ABSTRACT
The core architecture of the eukaryotic cell was established well over one billion years ago, and is largely retained in all extant lineages. However, eukaryotic cells also possess lineage-specific features, frequently keyed to specific functional requirements. One quintessential core eukaryotic structure is the nuclear pore complex (NPC), responsible for regulating exchange of macromolecules between the nucleus and cytoplasm as well as acting as a nuclear organizational hub. NPC architecture has been best documented in one eukaryotic supergroup, the Opisthokonts (e.g. Saccharomyces cerevisiae and Homo sapiens), which although compositionally similar, have significant variations in certain NPC subcomplex structures. The variation of NPC structure across other taxa in the eukaryotic kingdom however, remains poorly understood. We explored trypanosomes, highly divergent organisms, and mapped and assigned their NPC proteins to specific substructures to reveal their NPC architecture. We showed that the NPC central structural scaffold is conserved, likely across all eukaryotes, but more peripheral elements can exhibit very significant lineage-specific losses, duplications or other alterations in their components. Amazingly, trypanosomes lack the major components of the mRNA export platform that are asymmetrically localized within yeast and vertebrate NPCs. Concomitant with this, the trypanosome NPC is ALMOST completely symmetric with the nuclear basket being the only major source of asymmetry. We suggest these features point toward a stepwise evolution of the NPC in which a coating scaffold first stabilized the pore after which selective gating emerged and expanded, leading to the addition of peripheral remodeling machineries on the nucleoplasmic and cytoplasmic sides of the pore.
KEYWORDS: eukaryogenesis, molecular evolution, mRNA export, nuclear pore complex, Trypanosoma brucei
In the beginning……
The origin of the eukaryotic cell (eukaryogenesis) is a major evolutionary transition in the history of life,1 accompanied by the emergence of extensive intracellular compartmentalization. The most pronounced compartment is the nucleus, surrounded by a double-membrane nuclear envelope (NE) that is contiguous with the endoplasmic reticulum. The most prominent macromolecular assembly, present in all NEs, are nuclear pore complexes (NPCs - 8-fold symmetric cylindrical multiprotein structures (∼50 MDa in yeast) that mediate macromolecular exchange between the nucleus and the cytoplasm (Fig. 1). Since the NPC is uniquely and ubiquitously a eukaryotic feature, we can facilitate a reconstruction of evolutionary history, identify potential adaptive mechanisms, and look for the imprints that the transition from the first to the last eukaryote common ancestor (FECA to LECA) left at the structural and molecular levels through a detailed study and comparison of the NPC's components from key organisms across the eukaryotic lineage.
Figure 1.
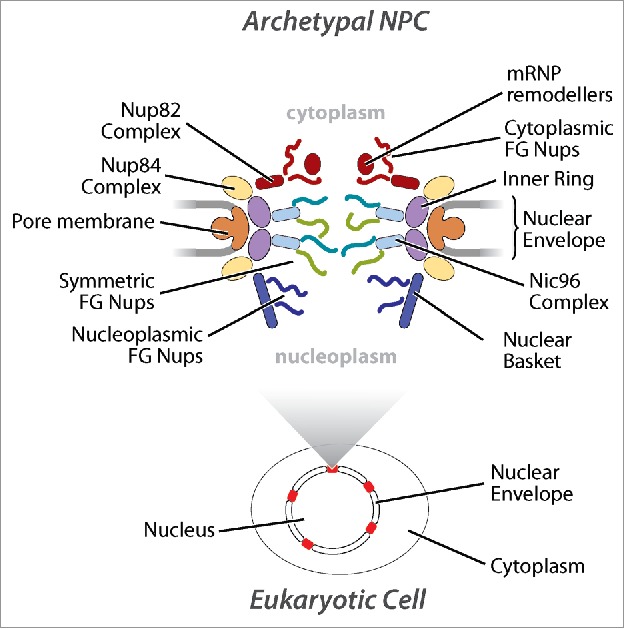
Schematic of opisthokont NPCs. A schematic of textbook NPCs highlighting each distinct nuclear pore subcomplex. The approximate location messenger ribonucleprotein (mRNP) remodeling factors required for mRNA export out of the NPC is also shown.
Ultrastructural studies hint at a high level of morphological NPC conservation across eukarya. However, morphological similarity, especially at comparatively low resolution, cannot define molecular structure or function that requires a significantly greater level of dissection. Until recently, detailed compositional, structural and functional information of the NPC was only available for Saccharomyces cerevisiae (Sc) and Homo sapiens (Hs), both members of the Opisthokont supergroup, and thus relatively, closely related.2-4 In the last 7 years, NPC components have been well cataloged in 2 further supergroups, Excavata (Trypanosoma brucei (Tb)) by us,4,5 and Archaeplastida (Arabidopsis thaliana (At)),6,7 resulting in remarkable initial insights into the structure, evolution and species-specific adaptations of the NPC.
Copy and paste: the NPC's scaffold arose through ancient duplications of a progenitor coating complex
NPCs are comprised of multiples of ∼30 different proteins termed nucleoporins (Nups) that are classified into 3 major subtypes; pore membrane (Poms), core scaffold, and phenylalanine-glycine (FG)-repeat containing Nups (FG-Nups) (Fig. 1).3-11 Poms and the core scaffold form the major structural framework while FG-Nups establish the permeability barrier of the NPC and facilitate nucleocytoplasmic transport through interactions with soluble transport factors (karyopherins), with directionality dependent on a Ran GTP/GDP gradient.12
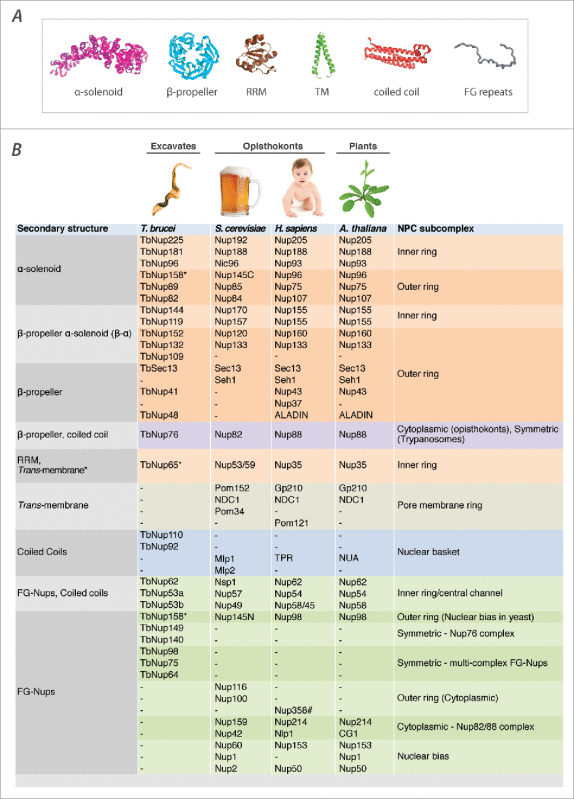
There is extremely low sequence similarity between excavate and opisthokont Nups.4 Despite this, trypanosome Nups share a remarkable architectural and domain organization with opisthokonts and plants, highlighting that structural considerations such as detailed fold arrangements are key to the function of these proteins (Table 1).4,5 The core scaffold of the NPC comprises the inner and outer rings that are composed almost exclusively of proteins comprised of β-propellers, α-solenoids, or an N-terminal β-propeller and C-terminal α-solenoid (β-α).13,14 These characteristics are shared with major classes of membrane interacting proteins, such as vesicle coat proteins (COPI, COPII, clathrin and tethering complexes) and intraflagellar complexes, suggestive of a common ancestry between the endomembrane trafficking system and the NPC, a theory known as the “protocoatomer hypothesis”.8,13,14 Moreover, the overall architecture of the NPC's core scaffold appears to consist of multiple kinds of structural modules that nevertheless resemble each other, both in composition and in arrangement of fold and domain types.8,15,16 This supports the idea that the elaborate architecture of the NPC arose through repeated duplication events from a simple progenitor coating complex.8,17,18
Table 1.
Protein folds and the components of the NPC in trypanosomes, plants and opisthokonts. (A) The NPC is principally comprised of proteins with the fold types highlighted. α-solenoids are stacked pairs of α (α) helices, composed of anti parallel repeating α subunits that in turn form a kind of “superhelix”,87 β-propeller proteins typically have between 4 to 8 blade-shaped β (β) sheets arranged in a toroidal fashion,88 RRM is RNA recognition motif, TM is trans membrane domain, coiled coils are 2 or more α helices that wind around each other to form larger helical bundles and FG-repeats are phenylalanine glycine repeats typically found in the intrinsically disordered protein domains of Nups that interact with transport factors in the nucleus. (B) A catalog of trypanosome Nups compared with those in other well-studied taxa. Orthologs of individual TbNups are based on interactome mapping and immunolocalization of NPC subcomplexes in the trypanosome NPC. Trypanosomes lack pore membrane Nups and cytoplasmic or nuclear biased Nups. Furthermore, several trypanosome FG-Nups and the nuclear basket Nups are not orthologous to those in opisthokonts and plants. Trypanosome nuclear basket Nups are half the size of those in other eukaryotes studied to date and appear to have arisen independently through evolution.38,39 The RNA-recognition motif (RRM) containing TbNup65 has a trans-membrane (TM) domain instead of an amphipathic lipid-sensing (ALPS) motif30,31 that is found in the equivalent opisthokont and plant orthologs, and maybe the way trypanosomes compensate for a lack of pore membrane (POMs) Nups. TbNup158* lacks the catalytic residues required to undergo autoproteolysis that generates 2 individual proteins such as yeast Nup145N/Nup145C and verterbrate Nup98/Nup964,5, and as such remains a single N-terminal FG-domain/C-terminal α-solenoid FG-Nup.4,5 Nup358# is a metazoan specific multi-domain protein that contains FG-Nups, Ran binding domains, multiple Zinc finger domains, an E3-SUMO ligase domain and a cylophilin domain.89
 |
Variations on a theme: using the same building blocks to assemble different NPC structures
At the heart of the NPC there appears to be a remarkably conserved core subcomplex. This is the inner ring complex, almost identical in protein composition and arrangement between all eukaryotes studied, including trypanosomes.5,7,8,16,19,20 This structure must already have been present in LECA, with relatively few post-LECA innovations in any organism. Surely, this forms a keystone to hold the NPC together.
However, the modular scaffold architecture of the NPC – a scaffold of similar building blocks – can open a simple path to the evolutionary elaboration and diversification of the NPC, through altering the number or design of each building block. And indeed, there is a compositional flexibility in the outer ring complex, which revolves around the presence or absence of a subset of proteins in different organisms (Table 1). Most variable is the complement of β-propeller proteins, as only Sec 13 is present in all characterized versions of this complex. Sec 13 is also present in COPII complexes, further underscoring the evolutionary relationship between the NPC and coating complexes.13,14 Nonetheless, what appear to be minor species-specific variations of the outer ring complex in opisthokonts have revealed an interesting architectural divergence between yeast and metazoa. The most compelling example is the metazoan outer ring, which despite near identical compositional and architectural similarity with yeast, comprises 2 reticulated rings,11,21 in contrast to a single ring in yeast.8,15,22 Thus, architectural and functional similarities cannot be assumed simply from a similar list of components. The presence or absence of a “few small β-propeller proteins” may disguise previously unsuspected and significant species-specific elaborations of this outer ring in other eukaryotic lineages. Indeed, the major difference we discovered through interactomics between trypanosomes and other well studied taxa is the presence of 3 β-α Nups in the outer ring complex, as opposed to just 2 in opisthokonts and plants (Table 1). This difference in composition may represent a completely unique outer ring structure in trypanosome NPCs (TbNPC). Discerning the stoichiometry, arrangement and morphology of TbNPC components is thus clearly essentially to fully understand the architecture of the trypanosome outer ring, and hence understand its function.
An extremely well conserved component of the outer ring complex with interesting properties is TbNup158 (HsNup98–96 or ScNup145N/C; see Table 1). It is easily identifiable as it encoded in most eukaryotic genomes as a single protein comprised of a large N-terminal FG-Nup domain and a large C-terminal α-solenoid separated by a β-sandwich that contains within it catalytic residues that lead to its autoproteolytic cleavage into 2 separate proteins.23 However, although the 2 proteins still associate with one another in the NPC, it is generally believed that cleavage into 2 different polypeptides allows the FG-Nup domain of this protein to be dynamic and have a role in gene regulation, while the α-solenoid portion remains a stable structural member of the outer ring complex.24,25 The tripeptide catalytic residues in the trypanosome ortholog (TbNup158) have altered during evolution to prevent cleavage, resulting in the protein being incorporated into the TbNPC outer ring as a single protein, possibly to eliminate this dynamism and negate a role in gene regulation which is unusual in trypanosomes (see below and Fig. 2). Once again, apparently minor deviations in the composition of the trypanosome NPC can lead to significant alterations in structure and presumably, function. This is not limited to trypanosomes. The same catalytic activity is lacking in another flagellated excavate Giardia lamblia,4 and autoproteolytic cleavage has been shown to be dispensable for cell growth in fission yeast.26 Additionally, the Apicomplexan parasite Plasmodium falciparum has an unusual fusion of a portion of the α solenoid domain of this protein (ScNup145C) with PfSec 1327. Thus, trypanosomes do not represent the exception – instead, they are part of the rule that the outer ring complex (comprising one-third of the NPC's mass) can apparently vary considerably in form and function between different organisms.
Figure 2.
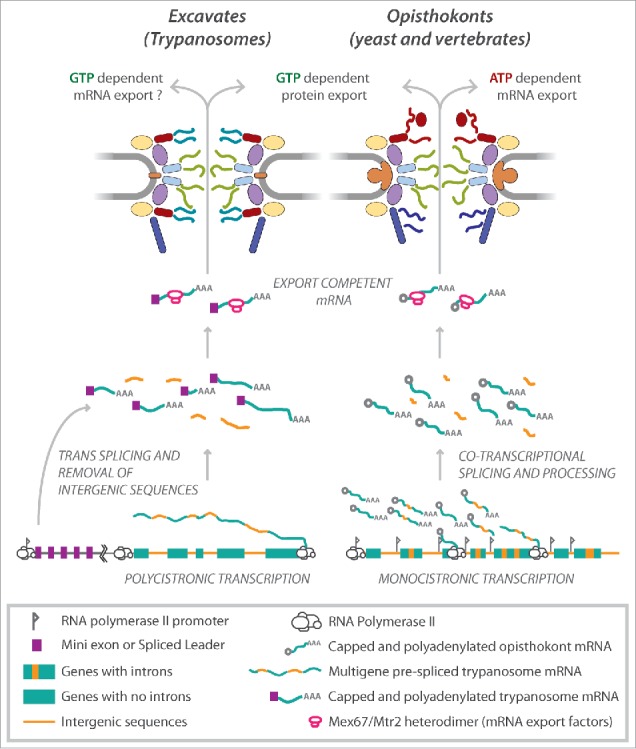
(A) comparison between Excavate and Opisthokont NPCs and transport through them. Trypanosomes (Excavates) have a symmetric NPC with the exception of the nuclear basket unlike in opisthokonts (yeast and humans). Furthermore, trypanosomes lack the cytoplasmic mRNA platform including mRNP (ribonucleoproteins) remodeling factors such as the ATP-dependent DEAD box helicase Dbp5 and Gle1 that are crucial for mRNA export in opisthokonts.15,41 Instead, mRNA export in trypanosomes appears to rely on the RanGDP/GTP gradient similar to protein export, as opposed to ATP in opisthokonts.5 This may be related to the unusual mechanisms trypanosomes use for controlling gene expression. Trypanosome protein-coding genes are intronless and each gene lacks an individual polymerase II promoter.58 Thus, trypanosome genes (green boxes) are transcribed into long multigene (polycistronic) transcripts that are resolved into single mRNAs by trans-splicing of a mini exon, also known as the spliced leader sequence (purple box) at the 5′ end of each gene, splicing out of intergenic (orange lines) sequences and polyadenylated (AAA). Processed and export competent mRNA are exported through the NPC by the Mex67/Mtr2 heterodimer (pink ovals). Protein coding genes in opisthokonts (and most other eukaryotes) have introns (orange boxes) and individual promoters (flags) and are transcribed as singly (monocistronic). Transcribed mRNAs are co-transcriptionally 5′-capped (gray circles), and introns spliced (orange lines) before export by Mex67/Mtr2 in conjunction with the actions of the ATP dependent helicase Dbp5.
TMs and ALPS: Multiple ways to tether a leviathan
Opisthokonts also have trans-membrane (TM) proteins that interact with core scaffold Nups to anchor the NPC to the pore.3,9 Although not well conserved,28 opisthokonts and plants all have orthologs of Gp210 (Pom152 in yeast, Gp210 in plants and humans) and Ndc1 (Table 1), pointing to ancient and possibly pre-LECA origins for both proteins, as plants - like trypanosomes - diverged early from the eukaryotic lineage. Remarkably, we found that both proteins are absent from the trypanosome NPC, suggestive of secondary loss. Instead, we found that trypanosomes rely on the ortholog of the RNA-recognition motif (RRM) containing HsNup35/ScNup53 to anchor the NPC.5 Nup35 is crucial for NPC biogenesis in opisthokonts.29 Furthermore, it connects components of the inner ring in opisthokonts with the pore membrane where it interacts via an amphipathic lipid–packing sensor (ALPS) motif,30-32 and is a key nucleator for in vitro assembly of an inner ring complex, raising the possibility that it is key in vivo for directing assembly of the inner ring, and so whole NPC, from the pore membrane inwards.33 By contrast, TbNup65 (Table 1), the trypanosome ortholog of Nup35, instead interacts with the pore membrane through a canonical trans-membrane (TM) domain. Somehow, ALPS and TM domains can be interchanged in evolution, and there is thus plasticity in the exact mechanism used for anchoring the NPC to the pore membrane. Indeed, all Poms in the Aspergillus nidulans NPC can be deleted without compromising the viability of the cells, in the presence of an intact outer ring complex.34 Thus, additional anchoring in trypanosomes may rely heavily on potential ALPS motifs that have been identified on the β-propellers of several outer ring β-α Nups in opisthokonts 32 or be provided by the lamin analog NUP-1 that co-purifies with members of the trypanosome outer ring complex.5,35 Unfortunately, little is known about how the NPC assembles outside of opisthokonts, so the role of these motifs in NPC assembly or function remains obscure.
Through the looking glass: FG-Nups are symmetrically distributed within the TbNPC
FG-Nups are proteins carrying significant intrinsically disordered domains that contain multiple repeats of degenerate phenylalanine-glycine (FG) motifs. These domains collectively fill the central channel of the NPC, and facilitate nucleocytoplasmic transport through interactions with soluble cargo-carrying transport factors.12 Approximately one third of FG-Nups in opisthokonts exhibit a biased localization within the NPC, being present either on the cytoplasmic side or the nucleoplasmic side of the pore.9,36 This asymmetry is required for aspects of NPC function, especially mRNA export, which relies on the cytoplasmically biased Nup82 complex for messenger ribonucleoproteins (mRNPs) remodeling before release into the cytoplasm for translation by ribosomes.15,37
There are significant differences between opisthokont and trypanosome FG-motif domain sequences, making it near impossible to identify orthologs of opisthokont FG-Nups in trypanosomes by in silico means alone. However, through affinity capture, we were able to identify orthologs of FG-Nups, based on the core scaffold structure with which they associate.5 We identified inner ring (central channel) FG-Nups, outer ring FG-Nups, a group that associates with both inner and outer ring complexes (multi-complex) and a third complex which we termed the TbNup76 complex, which consists of TbNup76 (an ortholog of ScNup82/HsNup88), and TbNup140 and TbNup149, the 2 largest FG-Nups in the TbNPC (see Table 1). One might have thought that, having identified the associated complex, we would then know where the FG-Nups were placed in the TbNPC. But this was not the case. Rather, we discovered via immuno-electron microscopy (iEM) localization that, with the exception of the nuclear basket,5,38,39 all Nup classes and subcomplexes were equally and symmetrically distributed between the nuclear and cytoplasmic faces of the NPC.5 Astonishingly, this suggests that the trypanosome NPC lacks a clear nucleoplasmic or cytoplasmic biased localization of FG-Nups, in complete contrast to opisthokonts.9,36 The symmetric arrangement in trypanosomes is consistent with the hypothesis that inherent NPC asymmetry is not necessary for basic nucleocytoplasmic transport.40
A major source of Nup asymmetry in opsithokonts is the exclusively cytoplasmic ScNup82/HsNup88 subcomplex, tethering specialized FG-Nups that provide the interaction platform for factors critical for mRNA export.15,37 However, TbNup76, the presumed trypanosome ortholog of ScNup82, is located on both the cytoplasmic and nucleoplasmic faces of the trypanosome NPC.5 Herein lies another cautionary tale of making sure that one does not assume that, just because proteins are orthologous, they function in the same way in organisms that are evolutionarily distant. After all, a bat's wing is orthologous to our hands! Furthermore, the FG-Nups that associate with TbNup76 do not bear any resemblance to the equivalent Nup82/Nup88 complex FG-Nups in opisthokonts and plants which importantly, contain N-terminal β-propeller domains required to mediate interactions with the ATP-dependent DEAD box RNA helicase Dbp5 and the RNA export mediator Gle1 with its cofactor IP6 (inositol hexakisphosphate) to form a remodeling factory to process messenger ribonucleoproteins (mRNPs) before nuclear exit.41-43 In fact, no single FG-Nup in the TbNPC contains a N-terminal β-propeller domain.
Rules of the road: 2 major FG-Nup flavors, 2 modes of transport?
Interestingly, despite the fact that FG-Nups are symmetrically distributed in the TbNPC, there still are different FG “flavors” (Supplementary Figure 4 in Obado et al, 2016). A perusal of the flavors used by trypanosomes compared with opisthokonts suggests that these flavor types are roughly preserved, even though their nucleocytoplasmic asymmetry is not. The more centrally localized trypanosome FG Nups are enriched predominantly in “GGFG” motifs; the multi-complex FG-Nups, which seem to be more peripheral on the NPC, are enriched instead in “FSFG,” “FG,” “SVFG” and “PAFG” repeats. In opisthokonts, there are also only a few major FG repeat flavors, the majority of which fall into the category of either “GLFG”-like or “FxFG”-like.44,45 We also know that there are 2 major kinds of transport factor: karyopherin-like, and non-karyopherin-like, with very different preferences for FG repeat flavors.46,47 Certainly, trypanosomes show us that FG flavor may not necessarily be all about nucleocytoplasmic positioning or imparting a directionality to nuclear transport. Perhaps instead, these 2 major FG repeat flavors delineate specific transport conduits for the trafficking pathways across the NPC mediated by the 2 kinds of transport factor?
Nip and tuck: Sculpting the NPC to reflect biology
So how does the trypanosome mRNA export machinery function in the absence of FG-Nup asymmetry? Unlike protein transport, mRNA (poly-adenylated RNA) export in opisthokonts is Ran-independent, being powered in an ATP-dependent manner by Dbp5 and is mediated by non-karyopherin RNA export factors (the Mex67:Mtr2 heterodimer in yeast, termed TAP:p15 in humans),41,48-50 Fortunately, transport factors (karyopherin and non-karyopherin) generally are extremely well conserved across the eukaryotic kingdom, including even in trypanosomes, which have clear orthologs of the RNA export factors Mex67 and Mtr2.51,52
We tagged and affinity captured TbMex67 to identify the composition of any potential RNA processing and export factors in trypanosomes. As well as its partner TbMtr2,51 TbMex67 associates strongly with the TbNup76 complex, even though this is found on both sides of the NPC; in opisthokonts, Mex67 associates strongly with the Nup82/Nup88 complex, as one might expect.41,50 However, no potential orthologs of Dbp5 and Gle1 co-isolated with TbMex67, or even found in the genome.5 So, although at least some aspect of the function of this complex – binding to RNA export factors – seems to be conserved in trypanosomes, it does so in a very different spatial context and by a different mechanism that lacks the anticipated helicase processing factory. In addition, TbMex67 bound strongly to the GTPase Ran, to Ran binding protein 1 (RBP1) and to a GTPase activating protein (GAP). In opisthokonts, Ran, RBP1 and RanGAP work in synchrony to expedite the hydrolysis of RanGTP to RanGDP in the cytoplasm to facilitate the release of protein cargo from a RanGTP-karyopherin complex.53-55 Therefore, the co-isolation of Ran, RBP1 and the GAP with the non-karyopherin transporters Mex67-Mtr2 is extremely unusual and has not been previously observed in other organisms. This suggests that unlike in opisthokonts and plants, GTP and not ATP powers mRNA export in trypanosomes. Interestingly, non protein coding RNA in opisthokonts, such as tRNAs, small nuclear RNAs, micro RNAs and pre-ribosomal subunits, are exported out of the nucleus on a Ran gradient by karyopherins, similar to protein export (reviewed in 47,56). Additionally, a small subset of mRNA export is dependent on the karyopherin, Exportin 1 (Crm1).57 Thus trypanosomes appear to have modified their NPC and the Ran-mediated export pathways, a system that already has enormous flexibility, expanding it to include all transport, including mRNA export. Why have they done this? We don't know; we have speculated it may be linked to their rather unusual mechanisms for controlling gene expression. Trypanosome protein-coding genes are intron-less, lack individual RNA polymerase II promoters and are organized into large polycistronic transcription units (Fig. 2).58,59 Thus, the regulation of gene expression relies mainly on mRNA turnover and translation rates.60,61 Hence, perhaps gene expression is controlled at the point of nucleo-cytoplasmic export through differential export in a highly regulated manner. In addition, the exclusive trans-splicing of protein-coding mRNAs in trypanosomes (Fig. 2) may also relax requirements for extensive chaperoning during nuclear export. Moreover, these results indicate that the asymmetric localization of many FG Nups in opisthokont NPCs is at least in part strongly associated with the co-transcriptionally linked mRNA processing and export pathway, with the final stages of mRNA processing embedded in the NPC in opisthokonts, and asymmetrically disposed FG Nups providing the necessary docking sites. Thus, quality control occurs at the nuclear basket and nucleoplasmic FG-Nups and then the remodeling of mRNPs to facilitate export occurring at the cytoplasmic end of the NPC.15,62
In any case, all this underscores the intimate link between the NPC and gene expression, as well as the incredible flexibility of the NPC, which although probably fully formed at LECA has the capacity to mold and adapt its structure to the demands of individual lineages by acting as an interaction platform for several nuclear processes while still maintaining its structural core and primary function as a transport hub.
What do we now know about the evolution of the NPC?
The origin of eukaryotes and the events surrounding the transition from prokaryotes to eukaryotes remain obscure despite several attempts at reconstructing the pathways involved. Trypanosomes diverged early in eukaryotic evolution and can provide important evolutionary insights into fundamental cell biologic processes shared by all eukaryotes. Indeed, several key features of molecular biology have first been identified in trypanosomes. Examples include antigenic variation, GPI-anchored proteins, RNA editing, polycistronic transcription and trans-splicing.63-67 Originally considered a “quirky” character of these parasites, these fundamental biologic processes have now been found to be common in other eukaryotes (including RNA editing in mammals,68,69 polycistronic transcription and trans-splicing in the nematode Caenorhabditis elegans).70,71 Now, the trypanosome is performing this function again, providing insights into the structure and evolution of the NPC and mechanisms of nucleocytoplasmic transport.
The shared evolutionary relationship between the NPC core scaffold and the endomembrane system is well documented.18 Furthermore, the 2 large yeast inner ring α-solenoid ScNups188/192 (Table 1) share architectural features with karyopherins (also α-solenoid proteins), suggestive of a co-evolution of transport factors, structural components of the NPC, and the endomembrane trafficking system.72,73 Indeed, it is noteworthy that the assembly of vesicle coats and the translocation of proteins through membranes are controlled by Ras-like GTPases termed Rabs,74 just as the translocation of cargo in the NPC is controlled by the Ras-like GTPase, Ran.
The symmetry of the TbNPC alludes to our previous theory that there may have been a stepwise acquisition of complexity in the NPC in the FECA to LECA transition in which an early non-specific pore comprised of coating proteins but lacking a gating function (Fig. 3).17,18 Later, the evolution of FG-Nups led to a more sophisticated gating system, that was further elaborated to include cytoplasmic and nucleoplasm biased FG-Nups and to include the elaborate mRNA export machinery in opisthokonts (Fig. 3).
Figure 3.
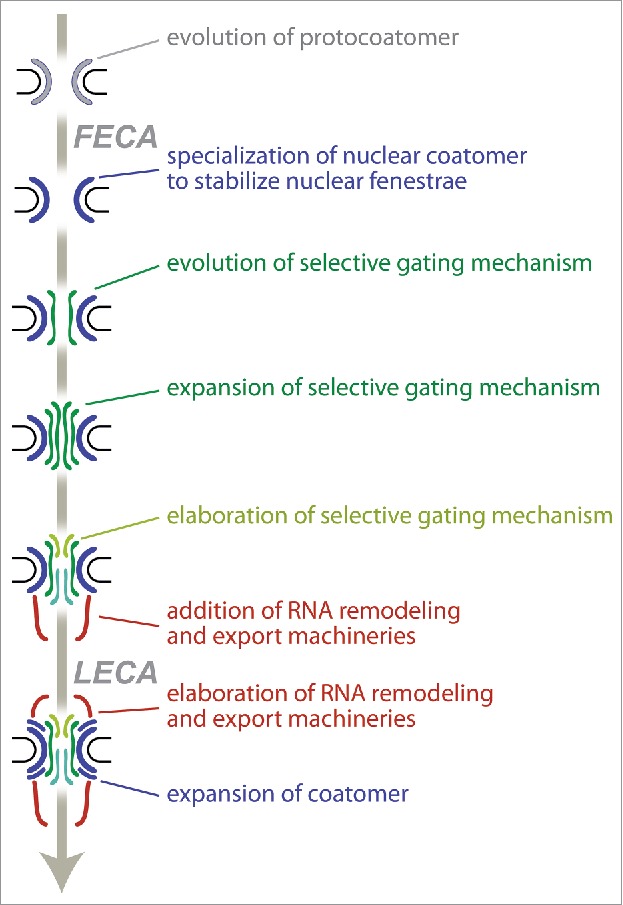
Evolution of the NPC. We propose a model in which the NPC evolved gating functions in a stepwise manner starting with a simple coat that acquired complexity through a series of duplications as observed in endomembrane trafficking.17,18 This then led to the evolution of FG-Nups and then further diversification into the current metazoan type NPC with a nuclear basket and cytoplasmic filaments.
Trypanosomes have no cytoplasmic or nucleoplasmic biased FG-Nups. However, they have retained a level of asymmetry through the addition of the nuclear basket, which interestingly appears to have a distinct evolutionary history to that of opisthokonts.38,39 In the opisthokonts and plants, the nuclear basket is composed of large (∼200–250 kDa) coiled coil proteins, that associate with and coordinate several nuclear peripheral processes, thereby extending NPC functionality.36,62,75-78 Although the 2 nuclear basket proteins are much smaller (∼100kDa) in trypanosomes, our study showed by iEM, that they extend an average of 36nm into the nucleoplasm from the center of the TbNPC.5 Additionally, they have retained similar functions to those in other eukarya, by creating heterochromatin free zones around nuclear pores that are evident by electron microscopy in all eukaryotic nuclei, as well as recapitulating interactions with the spindle organizer.4,38,79-81 Thus, in conjunction with divergent lamin-like proteins,35,82 and unconventional kinetochores,83 it seems that the trypanosome nucleus uses unique protein complexes in parallel with conserved core elements such as the spindle microtubules and outer kinetochore components, Ndc80/Nuf2 to facilitate trypanosome biology.84,85
Did trypanosomes retain an ancient symmetric assembly or reconfigure their molecular biology through the loss of introns and individual gene promoters (Fig. 2) and thereby dispense with the need to utilize ATP as an energy source for separately powering mRNA export? Perhaps investigations of the nuclear transport machineries in other divergent eukaryotes will help answer this question. Indeed, a recent study has shown that mRNA export pathways in Apicomplexa also appears to be divergent86; in addition, so far investigators have been unable to identify any Mex67 orthologs in Toxoplasma gondii, indicating that this organism may be even more divergent than trypanosomes in its nucleocytoplasmic transport machineries.86 In fact, very little is known about mRNA processing and export in most protists. Thus our study highlights and reinforces the need to sample and study a broad distribution of eukaryotic taxa to gain insight into evolutionary origins of function and mechanism at the nuclear envelope – as well as of course in many other cellular processes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Brian T. Chait for enormous contributions to proteomics, Javier Fernandez Martinez for critical reading of the manuscript.
Funding
This work described above was supported in part by the Medical Research Council (project grant MR/N010558/1 to MCF), The National Institutes of Health (NIAID Exploratory/Developmental Research Grant 1R21AI096069 to MPR, NIGMS GM103314 to BTC, GM103511 and GM109824 to MPR and BTC) and the Wellcome Trust (082813/Z/07/Z to MCF and MPR).
References
- [1].Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biology letters 2010; 6:342-5; PMID:20031978; https://doi.org/ 10.1098/rsbl.2009.0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al.. The revised classification of eukaryotes. J Eukaryot Microbiol 2012; 59:429-93; PMID:23020233; https://doi.org/ 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158:915-27; PMID:12196509; https://doi.org/ 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics : MCP 2009; 8:2119-30; https://doi.org/ 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol 2016; 14:e1002365; PMID:26891179; https://doi.org/ 10.1371/journal.pbio.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 2010; 22:4084-97; PMID:21189294; https://doi.org/ 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tamura K, Hara-Nishimura I. The molecular architecture of the plant nuclear pore complex. J Exp Bot 2013; 64:823-32; PMID:22987840; https://doi.org/ 10.1093/jxb/ers258 [DOI] [PubMed] [Google Scholar]
- [8].Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al.. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695-701; PMID:18046406; https://doi.org/ 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- [9].Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 2000; 148:635-51; PMID:10684247; https://doi.org/ 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schwartz TU. The Structure Inventory of the Nuclear Pore Complex. J Mol Biol 2016; 428:1986-2000; PMID:27016207; https://doi.org/ 10.1016/j.jmb.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al.. In situ structural analysis of the human nuclear pore complex. Nature 2015; 526:140-3; PMID:26416747; https://doi.org/ 10.1038/nature15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wente SR, Rout MP. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb Perspect Biol 2010; 2(10):a000562; PMID:20630994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2004; 2:e380; PMID:15523559; https://doi.org/ 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A 2006; 103:2172-7; PMID:16461911; https://doi.org/ 10.1073/pnas.0506345103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, et al.. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016; 167:1215-28 e25; PMID:PMID:27839866; https://doi.org/ 10.1016/j.cell.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al.. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 2016; 352:363-5; PMID:27081072; https://doi.org/ 10.1126/science.aaf0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Field MC, Koreny L, Rout MP. Enriching the pore: splendid complexity from humble origins. Traffic 2014; 15:141-56; PMID:24279500; https://doi.org/ 10.1111/tra.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Field MC, Sali A, Rout MP. Evolution: On a bender–BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol 2011; 193:963-72; PMID:21670211; https://doi.org/ 10.1083/jcb.201102042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011; 146:277-89; PMID:21784248; https://doi.org/ 10.1016/j.cell.2011.06.039 [DOI] [PubMed] [Google Scholar]
- [20].Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 2010; 189:1129-42; PMID:PMID:20566687; https://doi.org/ 10.1083/jcb.200912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, et al.. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013; 155:1233-43; PMID:24315095; https://doi.org/ 10.1016/j.cell.2013.10.055 [DOI] [PubMed] [Google Scholar]
- [22].Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, et al.. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol 2012; 196:419-34; PMID:22331846; https://doi.org/ 10.1083/jcb.201109008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol Cell 2002; 10:347-58; PMID:12191480; https://doi.org/ 10.1016/S1097-2765(02)00589-0 [DOI] [PubMed] [Google Scholar]
- [24].Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 2002; 13:1282-97; PMID:11950939; https://doi.org/ 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genetics 2013; 9:e1003308; PMID:23468646; https://doi.org/ 10.1371/journal.pgen.1003308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Asakawa H, Mori C, Ohtsuki C, Iwamoto M, Hiraoka Y, Haraguchi T. Uncleavable Nup98-Nup96 is functional in the fission yeast Schizosaccharomyces pombe. FEBS Open Bio 2015; 5:508-14; PMID:26137436; https://doi.org/ 10.1016/j.fob.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe'er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, et al.. PfSec 13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J Cell Sci 2013; 126:3055-69; PMID:23687383; https://doi.org/ 10.1242/jcs.122119 [DOI] [PubMed] [Google Scholar]
- [28].Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PloS One 2010; 5:e13241; PMID:20949036; https://doi.org/ 10.1371/journal.pone.0013241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Molecular biology of the cell 2008; 19:1753-62; PMID:18256286; https://doi.org/ 10.1091/mbc.E07-08-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Doucet CM, Esmery N, de Saint-Jean M, Antonny B. Membrane Curvature Sensing by Amphipathic Helices Is Modulated by the Surrounding Protein Backbone. PloS One 2015; 10:e0137965; PMID:26366573; https://doi.org/ 10.1371/journal.pone.0137965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol 2007; 14:138-46; PMID:17220896; https://doi.org/ 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- [32].Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, et al.. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Molecular & cellular proteomics : MCP 2014; 13:2911-26; https://doi.org/ 10.1074/mcp.M114.040915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat Struct Mol Biol 2015; 22:774-81; PMID:26344569; https://doi.org/ 10.1038/nsmb.3084 [DOI] [PubMed] [Google Scholar]
- [34].Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell 2009; 20:616-30; PMID:19019988; https://doi.org/ 10.1091/mbc.E08-06-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, et al.. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS biology 2012; 10:e1001287; PMID:22479148; https://doi.org/ 10.1371/journal.pbio.1001287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 2004; 15:4261-77; PMID:15229283; https://doi.org/ 10.1091/mbc.E04-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adams RL, Terry LJ, Wente SR. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics 2014; 197:1213-24; PMID:24931410; https://doi.org/ 10.1534/genetics.114.164012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Holden JM, Koreny L, Obado S, Ratushny AV, Chen WM, Chiang JH, Kelly S, Chait BT, Aitchison JD, Rout MP, et al.. Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol Biol Cell 2014; 25:1421-36; PMID:24600046; https://doi.org/ 10.1091/mbc.E13-12-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Holden JM, Koreny L, Kelly S, Chait BT, Rout MP, Field MC, Obado SO. Touching from a distance. Nucleus 2014; 5:304-10; PMID:25482119; https://doi.org/ 10.4161/nucl.32228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zeitler B, Weis K. The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo. J Cell Biol 2004; 167:583-90; PMID:15557115; https://doi.org/ 10.1083/jcb.200407156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus 2011; 2:540-8; PMID:22064466; https://doi.org/ 10.4161/nucl.2.6.17881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell 2004; 16:749-60; PMID:15574330; https://doi.org/ 10.1016/j.molcel.2004.10.032 [DOI] [PubMed] [Google Scholar]
- [43].Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nature Cell Biol 2006; 8:668-76; PMID:16783364; https://doi.org/ 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- [44].Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 2007; 129:83-96; PMID:17418788; https://doi.org/ 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- [45].Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al.. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics : MCP 2010; 9:2205-24; https://doi.org/ 10.1074/mcp.M000035-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Christie M, Chang CW, Rona G, Smith KM, Stewart AG, Takeda AA, Fontes MR, Stewart M, Vértessy BG, Forwood JK, et al.. Structural Biology and Regulation of Protein Import into the Nucleus. J Mol Biol 2016; 428:2060-90; PMID:26523678; https://doi.org/ 10.1016/j.jmb.2015.10.023 [DOI] [PubMed] [Google Scholar]
- [47].Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 2007; 8:761-73; PMID:17786152; https://doi.org/ 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- [48].Clouse KN, Luo MJ, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol 2001; 3:97-9; PMID:11146633; https://doi.org/ 10.1038/35050625 [DOI] [PubMed] [Google Scholar]
- [49].Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J 1999; 18:2593-609; PMID:10228171; https://doi.org/ 10.1093/emboj/18.9.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. The EMBO journal 1997; 16:3256-71; PMID:9214641; https://doi.org/ 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dostalova A, Kaser S, Cristodero M, Schimanski B. The nuclear mRNA export receptor Mex67-Mtr2 of Trypanosoma brucei contains a unique and essential zinc finger motif. Mol Microbiol 2013; 88:728-39; PMID:23560737; https://doi.org/ 10.1111/mmi.12217 [DOI] [PubMed] [Google Scholar]
- [52].O'Reilly AJ, Dacks JB, Field MC. Evolution of the karyopherin-beta family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PloS One 2011; 6:e19308; PMID:21556326; https://doi.org/ 10.1371/journal.pone.0019308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J 1995; 14:705-15; PMID:7882974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Seewald MJ, Korner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature 2002; 415:662-6; PMID:11832950; https://doi.org/ 10.1038/415662a [DOI] [PubMed] [Google Scholar]
- [55].Seewald MJ, Kraemer A, Farkasovsky M, Korner C, Wittinghofer A, Vetter IR. Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol Cell Biol 2003; 23:8124-36; PMID:14585972; https://doi.org/ 10.1128/MCB.23.22.8124-8136.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Okamura M, Inose H, Masuda S. RNA Export through the NPC in Eukaryotes. Genes (Basel) 2015; 6:124-49; PMID:25802992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Delaleau M, Borden KL. Multiple Export Mechanisms for mRNAs. Cells 2015; 4:452-73; PMID:26343730; https://doi.org/ 10.3390/cells4030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al.. The genome of the African trypanosome Trypanosoma brucei. Science 2005; 309:416-22; PMID:16020726; https://doi.org/ 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- [59].Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J 2002; 21:1881-8; PMID:11953307; https://doi.org/ 10.1093/emboj/21.8.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathogens 2010; 6:e1001090; PMID:20838601; https://doi.org/ 10.1371/journal.ppat.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vasquez JJ, Hon CC, Vanselow JT, Schlosser A, Siegel TN. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res 2014; 42:3623-37; PMID:24442674; https://doi.org/ 10.1093/nar/gkt1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 2010; 11:490-501; PMID:20571586; https://doi.org/ 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- [63].Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986; 46:819-26; PMID:3019552; https://doi.org/ 10.1016/0092-8674(86)90063-2 [DOI] [PubMed] [Google Scholar]
- [64].Feagin JE, Abraham JM, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 1988; 53:413-22; PMID:2452697; https://doi.org/ 10.1016/0092-8674(88)90161-4 [DOI] [PubMed] [Google Scholar]
- [65].Hoeijmakers JH, Frasch AC, Bernards A, Borst P, Cross GA. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature 1980; 284:78-80; PMID:6243753; https://doi.org/ 10.1038/284078a0 [DOI] [PubMed] [Google Scholar]
- [66].Imboden MA, Laird PW, Affolter M, Seebeck T. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res 1987; 15:7357-68; PMID:3658696; https://doi.org/ 10.1093/nar/15.18.7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sather S, Agabian N. A 5′ spliced leader is added in trans to both alpha- and beta-tubulin transcripts in Trypanosoma brucei. Proc Natl Acad Sci U S A 1985; 82:5695-9; PMID:2994042; https://doi.org/ 10.1073/pnas.82.17.5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Anant S, Blanc V, Davidson NO. Molecular regulation, evolutionary, and functional adaptations associated with C to U editing of mammalian apolipoproteinB mRNA. Prog Nucleic Acid Res Mol Biol 2003; 75:1-41; PMID:14604008 [DOI] [PubMed] [Google Scholar]
- [69].Blanc V, Davidson NO. C-to-U RNA editing: mechanisms leading to genetic diversity. J Biol Chem 2003; 278:1395-8; PMID:12446660; https://doi.org/ 10.1074/jbc.R200024200 [DOI] [PubMed] [Google Scholar]
- [70].Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet 1995; 11:132-6; PMID:7732590; https://doi.org/ 10.1016/S0168-9525(00)89026-5 [DOI] [PubMed] [Google Scholar]
- [71].Blumenthal T. Trans-splicing and operons in C. elegans. WormBook 2012:1-11; PMID:23175478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife 2013; 2:e00745; PMID:23795296; https://doi.org/ 10.7554/eLife.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, et al.. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 2013; 21:560-71; PMID:23499021; https://doi.org/ 10.1016/j.str.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol 2014; 6:a022616; PMID:25341920; https://doi.org/ 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol 2002; 156:617-30; PMID:11839768; https://doi.org/ 10.1083/jcb.200106046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell 2013; 24:3920-38; PMID:24152732; https://doi.org/ 10.1091/mbc.E13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ptak C, Aitchison JD, Wozniak RW. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol 2014; 28C:46-53; https://doi.org/ 10.1016/j.ceb.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 2007; 19:1537-48; PMID:17513499; https://doi.org/ 10.1105/tpc.106.049239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 2010; 29:1659-73; PMID:20407419; https://doi.org/ 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol 2005; 170:225-35; PMID:16027220; https://doi.org/ 10.1083/jcb.200504140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al.. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell 2004; 15:4854-65; PMID:15356261; https://doi.org/ 10.1091/mbc.E04-07-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Maishman L, Obado SO, Alsford S, Bart JM, Chen WM, Ratushny AV, Navarro M, Horn D, Aitchison JD, Chait BT, et al.. Co-dependence between trypanosome nuclear lamina components in nuclear stability and control of gene expression. Nucleic Acids Res 2016; 44:10554-70; PMID:27625397; https://doi.org/ 10.1093/nar/gkw751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Akiyoshi B, Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell 2014; 156:1247-58; PMID:24582333; https://doi.org/ 10.1016/j.cell.2014.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].D'Archivio S, Wickstead B. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J Cell Biol 2016; PMID:28034897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ogbadoyi E, Ersfeld K, Robinson D, Sherwin T, Gull K. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 2000; 108:501-13; PMID:10794572; https://doi.org/ 10.1007/s004120050402 [DOI] [PubMed] [Google Scholar]
- [86].Serpeloni M, Jimenez-Ruiz E, Vidal NM, Kroeber C, Andenmatten N, Lemgruber L, Mörking P, Pall GS, Meissner M, Ávila AR. UAP56 is a conserved crucial component of a divergent mRNA export pathway in Toxoplasma gondii. Mol Microbiol 2016; 102(4):672-689; PMID:27542978; https://doi.org/ 10.1111/mmi.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Fournier D, Palidwor GA, Shcherbinin S, Szengel A, Schaefer MH, Perez-Iratxeta C, Andrade-Navarro MA. Functional and genomic analyses of alpha-solenoid proteins. PloS One 2013; 8:e79894; PMID:24278209; https://doi.org/ 10.1371/journal.pone.0079894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 1999; 24:181-5; PMID:10322433; https://doi.org/ 10.1016/S0968-0004(99)01384-5 [DOI] [PubMed] [Google Scholar]
- [89].Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 1995; 270:14209-13; PMID:7775481; https://doi.org/ 10.1074/jbc.270.23.14209 [DOI] [PubMed] [Google Scholar]


