Abstract
While many are familiar with actin as a well-conserved component of the eukaryotic cytoskeleton, it is less often appreciated that actin is a member of a large superfamily of structurally related protein families found throughout the tree of life. Actin-related proteins include chaperones, carbohydrate kinases, and other enzymes, as well as a staggeringly diverse set of proteins that use the energy from ATP hydrolysis to form dynamic, linear polymers. Despite differing widely from one another in filament structure and dynamics, these polymers play important roles in ordering cell space in bacteria, archaea, and eukaryotes. It is not known whether these polymers descended from a single ancestral polymer or arose multiple times by convergent evolution from monomeric actin-like proteins. In this work, we provide an overview of the structures, dynamics, and functions of this diverse set. Then, using a phylogenetic analysis to examine actin evolution, we show that the actin-related protein families that form polymers are more closely related to one another than they are to other nonpolymerizing members of the actin superfamily. Thus all the known actin-like polymers are likely to be the descendants of a single, ancestral, polymer-forming actin-like protein.
BACKGROUND
Actin was one of the first biological polymers to be biochemically characterized. It was first extracted from skeletal muscle tissue by Halliburton in 1887, who referred to it as “myosin ferment” due to its ability to coagulate in the presence of the motor protein, myosin (Halliburton, 1887). In 1942, Straub purified monomeric actin, opening up the possibility of studying polymer behavior in vitro (Straub, 1942). Later, Huxley and Hanson showed that actin filaments are one of the major components of contractile muscle sarcomeres (Hanson and Huxley, 1953), and Pollard identified actin as the thin filaments present in the contractile cytoplasm of Acanthamoeba (Pollard et al., 1970). Since then, work by many researchers has elucidated the ultrastructure, regulation, dynamics, and function of actin filaments from a wide range of eukaryotes, in which it functions in concert with other cytoskeletal filaments to control cellular organization. Although this work revealed the ubiquity of eukaryotic actin, actin’s place as one of the distinguishing features of eukaryotes changed with the discovery of similar filaments within bacteria, archaea, and mobile genetic elements (Bork et al., 1992; Møller-Jensen et al., 2002). As this work made clear, actin is in fact a single member of a much larger superfamily of proteins that are present in organisms across the tree of life. Here we refer to members of this superfamily as “actins” if they cluster with canonical eukaryotic actins in phylogenetic analyses, while we call other proteins in the superfamily “actin-related” polymers or ”actin-related” proteins.
Strikingly, many members of the superfamily have been shown to form dynamic polymers. Despite differing widely in their higher-order structures and assembly kinetics, these polymers carry out a number of conserved functions across the domains of life: they control the shape of both eukaryotic and many prokaryotic cells; are common, conserved components of the cell division and chromosome segregation machinery (Mishra et al., 2014); and control the dynamic internal organization of the cytoplasm in eukaryotes and some prokaryotes. In addition, the actin superfamily also includes a number of other proteins that do not form polymers, including chaperones such as Hsp70/DnaK and enzymes such as hexokinase and glutamate mutase (Hurley, 1996).
The structural similarity between these diverse proteins raises a number of questions. How are the actin-like polymers and nonpolymerizing actin-like enzymes related? How likely is it that a diverse set of actin-related enzymes form polymers? Did polymerization arise once, or multiple times, during evolution? What common features do these evolutionarily distant proteins share that enables them to form polymers, and what features are modified across evolution to change their filament superstructure, polymerization dynamics, and cellular function? In this review, we look at the evolution of actin-like filament structure and function across the tree of life. In doing so, we consider two reciprocal hypotheses: 1) that the self-assembly of actin filaments arose only once in evolution—that is, all extant actin-like filaments are descended from a single ancestral polymer, or 2) polymerization arose multiple times within the actin superfamily as the result of modifications in the structure of one or more monomeric ATPases with an actin-like fold.
In exploring the evolution of actin-like proteins, we begin by examining their structure and functions across systems. We then look at the phylogenetic tree of actins and related proteins to test these alternative hypotheses and determine the origin of polymerization within the actin superfamily.
THE STRUCTURE OF ACTIN AND ACTIN-LIKE FILAMENTS
Actin is one of the most ubiquitous and highly conserved proteins in eukaryotes, being 91% identical in sequence between Caenorhabditis elegans and Homo sapiens. It is also among the set of proteins that appear to have been inherited from the last common ancestor of all eukaryotes. Nevertheless, the sequences of actins differ more widely in some eukaryotic lineages, for example, in plants (McDowell et al., 1996). Thus the high level of sequence conservation between actins across eukaryotes may not reflect a fundamental limit in actin’s ability to tolerate sequence changes while still forming dynamic filaments. Instead, this conservation may reflect other complex constraints, for example, those imposed by the need to preserve interactions with a wide set of actin-binding proteins, or those due to actin’s high levels of expression in eukaryotes (Pál et al., 2001). In addition, when we consider the prokaryotic members of the actin family, the degree of variation that can be tolerated within the context of an actin-like polymer becomes clear. These distant relatives have sequences so divergent that they were only first identified using a motif built from an alignment of actin, Hsp70, and hexokinase (Bork et al., 1992).
The first low-resolution structure of the actin monomer was obtained by Kabsch, Mannherz, and Suck in 1985, and the same team produced the first atomic model in 1990 with the help of Pai and Holmes (Kabsch et al., 1985, 1990). With the structure in hand, it became immediately apparent that, despite their low sequence similarity, actin and hexokinases shared the same fold (Steitz et al., 1978). At the heart of actin’s two globular domains is a nucleotide-binding pocket that binds and hydrolyzes ATP. Importantly, the structure of the monomer is profoundly changed by nucleotide binding and subtly altered by nucleotide hydrolysis from ATP to ADP. As a result, ATP hydrolysis and subsequent phosphate release lead to movement of the two domains of the protein relative to one another (Korn et al., 1987; Orlova and Egelman, 1992; Murakami et al., 2010). It is this link between nucleotide binding, ATP hydrolysis, and changes in monomer structure that are likely to make actin-related enzymes a good starting point for the formation of actin-like filaments.
Allostery and monomer contacts within an actin-like filament
When embedded within a filament, actin-related monomers make extensive contacts with one another. These include head-to-tail contacts between monomers within individual protofilaments and contacts between the two filament strands (Figure 1). In most actin-like polymers, the location of the head-to-tail monomer contacts within the protofilaments are conserved: subdomain Ia in one monomer interacts with residues in subdomain Ib in the next monomer in the series, while subdomain IIa interacts with subdomain IIb in a similar way (Figure 1C). Even in FtsA, which is missing subdomain Ib, the longitudinal contacts appear to be conserved; an insertion into subdomain Ia takes the place of subdomain Ib within a presumed polymer (Szwedziak et al., 2012; Ozyamak et al., 2013b). Thus these contacts may be essential across actin-related proteins to enable the formation of dynamic filaments, because nucleotide binding shortens the distance separating subdomains Ib and IIb, matching the distance between subdomain Ia and IIa (van den Ent et al., 2014) (Figure 1).
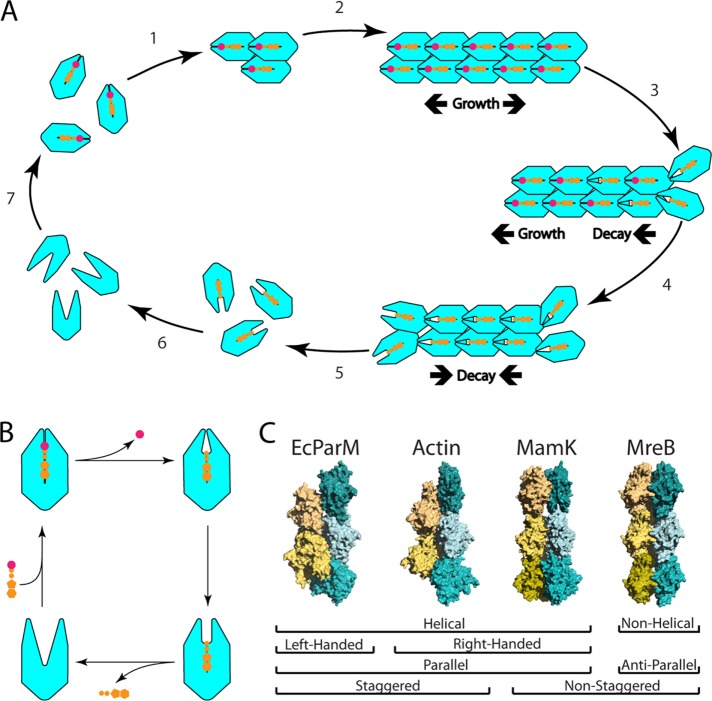
FIGURE 1:
Polymer formation is a repeated feature within the actin superfamily. (A) Polymerization of actin homologues requires the formation of a filament nucleus (1). Once a nucleus has formed, filament elongation is rapid (2). Once monomers incorporate into a growing filament, they begin to hydrolyze ATP (3) until the filament end(s) is(are) composed of ADP-bound monomers (4). Filaments initially grow in both directions. Under certain conditions, some filaments can grow from one end and shrink from the other (a process referred to as treadmilling). Eventually, lower affinity between ADP-bound monomers allows for filament disassembly from the ends (5). ADP then dissociates from ADP-bound monomers (6), which then rapidly rebind ATP (7). (B) ATP hydrolysis, phosphate release, ADP/ATP exchange are associated with changes in monomer conformation that influence filament architecture and actin on/off rates. (C) Among actin-like filaments, the contacts within individual protofilaments vary little. By varying lateral contacts between protofilaments, different filament structures with different properties and behaviors can be generated. A subset of these structures is displayed here. E. coli ParM is a parallel, left-handed, helical filament whose protofilaments are staggered (PDB ID: 5AEY) (Bharat et al., 2015). Actin is a parallel, helical filament whose protofilaments are staggered (PDB ID: 4A7N) (Behrman et al., 2012). MamK filaments are parallel, helical filaments whose protofilaments are not staggered (PDB ID: 5LJV) (Löwe et al., 2016). MreB is a nonhelical, antiparallel filament whose protofilaments are not staggered and that has an intrinsic curvature (PDB ID: 4CZJ) (van den Ent et al., 2014).
As a result of these contacts, changes in the conformation of individual monomers that result from ATP hydrolysis and phosphate release can be felt by neighboring monomers in the filament. As well as making actin an excellent chassis for use in the generation of cytomotive filaments (filaments that generate force via polymerization or depolymerization), this type of monomer–monomer communication may enable actin-related proteins to act as allosteric enzymes with high cooperativity. It is therefore formally possible that some members of the enzyme/chaperone actin subfamilies form polymers. This would not be without precedent, as other enzymes have been shown to assemble into filaments that regulate their activity: one of the best examples being the non–actin related protein, CTP synthetase (Barry et al., 2014). In this case, polymerization can either activate or inhibit CTP synthetase activity depending on the organism (Noree et al., 2010; Lynch and Kollman, 2016). Moreover, several metabolic enzymes in yeast have been shown to form pH-dependent polymers in response to starvation (Shen et al., 2016). For most actin-related enzymes, however, this hypothesis remains to be tested.
Variation in the structure of actin-related filaments
Even though all members of the actin superfamily appear similar at the monomer level, they differ greatly in their filament structure. Much of this variation comes from changes in the lateral contacts between protofilaments (van den Ent et al., 2014). This can alter helical pitch and filament handedness and can determine whether filament strands lie parallel or antiparallel to one another (Figure 1C). Because of this, filaments built from monomers with a similar fold can have very different properties.
In the case of eukaryotic actin, two parallel filament strands twist around one another like rope to form a right-handed helical filament that is both chiral and polar. Many of these structural features of the polymer are essential for actin’s function. For example, filaments must be polar to be used as a substrate for directed molecular motors, like myosin (Schuh, 2011). However, the protofilament polarity, pitch, handedness, and degree of stagger between protofilaments varies across polymers within the actin superfamily. Moreover, as highlighted in the following sections, these structural differences have significant consequences for polymer dynamics and function (see Figure 1).
MreB polymers are involved in bacterial cell wall synthesis and maintenance of prokaryotic rod shape. MreB forms two-stranded, antiparallel, nonhelical filaments, with no stagger between the subunits in each protofilament (van den Ent et al., 2014). The lack of a twist is essential to MreB function in two ways. First, because MreB associates with the membrane through a single face, the absence of filament twist enables adjacent monomers to form tight contacts with the membrane. Furthermore, because MreB lacks a helical twist, MreB filaments can sustain an intrinsic curvature—something that appears to be crucial for its function in the regulation of bacterial cell shape (Salje et al., 2011; van den Ent et al., 2014).
MamK organizes the cellular distribution of magnetic organelles (magnetosomes) within magnetotactic bacteria. MamK filaments form parallel helical filaments like actin but with no stagger between the subunits, leaving an open cavity between the protofilaments (Ozyamak et al., 2013a; Löwe et al., 2016; Bergeron et al., 2017). This raises some interesting questions about how these structural differences influence MamK polymer dynamics, the interaction of the polymer with magnetosomes, and how the lack of stagger affects the mechanical properties of MamK filaments.
ParM from the Escherichia coli R1 plasmid (EcParM) forms parallel, twisted filaments, but unlike actin, has a left-handed twist. In contrast, ParM from the Bacillus thuringiensis pBMB67 plasmid (BtParM), forms two-stranded, supercoiled, antiparallel, helical filaments. Interestingly, in the presence of its DNA adaptor protein (ParR), these filaments associate to form a four-stranded nanotubule with an open core; a structure reminiscent of microtubules (Jiang et al., 2016b). While this structure’s mechanical properties have not been studied, it is likely that these filaments are stiffer than two-stranded filaments, again reminiscent of the mechanical properties of microtubules or of stiff fascin–cross-linked actin bundles seen in many eukaryotic cells (Jansen et al., 2011; Takatsuki et al., 2014).
Variation in filament kinetics
Actin-like polymers also differ widely from one another in their dynamics. Changes in kinetic parameters (e.g., nucleation rate, monomer on/off rate, ATPase activity, phosphate-release kinetics) can yield polymers with different length distributions, stability, and polymerization and depolymerization rates. Importantly, however, because polymer dynamics are sensitive to changes in amino acid sequence that cause little or no change in secondary and tertiary structure, this may not be evident from a simple structural analysis of the polymer. Changes in kinetics can also be brought about by associated regulators. Thus, in eukaryotes, a small number of actins are used to perform a wide variety of cellular tasks through the action of a large number of actin-binding proteins that modify different kinetic and/or structural parameters of the monomer/polymer. This tunability enables cells to exert dynamic local control over the actin cytoskeleton to determine where actin filaments form, how fast they elongate, and for how long they persist (Pollard et al., 2001). Cross-links between actin filaments further expand this repertoire to enable networks with different filament organization (Mullins et al., 1998; Jacinto and Baum, 2003).
In eukaryotes, much of the control is exerted at the level of nucleation. This is because actin is a so-called nucleation-condensation polymer, that is, the affinity of a monomer for another monomer is much lower than its affinity for a growing filament or a filament nucleus (Lal et al., 1984; Mullins et al., 1998). This is both a natural consequence of monomers having multiple contacts in the context of a filament and a useful property, because it disfavors the spontaneous nucleation of filaments even in the presence of high concentrations of the monomer. As a result, nearly all actin-like polymers whose assembly dynamics have been elucidated have been found to be nucleation-limited. However, relative to eukaryotic actin, the nucleation rate tends to be much faster in the case of bacterial actins. For example, the nucleation rate of EcParM is 200 times faster than eukaryotic actin, even though filaments elongate at a similar rate (Garner et al., 2004). Similarly, the plasmid-segregating actin AlfA from Bacillus subtilis also assembles via a rapid, yet nucleation-limited pathway (Polka et al., 2009). In these cases, the ability to undergo spontaneous polymerization at a suitable site may do away with the need for a dedicated nucleator. Interestingly, there are eukaryotic actins within parasites such as Toxoplasma gondii that assemble isodesmically (the affinity of a monomer for another monomer is equal to the affinity between a monomer and a filament end), thus obviating the need for a nucleator (Skillman et al., 2013). These actins have been reported to exhibit atypical lateral filament contacts (Vahokoski et al., 2014) and form short microfilaments just beneath the cell membrane.
Following polymerization, the behavior of a filament will depend on the rates of ATP hydrolysis and phosphate release. As ATP-bound monomers are incorporated into actin filaments, bound ATP is hydrolyzed such that the ends of the filament tend to be composed of ATP-actin as long as the filament continues to grow (Carlier et al., 1984) (Figure 1). This in turn affects polymer dynamics because, for eukaryotic actin, the ADP-bound form has an off rate at the minus end that is much higher than that of ATP actin (Pollard and Borisy, 2003). This, together with asymmetries in the on/off rates at each end of the filament, can lead to treadmilling, whereby monomers flux through a filament of constant length as the filament grows via monomer addition at its (+) end at the same rate as it shrinks via monomer loss from its (−) end (Kirschner, 1980). Other actin-like polymers display this behavior. These include the plasmid segregators Alp7a and AlfA (Derman et al., 2009; Polka et al., 2009) and MamK, which treadmills to distribute magnetosomes between daughter cells (Toro-Nahuelpan et al., 2016).
Actin-related filaments can also exhibit dynamic instability, the rapid transitioning between states of polymerization and depolymerization—a key feature of microtubule dynamics (Mitchison and Kirschner, 1984). While eukaryotic actin does not exhibit dynamic instability (for a possible exception see Husson et al., 2011), it is observed in the plasmid-segregating polymers EcParM and Alp7a (Garner et al., 2004; Drew and Pogliano, 2011). Although there are exceptions to this rule, for example, AlfA (Polka et al., 2014), this suggests that evolution may have converged on a common solution that allows the efficient “search and capture” of DNA by cytoskeletal elements across different systems (Mitchison and Kirschner, 1984).
At the same time, there are many clades of structurally similar enzymes that do not appear to form polymers. These include the many “actin-related proteins” (Arps) in eukaryotes. Although, in the case of the Arp2/3 complex, the actin-related proteins in the complex have lost their ability to form filaments themselves, the conformational change associated with their ATPase activity is still used to cause allosteric changes required for the protein to function. The nucleotide state determines whether the dimer is in an open or closed state, which regulates its ability to seed new actin filaments (Goley et al., 2004; Dalhaimer et al., 2008) and its affinity for other factors (Ti et al., 2011). It remains to be seen whether there are similar nonpolymerizing proteins within the bacterial family of “actin-like proteins” (Alps) (Derman et al., 2009).
THE FUNCTIONS OF ACTIN-LIKE PROTEINS
Having examined the structure and dynamics of actin and actin-related proteins and polymers, we next explore the conserved roles of the different actin-like polymers: focusing on their roles in the regulation of cell shape, organization, and division across kingdoms.
Bridging long-length scales
One of the primary cellular roles of several different actin-like polymers is to bridge length scales. As both actin and ParM filaments have a persistence length of ∼10 microns (Gittes et al., 1993; Choi et al., 2008), cells beneath this size can use these relatively rigid filaments as a means to establish and/or read out long-range order (Theriot, 2013). To exert spatial control over greater distances, short filaments can be cross-linked together or coupled to stiffer cellular components (e.g., microtubules). In large eukaryotic cells, these actin tracks are used to traffic cellular components over long distances, a transport mediated by processive myosin motors moving along parallel actin filament bundles (Boldogh and Pon, 2006; Massarwa et al., 2009; Schuh, 2011; Rousso et al., 2013). Conversely, at the small length scales found in most prokaryotic cells, cargoes such as plasmids are trafficked to cell poles by associating with the growing filament ends. (Drew and Pogliano, 2011; Gayathri et al., 2012; Polka et al., 2014; Toro-Nahuelpan et al., 2016) This strategy obviates the need for linear stepping motors, molecules that have yet to be discovered in prokaryotes.
In this context, filaments can be used to organize cell space in several ways. When elongating within a confined space with a simple geometry (like a cylinder), long and stiff filaments will orient along the cell’s long axis, thus growing to the ends of the cell (Møller-Jensen et al., 2003; Garner et al., 2007). Likewise, in the filamentous fungus Ashbya gossypii, cell wall material is trafficked to the growing tips of the hypha along bundles of actin filaments (Schmitz et al., 2006), and in budding yeast, actin-mediated transport plays a critical role in moving material from the mother to the emerging bud (Yin et al., 2000; Rousso et al., 2013; Westermann, 2014; Knoblach and Rachubinski, 2015). Thus, actin filaments perform a vital function in cell shape control in both fungi and plant cells by defining the sites of cell wall insertion, while actin-like (e.g., MreB) filaments perform a similar role in generating the form of rod-shaped bacteria (see below) (Dye et al., 2005).
Interestingly, because animal cells have a flexible form, actin performs a similar role in the regulation of animal cell shape in a different way. Here actin filaments are often cross-linked to generate higher-order networks, whose material properties can be precisely tuned to set the physical properties of cells and tissues. Thus, in animal cells, the actin meshwork that underlies the plasma membrane provides the plasma membrane with vital structural support and with a capacity to undergo dynamic changes in shape, for example, to form protrusions, retraction fibers, and phagocytic cups (Clerc and Sansonetti, 1987; Cramer and Mitchson, 1993; Nemethova et al., 2008). Similarly, actin helps to define the material properties of both the nucleoplasm (Grosse and Vartiainen, 2013) and the cytoplasm (Gordon et al., 1976).
Force generation through polymerization dynamics
By leveraging the release of free energy associated with ATP hydrolysis, cells can use the polymerization of filaments to exert force. In the context of actin networks, force production has been posited to occur in a variety of ways (Dmitrieff and Nédélec, 2016). One of the simplest mechanisms is the Brownian ratchet (Theriot, 2000), where ATP hydrolysis within filaments holds the system far from equilibrium, and thus constantly polymerizing filament ends iteratively rectify fluctuations in the position of a load. Similarly, in EcParM, insertional polymerization at ParM filament ends causes directional plasmid movement (Garner et al., 2007). It has also been proposed that cells can use the depolymerization of actin filaments to generate pulling forces (Jégou et al., 2013). Moreover, in the presence of multivalent motors, like myosin II, which can bind multiple actin filaments at the same time, the forces exerted by the motors can cause an actin network to contract or expand (Huxley and Niedergerke, 1954). However, because individual filaments tend to buckle under compressive forces, this break in the symmetry usually ensures that cross-linked actin networks contract under the influence of myosin-based motors (Lenz, 2014).
Finally, actin-related filaments can also function to sense local curvature, for example, through their association with membranes. The best example of this is MreB. MreB filaments are highly curved (Salje et al., 2011; van den Ent et al., 2014). This may enable MreB filaments to read out the differences in the local curvature of the bacterial membrane, causing them to align along the circumference of the cell, perpendicular to the straight, long cell axis. This orientation would guide the associated cell wall synthetic machinery so that new cell wall material is laid down in hoops, ensuring that bacteria retain their rod shape against the internal pressure (Chang and Huang, 2014). MreB filaments have also been shown to show a small, but significant increased localization at points of negative Gaussian curvature (Ursell et al., 2014). Interestingly, an archaeal actin, crenactin, is present in rod-shaped crenarchaea, but is absent from many more pleomorphic archaea (which can change their shape in response to environmental cues), suggesting that it could perform a similar function in guiding archaeal cell shape (Ettema et al., 2011), even though the shape of these cells is supported by an external S-layer rather than a cell wall.
Cell division
Cell division is one of the most basic cellular processes to rely on force production and on long-range intracellular trafficking. Actin, along with myosin II, is an essential component of the cytokinetic ring in eukaryotes (Wong et al., 2002; Miller, 2011). The contractile forces generated by myosin II within the contractile ring cause actin filaments to slide past one another (Miller, 2011). At a larger scale, similar rings, encompassing many cells, provide the contractile forces required for wound healing and tissue morphogenesis (Jacinto and Baum, 2003). Importantly, in these cases, the tension generated around the ring can function to drive changes in cell/tissue shape and/or to provide a template with a simple uniform geometry that can then be used to guide and organize external processes such as cell wall synthesis in yeast or dorsal closure in fly embryos (Jacinto et al., 2002).
Interestingly, bacterial actins are also used in prokaryotic cell division. In this case, actin-related proteins aid division without the aid of a myosin-like motor. The bacterial actin FtsA polymerizes along with the tubulin homologue FtsZ at the septum of dividing cells (Ma et al., 1996). Both FtsA and FtsZ treadmill around the division plane (Loose and Mitchison, 2014; Bisson-Filho et al., 2017), and treadmilling controls the location and activity of the associated cell wall synthetic enzymes. It is unclear whether FtsA/FtsZ treadmilling is responsible for generating the forces for cell division, or whether the filaments themselves bend the membrane. Alternatively, the composite FtsA/FtsZ ring may simply act as a scaffold guiding the movement of enzymes that insert septal cell wall material (Egan and Vollmer, 2015)—as has been proposed for actomyosin rings in eukaryotes with a cell wall (Otegui and Staehelin, 2000; Chang, 2017), where actin filament turnover likely contributes to ring homeostasis (Chew et al., 2017).
During cell division, actin-related proteins also drive the segregation of large cellular objects (Jiang et al., 2016a). For example, MamK is responsible for distributing magnetosomes along the long cell axis (Komeili et al., 2006) and partitioning them into daughter cells (Toro-Nahuelpan et al., 2016), while bidirectionally growing ParM filament bundles segregate low-copy plasmids to ensure each daughter cell inherits a copy. Similar end-finding strategies have been observed for other actin-related plasmid segregators, each displaying subtle variation in its filament kinetics and regulation (Derman et al., 2009; Polka et al., 2009). Thus actin-related proteins are an essential component of the cell division machinery across kingdoms (Balasubramanian et al., 2012).
Macromolecular scaffolds
Finally, actin and actin-related filaments can serve as scaffolds for larger complexes. An example of this is the dynactin complex. This complex links vesicles to the motor dynein and contains short filaments of actin and ARP1 (Urnavicius et al., 2015). This filament helps to orient and adapt various organelles for microtubule-mediated transport (Gill et al., 1991; Plamann et al., 1994). The polarity of this actin-like filament ensures that dynein engages with its cargo in the proper orientation. This type of role for short filaments may be more widespread, as actin and ARPs have recently been suggested to play a similar function in the regulation of eukaryotic transcription (de Lanerolle, 2012).
THE EVOLUTION OF ACTIN-FAMILY PROTEINS
Structural similarity and shared ancestry among actin-like proteins
To what extent do the related structure and function of prokaryotic and eukaryotic actins reflect the action of selection, and to what extent do they reflect the constraints imposed by common ancestry? To answer this, we next take a look at the evolution of the actin-related proteins.
Because primary sequence evolves faster than tertiary structure (Illergård et al., 2009), structural similarities can preserve evidence of common evolutionary ancestry even after sequences have diverged beyond recognition. At the same time, proteins with different evolutionary origins may converge on (that is, evolve toward) similar structures in response to selection for a particular function. Given the diversity of functions performed by actin-like proteins across the tree of life, it therefore seems reasonable to ask whether their shared fold is the result of convergence or common ancestry.
Here, to provide a formal test of the common ancestry of all protein families sharing the actin fold, we built hidden Markov models (HMMs) from sequence alignments of each family and aligned these profiles using the hhalign program in the HH-suite package (Remmert et al., 2011). These comparisons are more sensitive than a standard BLAST search because they incorporate information about the evolutionary conservation of residues within each protein family. Because they are still based on comparisons of primary sequences, they also provide a way to test hypotheses of common ancestry or convergence. This is done by determining whether the levels of sequence identity between two families are better than those expected by chance, giving evidence for descent from a common ancestor or of a common structural constraint. The results of this analysis (Supplemental Figure S2) provide strong evidence of significant sequence similarity, and therefore common ancestry, among many of the actin-like protein families. This is consistent with their structural similarities and suggests that filament-forming and monomeric actin-like families share a common ancestor. Note that a lack of significant similarity does not rule out an evolutionary relationship between two families, particularly in light of the structural similarities among all these families.
Evolution of polymerization in the actin superfamily
Having established a likely common ancestry for all known actin-like proteins, we next inferred a phylogenetic tree to determine the relationships among the subfamilies and to assess the number of times that polymerization has evolved. A prerequisite for this phylogenetic analysis is sufficient similarity among sequences to permit alignment. Thus we limited our tree inference to the set of actin-like families for which all pairs show significant pairwise sequence similarity (with an E-value cutoff of < 10−5): a set that includes the polymers actin, MreB, FtsA, ParM, and PilM; and the enzymes DnaK, type II secretion protein L, diol dehydratase reactivase, peptidase M22, BcrAD-BadFG, glutamate mutase, 2-hydroxyglutaryl-CoA-reductase, EutA, FGGY carbohydrate kinase, AnmK, GDA1-CD39, and PPX GPPA phosphatase (Supplemental Tables S1 and S2). HMMs built from representative sequences for each of these subfamilies were aligned against one another using the E-INS-i mode in mafft (Katoh and Standley, 2013), and poorly aligning positions were identified and removed using trimAl (Capella-Gutiérrez et al., 2009). Bootstrapped maximum-likelihood phylogenies were inferred under the LG+C20+G+F model in IQ-Tree (Nguyen et al., 2015), providing a model that accommodates variation in exchange rates among amino acids (LG) (Le and Gascuel, 2008; Quang et al., 2008), variation in equilibrium composition frequencies across sites (C20) (Quang et al., 2008), and variation in evolutionary rates across sites (G, discrete gamma distribution) (Yang, 1994).
The maximum-likelihood phylogeny (Figure 2) resolves the filament-forming actin families into a single clan (Wilkinson et al., 2007) or lineage on the tree, so the split occurring between polymers and monomers receives good (93) bootstrap support. This result strongly suggests that polymerization evolved once during the evolution of actins and that all filament-forming actins are descended from an ancestral polymer-forming, actin-like protein.
FIGURE 2:
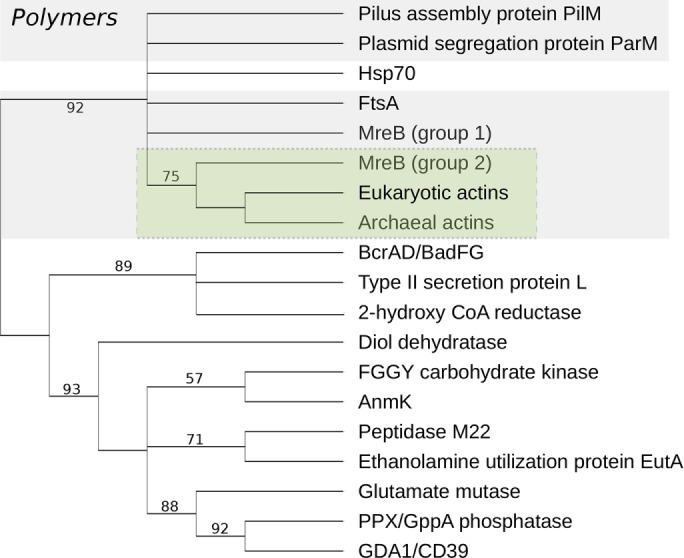
Evolutionary relationships among members of the actin superfamily. The phylogeny was inferred under the LG+C20+G+F model in IQ-Tree (Nguyen et al., 2015), and branch supports are maximum-likelihood bootstrap values. All polymer-forming actins cluster together in the tree, suggesting that the capacity to form filaments arose once during the evolution of the actin fold. Interestingly, the molecular chaperone DnaK/Hsp70 falls within the polymer lineage, suggesting that it may have evolved from an ancestral polymer-forming actin by loss of polymerization. The actin-like proteins of Crenarchaeota and the Asgard archaea (indicated as “Archaeal actins”) are the closest prokaryotic relatives of bona fide eukaryotic actins, consistent with a close relationship between the Asgard superphylum and the archaeal host cell for the mitochondrial endosymbiont (Braun et al., 2015; Spang et al., 2015; Zaremba-Niedzwiedzka et al., 2017). The archaeal actins and the eukaryotic actins together form a lineage that is most closely related to the cell shape–determining protein MreB, found in rod-shaped bacteria. Our phylogenetic analysis suggest that actins are nested within the diversity of MreB proteins (green box), although statistical support for the specific relationship is low (Supplemental Figure S1). We have depicted the actin tree as unrooted: the divergences between superfamily members are ancient, with some likely occurring before the time of the last universal common ancestor. It nonetheless seems reasonable to suppose that the polymer-forming actins evolved from an ancestral monomer, suggesting that the root may lie somewhere among the monomeric actins. Among modern actin-like proteins, only two proteins, both monomeric—benzoyl-CoA reductase (BcrAD/BadFG) and hydantoinase (not depicted in this tree, due to high levels of sequence divergence)—are broadly distributed in bacteria, archaea, and among some eukaryotic lineages and may represent good candidates for the oldest extant members of the superfamily; both also perform functions that may have been important during the evolution of early life. A complete version of this schematic tree is available in the Supplemental Material (Supplemental Figure S1).
Given the lack of an obvious outgroup, it is impossible to unambiguously root the tree of actins and actin-like proteins. It nonetheless seems reasonable to assume that polymers would evolve from monomers. Interestingly, however, the monomeric actins do not form a single lineage in our tree, rather, the Hsp70/DnaK family of molecular chaperones is found nested within the polymer clade, separated from the other monomeric actins with good bootstrap support. Although our unrooted tree does not enable us to exclude the possibility that the root of the entire actin family lies within one of these two groups, the high level of sequence similarity between Hsp70/DnaK and the filament-forming actins (Supplemental Figure S2) is, in our view, more readily explained by an origin of this molecular chaperone via a secondary loss of polymerization. Consistent with this view, it is interesting to note that, while none of the actin family polymers can be confidently mapped to the last universal common ancestor, some of the monomeric forms—including benzoyl-CoA reductase and hydantoinase—are broadly distributed in modern bacteria, archaea, and some eukaryotes (Finn et al., 2016) (Supplemental Table S1), indicating the radiation of the monomeric actins may predate the deepest splits in the tree of life.
The prokaryotic origins of eukaryotic-type actin
Given that eukaryotes appear to have arisen from the symbiosis of cells from both bacterial and archaeal lineages, both of which contain actin-like proteins, the origin of eukaryotic actin has long remained a question in the field. Fortunately, this mystery now appears to have been resolved. Recent discoveries have identified close homologues of the eukaryotic-type actins in the genomes of members of the TACK and “Asgard” superphyla (Braun et al., 2015; Spang et al., 2015; Zaremba-Niedzwiedzka et al., 2017), the lineages of archaea that appear to be most closely related to eukaryotes. Furthermore, functional studies of “crenactin” filaments from Pyrobaculum indicate that they are nearly identical to the filaments formed by eukaryotic actin (Izoré et al., 2014; Braun et al., 2015), and at least one gelsolin-like protein (arcadin-2) seems to act on crenactin polymers in a manner similar to the way in which gelsolin acts on eukaryotic actin (Izoré et al., 2016). While functional studies have not yet been performed on Asgard actins, both crenactins and Asgard actins cluster with the actins of canonical eukaryotes in our phylogenetic analysis (Figure 2). Thus direct evolutionary precursors of eukaryotic actin appear to have first evolved in the archaeal ancestors of eukaryotes. It is also interesting to note here that the metagenomic assembly of Lokiarchaeum, the first member of the Asgard group to be discovered, also encodes potential homologues of the actin modulators profilin and gelsolin. If these proteins prove to modulate the polymer kinetics of a Lokiarchaeal actin in vitro, it would suggest that at least some of the functions and regulatory mechanisms of eukaryotic actins evolved before the split between eukaryotes and archaea. While the cellular function of these archaeal actin homologues remains a matter of speculation, this is an exciting time for research into the origin of the eukaryotic cytoskeleton. It is clear that much more functional information is needed to answer the following questions: Do Asgard actins form actin-like filaments? If they do, what are the kinetic properties of archaeal actin polymers? Do the predicted actin modulators present in the Lokiarchaea modulate Lokiarchaeal actin filaments? Does crenactin perform similar cellular functions to that of eukaryotic actin?
CONCLUSIONS AND PERSPECTIVES
In this review, we have examined the similarities in the structure and function of actin-like proteins across the tree of life. Importantly, our evolutionary analysis suggests that the known polymer-forming actin-like proteins from bacteria, archaea, and eukaryotes have all arisen from a single ancestral polymer-forming protein. This makes actin and its relatives an ancient protein (Doolittle, 1995). Importantly, much of the structural variation that appears to underpin changes in the biological function of actin-related proteins across domains is based on changes in lateral contacts between protofilaments (van den Ent et al., 2014), yielding changes in twist, packing, curvature, and polarity. In addition, it is clear that there are wide differences in the dynamics of polymer formation and disassembly across the clade. More work needs to be done to survey the variations in kinetic properties of different actin-like subfamilies (MreB, actin, MamK, ParM, etc.) to understand how subtle differences in the context of a similar fold can lead to dramatically different behaviors. For the one well-studied case of ParM, a weakening of the cross-protofilament contacts occurs when filaments are ADP bound (Bharat et al., 2015). In addition, our analysis raises the question of whether Hsp70/DnaK can form polymers or has lost that capability during evolution.
It seems remarkable that so many of the cytomotive filaments used to generate intracellular forces, to order cellular organization, and to drive cell division across living systems are based on this single scaffold. This reflects the role of evolution as tinkerer (Jacob, 1977). It may also point to the special properties of the actin fold within the polymer-forming enzymes. Actin couples ATP hydrolysis and phosphate release to large-scale changes in conformation, altering monomer–monomer contacts in the context of a filament, giving rise to polymer dynamics and the capacity to do work. Of course, there are instances in which biological polymers appear to have evolved from nonpolymerizing proteins. These include CTP synthetase, sickle-cell hemoglobin, and MSP from C. elegans (Edelstein et al., 1973; Italiano et al., 2001; Ingerson-Mahar et al., 2010). However, sickle-cell hemoglobin forms long, static, polar filaments that distort cells, and CTP synthase and MSP form nonpolar filaments. CTP synthase polymerization appears to inhibit the enzyme activity in most cases, and MSP polymerization is not directly coupled to its own catalytic activity. As a result, its directed polymerization must be driven by the action of an external pH gradient. These polymers appear to lack the functional flexibility of actin.
In sum, actin is a remarkable protein family. Polymer-forming actin appears to be an ancient protein whose ability to generate long-range order and force has been used again and again in evolution to perform many of the tasks that help give cells their dynamic shape and internal organization.
Acknowledgments
We thank Mohan Balasubramanian, Scott Dawson, Gautam Dey, Thijs Ettema, Ann-Christin Lindås, Jan Löwe, Andrew Murray, and Tom Pollard for comments. This work was supported by Wellcome Trust grant 203276/Z/16/Z for B.B. and E.G. and National Institutes of Health grants DP2AI117923-01 for E.G. and F31GM116441 for P.R.S. T.A.W. is supported by a Royal Society University Research Fellowship.
Footnotes
REFERENCES
- Balasubramanian MK, Srinivasan R, Huang Y, Ng KH. Comparing contractile apparatus-driven cytokinesis mechanisms across kingdoms. Cytoskeleton (Hoboken) 2012;69:942–956. doi: 10.1002/cm.21082. [DOI] [PubMed] [Google Scholar]
- Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollman JM, Gitai Z. Large-scale filament formation inhibits the activity of CTP synthetase. Elife. 2014;3:e03638. doi: 10.7554/eLife.03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman E, Muller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JRC, Hutto R, Ozyamak E, Hom N, Hansen J, Draper O, Byrne ME, Keyhani S, Komeili A, Kollman JM. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. 2017;26:93–102. doi: 10.1002/pro.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TAM, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Orlova A, Valegård K, Lindås AC, Schröder GF, Egelman EH. Archaeal actin from a hyperthermophile forms a single-stranded filament. Proc Natl Acad Sci USA. 2015;112:9340–9345. doi: 10.1073/pnas.1509069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D, Korn ED. Evidence for an ATP cap at the ends of actin filaments and its regulation of the F-actin steady state. J Biol Chem. 1984;259:9983–9986. [PubMed] [Google Scholar]
- Chang F. Forces that shape fission yeast cells. Mol Biol Cell. 2017;28:1819–1824. doi: 10.1091/mbc.E16-09-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Huang KC. How and why cells grow as rods. BMC Biol. 2014;12:2102. doi: 10.1186/s12915-014-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew TG, Huang J, Palani S, Sommese R, Kamnev A, Hatano T, Gu Y, Oliferenko S, Sivaramakrishnan S, Balasubramanian MK. Actin turnover maintains actin filament homeostasis during cytokinetic ring contraction. J Cell Biol. 2017 doi: 10.1083/jcb.201701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CL, Claridge SA, Garner EC, Alivisatos AP, Mullins RD. Protein-nanocrystal conjugates support a single filament polymerization model in R1 plasmid segregation. J Biol Chem. 2008;283:28081–28086. doi: 10.1074/jbc.M803833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Sansonetti PJ. Entry of Shigella flexneri into HeLa cells: evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55:2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L, Mitchison TJ. Moving and stationary actin filaments are involved in spreading of postmitotic PtK2 cells. J Cell Biol. 1993;122:833–843. doi: 10.1083/jcb.122.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhaimer P, Pollard TD, Nolen BJ. Nucleotide-mediated conformational changes of monomeric actin and Arp3 studied by molecular dynamics simulations. J Mol Biol. 2008;376:166–183. doi: 10.1016/j.jmb.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle P. Nuclear actin and myosins at a glance. J Cell Sci. 2012;125:4945–4949. doi: 10.1242/jcs.099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieff S, Nédélec F. Amplification of actin polymerization forces. J Cell Biol. 2016;212:763–766. doi: 10.1083/jcb.201512019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF. The origins and evolution of eukaryotic proteins. Philos Trans R Soc Lond B Biol Sci. 1995;349:235–240. doi: 10.1098/rstb.1995.0107. [DOI] [PubMed] [Google Scholar]
- Drew KRP, Pogliano J. Dynamic instability-driven centering/segregating mechanism in bacteria. Proc Natl Acad Sci USA. 2011;108:11075–11080. doi: 10.1073/pnas.1018724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein SJ, Telford JN, Crepeau RH. Structure of fibers of sickle cell hemoglobin. Proc Natl Acad Sci USA. 1973;70:1104–1107. doi: 10.1073/pnas.70.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Vollmer W. The stoichiometric divisome: a hypothesis. Front Microbiol. 2015;6:455. doi: 10.3389/fmicb.2015.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJG, Lindås A-C, Bernander R. An actin-based cytoskeleton in archaea. Mol Microbiol. 2011;80:1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchel AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri P, Fujii T, Møller-Jensen J, van den Ent F, Namba K, Löwe J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338:1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Eisenberg E, Korn ED. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J Biol Chem. 1976;251:4778–4786. [PubMed] [Google Scholar]
- Grosse R, Vartiainen MK. To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol. 2013;14:693–697. doi: 10.1038/nrm3681. [DOI] [PubMed] [Google Scholar]
- Halliburton WD. On muscle-plasma. J Physiol (Lond.) 1887;8:133–202. doi: 10.1113/jphysiol.1887.sp000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Huxley HE. Structural basis of the cross-striations in muscle. Nature. 1953;172:530–532. doi: 10.1038/172530b0. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier M-F. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell. 2011;43:464–477. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Illergård K, Ardell DH, Elofsson A. Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins. 2009;77:499–508. doi: 10.1002/prot.22458. [DOI] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano JE, Stewart M, Roberts TM. How the assembly dynamics of the nematode major sperm protein generate amoeboid cell motility. Int Rev Cytol. 2001;202:1–34. doi: 10.1016/s0074-7696(01)02002-2. [DOI] [PubMed] [Google Scholar]
- Izoré T, Duman R, Kureisaite-Ciziene D, Löwe J. Crenactin from Pyrobaculum calidifontis is closely related to actin in structure and forms steep helical filaments. FEBS Lett. 2014;588:776–782. doi: 10.1016/j.febslet.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izoré T, Kureisaite-Ciziene D, McLaughlin SH, Löwe J. Crenactin forms actin-like double helical filaments regulated by arcadin-2. Elife. 2016;5:213. doi: 10.7554/eLife.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Baum B. Actin in development. Mech Dev. 2003;120:1337–1349. doi: 10.1016/j.mod.2003.06.006. [DOI] [PubMed] [Google Scholar]
- Jacinto A, Wood W, Woolner S, Hiley C, Turner L, Wilson C, Martinez-Arias A, Martin P. Dynamic analysis of actin cable function during Drosophila dorsal closure. Curr Biol. 2002;12:1245–1250. doi: 10.1016/s0960-9822(02)00955-7. [DOI] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. Mechanism of actin filament bundling by fascin. J Biol Chem. 2011;286:30087–30096. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou A, Carlier M-F, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- Jiang S, Ghoshdastider U, Narita A, Popp D, Robinson RC. Structural complexity of filaments formed from the actin and tubulin folds. Commun Integr Biol. 2016a;9:e1242538. doi: 10.1080/19420889.2016.1242538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Narita A, Popp D, Ghoshdastider U, Lee LJ, Srinivasan R, Balasubramanian MK, Oda T, Koh F, Larsson M, Robinson RC. Novel actin filaments from Bacillus thuringiensis form nanotubules for plasmid DNA segregation. Proc Natl Acad Sci USA. 2016b;113:E1200–E1205. doi: 10.1073/pnas.1600129113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D. Three-dimensional structure of the complex of actin and DNase I at 4.5 A resolution. EMBO J. 1985;4:2113–2118. doi: 10.1002/j.1460-2075.1985.tb03900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW. Implications of treadmilling for the stability and polarity of actin and tubulin polymers in vivo. J Cell Biol. 1980;86:330–334. doi: 10.1083/jcb.86.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA. Sharing the cell’s bounty—organelle inheritance in yeast. J Cell Sci. 2015;128:621–630. doi: 10.1242/jcs.151423. [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Korn E, Carlier M, Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Lal AA, Korn ED, Brenner SL. Rate constants for actin polymerization in ATP determined using cross-linked actin trimers as nuclei. J Biol Chem. 1984;259:8794–8800. [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lenz M. Geometrical origins of contractility in disordered actomyosin networks. Phys Rev X. 2014;4:041002. [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, He S, Scheres SHW, Savva CG. X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization. Proc Natl Acad Sci USA. 2016;113:13396–13401. doi: 10.1073/pnas.1612034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch E, Kollman J. The structural basis of enzyme regulation by CTP synthase metabolic filaments. Biophys J. 2016;110:26a–33. [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Schejter ED, Shilo B-Z. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Dev Cell. 2009;16:877–888. doi: 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics. 1996;142:587–602. doi: 10.1093/genetics/142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL. The contractile ring. Curr Biol. 2011;21:R976–R978. doi: 10.1016/j.cub.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Huang J, Balasubramanian MK. The yeast actin cytoskeleton. FEMS Microbiol Rev. 2014;38:213–227. doi: 10.1111/1574-6976.12064. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Møller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- Møller-Jensen J, Jensen RB, Löwe J, Gerdes K. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Yasunaga T, Noguchi TQP, Gomibuchi Y, Ngo KX, Uyeda TQP, Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Nemethova M, Auinger S, Small JV. Building the actin cytoskeleton: filopodia contribute to the construction of contractile bundles in the lamella. J Cell Biol. 2008;180:1233–1244. doi: 10.1083/jcb.200709134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A, Egelman EH. Structural basis for the destabilization of F-actin by phosphate release following ATP hydrolysis. J Mol Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Cytokinesis in flowering plants: more than one way to divide a cell. Curr Opin Plant Biol. 2000;3:493–502. doi: 10.1016/s1369-5266(00)00119-9. [DOI] [PubMed] [Google Scholar]
- Ozyamak E, Kollman JM, Agard DA, Komeili A. The bacterial actin MamK: in vitro assembly behavior and filament architecture. J Biol Chem. 2013a;288:4265–4277. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyamak E, Kollman JM, Komeili A. Bacterial actins and their diversity. Biochemistry. 2013b;52:6928–6939. doi: 10.1021/bi4010792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polka JK, Kollman JM, Mullins RD. Accessory factors promote AlfA-dependent plasmid segregation by regulating filament nucleation, disassembly, and bundling. Proc Natl Acad Sci USA. 2014;111:2176–2181. doi: 10.1073/pnas.1304127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Actin dynamics. J Cell Sci. 2001;114:3–4. doi: 10.1242/jcs.114.1.3. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Shelton E, Weihing RR, Korn ED. Ultrastructural characterization of F-actin isolated from Acanthamoeba castellanii and identification of cytoplasmic filaments as F-actin by reaction with rabbit heavy meromyosin. J Mol Biol. 1970;50:91–97. doi: 10.1016/0022-2836(70)90106-3. [DOI] [PubMed] [Google Scholar]
- Quang LS, Gascuel O, Lartillot N. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics. 2008;24:2317–2323. doi: 10.1093/bioinformatics/btn445. [DOI] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Rousso T, Shewan AM, Mostov KE, Schejter ED, Shilo B-Z. Apical targeting of the formin Diaphanous in Drosophila tubular epithelia. Elife. 2013;2:e00666. doi: 10.7554/eLife.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H-P, Kaufmann A, Köhli M, Laissue PP, Philippsen P. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol Biol Cell. 2006;17:130–145. doi: 10.1091/mbc.E05-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M. An actin-dependent mechanism for long-range vesicle transport. Nat Cell Biol. 2011;13:1431–1436. doi: 10.1038/ncb2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q-J, Kassim H, Huang Y, Li H, Zhang J, Li G, Wang PY, Yan J, Ye F, Liu J-L. Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J Genet Genomics. 2016;43:393–404. doi: 10.1016/j.jgg.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skillman KM, Ma CI, Fremont DH, Diraviyam K, Cooper JA, Sept D, Sibley LD. The unusual dynamics of parasite actin result from isodesmic polymerization. Nat Commun. 2013;4:2285. doi: 10.1038/ncomms3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Anderson CM, Stenkamp RE. Sequencing a protein by x-ray crystallography. II. refinement of yeast Hexokinase B co-ordinates and sequence at 2.1 angstroms resolution. J Mol Biol. 1978;123:15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- Straub FB. Actin. Stud Inst Med Chem Univ Szeged. 1942;2:3–15. [Google Scholar]
- Szwedziak P, Wang Q, Freund SMV, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki H, Bengtsson E, Månsson A. Persistence length of fascin-cross-linked actin filament bundles in solution and the in vitro motility assay. Biochim Biophys Acta. 2014;1840:1933–1942. doi: 10.1016/j.bbagen.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Theriot JA. The polymerization motor. Traffic. 2000;1:19–28. doi: 10.1034/j.1600-0854.2000.010104.x. [DOI] [PubMed] [Google Scholar]
- Theriot JA. Why are bacteria different from eukaryotes. BMC Biol. 2013;11:119. doi: 10.1186/1741-7007-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S-C, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro-Nahuelpan M, Müller FD, Klumpp S, Plitzko JM, Bramkamp M, Schüler D. Segregation of prokaryotic magnetosomes organelles is driven by treadmilling of a dynamic actin-like MamK filament. BMC Biol. 2016;14:88. doi: 10.1186/s12915-016-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial cell shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahokoski J, Bhargav SP, Desfosses A, Andreadaki M, Kumpula EP, Martinez SM, Ignatev A, Lepper S, Frischknecht F, Sidén-Kiamos I, et al. Structural differences explain diverse functions of Plasmodium actins. PLoS Pathog. 2014;10:e1004091. doi: 10.1371/journal.ppat.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. Elife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial inheritance in yeast. Biochim Biophys Acta. 2014;1837:1039–1046. doi: 10.1016/j.bbabio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, McInerney JO, Hirt RP, Foster PG, Embley TM. Of clades and clans: terms for phylogenetic relationships in unrooted trees. Trends Ecol Evol. 2007;22:114–115. doi: 10.1016/j.tree.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Wong KCY, D’Souza VM, Naqvi NI, Motegi F, Mabuchi I, Balasubramanian MK. Importance of a myosin II-containing progenitor for actomyosin ring assembly in fission yeast. Curr Biol. 2002;12:724–729. doi: 10.1016/s0960-9822(02)00790-x. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]