ABSTRACT
Genomic variation is a source of functional diversity that is typically studied in genic and non-coding regulatory regions. However, the extent of variation within noncoding portions of the human genome, particularly highly repetitive regions, and the functional consequences are not well understood. Satellite DNA, including α satellite DNA found at human centromeres, comprises up to 10% of the genome, but is difficult to study because its repetitive nature hinders contiguous sequence assemblies. We recently described variation within α satellite DNA that affects centromere function. On human chromosome 17 (HSA17), we showed that size and sequence polymorphisms within primary array D17Z1 are associated with chromosome aneuploidy and defective centromere architecture. However, HSA17 can counteract this instability by assembling the centromere at a second, “backup” array lacking variation. Here, we discuss our findings in a broader context of human centromere assembly, and highlight areas of future study to uncover links between genomic and epigenetic features of human centromeres.
KEYWORDS: CENP-A, chromosome, genome stability, kinetochore, repetitive DNA
The centromere is the chromosomal locus that is important in chromosome pairing and essential for chromosome segregation during cell division. It is the site of kinetochore formation, the multi-protein structure that attaches chromosomes to spindle microtubules for segregation during cell division. Despite this essential role in chromosome inheritance, the features of centromeres vary among organisms. Centromeres range in size from small point centromeres (∼125bp) in budding yeasts to large regional centromeres (100kb–5Mb) in humans and plants.1 Despite these genomic disparities, the proteins of eukaryotic centromeres are related, emphasizing the functional importance of the locus. Centromeres are defined by specialized nucleosomes containing the histone H3 variant CENP-A. CENP-A nucleosomes are interspersed with canonical H3 nucleosomes to create a unique type of chromatin that differentiates the centromere from the rest of the chromosome.2,3 Centromeric (CEN) chromatin also serves as the foundation of the kinetochore, interacting with CENP-C and other members of the constitutive centromere-associated network (CCAN) to assemble the protein network between the DNA and the microtubules.4
Assembly of CEN chromatin occurs on DNA sequences that differ among organisms and even within the same individual, suggesting general sequence independence for recruitment of CENP-A and other centromere proteins. The lack of sequence similarities at eukaryotic centromeres has encouraged the current view of centromere identity as an epigenetically defined process, with little contribution from the underlying genomic sequences. However, de novo engineered centromeres (i.e. human artificial chromosomes, HACs) have only been generated from specific sequences, and centromeres in most organisms are consistently maintained at the same genomic location, raising the possibility of sequence-dependent aspects of centromere specification.5-7 Few groups have studied large regional centromeres from a genomic perspective, primarily because centromeres are located within extensive and complex genomic regions enriched for repetitive DNA and retrotransposons.8,9 The centromeres of plants (maize, rice, potato, Arabidopsis thaliana) are among the most well-defined regional centromeres, and in fact, several recent studies have produced nearly complete assemblies of a few maize centromeres.10-13 These impressive advances notwithstanding, we have focused this commentary on human centromeres and our recent studies exploring links between α satellite genomic structure and centromere function.
Human centromeres are located at regions of A-T rich α satellite, a DNA repeat based on a 171 bp monomer. Monomers are 50–70% identical, and a defined number of monomers are arranged tandemly to create a higher order repeat (HOR) unit. The number of monomers and their order in the HOR confer chromosome specificity; that is, the centromeres of different human chromosomes are defined by HORs that are structurally distinct. For instance, the Homo sapiens chromosome X (HSAX) is defined by a HOR of 12 monomers (DXZ1, 12-mer), whereas the HSA8 is defined by a 7-mer HOR (D8Z2). The order and sequence of the monomers differ between chromosome-specific HORs, and thus, α satellite arrays on each chromosome can be distinguished. Despite these structural and organizational features that discriminate between α satellite arrays, the human genome assembly lacks contiguous α satellite sequences at centromeres. Monomers are readily identified by sequencing, but the HORs that define each chromosome-specific α satellite region are reiterated hundreds to thousands of times so that the highly homogeneous arrays (97–100% identical) extend over many megabases. This makes it difficult to accurately assemble long α satellite arrays, especially from short sequence reads.8 Without contiguous centromeric genome assemblies, it has been challenging to link specific features of α satellite organization to centromere function. Recent computational efforts have resulted in graphical models of human centromere sequences,14-16 a first step toward linear centromere maps. This approach has allowed assessment of genetic content within α satellite DNA, revealing some of the diversity in satellites within and among centromeres and reinforcing that many satellites are distinct among different chromosomes. A limitation of these “maps” is that they do not delineate the order of sequences within any given centromere, so the long-range organization of α satellite arrays in a single individual, much less the population, remains largely undetermined.
From these studies, we know that most human chromosomes have multiple HORs, that is more than one multi-megabase higher order α satellite array within the centromere region. Chromosomes like HSA1, HSA5, HSA7, HSA15, and HSA18 have 2 (or more) independent chromosome-specific α satellite arrays. Moreover, HSA1, HSA5, and HSA19, share at least one array (D1Z5), further emphasizing the structural complexity of centromere regions.17,18 The multi-array organization of human centromere regions presents a new view of normal chromosome biology. Structurally, endogenous human chromosomes closely resemble dicentric or tricentric chromosomes that have been thought to arise primarily through genome rearrangements that fuse 2 or more different chromosomes. Multi-array endogenous chromosomes are generally stable. Conversely, dicentric chromosomes were originally described by Barbara McClintock as unstable chromosomes that were poorly tolerated by the genome.19-21 Dicentrics that arise by genome rearrangement occur frequently in humans (1 in 1000 individuals) and are stable through mitosis and meiosis.22 This has been thought to be due to the poorly understood phenomenon of centromere inactivation, a process by which one centromere loses its identity and function. Human chromosomes, even multi-array chromosomes, have only one site of centromere and kinetochore formation, so, in a sense, they are normal models for studying active and inactive α satellite arrays. The fact that most endogenous human chromosomes possess more than one α satellite array suggests that the humans may be inherently more tolerant of dicentrics caused by genome rearrangement. Interestingly, when the same acquired dicentric chromosome occurs in multiple individuals, a specific centromere is often inactivated.23 Results like these have led to models describing differences in the functional potential (i.e., “centromere strength”) of distinct α satellite arrays. It is conceivable that the independent arrays on endogenous chromosomes may also exhibit variable strength or functional capabilities.
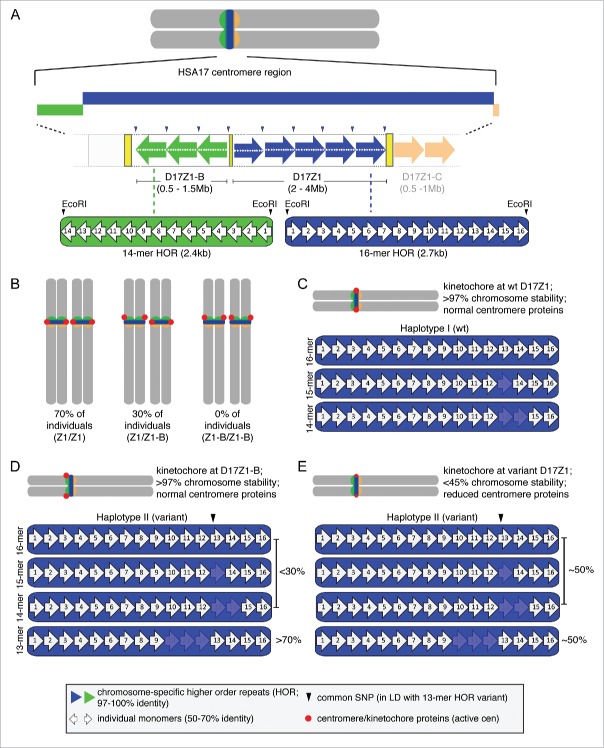
HSA17 has 3 α satellite arrays D17Z1, D17Z1-B and D17Z1-C, and is essentially structurally tricentric24-26 (Fig. 1A). These α satellite arrays (and other DNA segments in the human genome) were named according to established gene nomenclature [D17Z1: DNA segment (D), chromosomal assignment (17), complexity of DNA (Z for repetitive), and sequential number (1, 2, 3…) to confer uniqueness of DNA segment].27,28 We previously showed that within the population, 70% of individuals carry 2 HSA17s in which D17Z1 is the site of centromere and kinetochore assembly, based on the presence of inner and outer kinetochore proteins29 (Fig. 1B). About 30% of the population has a heterozygous centromere configuration in which D17Z1 is the active centromere and the location of kinetochore assembly on one homolog, while D17Z1-B is the active centromere on the other (Fig. 1B). Such flexibility in centromere location, termed centromeric epialleles, is mitotically and meiotically stable.29 The molecular basis of centromeric epialleles, that is, the ability to switch the position of centromere and kinetochore assembly between α satellite arrays, is not understood. Furthermore, they are not exclusive to HSA17. Our recent studies have identified centromeric epialleles on HSA1 and HSA7 (S McNulty, J Ross, and B Sullivan, unpublished observations), indicating that flexibility in centromere location is an intrinsic property of human chromosomes. In the case of HSA17, D17Z1 appears to be the major site of centromere and kinetochore assembly, suggesting that it is a dominant centromere. Collectively, these observations raise several interesting questions. What would make one centromere stronger than the other? Why might one α satellite array be a preferred centromere? How is the site of centromere assembly chosen when 2 or more competent arrays are available?
Figure 1.
α satellite variation and the molecular basis of centromeric epialleles in humans. (A) The centromere region of human chromosome 17 (HSA17) contains 3 α satellite arrays that are each defined by a different higher order repeat (HOR) unit. α satellite DNA is composed of 171bp monomers (white arrows) that are 50–70% identical. A defined number of monomers are tandemly arranged to create a HOR that is chromosome-specific. D17Z1 (blue), the predominant array is defined by a canonical 16-monomer HOR; EcoRI restriction sites demarcate the first monomer of each HOR. D17Z1-B (green) and D17Z1-C (shaded orange) are each defined by different 14-mer HORs. The monomers are numbered by their order in the HOR, and do not necessarily indicate sequence identity at the same monomer position between HORs of different arrays. (B) In the population, 70% of individuals carry 2 HSA17s that assemble the centromere and kinetochore (red dot) at D17Z1 (Z1/Z1). In 30% of individuals (Z1/Z1-B), D17Z1-B is the active centromere on one HSA17 homolog and D17Z1 is the centromere on the other homolog. No individuals have been identified yet that assemble both HSA17 centromeres at D17Z1-B (Z1-B/Z1-B). (C) D17Z1 is a polymorphic array. Single and multiple monomeric deletions produce HOR variants, including 15-mers, 14-mers, 13-mers, as well as 12-mers and 11-mers (not shown). (D) Some monomers also carry a common SNP in monomer 13 (black arrowhead) that creates an EcoRI site. This SNP is in linkage disequilibrium (LD) with the 13-mer HOR. Arrays containing specific HORs and the SNPs exist as distinct haplotypes in humans. Wild-type haplotype (I) occurs in most individuals and is defined by the canonical 16-mer HOR, as well as rarer 15- and 14-mers (C). Wild-type D17Z1 arrays are usually the site of centromere and kinetochore assembly (red circles) on mitotically stable HSA17. Haplotype II is defined by HOR variants that include a high proportion of 13-mers, many of which contain the SNP. D17Z1 arrays that have a high proportion of variant HORs are less likely to be the site of centromere assembly. Instead, the centromere is formed at “backup” array D17Z1-B and the HSA17 is extremely stable. (E) In a subset of Haplotype II individuals, the proportion of wild-type to variant HORs within the multi-megabase D17Z1 array is nearly equivalent. In these instances, if the centromere forms at D17Z1, the HSA17 is extremely unstable in mitosis due to a deficiency in centromere and kinetochore proteins (small red circles) and abnormal kinetochore architecture.
In answering these questions, one can consider models of gene expression and genetic variation. Gene function can be affected by genomic variation within the gene body or regulatory regions that alter promoter activity, splicing, enhancer activity, or transcription factor binding. Early studies of α satellite arrays uncovered substantial genomic variation at centromeres.30-32 Within the same chromosome-specific array, this variation exists in multiples forms: as HOR size variation, single nucleotide polymorphisms (SNPs) within HORs, and differences in total array size between homologues and among individuals. HSA17 exhibits extensive α satellite variation.31,33-35 The major array D17Z1 is classically defined by a HOR of 16 monomers (16-mer) (Fig. 1A, C), however, monomeric deletions have produced variant D17Z1 HORs that contain 15-, 14-, 13-, 12-, and 11-mers.31,34 These deletions have not swept through entire arrays, but are typically present as a fraction of an array, so that 35% of the human population has hybrid D17Z1 arrays containing both wild-type (16-mer) HORs and variant HORs. D17Z1-B and D17Z1-C appear to be homogenous arrays; both are defined by different, but related, 14-mer HORs.16,24
In our recent study, we explored the role of α satellite variation in controlling centromeric epialleles.36 We specifically tested the role of total α satellite array size in determining the location of the centromere on HSA17. D17Z1 and D17Z1-B arrays were molecularly sized from multiple individuals whose HSA17 centromeres had been functionally characterized.29 D17Z1 is overall a much larger array, ranging from 2–4 Mb between HSA17s homologues and among different individuals.36-38 Total array size for D17Z1-B and D17Z1-C is smaller, ranging 0.3–1.5 Mb24,29,36 (K Chew and B Sullivan, unpublished observations). We found that large D17Z1 arrays (≥ 3 Mb) tended to be the site of centromere and kinetochore assembly. However, when D17Z1 and D17Z1-B were closer in size, D17Z1-B tended to be active. Although array length cannot absolutely predict the site of centromere assembly, our findings imply that a large D17Z1 array may recruit or retain an increased critical mass of centromere proteins, giving it an advantage over a smaller D17Z1-B array.
About 70% of the population has 2 HSA17s in which D17Z1 is the centromere on both homologs, and ∼30% of individuals carry one homolog in which D17Z1-B is the active centromere.29,36 We were particularly interested in the fraction of HSA17s in which D17Z1 is not the active centromere, and delved deeper into the genomic structure of D17Z1 arrays on these chromosomes. The D17Z1 canonical 16-mer HOR is operationally defined by EcoRI sites that designate monomer one of the HOR and define the boundary between individual HORs.26 D17Z1 has 2 major polymorphisms. First, there is a size polymorphism caused by deletion of 3 monomers, yielding a 13-mer HOR31,39 (Fig. 1C–E). As already mentioned, additional monomeric deletions have produced HOR variants ranging from 15-mer to 11-mer.31,34 In addition, a subset of HORs contain a SNP that has introduced an EcoRI site within monomer 13 (Fig. 1D, E).33 HOR size variants and the presence or absence of the common SNP define distinct haplotypes.34 Haplotype I (wild-type) contains 16-, 15-, and 14-mer HORs and is present in 65% of HSA17s in the population. Haplotype II (variant) is present in 35% of HSA17s in the population. It is defined by the 13-mer HOR that is in linkage disequilibrium with the SNP. Interestingly, the frequencies of wild-type to variant D17Z1 haplotypes (65%:35%) resembles that of active to inactive D17Z1 arrays (70:30).
We also measured D17Z1 variation in the context of centromere location among various individuals and within a multigenerational family. Individuals carrying HSA17 centromeric epialleles exhibited substantial D17Z1 variation. Variant D17Z1 arrays defined by Haplotype II (i.e., containing the 13-mer HOR and the EcoRI SNP) were negatively associated with centromere function. That is, centromere and kinetochore assembly was more likely to be occur at D17Z1-B if D17Z1 was variant. Overall, our findings indicated that a large D17Z1 array composed of > 50% wild-type HORs will typically be the site of centromere assembly (Fig. 1C). However, smaller D17Z1 arrays (< 3Mb) that contain more (> 80%) variant HORs are more likely to be inactive and instead, the centromere will be assembled at D17Z1-B (Fig. 1D). Our studies suggest that D17Z1-B serves as a “backup” array, when the amount of variation within D17Z1 exceeds 80%.
Intriguingly, several D17Z1 arrays exhibiting 50–70% variation were chosen as the site of centromere assembly (Fig. 1E). We found that these HSA17s were highly unstable and showed increased aneuploidy over time. Notably, HSA17s with active, invariant D17Z1 or active D17Z1-B arrays did not exhibit appreciable instability. The HSA17 mutants allowed us to test the causal relationship between D17Z1 variation and HSA17 instability. We measured key centromere proteins on active variant D17Z1 arrays and compared them to active wild-type D17Z1 or D17Z1-B arrays. CENP-A is a variant of histone H3 that creates a unique type of chromatin exclusive to the centromere.2,3,40 CENP-C is a member of the CCAN that links the inner and outer kinetochore and is important for CENP-A recruitment and kinetochore maturation.41-43 We observed reduced amounts of CENP-A and CENP-C on variant D17Z1 arrays, but not wild-type D17Z1 or D17Z1-B arrays. These results suggested that the molecular basis for instability of HSA17s with active, variant arrays is an architectural kinetochore defect. Our studies also suggest that there is a critical molecular threshold for α satellite variation and centromere formation. In our data set, centromere formation occurred on D17Z1 arrays with moderate (50–70%) variation at the cost of decreased chromosome stability. It is not clear why a chromosome would continue to assemble the kinetochore at a mutated array. Determining if centromere function on these “threshold” arrays eventually switches to the backup D17Z1-B array to correct HSA17 instability and identifying molecular triggers that stimulate the shift are important next steps in the study of centromeric epialleles.
Overall, our findings indicate that different α satellite sequences do not have equal functional potential and that variation within α satellite organization negatively affects centromere assembly and function. The relationship between long-range organization of α satellite, i.e., where wild-type versus variant HORs are situated across a 4Mb array, is not known. A prior study of 3 HSA17s intimated that HOR variants within D17Z1 are clustered into domains.44 We do not know how HOR size variants are organized in active vs. inactive D17Z1 arrays in our functionally characterized data set. A few models could explain how α satellite organization affects centromere assembly and function. Inactive D17Z1 arrays in our study exhibited > 80% variation and had 30–60 times more HOR size/SNP variation compared with HSA17s with active D17Z1 arrays. These inactive arrays are homogenously variant, so the large number of variant HORs dispersed across the entire array may prevent or disfavor CENP recruitment/maintenance, skewing centromere assembly toward D17Z1-B. However, several of the D17Z1 arrays in our data set exhibited an intermediate range of variation and centromere function. The arrays were 50–70% variant and were often chosen as the site of centromere assembly, but the HSA17s were unstable. On variant arrays containing an equal amount of wild-type and variant HORs, the long-range organization of the entire α satellite region and where centromeric chromatin is located may be crucial. α satellite organization in which variant HORs are clustered at one end of the array may be less detrimental because centromere assembly can occur on wild-type HORs concentrated at the opposite end of the array. However, if variant and wild-type HORs are interspersed across the entire array, the irregularity might disrupt structural requirements for kinetochore architecture, such as CENP-C-mediated bridging between nucleosomes.4
CENP-B is another constitutive centromere protein with α satellite DNA binding properties. It recognizes the CENP-B box, a 17-bp sequence motif found in a subset of α satellite monomers on all human chromosomes except the Y.45-47 CENP-B is thought to position CENP-A nucleosomes and to stabilize CENP-A and CENP-C, based on the position of the CENP-B box within the DNA that is wrapped around the nucleosome.48-53 The number and location of CENP-B boxes might correlate with the ability of an array to achieve the proper higher order structure required for centromere function. Monomeric deletions that gave rise to variant HORs may have altered the number of available binding sites for CENP-B which could destabilize the interactions between CENP-A nucleosomes, CENP-C, and other CCAN proteins. Our rough calculations of the number of CENP-B boxes in highly variant arrays indicate that they have 25–50% fewer CENP-B boxes than wild-type arrays (NG Peterson and BA Sullivan, unpublished observations). Nevertheless, variant D17Z1 arrays have 5–6 times more CENP-B boxes than D17Z1-B arrays, suggesting that a factor other than the overall number of CENP-B boxes affects functional potential of variant arrays. CENP-B boxes within variant arrays could be mutated so that they are not recognized by CENP-B, but without extensive sequence information we cannot test this hypothesis. Alternatively, HORs containing 13-mers are shorter than wild-type (16-mer) HORs and the decreased HOR length might alter positioning of nucleosomes and centromere protein complexes across a variant D17Z1 array.49,53-55
Centromeric transcription is an integral part of kinetochore assembly and mitosis.56-63 In humans, each α satellite array produces a unique set of array-specific, long non-coding transcripts (SM McNulty and BA Sullivan, unpublished observations). We speculate that transcription of wild-type vs. variant α satellite HORs are correlated with distinct differences in centromere assembly. Genomic variation within D17Z1 may alter the abundance, stability, and/or structure of long, non-coding α satellite RNAs so that variant transcripts are less stable or cannot interact properly with centromere proteins. Additional studies are needed to distinguish transcription at variant and wild-type D17Z1 arrays and to capture the interaction of these transcripts with centromere protein complexes.
The extent of genomic variation within D17Z1 beyond what we have studied is not known, and much less so are the types and frequency of variation in other α satellite arrays located on different human chromosome. Concerted efforts to expand genomic studies of highly repetitive sequences will allow us to fully uncover links between α satellite DNA organization and centromere function.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The research described here was supported by March of Dimes grant 1FY13–517 and National Institutes of Health grant 5R01-GM098500.
References
- [1].Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 2008; 9:923-37; PMID:19002142; https://doi.org/ 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2002; 2:319-30; PMID:11879637; https://doi.org/ 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Structural Mol Biol 2004; 11:1076-83; https://doi.org/ 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Musacchio A, Desai A. A molecular view of Kinetochore assembly and function. Biology (Basel) 2017; 6(1):5; PMID:28125021; https://doi.org/ 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grimes B, Rhoades A, Willard H. Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 2002; 5:798-805; PMID:12027565; https://doi.org/ 10.1006/mthe.2002.0612 [DOI] [PubMed] [Google Scholar]
- [6].Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 1998; 16:431-9; PMID:9592390; https://doi.org/ 10.1038/nbt0598-431 [DOI] [PubMed] [Google Scholar]
- [7].Harrington J, Van Bokkelen G, Mays R, Gustashaw K, Willard H. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 1997; 15:345-55; PMID:9090378; https://doi.org/ 10.1038/ng0497-345 [DOI] [PubMed] [Google Scholar]
- [8].Miga KH. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res 2015; 23:421-6; PMID:26363799; https://doi.org/ 10.1007/s10577-015-9488-2 [DOI] [PubMed] [Google Scholar]
- [9].Plohl M, Mestrovic N, Mravinac B. Centromere identity from the DNA point of view. Chromosoma 2014; 123:313-25; PMID:24763964; https://doi.org/ 10.1007/s00412-014-0462-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo DH, Shi J, Gao Z, Han F, Lee H, Xu R, et al.. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet 2009; 5:e1000743; PMID:19956743; https://doi.org/ 10.1371/journal.pgen.1000743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gong Z, Wu Y, Koblizkova A, Torres GA, Wang K, Iovene M, Neumann P, Zhang W, Novák P, Buell CR, et al.. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 2012; 24:3559-74; PMID:22968715; https://doi.org/ 10.1105/tpc.112.100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat Genet 2004; 36:138-45; PMID:14716315; https://doi.org/ 10.1038/ng1289 [DOI] [PubMed] [Google Scholar]
- [13].Maheshwari S, Ishii T, Brown CT, Houben A, Comai L. Centromere location in Arabidopsis is unaltered by extreme divergence in CENH3 protein sequence. Genome Res 2017; 27:471-8; PMID:28223399; https://doi.org/ 10.1101/gr.214619.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hayden KE. Human centromere genomics: now it's personal. Chromosome Res 2012; 20:621-33; PMID:22801774; https://doi.org/ 10.1007/s10577-012-9295-y [DOI] [PubMed] [Google Scholar]
- [15].Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 2014; 24:697-707; PMID:24501022; https://doi.org/ 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al.. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 2015; 43:D670-81; PMID:25428374; https://doi.org/ 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pironon N, Puechberty J, Roizes G. Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21. BMC Genomics 2010; 11:195; PMID:20331851; https://doi.org/ 10.1186/1471-2164-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shepelev VA, Uralsky LI, Alexandrov AA, Yurov YB, Rogaev EI, Alexandrov IA. Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom Data 2015; 5:139-46; PMID:26167452; https://doi.org/ 10.1016/j.gdata.2015.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McClintock B. The behaviour of successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA 1939; 25:405-16; PMID:16577924; https://doi.org/ 10.1073/pnas.25.8.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics 1941; 26:234-82; PMID:17247004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc Natl Acad Sci U S A 1942; 28:458-63; PMID:16578057; https://doi.org/ 10.1073/pnas.28.11.458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stimpson KM, Matheny JE, Sullivan BA. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res 2012; 20:595-605; PMID:22801777; https://doi.org/ 10.1007/s10577-012-9302-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sullivan BA, Wolff DJ, Schwartz S. Analysis of centromeric activity in Robertsonian translocations: implications for a functional acrocentric hierarchy. Chromosoma 1994; 103:459-67; PMID:7720412; https://doi.org/ 10.1007/BF00337384 [DOI] [PubMed] [Google Scholar]
- [24].Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet 2009; 5:e1000641; PMID:19749981; https://doi.org/ 10.1371/journal.pgen.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rudd M, Willard H. Analysis of the centromeric regions of the human genome assembly. Trends Genet 2004; 20:529-33; PMID:15475110; https://doi.org/ 10.1016/j.tig.2004.08.008 [DOI] [PubMed] [Google Scholar]
- [26].Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol 1986; 6:3156-65; PMID:3785225; https://doi.org/ 10.1128/MCB.6.9.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shows TB, McAlpine PJ, Boucheix C, Collins FS, Conneally PM, Frezal J, Gershowitz H, Goodfellow PN, Hall JG, Issitt P, et al.. Guidelines for human gene nomenclature. An international system for human gene nomenclature (ISGN, 1987). Cytogenet Cell Genet 1987; 46:11-28; PMID:3507270; https://doi.org/ 10.1159/000132471 [DOI] [PubMed] [Google Scholar]
- [28].Willard HF, Skolnick MH, Pearson PL, Mandel JL. Report of the committee on Human Gene Mapping by Recombinant DNA techniques. Cytogenet Cell Genet 1985; 40:360-489; PMID:3864601; https://doi.org/ 10.1159/000132180 [DOI] [PubMed] [Google Scholar]
- [29].Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA. Functional epialleles at an endogenous human centromere. Proc Natl Acad Sci U S A 2012; 109:13704-9; PMID:22847449; https://doi.org/ 10.1073/pnas.1203126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Warburton PE, Willard HF. PCR amplification of tandemly repeated DNA: analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human alpha satellite DNA. Nucleic Acids Res 1992; 20:6033-42; PMID:1461735; https://doi.org/ 10.1093/nar/20.22.6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Waye JS, Willard HF. Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res 1986; 14:6915-27; PMID:3763396; https://doi.org/ 10.1093/nar/14.17.6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 1987; 25:207-14; PMID:2822935; https://doi.org/ 10.1007/BF02100014 [DOI] [PubMed] [Google Scholar]
- [33].Warburton P, Greig G, Haaf T, Willard H. PCR amplification of chromosome-specific alpha satellite DNA: Definition of centromeric STS markers and polymorphic analysis. Genomics 1991; 11:324-33; PMID:1685138; https://doi.org/ 10.1016/0888-7543(91)90139-6 [DOI] [PubMed] [Google Scholar]
- [34].Warburton PE, Willard HF. Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: evidence for concerted evolution along haplotypic lineages. J Mol Evol 1995; 41:1006-15; PMID:8587099; https://doi.org/ 10.1007/BF00173182 [DOI] [PubMed] [Google Scholar]
- [35].Willard HF, Greig GM, Powers VE, Waye JS. Molecular organization and haplotype analysis of centromeric DNA from human chromosome 17: implications for linkage in neurofibromatosis. Genomics 1987; 1:368-73; PMID:2896633; https://doi.org/ 10.1016/0888-7543(87)90041-3 [DOI] [PubMed] [Google Scholar]
- [36].Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res 2016; 26:1301-11; PMID:27510565; https://doi.org/ 10.1101/gr.206706.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wevrick R, Willard HF. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci U S A 1989; 86:9394-8; PMID:2594775; https://doi.org/ 10.1073/pnas.86.23.9394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wevrick R, Earnshaw WC, Howard-Peebles PN, Willard HF. Partial deletion of alpha satellite DNA associated with reduced amounts of the centromere protein CENP-B in a mitotically stable human chromosome rearrangement. Mol Cell Biol 1990; 10:6374-80; PMID:2247061; https://doi.org/ 10.1128/MCB.10.12.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Warburton PE, Waye JS, Willard HF. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 1993; 13:6520-9; PMID:8413251; https://doi.org/ 10.1128/MCB.13.10.6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A 1991; 88:3734-8; PMID:2023923; https://doi.org/ 10.1073/pnas.88.9.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 2013; 200:45-60; PMID:23277427; https://doi.org/ 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol 2015; 210:11-22; PMID:26124289; https://doi.org/ 10.1083/jcb.201412028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nagpal H, Hori T, Furukawa A, Sugase K, Osakabe A, Kurumizaka H, Fukagawa T. Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol Biol Cell 2015; 26:3768-76; PMID:26354420; https://doi.org/ 10.1091/mbc.E15-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Warburton P, Willard HF. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol 1990; 216:3-16; PMID:2122000; https://doi.org/ 10.1016/S0022-2836(05)80056-7 [DOI] [PubMed] [Google Scholar]
- [45].Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genetics 1994; 3:1245-57; PMID:7987298; https://doi.org/ 10.1093/hmg/3.8.1245 [DOI] [PubMed] [Google Scholar]
- [46].Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 1992; 116:585-96; PMID:1730770; https://doi.org/ 10.1083/jcb.116.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Haaf T, Ward D. Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum Mol Genet 1994; 3:697; PMID:8081355; https://doi.org/ 10.1093/hmg/3.5.697 [DOI] [PubMed] [Google Scholar]
- [48].Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell 2007; 131:1287-300; PMID:18160038; https://doi.org/ 10.1016/j.cell.2007.10.045 [DOI] [PubMed] [Google Scholar]
- [49].Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/alpha-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells 1998; 3:533-48; PMID:9797455; https://doi.org/ 10.1046/j.1365-2443.1998.00210.x [DOI] [PubMed] [Google Scholar]
- [50].Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Structural Mol Biol 2013; 20:687-95; https://doi.org/ 10.1038/nsmb.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B enhances the Fidelity of Human Centromere Function. Dev Cell 2015; 33:314-27; PMID:25942623; https://doi.org/ 10.1016/j.devcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fujita R, Otake K, Arimura Y, Horikoshi N, Miya Y, Shiga T, Osakabe A, Tachiwana H, Ohzeki J, Larionov V, et al.. Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res 2015; 43:4909-22; PMID:25916850; https://doi.org/ 10.1093/nar/gkv405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, Yokoyama S. Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. J Biol Chem 2005; 280:41609-18; PMID:16183641; https://doi.org/ 10.1074/jbc.M509666200 [DOI] [PubMed] [Google Scholar]
- [54].Henikoff JG, Thakur J, Kasinathan S, Henikoff S. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci Adv 2015; 1(1):e1400234; https://doi.org/ 10.1126/sciadv.1400234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Thakur J, Henikoff S. CENPT bridges adjacent CENPA nucleosomes on young human alpha-satellite dimers. Genome Res 2016; 26:1178-87; PMID:27384170; https://doi.org/ 10.1101/gr.204784.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O'Neill MJ, et al.. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 2009; 118:113-25; PMID:18839199; https://doi.org/ 10.1007/s00412-008-0181-5 [DOI] [PubMed] [Google Scholar]
- [57].Hall LE, Mitchell SE, O'Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res 2012; 20:535-46; PMID:22760449; https://doi.org/ 10.1007/s10577-012-9297-9 [DOI] [PubMed] [Google Scholar]
- [58].Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 2007; 179:411-21; PMID:17984319; https://doi.org/ 10.1083/jcb.200706176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, Wong LH. LINE Retrotransposon RNA is an essential structural and functional Epigenetic component of a Core Neocentromeric Chromatin. PLoS Genet 2009; 5:e1000354; PMID:19180186; https://doi.org/ 10.1371/journal.pgen.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rosic S, Kohler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 2014; 207:335-49; PMID:25365994; https://doi.org/ 10.1083/jcb.201404097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci U S A 2004; 101:15986-91; PMID:15514020; https://doi.org/ 10.1073/pnas.0407154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 2005; 19:2301-6; PMID:16204182; https://doi.org/ 10.1101/gad.344205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Blower MD. Centromeric Transcription Regulates Aurora-B localization and activation. Cell Rep 2016; 15:1624-33; PMID:27184843; https://doi.org/ 10.1016/j.celrep.2016.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]