are soluble PML isoforms that are inherited from mother to daughter cells (but not as CyPNs) and newly synthesized PML proteins. The relative contribution of soluble and CyPN-associated PML to the assembly of PML NBs is not clear.
The analysis of 20 different NUPs, which represent distinct NPC subcomplexes, revealed a clear preference for incorporation of peripheral NUPs over scaffold NUPs into CyPNs. This is in contrast to annulate lamellae that recruits most NUPs, including peripheral and scaffold NUPs, to form NPCs.45,46 In addition, this result shows that CyPNs primarily recruit NUPs that have a role in nuclear transport and that mediate contact with nuclear import complexes as they pass through the NPC. Our data further indicate that the assembly of CyPNs is dependent on the same protein-protein interactions (the NLS-karyopherin-FG-NUP axis) that are known to be essential for nuclear import.15,19,20 It is well established that nuclear import is switched off during mitosis (due to lack of a nuclear membrane) and turned on again shortly after mitosis has been completed. Notably, CyPN formation coincides with nuclear import reactivation after mitosis. It is also clear from the present study that detection of CyPNs between 20 and 30 minutes after anaphase onset coincides with the detection of progeny PML NBs in the nucleus. Thus the experimental approach presented in the present paper may prove useful for studies on post-mitotic reactivation of nuclear import.
Although protein complexes consisting of protein cargoes, import receptors and NUPs readily can be detected on the surface of CyPNs, it is not clear if these protein-protein interactions persist during transport from CyPNs to the nuclear pores. An alternative possibility is that these interactions become disrupted concomitant with disassembly of CyPNs, suggesting they will have to form a second time during translocation through the nuclear pores. Another question that needs to be answered is to what extent nuclear import factors, including KPNB1 and NUPs, contribute directly in CyPN disassembly before nuclear import. Our observation that the ratio of cytoplasmic PML foci per cell increases following KPNB1-depletion indicate that import factors facilitate transition of CyPNs into units that are small enough in size to be translocated through the nuclear pores. However, further investigations are required to elucidate the detailed process of CyPN disassembly and to determine the protein composition, as well as the function, of complexes that are transported from CyPNs to NPCs.
While previous models for nuclear import suggest that interactions between protein cargo-karyopherin complexes and FG-containing NUPs are strictly NPC-coupled events, the present study suggests that these interactions also take place at NPC distal sites. Particularly, the concept that peripheral NUPs play an interactive role together with karyopherins at protein cargo retrieval sites, before nuclear import, complements a publication by Rabut et al. showing that several NUPs have a relatively short NPC residence time.14 While others have linked off-pore functions of NUPs to nuclear processes such as transcription regulation and genome maintenance27,28,30-32 we provide data that are consistent with a possible role for soluble NUPs in protein transport. The concept that nuclear cargo proteins and FG-NUPs are pre-assembled before entry into the pore channel does not appear to be in conflict with well established models for nucleo-cytoplasmic transport, such as the selective phase model.21-23 On the contrary, assembly of FG-NUPs and the nuclear cargo before import may lead to a further increase in affinity between import complexes and the pore channel environment, thus facilitating interactions between incoming cargo and NPCs. It is also possible that the presence of FG-NUPs facilitates solubilization and prevents reaggregation of aggregation prone nuclear proteins during cargo retrieval in the cytoplasm. In agreement with this, several import receptors, including IMP4, IMP9 and IMPβ, have been shown to exert a solubilizing effect on cargo proteins.50
In our analysis of NUP subclasses, we identified 3 exceptions that deviated from the general pattern of CyPN recruitment. 1) NUP50 and NUP153 are classified as FG-containing proteins, but were not observed to colocalize with PML in the cytoplasm. These NUPs are primarily associated with the nuclear basket and the absence of these proteins in CyPNs may be related to their nucleoplasmic rather than cytoplasmic location.51,52 2) RAE1 accumulates in the CyPNs although it does not contain annotated FG-repeats. However, this protein has been shown to bind directly to the FG-repeat-containing NUP98, suggesting it may be recruited through its ability to interact with this protein.53,54 3) NUP53 was detected in CyPNs even though this protein is regarded as an inner ring scaffold NUP. It should be noted, however, that the signal produced by mNG-tagged NUP53 within CyPNs was observed to be considerably weaker compared with other NUPs. Indeed, the inner ring localization of NUP53 and its small number of FG motifs (only 3 annotations) might explain the weak accumulation of this protein at CyPNs.
Although karyopherin import receptors are not generally classified as NUP family members, IF analysis of subcellular distribution of KPNB1 reveals a high degree of overlap between this protein and most NUPs. Interestingly, we observe small, mobile cytoplasmic foci containing both KPNB1 and FG-NUPs, which are formed independently of PML. Although this observation underscores the interactive relationship between import receptors and NUPs both in the cytoplasm and in the nuclear membrane, it is not clear from the present study if these smaller foci are functionally equivalent to CyPNs. In addition, although we cannot rule out the possibility that the small PML-independent foci are not related to annulate lamellae, this seems unlikely since we do not detect any annulate lamellae by electron microscopy in HaCaT cells. Furthermore, primary cells such as MEFs generally do not have detectable annulate lamellae.55 Another possible reason why these structures are so poorly characterized (but yet so abundant) may be that they are protein aggregates and therefore difficult to detect by electron microscopy. Notably, KPNB1 has also been implicated in post-mitotic NPC assembly, suggesting that it might function as a chaperone for newly synthesized FG-NUPs.21,56,57
In summary, the present study demonstrates that NUPs and KPNB1 are recruited to CyPNs concomitant with CyPN disassembly and PML nuclear import. The schematic in Fig. 6 illustrates our proposed model for post-mitotic nuclear import of PML. Thanks to the unique ability of PML to form protein aggregates that persist in the cytoplasm as the cell turns from a nuclear import inactive state to a nuclear import active state, the dynamics of PML nuclear import complex assembly can be visualized and studied in living cells. The data challenge the assumption that the role of NUPs during nucleo-cytoplasmic transport is restricted within NPCs and suggest a more interactive role of import receptors and NUPs at nuclear cargo retrieval sites in the cytoplasm. Further studies should clarify if this represents a general mechanism in nuclear import that also applies for proteins other than PML.
Figure 6.
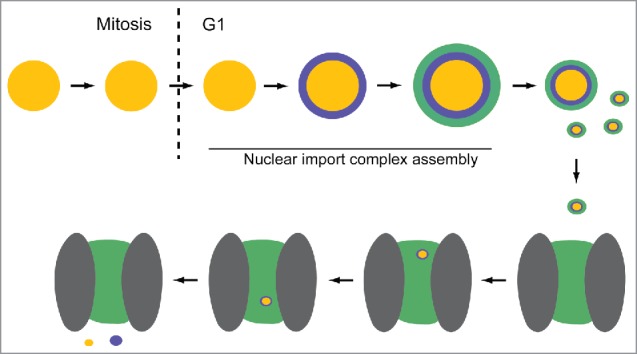
Model for post-mitotic nuclear import of PML. PML within a single MAPP is depicted as yellow spheres. KPNB1 (purple) and FG-NUPs (green) are recruited successively during G1 phase to form a CyPN. The CyPN dissociates into smaller components (illustrated by smaller spheres) that can translocate through a nuclear pore. We propose that these smaller nuclear cargo complexes may retain an outer layer of FG-repeat proteins that have the potential to facilitate interactions with the nuclear pore environment (gray ovals = scaffold NUPs; green area = hydrogel-like web of FG-repeats). On the nucleoplasmic side, PML and KPNB1 dissociate.
Materials and methods
Additional material and methods can be found in Supplementary Information (online).
Cells
HaCaT (immortalized human keratinocyte) and U2OS (human osteosarcoma) cells were grown in Iscove's Modified Dulbecco's Medium (IMDM, Lonza) supplemented with 10% fetal bovine serum (FBS, Thermo Scientific), 90 U/ml penicillin and 90 U/ml streptomycin (Lonza). All cell lines used were tested for mycoplasma using a commercially available kit (LT07–118, Lonza). MEFs (mouse embryonic fibroblasts) were isolated from wild type and Pml−/- C57BL/6 mice and maintained in Dulbecco's Modified Eagle's Medium (DMEM, Lonza) supplemented with 10% FBS and 90 U/ml penicillin and 90 U/ml streptomycin.
In experiments where ATO was used, cells were grown in the presence of 0.25 µM ATO (202673, Sigma) for 24 hours (h).
Antibodies
Commercial primary antibodies used for immunofluorescence stainings were; rabbit anti-PML (NB100–59787, Novus Biologicals), mouse anti-PML (sc-966, Santa Cruz Biotechnology), mouse anti-Nuclear pore complex proteins mAb414 (ab24609, Abcam), mouse anti-KPNB1 (NB120–2811, Novus Biologicals), rabbit anti-NUP214 (ab70497, Abcam), mouse anti-NUP153 [SA1] (ab96462, Abcam), mouse anti-NUP62 (610498, BD Transduction Laboratories), rat anti-NUP98 (ab50610, Abcam), rabbit anti-TPR (ab84516, Abcam), rabbit anti-NUP155 (ab199528, Abcam), rabbit anti-NUP358/RanBP2 (ab64276, Abcam), rabbit anti-CRM1 (ab24189, Abcam), and mouse anti-Transportin/karyopherin β2 [D45] (ab10303, Abcam). Guinea pig IgG antibodies against NUP96, NUP107 and SEH1 have been described previously and were kindly provided by Dr. Volker Cordes at Max Planck Institute for Biophysical Chemistry (MPI-BPC), Germany.58-60 Secondary antibodies used were; Alexa Fluor® 488 goat anti-mouse IgG, Alexa Fluor® 488 goat anti-rabbit IgG, Alexa Fluor® 555 goat anti-rabbit IgG, Alexa Fluor® 555 goat anti-rat IgG, Alexa Fluor® 594 donkey anti-mouse IgG, and Alexa Fluor® 594 goat anti-rabbit IgG from Life Technologies, and Alexa Fluor® 594 donkey anti-guinea pig IgG from Jackson Laboratories.
For 2-color STED imaging we used Chromeo 505 goat anti-mouse IgG (15050, Active motif) in combination with Biotin-xx Fab anti-rabbit IgG (B-21078, Life Technologies), and V500-Streptavidin (561419, BD Biosciences/Horizon).
Plasmids and lentiviruses
Lentivirus vectors expressing the EYFP-PML I, EYFP-PML II isoforms and PML shRNAs have been described previously.61 The EYFP-PML IInls mutant, containing a non-functional central NLS due to alanine substitutions for arginine and lysine at position 486 and 487 respectively, was obtained by site-directed mutagenesis using the following QuickChange primers; GCCAGSCCCSGTGCCCCGCGGCGGTCATCAAGATGGAGT (forward), ACTCCATCCTGATGACCGCCGCGGGGCACTGGGTCTGGC (reverse). EYFP-positive HaCaT cells were sorted by FACSAriaII (Becton Dickinson) using a 100 µm nozzle. Before each sorting, new 35 µm filter was set up on the end of the sample line to avoid clumps of cells in the samples. Cell sorting gates were set up using histograms for green fluorescence (FL1). Non-transduced HaCaT cells were used as a negative control. For each of the EYFP-PML positive populations, 1 × 104 to 5 × 104 cells were sorted with a 70–90% sorting purity.
For expression of RFP-PML I, mNG-KPNB1, mNG-NUP58, mNG-NUP88, mNG-NUP62, mNG-NUP50, mNG-NUP53, mNG-NUP54, mNG-NUP37, mNG-NUP85, mNG-ALADIN, mNG-NDC1, mNG-RAE1, mNG-NUP62ΔFG, mNG-NUP62-NTD, mNG-NUP88ΔFG, and mNG-NUP58ΔFG we used the bicistronic lentivirus vectors pCDH-EF1-MCS-IRES-Puro or pCDH-EF1-MCS-IRES-Neo (CD532A-2 and CD533A-2, respectively; System Biosciences). All cDNA inserts were de novo synthesized, cloned, and sequence verified by GenScript. Supplementary Table S1 sums up the plasmids synthesized for this study in annotated GenBank format.
Production of infectious lentiviruses and stable transduction of HaCaT cells were performed as described previously.43 For expression of GST-tagged PML NLS, the PMLnls-mTQ2 sequence from pCHD_EF1_PMLnls_linker_mTQ2_IRES_Neo was cloned into the pETM30 vector (Table S1).
siRNA transfection, western blotting and flow cytometry
Control or KPNB1-targeted siRNAs (AllStars Negative Control siRNA (1027280) or FlexiTube GeneSolution GS3837, Qiagen), at a total concentration of 60 nM, were introduced into U2OS cells using oligofectamine transfection reagent (Life Technologies). One day after transfection, cells were either harvested for analysis or left to grow for one or 2 additional days in fresh IMDM growth medium. Whole cell lysates were prepared from U2OS cells at 3 consecutive days after transfection and analyzed by western blotting as described previously.43 Protein expression was detected using mouse anti-KPNB1 (05–1530, Millipore) and mouse anti-β-Actin (AC-15; Santa Cruz Biotechnology) antibodies.
To determine the cell cycle profile after transfection with control and KPNB1 siRNAs, cells were collected by trypsination, fixed in ice-cold 70% ethanol and permeabilized using 0.25% Triton X-100. Subsequently, cells were incubated with an anti-Histone H3 (phospho S10) antibody (ab5176, Abcam) for 1 h at room temperature (RT), washed with PBS and incubated with secondary antibody Alexa Flour 488 goat anti-rabbit IgG for 30 minutes (min) at RT. Cells were further resuspended in PBS containing 10 µg/ml RNase A (Qiagen) and 20 µg/ml propidium iodide (Sigma). All samples were analyzed using the Accuri C6 flow cytometer (BD Biosciences).
Immunofluorescence (IF) staining
Cells were grown on 12 mm or 18 mm (0.17 mm size selected, Assistent) glass coverslips, for analysis by confocal, or SIM and gSTED microscopy respectively. Samples were processed for IF as described previously62 and mounted in Vectashield mounting media with DAPI (Vector Laboratories). Cells subject to gSTED analysis were incubated with secondary antibodies Chromeo 505 goat anti-mouse IgG (15050, Active motif) and biotin-XX F(ab′)2 fragment of goat anti-rabbit IgG (H+L) (B21078, Life Technologies) in 0.5% BSA in PBS for 1 h at 37 °C, followed by incubation with V500-streptavidin (561419, BD Horizon™) in 0.5% BSA in PBS for 30 min at 37 °C. Finally, cells were mounted in ProLong Gold antifade mounting media (Life Technologies).
Confocal microscopy
Analysis of siRNA transfected U2OS cells, and characterization of NUP expression in HaCaT and MEF cells, was performed using a Leica TCS SP8 confocal microscope equipped with a 40× 1.30 NA oil immersion lens, a UV (405 nm) laser and a CW (Continuous Wavelength) white light laser. Z-stack images were acquired. The ratio of NUP-positive PML foci in U2OS cells was determined by manual inspection of more than 300 cells/sample using the Fiji Image J software.
Gated Stimulated Emission Depletion (gSTED) Microscopy
gSTED images of labeled HaCaT cells, on 0.17 mm glass coverslips (size selected, Assistent), were acquired using the Leica TCS SP8 gated STED microscope equipped with a 100× 1.40 NA oil immersion lens, a CW white light laser and 592 nm depletion beam. Images were subject to deconvolution using the Huygens Professional software (Scientific Volume Imaging).
Structured Illumination Microscopy (SIM)
SIM imaging was performed on a Deltavision OMX V4 microscope (GE Healthcare) equipped with 3 PCO.edge sCMOS cameras, 405 nm, 488 nm, 568 nm and 642 nm laser lines and a 60× 1.42 NA Plan Apochromat lense (Olympus). Z-stacks covering the whole cell, with sections spaced 0.125 µm apart, were acquired. For each Z-section, 15 raw images (3 rotations with 5 phases each) were recorded. Super-resolution images were reconstructed and aligned using the Softworx software (GE Healthcare).
Correlative Light Electron Microscopy (CLEM)
HaCaT cells expressing EYFP-PML I were grown on gridded coverslips (EMS), treated with 0.25 µM ATO for 24 h and fixed in 4% formaldehyde and 0.1% glutaraldehyde in 0.1 M PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, pH 6.9) overnight. For fluorescence microscopy, the coverslips were mounted on coverglass with Mowiol and Hoechst for nuclear staining. Fluorescence microscopy was performed using a 20x and 63x objectives on Zeiss LSM 780 and Zeiss LSM 710 confocal microscopes (Zeiss) with the appropriate channel. For recognition of cells with respect to the grid on coverslips, we used differential interference contrast (DIC) in combination with the 20x objective. After confocal microscopy, cells were further processed for CLEM as described earlier.63 Serial sections of approximately 100 nm were cut on a Leica Ultracut (Leica Microsystems GmbH), observed in JEOL-JEM 1230 at 80 kV and micrographs recorded with a Morada CCD camera (Olympus). Electron micrographs (EM) were overlaid with the fluorescence images of the same cell using Adobe Photoshop software. We used single confocal slides from whole stacks (approximately 700 nm thickness) and aligned these with relevant EM sections using the cell localization and the shape of the nucleus as guidance.
Live cell imaging and PML body tracking
HaCaT cells stably expressing RFP and mNG–tagged proteins were imaged using a Leica TCS SP8 confocal microscope equipped with a 40x oil 1.30 NA immersion lens. During imaging cells were maintained in a stage-top incubation system (37 °C, 5% CO2) on glass bottom dishes from MatTek (P35G-1.5–14-C). Z-stacks of 10–15 confocal sections (spaced 1 µm apart) were acquired simultaneously in the red and green channel, using Leica HyD hybrid detectors at 2 min intervals between acquisition start points. To generate PML body tracking plots, z-stacks of the entire time series were first projected using a maximal intensity projection algorithm (Image J; http://imagej.nih.gov/ij/). An Image J plugin, PMLBuddy, was designed that automatically tracked the RFP-PML I signal and subsequently plotted the red and green channels as separate and merged images. Source code and additional information about the plugin is available at: https://github.com/Oftatkofta/ImageJ-plugins. For quantitative measurements of KPNB1 and NUP recruitment to PML bodies after mitosis, the RFP signal from individual PML bodies were tracked manually in 3 dimensions (without projecting the stacks) and the mNG intensity at the center of the tracked body was measured.
Investigation of the nuclear size growth after anaphase onset was performed by live cell imaging of HaCaT GFP-H2B cells, using a Leica TCS SP8 confocal microscope equipped with a 40x oil 1.30 NA immersion lens and a stage-top incubation system (37 °C, 5% CO2). Cells were grown on glass bottom dishes (MatTek). Z-stacks of 9–13 confocal sections were acquired using Leica HyD hybrid detectors at 4 min intervals between acquisition start points, for a total imaging period of 4–5 h. The nuclear size of 50 daughter cell pairs was determined by calculating the area after z-stack projection using Fiji Image J software.
Statistical analysis
Data presented in Fig. 2 are expressed as mean ± standard deviation (SD) quantified from 3 independent experiments, and normalized to the control. Statistical significance was determined by the use of 2-tailed Student's t-test. Differences were considered significant when p was less than 0.05.
Statistical analysis of NUP recruitment kinetics (Fig. 4) was performed by calculating the inflection point (the point where the curve has risen to half of its maximum value) of fitted sigmoid curves derived from individual CyPNs. Data for KPNB1 were compared with the respective NUPs using a Wilcoxon Rank sum/Mann-Whitney test, with Bonferroni multiple testing correction.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Marianne Smestad, at the Department of Molecular Cell Biology, Oslo University Hospital, for technical assistance. We also thank Prof. Magnar Bjørås and Rajikala Suganthan, at the Department of Microbiology, Oslo University Hospital, for handling the mice and producing MEF cells. Dr. Volker Cordes, at Max Planck Institute for Biophysical Chemistry (MPI-BPC), Germany, kindly helped with reading and commenting on the manuscript. Finally, we appreciate the help we got from Dr. Alexander D. Rowe, at the Department of Medical Biochemistry, Oslo University Hospital, when it comes to statistical analysis.
Funding
The research was supported by the Norwegian Cancer Society (A.L. and S.O.B.) and the South-Eastern Norway Regional Health Authority (J.E., E.L. and A.P.).
Notes on contributors
A.L. designed experiments, performed experiments and wrote the paper. S.O.B. designed experiments and wrote the paper. J.E. designed and performed experiments. E.L., A.P., A.B., P.B., and B.D. performed experiments. K.O.S. provided expertise and assistance with SIM experiments. All authors reviewed the manuscript.
References
- [1].Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 2014; 111:E2453-61; PMID:24927568; https://doi.org/ 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Löschberger A, van de Linde S, Dabauvalle MC, Rieger B, Heilemann M, Krohne G, Sauer M. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci 2012; 125:570-5; PMID:22389396; https://doi.org/ 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- [3].Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. Nuclear pores. Architecture of the nuclear pore complex coat. Science (New York, NY) 2015; 347:1148-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Szymborska A, de Marco A, Daigle N, Cordes VC, Briggs JA, Ellenberg J. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science (New York, NY) 2013; 341:655-8; https://doi.org/ 10.1126/science.1240672 [DOI] [PubMed] [Google Scholar]
- [5].von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al.. In situ structural analysis of the human nuclear pore complex. Nature 2015; 526:140-3; PMID:26416747; https://doi.org/ 10.1038/nature15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al.. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science (New York, NY) 2016; 352:363-5; https://doi.org/ 10.1126/science.aaf0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature 1995; 377:246-8; PMID:7675110; https://doi.org/ 10.1038/377246a0 [DOI] [PubMed] [Google Scholar]
- [8].Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev 2001; 65:570-94, table of contents; PMID:11729264; https://doi.org/ 10.1128/MMBR.65.4.570-594.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci U S A 1995; 92:1769-73; PMID:7878057; https://doi.org/ 10.1073/pnas.92.5.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 1995; 83:683-92; PMID:8521485; https://doi.org/ 10.1016/0092-8674(95)90181-7 [DOI] [PubMed] [Google Scholar]
- [11].Soniat M, Chook YM. Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J 2015; 468:353-62; PMID:26173234; https://doi.org/ 10.1042/BJ20150368 [DOI] [PubMed] [Google Scholar]
- [12].Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell 2002; 10:1345-53; PMID:12504010; https://doi.org/ 10.1016/S1097-2765(02)00727-X [DOI] [PubMed] [Google Scholar]
- [13].Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, et al.. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science (New York, NY) 2003; 302:1571-5; https://doi.org/ 10.1126/science.1088372 [DOI] [PubMed] [Google Scholar]
- [14].Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004; 6:1114-21; PMID:15502822; https://doi.org/ 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- [15].Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 2007; 8:195-208; PMID:17287812; https://doi.org/ 10.1038/nrm2114 [DOI] [PubMed] [Google Scholar]
- [16].Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013; 154:971-82; PMID:23993091; https://doi.org/ 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A 2003; 100:2450-5; PMID:12604785; https://doi.org/ 10.1073/pnas.0437902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J 2002; 21:2843-53; PMID:12065398; https://doi.org/ 10.1093/emboj/cdf305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 2000; 102:99-108; PMID:10929717; https://doi.org/ 10.1016/S0092-8674(00)00014-3 [DOI] [PubMed] [Google Scholar]
- [20].Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 1995; 81:215-22; PMID:7736573; https://doi.org/ 10.1016/0092-8674(95)90331-3 [DOI] [PubMed] [Google Scholar]
- [21].Frey S, Görlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007; 130:512-23; PMID:17693259; https://doi.org/ 10.1016/j.cell.2007.06.024 [DOI] [PubMed] [Google Scholar]
- [22].Frey S, Richter RP, Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science (New York, NY) 2006; 314:815-7; https://doi.org/ 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- [23].Hülsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 2012; 150:738-51; PMID:22901806; https://doi.org/ 10.1016/j.cell.2012.07.019 [DOI] [PubMed] [Google Scholar]
- [24].Lim RY, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science (New York, NY) 2007; 318:640-3; https://doi.org/ 10.1126/science.1145980 [DOI] [PubMed] [Google Scholar]
- [25].Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 2003; 13:622-8; PMID:14624840; https://doi.org/ 10.1016/j.tcb.2003.10.007 [DOI] [PubMed] [Google Scholar]
- [26].Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev 2015; 29:337-49; PMID:25691464; https://doi.org/ 10.1101/gad.256495.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Buchwalter AL, Liang Y, Hetzer MW. Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol Biol Cell 2014; 25:2472-84; PMID:24943837; https://doi.org/ 10.1091/mbc.E14-04-0865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372-83; PMID:20144761; https://doi.org/ 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chatel G, Fahrenkrog B. Dynamics and diverse functions of nuclear pore complex proteins. Nucleus (Austin, Tex) 2012; 3:162-71; PMID:22555605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 2002; 13:1282-97; PMID:11950939; https://doi.org/ 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010; 140:360-71; PMID:20144760; https://doi.org/ 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- [32].Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet 2013; 9:e1003308; PMID:23468646; https://doi.org/ 10.1371/journal.pgen.1003308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007; 8:1006-16; PMID:17928811; https://doi.org/ 10.1038/nrm2277 [DOI] [PubMed] [Google Scholar]
- [34].Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000661; PMID:20452955; https://doi.org/ 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene 2001; 20:7223-33; PMID:11704850; https://doi.org/ 10.1038/sj.onc.1204765 [DOI] [PubMed] [Google Scholar]
- [36].Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J 1995; 14:1532-41; PMID:7729428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Borden KL, Lally JM, Martin SR, O'Reilly NJ, Solomon E, Freemont PS. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML. Proc Natl Acad Sci U S A 1996; 93:1601-6; PMID:8643677; https://doi.org/ 10.1073/pnas.93.4.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al.. The tripartite motif family identifies cell compartments. EMBO J 2001; 20:2140-51; PMID:11331580; https://doi.org/ 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Å Jul-Larsen, Grudic A, Bjerkvig R, Bøe SO. Subcellular distribution of nuclear import-defective isoforms of the promyelocytic leukemia protein. BMC Mol Biol 2010; 11:89; PMID:21092142; https://doi.org/ 10.1186/1471-2199-11-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci 2006; 119:1034-42; PMID:16492707; https://doi.org/ 10.1242/jcs.02817 [DOI] [PubMed] [Google Scholar]
- [41].Chen YC, Kappel C, Beaudouin J, Eils R, Spector DL. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol Biol Cell 2008; 19:3147-62; PMID:18480407; https://doi.org/ 10.1091/mbc.E08-01-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Å Jul-Larsen, Grudic A, Bjerkvig R, Bøe SO. Cell-cycle regulation and dynamics of cytoplasmic compartments containing the promyelocytic leukemia protein and nucleoporins. J Cell Sci 2009; 122:1201-10; PMID:19339552; https://doi.org/ 10.1242/jcs.040840 [DOI] [PubMed] [Google Scholar]
- [43].Lång E, Grudic A, Pankiv S, Bruserud O, Simonsen A, Bjerkvig R, Bjørås M, Bøe SO. The arsenic-based cure of acute promyelocytic leukemia promotes cytoplasmic sequestration of PML and PML/RARA through inhibition of PML body recycling. Blood 2012; 120:847-57; PMID:22692509; https://doi.org/ 10.1182/blood-2011-10-388496 [DOI] [PubMed] [Google Scholar]
- [44].Lang M, Jegou T, Chung I, Richter K, Münch S, Udvarhelyi A, Cremer C, Hemmerich P, Engelhardt J, Hell SW, et al.. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci 2010; 123:392-400; PMID:20130140; https://doi.org/ 10.1242/jcs.053496 [DOI] [PubMed] [Google Scholar]
- [45].Cordes VC, Rackwitz HR, Reidenbach S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp Cell Res 1997; 237:419-33; PMID:9434638; https://doi.org/ 10.1006/excr.1997.3806 [DOI] [PubMed] [Google Scholar]
- [46].Miller BR, Forbes DJ. Purification of the vertebrate nuclear pore complex by biochemical criteria. Traffic 2000; 1:941-51; PMID:11208084 [PMC free article] [PubMed] [Google Scholar]
- [47].Rawe VY, Olmedo SB, Nodar FN, Ponzio R, Sutovsky P. Abnormal assembly of annulate lamellae and nuclear pore complexes coincides with fertilization arrest at the pronuclear stage of human zygotic development. Hum Reprod 2003; 18:576-82; PMID:12615828; https://doi.org/ 10.1093/humrep/deg114 [DOI] [PubMed] [Google Scholar]
- [48].Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al.. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 2013; 10:407-9; PMID:23524392; https://doi.org/ 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 1997; 16:807-16; PMID:9049309; https://doi.org/ 10.1093/emboj/16.4.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jäkel S, Mingot JM, Schwarzmaier P, Hartmann E, Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 2002; 21:377-86; PMID:11823430; https://doi.org/ 10.1093/emboj/21.3.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol 2000; 20:5619-30; PMID:10891499; https://doi.org/ 10.1128/MCB.20.15.5619-5630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 1993; 72:29-38; PMID:8422679; https://doi.org/ 10.1016/0092-8674(93)90047-T [DOI] [PubMed] [Google Scholar]
- [53].Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol 1999; 145:237-54; PMID:10209021; https://doi.org/ 10.1083/jcb.145.2.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 2005; 121:223-34; PMID:15851029; https://doi.org/ 10.1016/j.cell.2005.02.016 [DOI] [PubMed] [Google Scholar]
- [55].Kessel RG. Annulate lamellae: a last frontier in cellular organelles In: Jeon KW, Friedlander M, eds. International Review of Cytology: Academic Press, Inc, 1992:43-120 [DOI] [PubMed] [Google Scholar]
- [56].Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell 2003; 14:4387-96; PMID:14551248; https://doi.org/ 10.1091/mbc.E03-05-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature 2003; 424:689-94; PMID:12894213; https://doi.org/ 10.1038/nature01898 [DOI] [PubMed] [Google Scholar]
- [58].Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 2003; 14:1923-40; PMID:12802065; https://doi.org/ 10.1091/mbc.E02-09-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 2010; 29:1659-73; PMID:20407419; https://doi.org/ 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Patre M, Tabbert A, Hermann D, Walczak H, Rackwitz HR, Cordes VC, Ferrando-May E. Caspases target only two architectural components within the core structure of the nuclear pore complex. J Biol Chem 2006; 281:1296-304; PMID:16286466; https://doi.org/ 10.1074/jbc.M511717200 [DOI] [PubMed] [Google Scholar]
- [61].Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci 2011; 124:280-91; PMID:21172801; https://doi.org/ 10.1242/jcs.075390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lonning PE, Boe SO. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp Cell Res 2004; 298:58-73; PMID:15242762; https://doi.org/ 10.1016/j.yexcr.2004.03.045 [DOI] [PubMed] [Google Scholar]
- [63].Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al.. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 2013; 126:939-52; PMID:23239026; https://doi.org/ 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- [64].Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 2012; 13:687-99; PMID:23090414; https://doi.org/ 10.1038/nrm3461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.