The actin assembly machinery coordinates a variety of membrane-remodeling processes. This paper shows that an inherited mutation in the WHAMM gene causes severe defects in autophagy and that the WHAMM protein normally binds to PI(3)P and nucleates actin to promote LC3 incorporation during autophagosome biogenesis.
Abstract
Actin nucleation factors function to organize, shape, and move membrane-bound organelles, yet they remain poorly defined in relation to disease. Galloway-Mowat syndrome (GMS) is an inherited disorder characterized by microcephaly and nephrosis resulting from mutations in the WDR73 gene. This core clinical phenotype appears frequently in the Amish, where virtually all affected individuals harbor homozygous founder mutations in WDR73 as well as the closely linked WHAMM gene, which encodes a nucleation factor. Here we show that patient cells with both mutations exhibit cytoskeletal irregularities and severe defects in autophagy. Reintroduction of wild-type WHAMM restored autophagosomal biogenesis to patient cells, while inactivation of WHAMM in healthy cell lines inhibited lipidation of the autophagosomal protein LC3 and clearance of ubiquitinated protein aggregates. Normal WHAMM function involved binding to the phospholipid PI(3)P and promoting actin nucleation at nascent autophagosomes. These results reveal a cytoskeletal pathway controlling autophagosomal remodeling and illustrate several molecular processes that are perturbed in Amish GMS patients.
INTRODUCTION
Actin is an abundant and essential molecule in eukaryotic organisms, in which the activities of nucleator proteins direct the polymerization of actin monomers into filaments during a variety of cellular processes (Pollard and Cooper, 2009). In human cells, branched actin filament networks are nucleated by the Arp2/3 complex (Rotty et al., 2013), which acts in collaboration with nucleation-promoting factors from the Wiskott-Aldrich syndrome protein (WASP) family (Rottner et al., 2010). Each of the 8 WASP-family members is thought to have a distinct membrane remodeling function in cells and to play a key role in development (Campellone and Welch, 2010), although only mutations in WASP itself have been directly associated with human illnesses (Moulding et al., 2013).
WASP promotes actin rearrangements in hematopoetic cells and is so named due to its involvement in Wiskott-Aldrich syndrome and related immune disorders (Derry et al., 1994). Other WASP-family members—N-WASP, WAVE1, WAVE2, WAVE3, WASH, WHAMM, and JMY—are expressed more broadly and perform a variety of cellular functions. As examples, N-WASP facilitates endocytosis and vesicle motility, WAVE1–3 drive protrusions of the plasma membrane during cell migration, and WASH controls endosome shape and trafficking (Campellone and Welch, 2010). Like the WAVE proteins, JMY can be recruited to the plasma membrane and enhances epithelial cell motility (Zuchero et al., 2009), but it also influences other intracellular functions such as transport from the trans-Golgi (Schluter et al., 2014). Finally, WHAMM is found on membranes of the secretory pathway, including the endoplasmic reticulum (ER), cis-Golgi, and ER–Golgi intermediate compartment (ERGIC), where it promotes membrane tubulation and trafficking (Campellone et al., 2008; Blom et al., 2015; Russo et al., 2016). WASP-family proteins appear to be crucial for mammalian development, because targeted disruptions of the mouse genes encoding N-WASP, WAVE2, or WASH are embryonic lethal (Campellone and Welch, 2010; Gomez et al., 2012). Whether the WHAMM and JMY proteins, which are 45% similar to one another, are also essential has yet to be determined.
Recently WHAMM and JMY have been implicated in macroautophagy (autophagy) (Coutts and La Thangue, 2015; Kast et al., 2015). Autophagy is a mechanism of cytoplasmic digestion involving double membrane–bound compartments called autophagosomes that fuse with lysosomes and is crucial in cellular homeostasis, stress responses, and development (Mizushima and Levine, 2010; Yang and Klionsky, 2010; Choi et al., 2013; Carlsson and Simonsen, 2015). Roles for actin and the Arp2/3 complex in autophagy in mammalian cells were obscure until actin and Cortactin, a stabilizer of Arp2/3 branchpoints, were found to influence this process (Lee et al., 2010; Aguilera et al., 2012). Functions for WASP-family members have also emerged. WASH can exert positive or negative effects on autophagy (Xia et al., 2013; Zavodszky et al., 2014), perhaps due to its participation in trafficking multiple types of endosomal cargo. Moreover, WHAMM-mediated Arp2/3 activation can drive autophagosomal rocketing (Kast et al., 2015), while JMY can interact with the autophagosomal protein LC3 and affect its expression and localization (Coutts and La Thangue, 2015). Interestingly, actin assembly was also recently revealed to take place at the interior of forming autophagosomes, and to be dependent on the phospholipid phosphatidylinositol 3-phosphate (PI(3)P; Mi et al., 2015). However, the connections between PI(3)P, actin nucleation factors, and autophagosomal biogenesis, as well as the physiological importance of actin assembly in autophagy, have not come into focus.
In the current study, we investigated the effects of a frameshift deletion in WHAMM that occurs alongside a WDR73 mutation in Amish patients with Galloway-Mowat syndrome (GMS; also called nephrocerebellar syndrome) (Jinks et al., 2015). Both founder mutations are coinherited in a homozygous manner in almost all cases due to their close proximity on chromosome 15. Given that preliminary characterizations of WDR73 have been undertaken (Jinks et al., 2015), the current study aimed to define the outcome of the WHAMM mutation in Amish GMS patient cells and to uncover the mechanisms underlying WHAMM function during autophagy.
RESULTS
Clinical features and genetic basis of Amish GMS
Amish GMS is an autosomal-recessive condition that was initially clinically delineated in 27 individuals (Jinks et al., 2015) and is phenotypically consistent with WDR73-associated GMS that occurs outside the Amish community (Colin et al., 2014; Ben-Omran et al., 2015; Vodopiutz et al., 2015). Shortly after birth, affected Amish children typically presented with irritability and roving eye movements. Psychomotor development was severely delayed. Common associations included progressive microcephaly with cerebellar atrophy, hypotonia, extrapyramidal movements, visual impairment with optic atrophy, multifocal seizures, and steroid-resistant nephrosis. Premature death in childhood was common, most often from complications from renal failure. Genome-wide single nucleotide polymorphism genotyping mapped the responsible locus to a small autozygous region on chromosome 15q25.2-q25.3, and parallel whole-exome sequencing revealed two novel homozygous variants within the region: a 1 base pair deletion (c.888delT; p.Phe296Leufs*26) in the last exon of WDR73 and a 7 base pair deletion (c.1264_1270delATAAAAG) in exon 5 of WHAMM (Jinks et al., 2015). Both variants cosegregated according to autosomal-recessive disease status in 26 affected patients and extended family members, with only one affected patient homozygous for the WDR73 variant and heterozygous for the WHAMM variant. This individual displayed the cardinal neurological features of GMS but died of a nonrenal cause, and no data on kidney involvement were available. This case provides evidence that homozygosity for the Amish WDR73 mutation is primarily responsible for the clinical presentation in this cohort (Jinks et al., 2015). However, the molecular and cellular phenotypes arising from the WHAMM mutation have not been explored.
Cells from Amish GMS patients are deficient in WHAMM expression
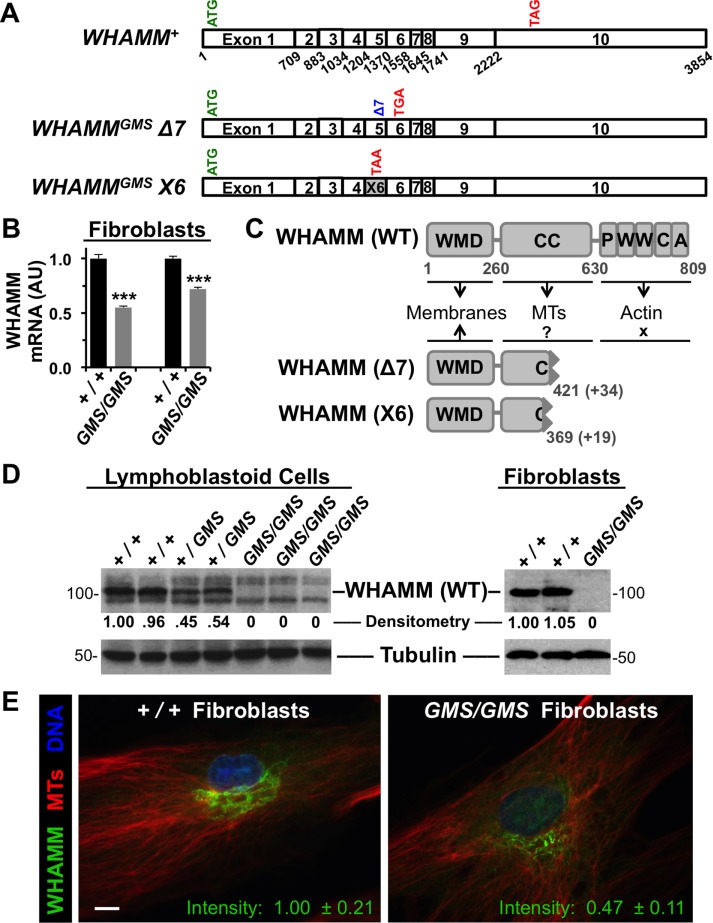
The canonical WHAMM gene is composed of 10 coding exons giving rise to a 3.8 kb transcript (Figure 1A). To examine whether the 7 base pair deletion at the 3′ end of exon 5 alters WHAMM transcript levels, we cultured primary dermal fibroblasts from Amish GMS patients and healthy Amish individuals, isolated RNA from the samples, and performed reverse transcription-PCR (RT-PCR). WHAMM mRNA levels in homozygous GMS/GMS mutant cells were present at 55–70% of the levels in +/+ normal cells (Figure 1B), suggesting that the WHAMMGMS variant encodes a less stable transcript.
FIGURE 1:
Cells from Amish GMS patients encode truncated WHAMM variants. (A) Diagrams of the exon organization in wild-type WHAMM cDNA and in the Δ7 and X6 variants are shown. Start and stop codons are indicated in green and red, respectively. (B) RNA was isolated from normal (+/+) or Amish GMS patient (GMS/GMS) fibroblasts and subjected to RT-PCR with primers to exons 9–10 of WHAMM or to β-actin. Relative levels of WHAMM-specific products (mean + SEM from three to four experiments) are shown. ***, p < 0.001 (t tests). (C) The 809-residue WHAMM(WT) protein includes a WMD that interacts with membranes, a CC region that binds microtubules (MTs), and a C-terminal PWWCA segment that promotes actin nucleation. The GMS Δ7 and X6 variants include the N-terminal 421 or 369 amino acids of WHAMM followed by 34 or 19 additional residues after the respective frameshifts. (D) Lymphoblastoid cell lines and skin fibroblasts from homozygous unaffected (+/+), heterozygous (+/GMS), or homozygous affected (GMS/GMS) individuals were subjected to immunoblotting with anti-WHAMM (left), anti-WWCA (right), and anti-tubulin antibodies. Relative levels of WHAMM(WT) were determined by densitometry of a representative blot. (E) Wild-type (+/+) or GMS patient (GMS/GMS) fibroblasts were stained with antibodies to visualize WHAMM (green) and microtubules (red), and with DAPI to label DNA (blue). The fluorescence intensity of WHAMM staining (mean ± SEM from 10–12 cells per genotype) was lower in patient cells, p < 0.001 (t test). Scale bar: 10 μm.
Given the position of the 7 base pair deletion, it may destabilize WHAMM mRNA by several mechanisms. As examples, a simple frameshift would result in a premature stop codon and possible nonsense-mediated decay, while a defect in splicing might also result in transcript degradation. To explore the effect of the Amish WHAMMGMS mutation on the gene transcript, we used several primer pairs to amplify portions of exons 4–8 using cDNA from Amish +/+ and GMS/GMS fibroblasts (Supplemental Figure S1A). With a plasmid control and +/+ cDNA sample, all primer pairs yielded PCR products corresponding to the predicted length of a WHAMM+ mRNA (Supplemental Figure S1B). With an Amish GMS patient sample, PCR products spanning exons 4–5, 4–7, 5–7, and 6–8 were similar in size to those from the control cells, consistent with a simple 7 base pair deletion (Supplemental Figure S1B). In contrast, a primer pair in exons 4 and 6 produced a smaller-than-expected PCR product specifically with the GMS sample (Supplemental Figure S1B). Sequencing of this product indicated that exon 4 was followed by an alternative exon from a predicted WHAMM RNA variant named X6 (XM_011521233). Incorporation of this cryptic exon also results in a frameshift and premature termination codon. We have thus termed the deletion and alternatively spliced transcripts WHAMMGMSΔ7 and WHAMMGMSX6, respectively (Figure 1A).
Wild-type WHAMM mRNA encodes an 809 amino acid protein consisting of a WHAMM membrane-interaction domain (WMD), a microtubule-binding coiled-coil (CC) region, and a polyproline-WH2-WH2-connector-acidic (PWWCA) segment that promotes actin nucleation in conjunction with the Arp2/3 complex (Figure 1C). In contrast, WHAMMGMSΔ7 and WHAMMGMSX6, if translated, would lead to truncated protein products that possess part of the CC region but none of the PWWCA motifs (Figure 1C and Supplemental Figure S1C). To examine WHAMM protein expression, we generated lymphoblastoid cell lines (LCLs) from control Amish individuals, heterozygous carriers of the WHAMMGMS and WDR73GMS variants, and GMS patients homozygous for both mutations. Consistent with expectations based on gene dosage, immunoblots using antibodies that recognize the C-terminal WWCA domain indicated that LCLs from carriers expressed approximately half the amount of full-length WHAMM compared with LCLs from wild-type individuals (Figure 1D). Moreover, no full-length WHAMM was detected in LCLs or fibroblasts from Amish GMS patients (Figure 1D).
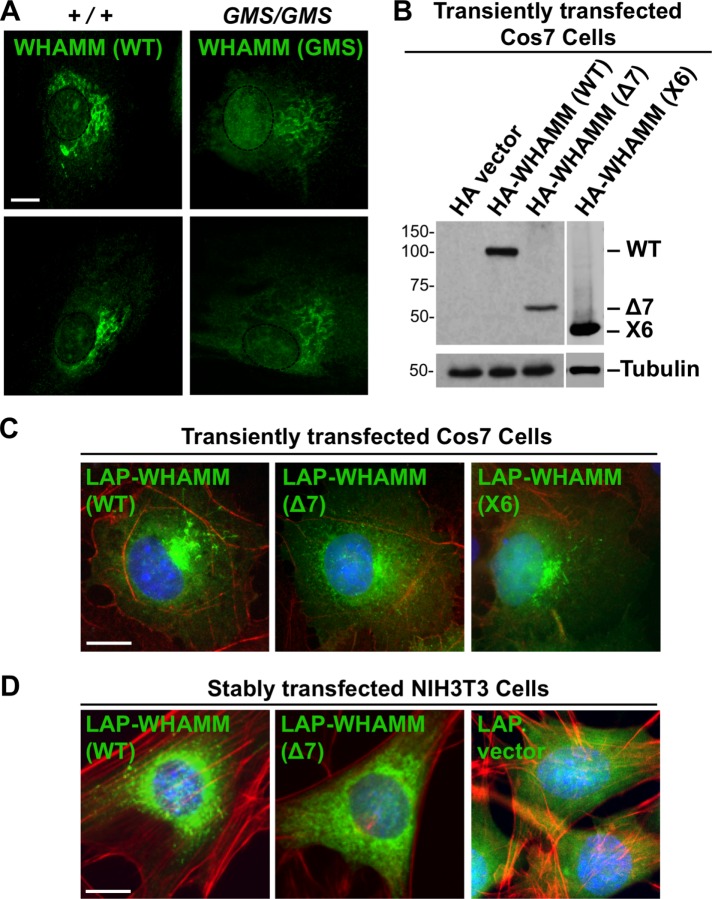
To explore whether truncated WHAMM proteins might be expressed from the WHAMMGMSΔ7 or X6 transcripts, we examined Amish GMS patient fibroblasts by immunofluorescence using antibodies to the WHAMM CC domain. While control cells exhibited a prominent Golgi-like staining pattern as expected, cells from GMS patients stained less intensely in the Golgi region and displayed a more dispersed reticular signal (Figure 1E). Total WHAMM fluorescence in GMS fibroblasts averaged less than half the level found in normal cells (Figure 1E), despite an apparent increase in nuclear fluorescence (Figure 2A). It is not clear whether the residual WHAMM staining is due to expression of the WHAMM(Δ7), WHAMM(X6), and/or other proteins derived from alternative splicing, as any such truncations would retain nearly half of the CC region that is recognized by the fluorescently labeled antibody (Campellone et al., 2008). Nevertheless, HA (hemagglutinin)- and LAP (localization and affinity purification)-tagged versions of WHAMM(Δ7) and/or WHAMM(X6) were capable of transient expression in Cos7 cells and stable expression in NIH3T3 cells following cDNA transfections (Figure 2, B–D). Consistent with the staining pattern of endogenous WHAMM in GMS patient cells, the tagged mutants localized more diffusely than tagged WHAMM(WT) in the transfected cell lines (Figure 2, C and D). Together these data indicate that Amish GMS patient cells lack the full-length WHAMM protein but appear to harbor a residual amount of at least one truncated WHAMM variant.
FIGURE 2:
Truncated WHAMM proteins exhibit altered properties in cells. (A) Fibroblasts from two healthy (+/+) Amish individuals or Amish GMS (GMS/GMS) patients were fixed and stained with anti-CC antibodies to visualize WHAMM (green). Nuclei, based on DAPI staining, were denoted with dashed ovals. (B) Cos7 cells were transfected with plasmids encoding a vector control, HA-WHAMM(WT), (Δ7), or (X6) and blotted with anti-HA or anti-tubulin antibodies. (C) Cos7 cells transiently expressing LAP-WHAMM(WT), (Δ7), or (X6) (green due to the GFP tag) were fixed and stained with phalloidin to visualize F-actin (red) and with DAPI to label DNA (blue). (D) NIH3T3 cells stably transfected with LAP-WHAMM(WT) or (Δ7) or a LAP vector control were fixed and stained as in C. All scale bars: 10 μm.
Amish WHAMM truncations do not promote actin nucleation but associate with microtubules
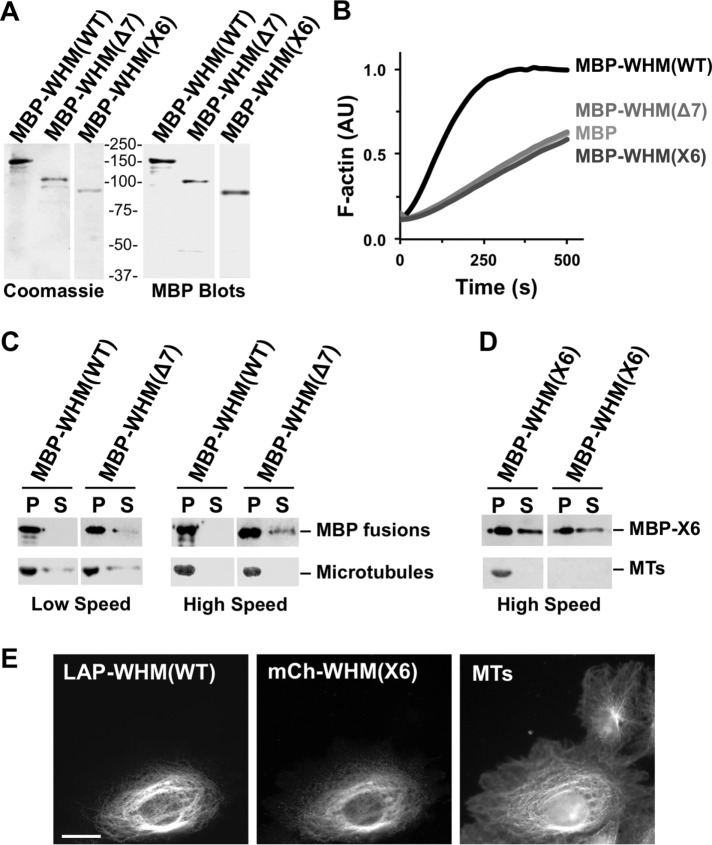
The two best-characterized molecular functions of WHAMM are Arp2/3-dependent actin nucleation via its PWWCA segment (Campellone et al., 2008) and microtubule binding via its CC region (Shen et al., 2012). The WHAMM(Δ7) variant is predicted to contain the first 421 amino acids of WHAMM followed by 34 inappropriate residues after the frameshift, while WHAMM(X6) is predicted to contain the first 369 amino acids of WHAMM plus 19 additional residues (Figure 1C). Therefore both truncated WHAMM derivatives lack roughly half of the microtubule-binding region and are completely devoid of the motifs required for actin nucleation. To determine how cytoskeletal interactions are affected in each WHAMM variant, we purified maltose-binding protein (MBP)-tagged versions of WHAMM(Δ7) and WHAMM(X6) in parallel to MBP-WHAMM(WT) (Figure 3A), and compared their abilities to promote actin assembly and bind microtubules in vitro.
FIGURE 3:
Amish GMS WHAMM truncations do not promote actin nucleation but can associate with microtubules. (A) Purified MBP-WHAMM(WT), (Δ7), and (X6) were analyzed by SDS–PAGE and Coomassie blue staining or anti-MBP immunoblotting. WHAMM is abbreviated WHM. (B) Actin was polymerized in the presence of the Arp2/3 complex and either MBP, MBP-WHAMM(WT), (Δ7), or (X6). The amount of F-actin, in arbitrary fluorescent units (AU), formed over time is shown. (C) Microtubules were incubated in the presence of MBP-WHAMM(WT) or (Δ7), and collected by centrifugation. Proteins found in pellet (P) and supernatant (S) samples were separated by SDS–PAGE. Tubulin was visualized by Ponceau S staining, while WHAMM fusions were detected by anti-MBP blotting. (D) MBP-WHAMM(X6) was incubated in the presence or absence of microtubules and processed as in C. (E) Cos7 cells transiently coexpressing LAP-WHAMM(WT) and mCherry-WHAMM(X6) were fixed and stained with antibodies to visualize microtubules.
For assessing actin assembly, we employed pyrene-actin polymerization assays, which measure the bulk formation of fluorescent actin filaments over time (Campellone et al., 2008). As expected, the full-length MBP-WHAMM(WT) protein accelerated actin polymerization in conjunction with the Arp2/3 complex (Figure 3B). Conversely, neither MBP-WHAMM(Δ7) nor MBP-WHAMM(X6) increased the rate of actin assembly (Figure 3B), confirming that, in the absence of its known C-terminal actin- and Arp2/3-binding motifs, WHAMM does not promote actin nucleation. For testing microtubule binding, we polymerized tubulin in vitro and performed low-speed and high-speed microtubule cosedimentation assays (Shen et al., 2012) with MBP-WHAMM(WT), MBP-WHAMM(Δ7), and MBP-WHAMM(X6). Despite the fact that the Δ7 mutant was missing part of its CC region, it copelleted with microtubules almost as well as wild-type WHAMM (Figure 3C), indicating that this construct retains significant microtubule-binding activity. Unfortunately, we were unable to assess whether the X6 mutant bound to microtubules, as this protein largely pelleted by itself (Figure 3D), suggesting that the purified X6 variant is more prone to self-aggregation than the Δ7 protein in vitro.
Overexpression of wild-type WHAMM at high levels in Cos7 cells is known to cause microtubule bundling (Campellone et al., 2008), so to explore whether the X6 mutant can interact with microtubules in cells, we coexpressed LAP-WHAMM(WT) with mCherry-tagged WHAMM(X6). When coexpressed with wild-type WHAMM, the X6 mutant localized to microtubule bundles (Figure 3E), suggesting that it is capable of associating with microtubules. Thus the two possible WHAMM truncations expressed by Amish GMS patients retain some ability to interact directly or indirectly with microtubules, but both completely lack the actin nucleation-promoting activity that is inherent to full-length WHAMM.
Amish GMS fibroblasts have altered F-actin organization and ERGIC morphology
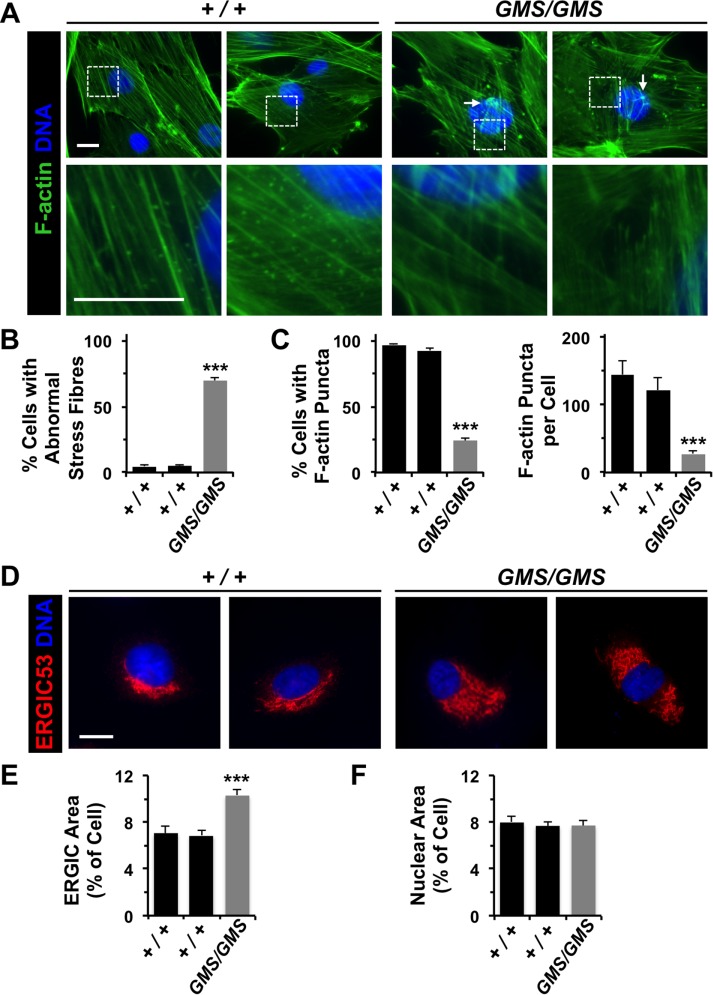
WHAMM is the only WASP-family protein known to function at membranes of the early secretory system, as depletion and overexpression approaches indicate that WHAMM helps localize the cis-Golgi near the centrosome and promotes tubulation of ERGIC-like membranes in an actin- and microtubule-dependent manner (Campellone et al., 2008; Russo et al., 2016). To determine whether patient cells display alterations in cytoskeletal or membrane organization, we stained cultured fibroblasts with probes to visualize microtubules and actin, as well as antibodies to detect organelles of the conventional secretory pathway. While Amish GMS patient fibroblasts exhibited normal-looking microtubule arrays (Figure 1E and Supplemental Figure S2), many contained unusually thick or stellate F-actin bundles (Figure 4, A and B), similar to the changes in stress fiber organization that were previously observed in some cells treated with siRNAs to WHAMM (Gad et al., 2012). GMS fibroblasts also harbored significantly fewer punctate, possibly vesicle-associated, F-actin structures than control cells (Figure 4, A and C). Each of these alterations in actin is consistent with a loss of nucleation activity resulting from the homozygous WHAMMGMS mutation. In accordance with the role of WHAMM in organizing membranes of the intermediate compartment and Golgi (Blom et al., 2015; Campellone et al., 2008), the ERGIC (Figure 4, D and E) and cis-Golgi (Supplemental Figure S2) appeared significantly larger in patient cells when compared with control cells. In contrast, nuclear size was unchanged (Figure 4F), as was ER morphology, ER exit-site distribution, and lysosome organization (Supplemental Figure S2). Collectively these results expand upon previous transient RNAi studies of WHAMM and highlight several actin-associated and organelle-organizing activities that are disrupted in patient fibroblasts.
FIGURE 4:
Amish GMS fibroblasts have altered F-actin organization and ERGIC morphology. (A) Fibroblasts from two healthy (+/+) Amish individuals or Amish GMS (GMS/GMS) patients were stained with phalloidin to visualize F-actin (green) and with DAPI to label DNA (blue). Unusual orthogonal bundles of F-actin fibers and aggregates are highlighted with arrows. A lack of cytoplasmic F-actin puncta in GMS cells is exemplified in the row of magnified images. (B) The percentage of cells with aberrant (orthogonal, aggregated, or stellate) F-actin stress fibers was quantified. Each bar represents the mean (+ SEM) from analyses of 350 cells. ***, p < 0.001 (Fisher’s exact test). (C) The percentage of cells with prominent cytoplasmic F-actin puncta in the juxtanuclear region was quantified. Each bar represents the mean (+ SEM) from analyses of 350 cells. For cells with puncta, the number per cell was also quantified. Each bar represents the mean (+ SEM) from analyses of 12 cells. ***, p < 0.001 (Fisher’s exact test, ANOVA). (D) Wild-type or GMS fibroblasts were stained with antibodies to detect ERGIC-53 (red) and with DAPI to label DNA (blue). (E, F) The areas occupied by the ERGIC, nucleus, and entire cell were calculated in ImageJ and converted to a ratio of organelle area/cell area. Each bar represents the mean (+ SEM) from analyses of 25 cells. ***, p < 0.001 (ANOVA). All scale bars: 10 μm.
Amish GMS cells have defects in autophagosomal biogenesis due to a loss in WHAMM function
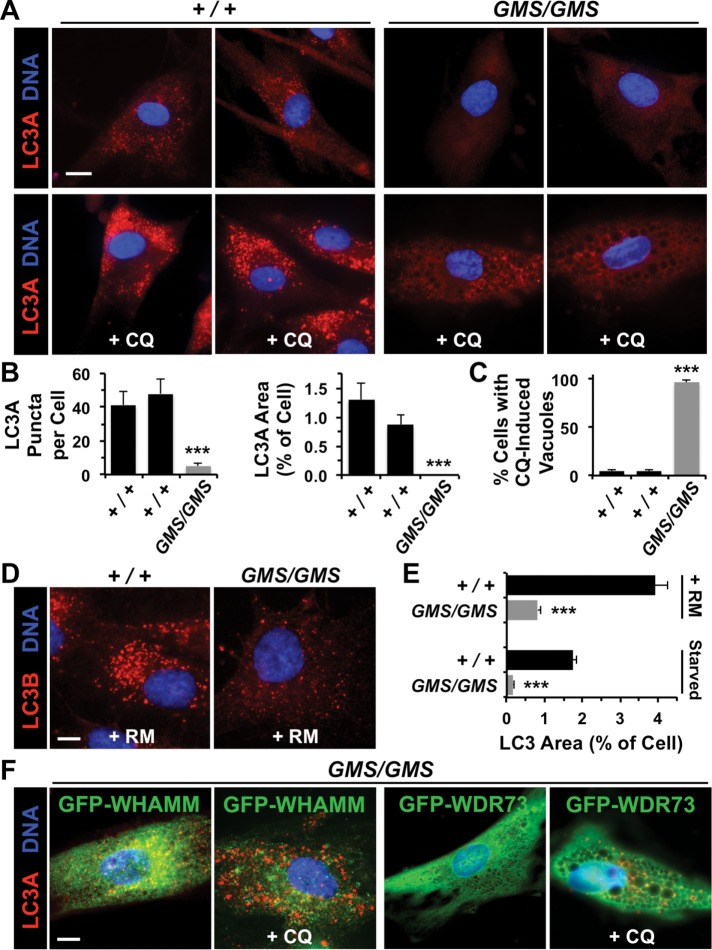
Given the clinical severity of Amish GMS, we investigated whether aspects of membrane remodeling beyond ERGIC morphology were altered in patient fibroblasts. One function requiring dramatic membrane shape changes is autophagy, a catabolic process in which cytoplasmic components are enveloped in double membrane–bound autophagosomes that can fuse with lysosomes, leading to the degradation of autolysosomal contents (Mizushima and Levine, 2010; Yang and Klionsky, 2010; Choi et al., 2013; Carlsson and Simonsen, 2015). While WHAMM was recently shown to localize to autophagosomes and promote their actin-based motility (Kast et al., 2015), the physiological importance of WHAMM in autophagy is not known. We examined patient fibroblasts for autophagosomes by staining the cells with antibodies to LC3, the standard marker of these organelles (Kabeya et al., 2000). Interestingly, while cells from healthy individuals contained LC3-positive preautophagosomal puncta under normal growth conditions, patient fibroblasts were virtually devoid of LC3-associated structures (Figure 5, A and B). To test whether patient cells could form autophagosomes under conditions in which lysosome-mediated degradation was blocked, we treated fibroblasts with chloroquine, a drug that inhibits vesicle acidification and causes autophagosomes to accumulate. While control cells exhibited the expected buildup of LC3-positive autophagosomes under these conditions (Figure 5A), many patient cells lost adherence, with the remaining cells exhibiting a diffuse LC3 staining pattern and a vacuole-filled cytoplasm (Figure 5, A and C). Thus patient fibroblasts display striking defects in autophagosomal biogenesis and trafficking that are exacerbated when lysosome-mediated degradation is inhibited.
FIGURE 5:
Amish GMS fibroblasts exhibit defects in autophagosomal biogenesis due to a loss in WHAMM function. (A) Fibroblasts from two healthy (+/+) Amish individuals or Amish GMS (GMS/GMS) patients were grown in the absence or presence of chloroquine (CQ) for 5 h and stained with antibodies to detect LC3A (red) and with DAPI to label DNA (blue). (B) The number of LC3A puncta per cell, and the relative area within each cell occupied by LC3+ structures was quantified using ImageJ. Each bar represents the mean (+ SEM) from analyses of 12 cells. ***, p < 0.001 (ANOVAs). (C) The percentage of cells with large cytoplasmic vacuoles was quantified. Each bar represents the mean (+ SEM) from analyses of 250 cells. ***, p < 0.001 (Fisher’s exact test). (D) Fibroblasts from a healthy individual or GMS patient were grown in the presence of rapamycin (RM) for 16 h and stained with antibodies to detect LC3B (red) and with DAPI to label DNA (blue). (E) Fibroblasts were treated with rapamycin for 16 h or starved for 90 min and stained as in D. The relative area within each cell occupied by LC3+ structures was quantified as in B. ***, p < 0.001 (t test). (F) Fibroblasts from a GMS patient were transfected with plasmids encoding GFP-WHAMM or GFP-WDR73, grown in the absence or presence of CQ, and stained as in A. Normal LC3 staining was observed in 29/35 GFP-WHAMM-expressing cells and 5/37 GFP-WDR73-expressing cells. All scale bars: 10 μm.
To determine whether patient fibroblasts could form autophagosomes under autophagy-inducing conditions, we treated cells with rapamycin or subjected them to starvation before staining for LC3. While fibroblasts from an Amish GMS patient contained some LC3-positive autophagosomal structures under both types of autophagy-inducing conditions, cells from a healthy individual contained significantly more (Figure 5, D and E). When measured as a fraction of the cytoplasm, LC3 staining was 5- to 10-fold less abundant in patient cells (Figure 5E). On average, LC3 puncta were ∼ 56% smaller in patient cells versus control cells following rapamycin treatment (0.87 μm2, n = 1158 vs. 0.38 μm2, n = 892; p < 0.001). Therefore patient fibroblasts have an autophagosomal biogenesis deficiency even when grown under pharmacological or nutritional conditions that ordinarily induce autophagy.
As Amish GMS patients have mutations in both WHAMM and WDR73, we sought to determine whether reexpression of either wild-type protein could restore autophagosomal formation to fibroblasts. Cells were transfected with plasmids encoding green fluorescent protein (GFP)-tagged WHAMM(WT) or WDR73(WT), and again stained with antibodies to LC3. In contrast to untransfected or GFP-WDR73–expressing cells, GFP-WHAMM–expressing cells harbored many LC3 puncta under normal growth conditions and accumulated autophagosomes after treatment with chloroquine (Figure 5F). These results indicate that WHAMM is sufficient to reestablish autophagosomal biogenesis in Amish GMS patient fibroblasts.
To assess autophagic digestion in patient samples, we used LCLs for measuring the levels of p62/SQSTM1, a protein that links polyubiquitinated proteins to LC3 in autophagosomes, and whose degradation serves as an indicator of autophagy (Klionsky et al., 2016). By immunoblotting LCL extracts for p62 and quantifying its abundance relative to tubulin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) loading controls (Figure 6A), we found that p62 levels were generally 50% higher in Amish GMS patient cells compared with cells from individuals with full-length WHAMM (Figure 6B). Notably, although LCLs express the wild-type WHAMM protein in a gene dose–dependent manner (Figure 1D), we have been unable to detect WDR73 by immunoblotting in any of the LCL extracts, implying that these immortalized B-cells express little, if any, WDR73 protein. Thus the accumulation of p62 is consistent with the idea that autophagic degradation is inefficient in GMS patient LCLs primarily because of the homozygous WHAMM mutation.
FIGURE 6:
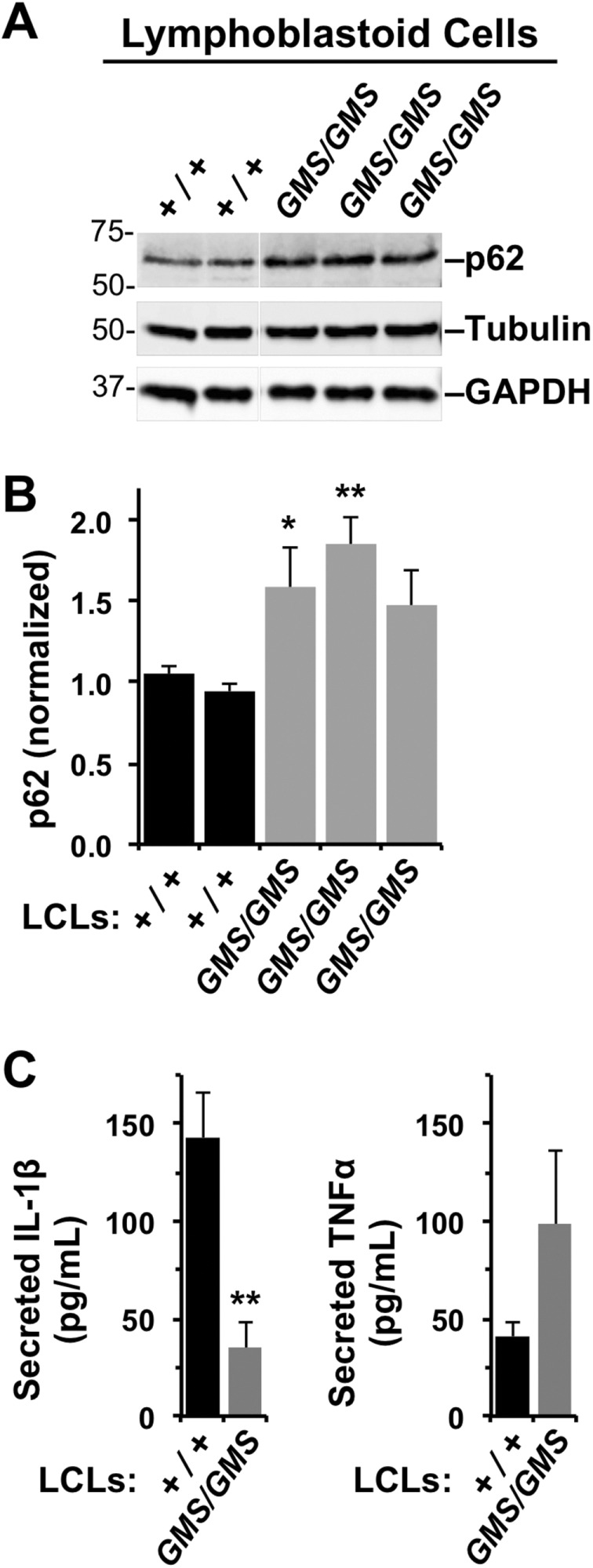
Lymphoblastoid cell lines from Amish GMS patients exhibit defects in autophagy-related processes. (A) LCLs from two healthy (+/+) Amish individuals or three Amish GMS (GMS/GMS) patients were cultured under normal growth conditions, collected, and subjected to immunoblotting with anti-p62/SQSTM1, anti-tubulin, and anti-GAPDH antibodies. (B) p62 band intensities were quantified relative to tubulin. Each bar represents the mean (+ SD) from analyses of three experiments. *, p < 0.05; **, p < 0.01 (ANOVA). (C) LCLs from several healthy (+/+) or Amish GMS (GMS/GMS) patients were stimulated with E. coli, and the levels of IL-1β and TNFα in culture supernatants were measured by ELISA. Each bar represents the mean (+ SEM) from four to six samples across two to three experiments. Data represent two to three patients per genotype. **, p < 0.01 (ANOVA).
As an independent measure of an autophagy-related process in LCLs, we examined unconventional secretion of interleukin-1β (IL-1β). This cytokine is expressed in immune cells in response to microbial products, and autophagosomal structures facilitate its export (Dupont et al., 2011; Zhang et al., 2015). For inducing IL-1β production, LCLs from Amish individuals with wild-type WHAMM and patients with GMS were treated with Escherichia coli, and the amount of IL-1β released into culture supernatants was measured by enzyme-linked immunosorbent assay (ELISA). Compared with normal LCLs, LCLs from GMS patients secreted less IL-1β (Figure 6C). In contrast, levels of tumor necrosis factor α (TNFα), a cytokine exported via the conventional secretory apparatus, did not differ significantly (Figure 6C). Hence Amish GMS patients possess fibroblasts with defects in autophagosomal biogenesis and immune cells with impaired autophagic functions.
Depletion or inactivation of WHAMM inhibits LC3 lipidation and clearance of protein aggregates
Given the autophagy-related defects in cells of Amish GMS patients, we next examined whether WHAMM and/or WDR73 might play a key role in autophagy in other cell lines. HeLa cells, when treated with a pool of siRNAs to WHAMM, were recently shown to contain smaller autophagosomes that lack actin tails (Kast et al., 2015). We also exposed HeLa cells to siRNAs for depleting WHAMM or WDR73 (Supplemental Figure S3, A and B). After starving the cells to induce autophagy and stalling flux with chloroquine, we assessed autophagosomal biogenesis by immunoblotting cell extracts for LC3 and measuring its conversion from a slower-migrating immature form (LC3-I) to a faster-migrating lipidated form (LC3-II) that associates with autophagosomal membranes and correlates with autophagosomal abundance (Klionsky et al., 2016). Comparisons of cells grown in normal media versus buffer plus chloroquine revealed that LC3-II levels increased in control and WDR73-depleted cells but not in WHAMM-depleted cells (Supplemental Figure S3, C and D).
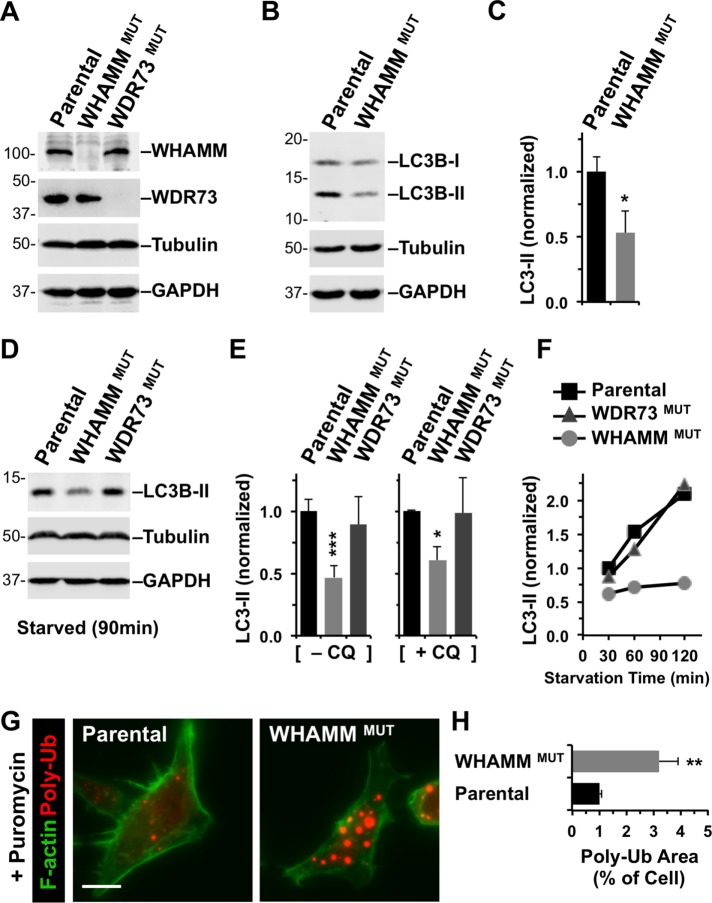
To further explore the role of WHAMM and WDR73 in autophagy using a genetically stable cellular system, we used clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated genome editing for creating WHAMM- or WDR73-deficient cells. To generate human cell lines that lack WHAMM or WDR73 function, we made derivatives of eHAP cells, a fully haploid fibroblast-like cell line (Essletzbichler et al., 2014). Frameshift mutations were introduced into exon 2 of WHAMM or exon 1 of WDR73, and immunoblotting indicated that expression of full-length WHAMM and WDR73 was lost in the respective WHAMMMUT and WDR73MUT cell lines (Figure 7A). Consistent with a deficiency in basal autophagy under standard growth conditions, the WHAMMMUT cell line displayed approximately half the amount of LC3-II as the parental cell line (Figure 7, B and C).
FIGURE 7:
Targeted inactivation of WHAMM inhibits lipidation of LC3 and clearance of protein aggregates. (A) Parental eHAP cells or cells with mutations engineered in WHAMM or WDR73 were analyzed by immunoblotting for WHAMM, WDR73, tubulin, and GAPDH. (B) eHAP and WHAMMMUT cells were analyzed by immunoblotting for LC3B, tubulin, and GAPDH. LC3B was detected in its immature (LC3-I) and mature lipidated (LC3-II) forms. (C) LC3B-II band intensities were quantified relative to tubulin. Each bar represents the mean (+ SD) from analyses of three experiments. *, p < 0.05 (t test). (D, E) eHAP cell derivatives were starved for 90 min in the absence or presence of chloroquine (CQ), and LC3-II levels were measured as in B and C. Each bar represents the mean (+ SD) from analyses of three to five experiments. *, p < 0.05; ***, p < 0.001 (ANOVA). (F) eHAP cell derivatives were starved for the indicated times, and LC3-II levels were measured as in C. (G) Parental or WHAMM-deficient cells were treated with puromycin for 3.5 h and stained with antibodies to visualize ubiquitinated protein aggregates (red) and with phalloidin to visualize F-actin (green). Scale bar: 10 μm. (H) The areas occupied by ubiquitinated protein aggregates and the entire cell were calculated using ImageJ and converted to a ratio. Each bar represents the mean (+ SEM) from analyses of 30–35 cells. **, p < 0.01 (t test).
We next shifted cells from rich growth media to starvation conditions and quantified LC3 conversion on immunoblots. While parental eHAP cells and WDR73MUT cells showed similar levels of LC3-II after 90 min of starvation, WHAMMMUT cells generated only half of the amount of LC3-II as the other two cell lines (Figure 7, D and E). Blocking flux with chloroquine resulted in a similar pattern of LC3 conversion (Figure 7E), suggesting that the lipidation deficit in WHAMM-deficient cells is primarily due to less autophagosomal biogenesis instead of increased autophagosomal degradation. Moreover, a time course revealed that the rate of LC3 lipidation between 30 and 120 min of starvation was slower in WHAMMMUT cells than in parental or WDR73MUT cells (Figure 7F). Thus genetic inactivation demonstrates a key role for WHAMM in autophagosomal biogenesis.
To determine whether this slower autophagosomal assembly impacts the eventual digestion of cargo, we induced the production of truncated misfolded proteins in eHAP and WHAMMMUT cells using the translational inhibitor puromycin and stained the cells with antibodies to ubiquitin. The parental cell line contained many cytoplasmic puncta indicative of small aggregates of polyubiquitinated proteins (Figure 7G). WHAMMMUT cells also accumulated aggregates, but they appeared much larger (Figure 7G) and occupied significantly greater areas of the cytoplasm than in eHAP cells (Figure 7H). Collectively our transient and stable loss-of-function studies indicate that WHAMM is important for promoting LC3 lipidation and autophagosomal biogenesis so that cells may ultimately degrade cytoplasmic components.
WHAMM is recruited to nascent autophagosomes by binding to PI(3)P
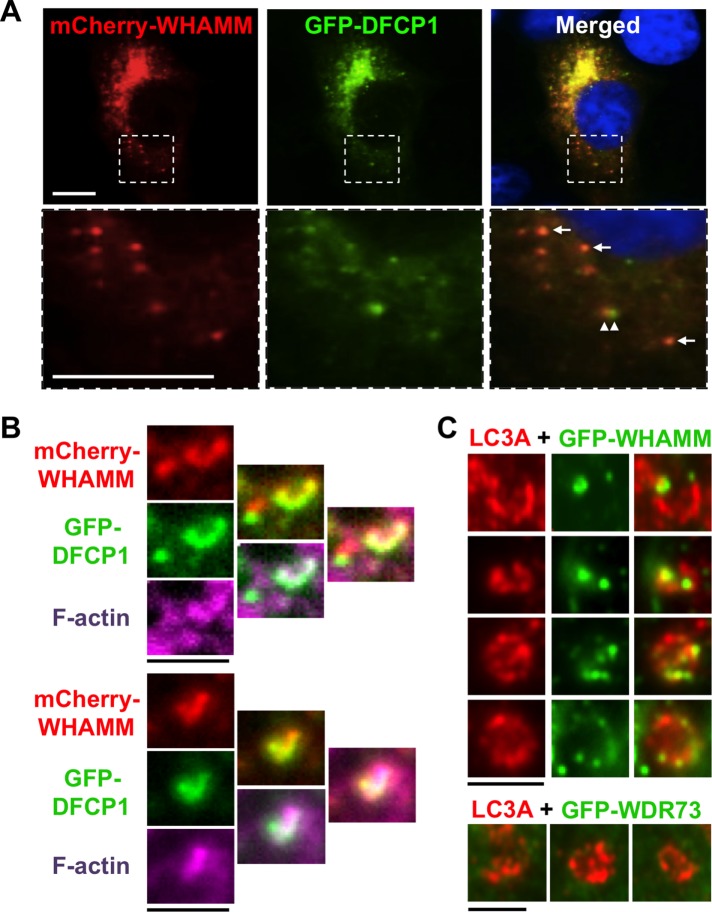
To investigate the mechanism underlying WHAMM’s normal function in autophagy, we first examined its localization. In most mammalian cells, the majority of endogenous and tagged WHAMM is associated with juxtanuclear ERGIC/cis-Golgi membranes (Campellone et al., 2008; Blom et al., 2015; Russo et al., 2016). In retinal epithelial cells, tagged murine WHAMM is also detected on puncta adjacent to LC3-positive vesicles and near regions of the ER called omegasomes (Kast et al., 2015). Omegasomes are transient hotspots for autophagosomal biogenesis marked by the protein DFCP1 (Axe et al., 2008). Consistent with these findings, when we starved fibroblasts, mCherry-WHAMM was located both at the Golgi region and adjacent to GFP-DFCP1 puncta (Figure 8A). We further found that mCherry-WHAMM and GFP-DFCP1 colocalized at some puncta (Figure 8A, arrows) and at omegasome-shaped structures that associated with actin filaments (Figure 8B), implying that WHAMM can promote actin nucleation at sites of autophagosomal biogenesis, not just at mature autophagosomes. Indeed, staining with antibodies to LC3 revealed that GFP-WHAMM localized to subdomains of developing (open) phagophores in addition to complete (closed) autophagosomes (Figure 8C). In contrast, WDR73 exhibited a diffuse nucleo-cytoplasmic distribution (Supplemental Figure S4) and was not enriched on autophagosomes (Figure 8C). The observations that WHAMM is recruited to omegasomes and phagophores point to a direct role for this protein in membrane remodeling before actin-based rocketing of mature autophagosomes.
FIGURE 8:
WHAMM localizes to omegasomes and subdomains of nascent autophagosomes. (A) Cos7 cells transiently expressing mCherry-WHAMM (red) and GFP-DFCP1 (green) were starved and stained with DAPI to label DNA (blue). Arrows indicate areas of WHAMM and DFCP1 colocalization; arrowheads highlight a region of WHAMM and DFCP1 juxtaposition. Scale bars: 10 μm. (B) Cos7 cells transiently expressing mCherry-WHAMM (red) and GFP-DFCP1 (green) were starved and stained with phalloidin to label F-actin (pink). Scale bars: 1.5 μm. (C) Cos7 cells transiently expressing GFP-WHAMM or GFP-WDR73 (green) were starved and stained with antibodies to label LC3 (red). Individual autophagosomal structures with punctate GFP-WHAMM localization to regions of membrane growth or curvature are highlighted in this composite. Scale bars: 1.5 μm.
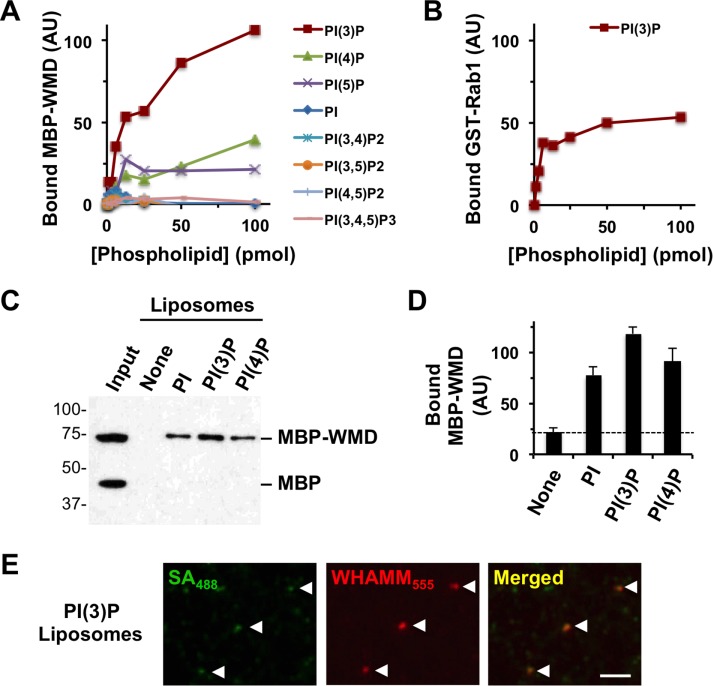
WHAMM localization to membranes of the secretory pathway is mediated by the WMD, which has been suggested to interact with multiple phospholipids in vitro (Campellone et al., 2008). Upon inspection of this domain, we noticed that residues 29–88 resemble a phospholipid-binding R-[Y/F]-xn-K-xn-R motif commonly present in PX domain proteins (Supplemental Figure S5A). Further examination of the WMD using the hhpred algorithm indicated that WHAMM residues 84–157 also share similarity with PX domains from many sorting nexin (SNX) proteins (Supplemental Figure S5B), which often bind to PI(3)P on endosomes (Teasdale and Collins, 2012). PI(3)P is also a crucial component of autophagosomal membranes (Roberts and Ktistakis, 2013) and was recently shown to be required for actin assembly at phagophores (Mi et al., 2015). Therefore we explored whether WHAMM could bind to this phospholipid by purifying an MBP-tagged version of the WMD and testing its capacity to bind to PI(3)P and 14 other lipids that were immobilized on nitrocellulose strips. After probing the strips with soluble MBP-WMD or MBP alone and detecting bound protein with anti-MBP antibodies, we found that the WMD bound strongly to PI(3)P and weakly to PI(4)P and PI(5)P (Supplemental Figure S5C). Similar phospholipid-binding patterns were observed for MBP-WHAMM(WT), MBP-WHAMM(Δ7), and MBP-WHAMM(X6) (Supplemental Figure S5C). To better gauge the affinity of the interactions between WHAMM and membranes, we used the MBP-WMD to probe arrays containing different concentrations of lipids. Again, the WMD bound to PI(3)P and, to a lesser extent, PI(4)P and PI(5)P (Figure 9A). At omegasomes, PI(3)P is responsible for binding DFCP1 and also for recruiting Rab1, a small G-protein that can positively regulate autophagosomal formation (Zoppino et al., 2010; Huang et al., 2011). Because Rab1 binds directly to WHAMM and recruits it to membranes (Russo et al., 2016), we also tested whether recombinant Rab1 could interact with PI(3)P. Indeed, glutathione S-transferase (GST)-tagged Rab1 also bound to PI(3)P (Figure 9B and Supplemental Figure S5D), suggesting that, in addition to binding to one another, WHAMM and Rab1 both bind to PI(3)P.
FIGURE 9:
WHAMM binds to PI(3)P in vitro. (A) Different quantities of phospholipids immobilized on a nitrocellulose membrane were probed with purified MBP-WMD. Bound protein was detected using antibodies to MBP and visualized by chemiluminescence. MBP-WMD binding was quantified by measuring the mean pixel intensity of protein-specific spots after normalizing to background staining. (B) Different concentrations of PI(3)P immobilized on a nitrocellulose membrane were probed with purified GST-Rab1A. Bound protein was detected using antibodies to GST and quantified as in A. (C) Synthetic liposomes containing 5% of the indicated phosphoinositides were incubated with MBP and MBP-WMD and collected by centrifugation. Liposome-associated proteins were subjected to SDS–PAGE and detected by immunoblotting for MBP. (D) The amount of MBP-WMD pulled down was determined by densitometry. Each bar represents the mean (+ SD) from three to four experiments. (E) PI(3)P-containing biotinylated liposomes were mixed with Alexa Fluor 555–labeled MBP-WHAMM and Alexa Fluor 488–labeled streptavidin and examined microscopically.
To assess whether the WMD was capable of binding to PI(3)P in a different context, we mixed MBP-WMD and MBP with synthetic liposomes containing PI, PI(3)P, or PI(4)P and performed vesicle–protein cosedimentation assays. Anti-MBP immunoblotting of pelleted samples indicated that MBP-WMD, but not the MBP control protein, interacted with all 3 phosphoinositide-containing liposomes (Figure 9C). Band quantification showed that slightly more MBP-WMD bound to PI(3)P than to PI (Figure 9D), suggesting that although the WMD acts as a promiscuous lipid-binding domain in vitro, it exhibits some preference for PI(3)P. Consistent with PI(3)P binding, when we mixed Alexa Fluor 555–labeled MBP-WHAMM(WT) with Alexa Fluor 488–labeled liposomes, WHAMM could be found on the PI(3)P-containing vesicle structures (Figure 9E).
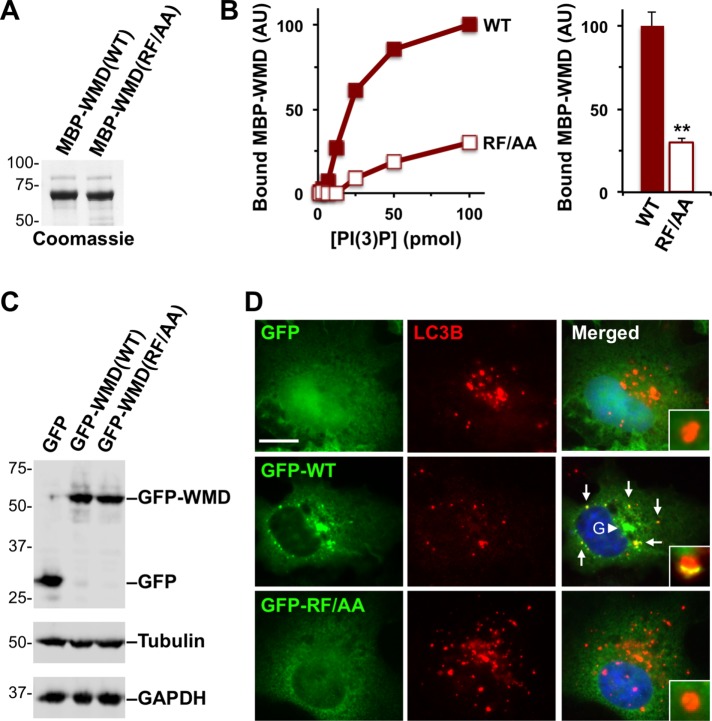
Because the WMD bears resemblance to PX domains, we finally tested whether residues predicted to be part of this domain were required for PI(3)P binding in vitro and for membrane localization in cells. We generated a R29A/F30A mutant version of the WMD fused to MBP and purified it alongside the wild-type MBP-WMD (Figure 10A). To compare the affinity of the WMD(RF/AA) mutant and WMD(WT) for PI(3)P, we used the MBP-tagged constructs to probe arrays containing different amounts of this phospholipid. As predicted, based on the conservation of R29 and F30 in the PX consensus sequence, mutation of these residues to alanine resulted in a dramatically lower degree of WMD binding to PI(3)P (Figure 10B). Even when PI(3)P was present at its highest amount (100 pmol), it bound approximately fourfold less WMD(RF/AA) than WMD(WT) (Figure 10B and Supplemental Figure S5E), further demonstrating that R29 and F30 are crucial for the interaction with PI(3)P.
FIGURE 10:
Point mutations in the WMD that disrupt binding to PI(3)P in vitro prevent localization to autophagosomes in cells. (A) Purified MBP-WMD(WT) and (RF/AA) were analyzed by SDS–PAGE and Coomassie blue staining. (B) Different quantities of phospholipids immobilized on a nitrocellulose membrane were probed with purified MBP-WMD(WT) or (RF/AA). Bound protein was detected using antibodies to MBP and quantified by measuring the intensity of protein-specific spots after normalizing to background staining. The bar graph represents the mean amounts (+ SEM) of MBP-WMD variants bound to 100 pmol of PI(3)P in three experiments. **, p < 0.01 (t test). (C) Cos7 cells were transiently transfected with plasmids encoding a GFP vector control, GFP-WMD(WT), or GFP-WMD(RF/AA) and blotted with anti-GFP, anti-tubulin, and anti-GAPDH antibodies. (D) Cos7 cells transiently expressing GFP, GFP-WMD(WT), or GFP-WMD(RF/AA) were starved and stained with antibodies to LC3B (red) and DAPI to label DNA (blue). Arrows indicate areas of WHAMM localization to LC3+ puncta; the arrowhead highlights the Golgi (G) region. Insets depict individual autophagosomes. Scale bar: 10 μm.
To determine the localization of the WMD(RF/AA) mutant in cells, we expressed it as a GFP-fusion protein in Cos7 fibroblasts in parallel to the wild-type GFP-WMD and GFP alone (Figure 10C). Under starvation conditions, GFP-WMD(WT) was found at the juxtanuclear Golgi region and at LC3-positive autophagosomes (Figure 10D). In contrast, GFP-WMD(RF/AA) exhibited a diffuse cytoplasmic localization and was not present on LC3-positive membranes (Figure 10D), indicating that residues R29 and F30 are required for WHAMM recruitment to autophagosomes. Collectively our results give rise to a model for autophagosomal biogenesis in healthy cells in which PI(3)P recruits WHAMM via its N-terminal PX-like WMD, and the WHAMM WCA domain drives actin assembly to promote membrane remodeling and LC3 incorporation.
DISCUSSION
For decades, WASP variants have been known to cause immunodeficiencies, but mutations in other actin nucleation factors have surprisingly not been characterized in relation to human disease. GMS is a genetic disorder featuring neurological abnormalities and progressive kidney dysfunction, and several variants of the WDR73 gene are thought to be responsible for its clinical hallmarks (Colin et al., 2014; Ben-Omran et al., 2015; Vodopiutz et al., 2015; Rosti et al., 2016). However, in parallel with those studies, we described coincident WDR73 and WHAMM mutations in all but one of 27 Amish individuals exhibiting clinical features consistent with GMS (Jinks et al., 2015). In the current work, we showed that cells from Amish children with GMS who are homozygous for a 7 base pair deletion in WHAMM lack a nucleation-proficient WHAMM protein and, as a result, have major defects in autophagy. Taken together with experiments using independent genome-edited cells and purified proteins, our results further revealed that WHAMM-mediated actin assembly is fundamentally important for autophagosomal biogenesis at a step that follows PI(3)P synthesis but precedes LC3 incorporation. In the absence of this PI(3)P-WHAMM-Arp2/3 pathway for autophagosomal remodeling, cells accumulate polyubiquitinated protein aggregates.
A major question arising from these studies is whether defects in autophagy or other WHAMM-associated functions modify the clinical outcomes in Amish GMS patients, as any resulting phenotypes may be concealed beneath a particularly severe clinical picture that arises through the coincident WDR73 mutation. Thus the direct medical relevance of findings using patient cells will likely remain unclear until individuals are identified who display bi-allelic WHAMM mutations in the absence of WDR73 mutations. Interestingly, it has been suggested that GMS is not a single gene disorder and that the clinical features of GMS can arise from mutations in genes other than WDR73 (Rosti et al., 2017). Moreover, since the initial description of Amish GMS, we have identified a second child in whom a recombination event led to heterozygosity for the WHAMM variant alongside the homozygous WDR73 mutation. This 3-year-old’s neurocognitive impairment is less significant than is typical for Amish children with GMS. Hypotonia remains the predominant clinical feature; he is able ride a tricycle, walk with assistance, gesture to indicate needs, and has not developed any renal impairment. How WHAMM heterozygosity may affect the clinical picture and/or cellular phenotypes in this patient or other Amish GMS patients remains to be determined.
Notably, the autophagy defects seen in Amish GMS patient fibroblasts were more severe than those in WHAMM-deficient HeLa or eHAP cells, as only the GMS cells vacuolated after treatment with chloroquine. This could be due to differences in cell type and tissue origin. However, it is also plausible that residual truncated WHAMM proteins exacerbate the autophagy defects in patient cells. Another possibility is that differences among the Amish, HeLa, and eHAP genomes influence autophagy or related membrane-trafficking processes. For example, aberrant WDR73 function in Amish GMS cells may worsen their autophagy defect. Although targeted loss-of-function and localization studies did not point to a direct role for WDR73 in autophagy, we have been unable to inactivate WDR73 in a WHAMMMUT background, implying that WDR73 is essential in WHAMM-deficient cells. Further studies are required to better understand the factors that impact autophagy in distinct cell lines.
In addition to the cellular phenotypes attributable to WHAMM function in Amish GMS patient cells, changes in WHAMM expression and WHAMM polymorphisms have been linked to several other conditions. WHAMM is one of 12 biomarkers that are differentially expressed in children with systemic onset juvenile idiopathic arthritis (Allantaz et al., 2007) and is one of 11 genes associated with risk for lupus-like diseases in dogs (Wilbe et al., 2015). The WHAMM and WDR73 genes are separated by only 1.7 Mb, and several humans with chromosome 15 microdeletions encompassing WHAMM or both WHAMM and WDR73 exhibit developmental delay or intellectual disability (Doelken et al., 2013; Burgess et al., 2014). Whether the specific loss of WHAMM and/or WDR73 function contributes to these illnesses has not been investigated. However, given that WHAMM is a microtubule-binding protein (Shen et al., 2012) and that WDR73 can localize to the mitotic spindle (Jinks et al., 2015), it is tempting to speculate that microtubule-dependent functions might be altered by mutations in WHAMM, WDR73, or both genes.
Apart from microtubule binding, the best-characterized biochemical activity of WHAMM is promoting actin nucleation with the Arp2/3 complex (Campellone et al., 2008). However, until recent years, little was known about actin assembly during autophagy in mammalian cells. The first nucleation factor studied in this process was Cortactin (Lee et al., 2010), which presumably influences filament branching at autophagosomes. The WASP-family member WASH is also involved in autophagy (Xia et al., 2013; Zavodszky et al., 2014), perhaps by controlling trafficking to autophagosomes. More recently, WHAMM was shown to promote Arp2/3-mediated autophagosomal rocketing (Kast et al., 2015), while JMY was shown to interact with LC3 and also nucleate actin at autophagosomes (Coutts and La Thangue, 2015). Finally, actin-capping protein was found to regulate actin polymerization at autophagosomes downstream of PI(3)P synthesis (Mi et al., 2015). Our current work both interconnects and expands on those studies.
JMY and WHAMM were proposed to influence autophagosomal maturation because a JMY siRNA reduced the amount of autophagosomes in U2OS epithelial cells (Coutts and La Thangue, 2015), while WHAMM siRNA treatment reduced autophagosomal size and movement in retinal epithelial cells (Kast et al., 2015). Expression of a WHAMM point mutant that should have a reduced affinity for Arp2/3 also decreased the number of LC3-positive autophagosomes in cells (Kast et al., 2015). Our results point to a crucial function for WHAMM early in biogenesis, between PI(3)P generation and LC3 lipidation. This is supported by our findings that WHAMM and Rab1 each bind to PI(3)P, and that WHAMM inactivation delays the conversion of LC3-I to LC3-II. How WHAMM facilitates LC3 conversion is not clear, but could be related to its ability to affect trafficking from the ERGIC, an important source of vesicle templates for LC3 lipidation (Ge et al., 2013, 2014). WHAMM-mediated actin assembly could also help shape the phagophore in a manner that promotes LC3 incorporation. Indeed, the punctate localization of WHAMM at omegasomes and autophagosomal membranes is consistent with WHAMM being the missing factor that promotes actin nucleation inside forming autophagosomes downstream of PI(3)P synthesis (Mi et al., 2015). Finally, we have highlighted the physiological importance of WHAMM through the use of Amish GMS patient cells, which lacked autophagosomal structures and possessed excess p62 under steady-state conditions, exhibited weak autophagic responses to rapamycin or starvation, and accumulated large vacuoles when lysosomal degradation was inhibited.
Autophagy is known to be up- or down-regulated in many disease states (Choi et al., 2013), but examples of genetic mutations that directly perturb this process to cause human illness are rare. In one example, recessive mutations in the epg5 gene, whose protein product appears to control autophagosome–lysosome fusion, give rise to a neurodevelopmental disorder called Vici syndrome (Byrne et al., 2016). A second example is a homozygous mutation in ATG5 that causes an E122D substitution and a neurodegenerative ataxia. ATG5 normally functions in conjugating phosphatidylethanolamine to LC3-I to create LC3-II during phagophore elongation, but cells from patients with the Atg5 variant exhibit less LC3 lipidation (Kim et al., 2016). Thus far, that is the only known mutation in a core component of the autophagy machinery with physiological and clinical consequences. However, based on our current results with Amish GMS patients, further studies on the molecular functions of WHAMM and WDR73 should also provide important insights into the mechanisms of autophagy that influence health and disease.
MATERIALS AND METHODS
Human genetics and cell culture
Studies leading to the definition of the genetic basis of clinical GMS in the Amish were described previously (Jinks et al., 2015). Primary patient cells were obtained from the blood and skin of human subjects as part of a previous study approved by the University of Arizona IRB (Project #10-0050-01, Genetic Studies in the Amish, IRB00005448). Subsequent cellular studies were approved by the University of Connecticut IRB (Protocol #H13-110, Cell Biology of a Neurodevelopmental Disorder, FWA00007125). Primary dermal fibroblasts isolated from two normal individuals and two Amish GMS patients were cultured in DMEM plus 20% fetal bovine serum (FBS) and antibiotic–antimycotic. Phenotype quantification was performed with one set of GMS fibroblasts only, because the second proliferated poorly and did not survive freeze–thaw cycles. LCLs (European Collection of Authenticated Cell Cultures) were derived from two normal Amish individuals, two heterozygous carriers, and three GMS patients, and grown in RPMI plus 10% FBS and antibiotic–antimycotic. Cos7, NIH3T3, and HeLa cells were cultured in DMEM plus 10% FBS and antibiotic–antimycotic. eHAP cells (Essletzbichler et al., 2014) and their derivatives were cultured in IMDM plus 10% FBS and penicillin–streptomycin. All mammalian cells were grown at 37°C in 5% CO2.
DNA, RNA, viruses, and insect cells
Key reagents are listed in Supplemental Table S1. Plasmids, bacmids, and baculoviruses encoding human WHAMM(WT) were described previously (Campellone et al., 2008; Shen et al., 2012; Russo et al., 2016). Plasmids encoding WHAMM(Δ7) were generated by site-directed mutagenesis on WHAMM(WT) using primers GACTCTCTTAAAAGACTTAAAAACAAGATGAAGTTG and CAACTTCATCTTGTTTTTAAGTCTTTTAAGAGAGTC. Plasmids encoding WHAMM(X6) were generated by replacing the appropriate region of WHAMM(WT) with a synthetic XhoI-SacI fragment of X6 (Integrated DNA Technologies). Recombinant baculoviruses were generated using the Bac-to-Bac system (Invitrogen). The plasmid for expression of GFP-WDR73 in mammalian cells was generated by PCR using a cDNA clone template (Open Biosystems), primers atcatcggtaccATGGATCCTGGGGACGACTGG and atcatcgcggccgcttaTCAGCGGGGGGCACAAAGGT, and cloned as a KpnI-NotI fragment into pKC-EGFP-C1 (Campellone et al., 2008). Plasmids for expression of MBP, MBP-WHAMM(WMD), and GST-Rab1 in insect cells were described elsewhere (Russo et al., 2016). Plasmids encoding the WMD R29A/F30A mutant were generated using a synthetic codon-optimized WMD(RF/AA) fragment flanked by KpnI-NotI sites (Integrated DNA Technologies). A synthetic WMD(WT) fragment was synthesized as a control (Integrated DNA Technologies). For quantification of human WHAMM mRNA, cDNA was prepared from skin fibroblasts with TRIzol reagent and the M-MLV reverse transcriptase (Invitrogen) and subsequently amplified using primers CTCCGTGCTCTGTCCTCATCCTCTCA (exon 9) and CTAACCATCCCACTGGCCAGGGTCCT (exon 10). The WHAMM PCR product corresponds to 456 nucleotides at the 3′ end of the open reading frame. WHAMM levels were normalized to β-actin, as described previously (Campellone et al., 2012). RT-PCR primers spanning exons 4–8 were used in Supplemental Figure S1. All bacteria were cultured using LB media with appropriate antibiotics at 37°C. Plasmids were maintained in E. coli XL-1 Blue and purified using standard kits (Machery-Nagel) before transfection. Sf9 insect cells were grown in ESF921 medium (Expression Systems) at 28°C and infected with baculoviruses encoding MBP-fusion proteins at a multiplicity of infection of ∼1 (Russo et al., 2016).
Transfections and chemical treatments
Standard mammalian cell transfections with DNA or RNA were performed using Lipofectamine-LTX or RNAiMAX (Invitrogen) (Russo et al., 2016). Dermal fibroblasts, however, were shifted to 0.60X DMEM, 0.25X RPMI medium, 0.25X F10 medium, 20% FBS, 1X nonessential amino acids, 1X Glutamax, and antibiotic–antimycotic (all from Invitrogen) at least 24 h before transfection. NIH3T3 clones stably encoding LAP, LAP-WHAMM(WT), or LAP-WHAMM(Δ7) were selected in media containing 690 µg/ml G418. For inducing LAP expression, 7.6 mM sodium butyrate was added for 16 h. Control siRNAs or Stealth siRNAs to WHAMM and WDR73 were purchased from Thermo Fisher Scientific. Treatments with Hank’s buffered saline solution were used in starvation experiments, chloroquine (Sigma) was used at 50 μM, rapamycin (Tocris) was used at 10 μM, and puromycin (Sigma) was used at 2 μg/ml.
Genome editing
HAP1 fibroblasts are a nearly haploid human cell line that contains an immortalizing BCR-ABL fusion and a single copy of all chromosomes, except for a heterozygous 30 Mb fragment of chromosome 15, which is integrated on the long arm of chromosome 19 (Carette et al., 2011). This diploid portion encompasses 330 genes, including WHAMM and WDR73. CRISPR/Cas9-engineered eHAP cells, which are fully haploid (Essletzbichler et al., 2014), were therefore used for mutagenesis of WHAMM and WDR73. Using CRISPR/Cas9-mediated recombination, a 10-nucleotide deletion was introduced into exon 2 of WHAMM, and a 10-nucleotide insertion was introduced into exon 1 of WDR73 (Haplogen, now Horizon Genomics). Mutations were confirmed by DNA sequencing (Haplogen, now Horizon Genomics). Using fluorescence in situ hybridization probes, we additionally verified the homozygosity and integrity of chromosomes 15 and 19 in eHAP derivatives (Supplemental Figure S6A). Briefly, whole-chromosome paint probes (Rainbow Scientific Cytocell) were hybridized and washed per the manufacturer’s protocol. Slides were mounted in a 1:6 dilution of 4′,6-diamidino-2-phenylindole (DAPI):Vectashield (Vector Laboratories). It is also important to note that, after editing, haploid cells tend to eventually revert to diploidy, a phenotype that we confirmed with karyotype analyses (Supplemental Figure S6B). Briefly, mitotic cells were arrested at metaphase via treatment with 0.1 μg/ml colcemid for 1 h, collected, swollen with a 0.075M KCl solution, and subsequently fixed with a modified Carnoy’s solution. Slides were G-banded with trypsin and Giemsa. All images were captured on an Olympus AX70 fluorescence microscope equipped with appropriate filters and analyzed with Leica Cytovision Imaging Software. Our attempts at introducing an inactivating mutation in exon 1 of WDR73 in the above WHAMM mutant background failed (Horizon Genomics). All WDR73 mutants were found to be in-frame, implying that WDR73 is an essential gene in this WHAMM-deficient background.
Immunoblotting and quantification
For preparation of protein extracts, cells were washed with phosphate-buffered saline (PBS) and lysed in 50 mM Tris-HCl (pH 7.6), 50 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin, leupeptin, pepstatin, and chymostatin, before being mixed with SDS–PAGE sample buffer. Protein samples were boiled and subjected to SDS–PAGE before being transferred to nitrocellulose membranes and stained with Ponceau S. Membranes were blocked in PBS with 5% milk before being probed with rabbit anti-WHAMM (Shen et al., 2012), chicken anti-WHAMM(WCA) (Campellone et al., 2008), rabbit anti-MBP (Shen et al., 2012), mouse anti–tubulin E7 (Developmental Studies Hybridoma Bank), mouse anti-GAPDH (Ambion), rabbit anti-LC3B or anti-p62/SQSTM1 (Cell Signaling Technology), rabbit anti-WDR73 (Santa Cruz Biotech), or mouse anti-HA (Covance) antibodies. Following washes, membranes were treated with IRDye-800 or horseradish peroxidase (HRP)-conjugated secondary antibodies for detection. Infrared or chemiluminescent bands were visualized with a Li-Cor Fc Imager, and their intensities were quantified using Li-Cor Image Studio software. For measurement of LC3 lipidation, LC3-II bands were normalized to tubulin and/or GAPDH bands in each lane. Statistical significance and p values were evaluated using an unpaired Student’s t test or one-way analysis of variance (ANOVA) using GraphPad Prism software.
Microscopy and quantification
Cells grown on 12 mm glass coverslips were fixed with 3.7% paraformaldehyde for 35 min. For WHAMM, microtubule, and LC3 staining, cells were treated with 100% methanol for 3.5 min at −20°C. We used the following primary antibodies: guinea pig anti-WHAMM(CC) or rabbit anti-GFP (Campellone et al., 2008), mouse anti–tubulin E7 (Developmental Studies Hybridoma Bank), mouse anti–ERGIC-53 (Alexis Biochemicals), rabbit anti-LC3A or anti-LC3B (Cell Signaling Technology), mouse anti–ubiquitin FK2 (Enzo Life Sciences), rabbit anti-WDR73 (Santa Cruz Biotechnology), rat anti-Grp94 (Novus Biologicals), mouse anti–Sec31A or anti-GM130 (BD Biosciences), and mouse anti-LAMP1 (Santa Cruz Biotechnology). We used secondary antibodies conjugated to Alexa Fluor 488, 555, or 647 (Invitrogen). DNA was visualized with 1 μg/ml DAPI, and F-actin was detected using 4U/ml Alexa Fluor–conjugated phalloidins (Invitrogen). Coverslips were mounted in ProLong Gold anti-fade (Invitrogen). Images were captured using 60× (1.40 NA) or 100× (1.45 NA) Plan-Apo objective lenses on a Nikon Ti-E microscope equipped with an Andor Clara-E camera, and Elements software. For measurement of fluorescence intensities, areas, and quantities, individual cells and/or organelles were outlined using ImageJ software. For intensity measurements, the mean pixel intensity of WHAMM staining in a cell was multiplied by its two-dimensional area and divided by the area of the cell. The intensity in control cells was set to 1. For organelle area measurements, an ERGIC, LC3, polyubiquitin, or DAPI signal was defined using a thresholding function in ImageJ and was divided by the area of the cell. Numbers of F-actin and LC3 puncta per cell were counted manually. For measuring the percentage of cells with F-actin puncta, abnormal stress fibers, or chloroquine-induced vacuoles, sample identities were coded, and randomly chosen cells were scored as 0 (negative) or 1 (positive) for the specific phenotype. Samples were decoded after quantification. Statistical significance and p values were evaluated using an unpaired Student’s t test, one-way ANOVA, or Fisher’s exact test using GraphPad Prism software.
Purified proteins
MBP, MBP-WHAMM(WT), MBP-WHAMM(WMD), GST, and GST-Rab1 preparations were described previously (Russo et al., 2016), and stored at −80°C in 20 mM MOPS (pH 7.0), 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM EDTA, and 10% glycerol. The Δ7 and X6 variants, as well as codon-optimized versions of WMD(WT) and WMD(RF/AA), were expressed and purified in an identical manner to other MBP fusions. MBP-WHAMM(WT) was covalently labeled using an Alexa Fluor 555 labeling kit (Invitrogen). All protein quantities were measured using Bradford assays (Bio-Rad), and purities were confirmed by SDS–PAGE analyses.
Actin assembly assays
Pyrene-actin assembly assays were performed essentially as described previously (Russo et al., 2016). Actin (Cytoskeleton) was resuspended in G-buffer (5 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.2 mM dithiothreitol (DTT), 0.2 mM ATP) and subjected to gel-filtration chromatography using a Superdex 200 column (GE Healthcare) to remove small filaments. Actin (2.0 µM; 7% pyrene labeled) was then polymerized in the presence of 20 nM Arp2/3 complex plus 200 nM MBP, MBP-WHAMM(WT), MBP-WHAMM(Δ7), or MBP-WHAMM(X6). Pyrene fluorescence (actin filaments) was measured using a Horiba Jobin Yvon Fluorolog-3 spectrofluorimeter capable of multiwavelength excitation/detection and equipped with a four-position sample changer. Similar results were obtained using different preparations of G-actin in up to four experiments for each MBP fusion protein.
Microtubule cosedimentation assays
Porcine brain tubulin was purified as described previously (Campellone et al., 2008) and diluted to 35 μM in BRB80 (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2) containing 20% glycerol, 1.25 mM GTP, and 5 mM DTT. Tubulin was then polymerized at 37°C and stabilized with stepwise additions of Taxol until reaching the final concentrations of 26 μM microtubules and 11 μM Taxol (Shen et al., 2012). Samples (22 μl) containing microtubules and fusion proteins were prepared by mixing BRB80, 100 mM KCl, 2 μM microtubules, and 500 nM precleared MBP-WHAMM(WT) or MBP-WHAMM(Δ7) at 25°C. (MBP-WHAMM(X6) pelleted by itself in the preclearance step.) After 10 min, 20 μl of each sample was overlaid onto BRB80 containing glycerol at 5% (low-speed assays) or 40% (high-speed assays). The samples were then centrifuged at 21,000 × g (low speed) in a benchtop microcentrifuge (Eppendorf) or at 40,000 × g (high speed) at 26°C for 21 min in a TLA100 rotor (Beckman) to sediment the microtubules. Supernatants were removed and precipitated with 10% trichloroacetic acid, and both the supernatant and pellet samples were mixed with SDS–PAGE sample buffer to equivalent volumes.
Lipid-binding assays
For protein overlay assays, PIP Strip and PIP Array nitrocellulose membranes spotted with 1–100 pmol of immobilized phospholipids (Echelon Biosciences) were hydrated in PBS, and blocked in blocking buffer (5% milk, 3% BSA, and 0.1% Tween-20 in PBS) for 1 h. PIP Strips were probed with 20 nM soluble MBP, MBP-WHAMM(WT), MBP-WHAMM(WMD), MBP-WHAMM(Δ7), MBP-WHAMM(X6), GST, or GST-Rab1 in blocking buffer for 2 h at room temperature. PIP Arrays were probed with 20 nM soluble MBP-WMD variants or 100 nM GST-Rab1. After washes, bound protein was detected using rabbit anti-MBP or rabbit anti-GST antibodies (Shen et al., 2012) in blocking buffer, followed by HRP-conjugated anti-rabbit antibodies in 5% milk, and visualized using enhanced chemiluminescence with film (GE Healthcare) or with a Li-Cor Fc Imager. For protein binding quantification, densitometry was performed by measuring the mean pixel intensity of protein-specific spots in ImageJ and normalizing these values to background staining, or by using Li-Cor Image Studio software. For liposome cosedimentation assays, 66 μM synthetic PolyPIPosomes (Echelon Biosciences) containing 65% polymerizable-PC, 29% polymerizable-PE, 5% polymerizable PI, PI(3)P, or PI(4)P, and 1% polymerizable biotin-PE were mixed with 1 μM MBP and 1 μM MBP-WMD at room temperature for 30 min in 10 mM Tris (pH 7.5), 1 mM MgCl2, 250 mM KCl, and 1% glycerol. Liposomes and proteins were centrifuged in a TLA100 rotor (Beckman) for 45 min at 55,000 rpm at 23°C. Pellet fractions were resuspended in sample buffer and analyzed by SDS–PAGE and immunoblotting with anti-MBP antibodies. For protein binding quantification, densitometry was performed by measuring the mean pixel intensity of MBP-WMD bands in ImageJ and normalizing these values to background staining. For microscopic visualization of WHAMM in association with liposomes, MBP-WHAMM(WT) was covalently labeled with Alexa Fluor 555 (Invitrogen), and PolyPIPosomes were detected with Alexa Fluor 488–streptavidin.
Cytokine secretion assays
Human LCLs were seeded at 5 × 105 cells/well and allowed to grow to 8 × 105 cells/well in 12-well plates before stimulation with 2 × 107 E. coli XL1-Blue for 3 h. The contents of each well were then transferred to microfuge tubes and placed on ice for 5 min before centrifugation at 635 × g for 5.5 min at 4°C. Supernatants were transferred to fresh tubes, recentrifuged, and again transferred to fresh tubes before being frozen at −20°C. Ready-SET-Go ELISA kits (eBioscience) were used to quantify levels of IL-1β and TNFα in once-thawed LCL supernatants. Briefly, 96-well Costar plates (Corning) were coated with capture antibodies in PBS for >24 h at 4°C, blocked at 23°C using 1% FBS in PBS for 1 h, and incubated with recombinant standards or LCL supernatants for 2 h; this was followed by cytokine detection antibodies for 1 h and Avidin-HRP for 30 min. Well contents were developed with tetramethylbenzidine for 5–15 min before quenching with 1 M H3PO4 and measurement of the colorimetric product at 450 nm on a SpectraMax M2 plate reader (Molecular Devices). IL-1β and TNFα concentrations were calculated from optical density values using SoftMax Pro version 4.8.
Statistical analyses
Statistical parameters and significance are reported in the figures and figure legends. Data were determined to be statistically significantly different when p < 0.05 by Student’s t test, ANOVA, or Fisher’s exact test in GraphPad Prism software as indicated in preceding sections.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01-GM107441 (K.G.C.), Medical Research Council grants G1002279 and G1001931 (A.H.C., E.B.), and the Newlife Foundation (A.H.C., E.B.). We thank Henry Chen (University of Connecticut) for assistance with ELISAs and Kevin Strauss (Clinic for Special Children) for communication of clinical and genetic data. We also thank the Amish families for their participation in these studies.
Abbreviations used:
- ANOVA
analysis of variance
- CC
coiled-coil
- DAPI
4′,6-diamidino-2-phenylindole
- DTT
dithiothreitol
- ELISA
enzyme-linked immunosorbent assay
- ER
endoplasmic reticulum
- ERGIC
ER–Golgi intermediate compartment
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GMS
Galloway-Mowat syndrome
- GST
glutathione S-transferase
- HA
hemagglutinin
- HRP
horseradish peroxidase
- IL-1β
interleukin-1β
- LAP
localization and affinity purification
- LCL
lymphoblastoid cell line
- MBP
maltose-binding protein
- PBS
phosphate-buffered saline
- PI(3)P
phosphatidylinositol 3-phosphate
- PWWCA
polyproline-WH2-WH2-connector-acidic
- RNAi
RNA interference
- RT-PCR
reverse transcription-PCR
- siRNA
small interfering RNA
- SNX
sorting nexin
- TNFα
tumor necrosis factor α
- WASP
Wiskott-Aldrich syndrome protein
- WMD
WHAMM membrane-interaction domain
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0022) on July 18, 2017.
REFERENCES
- Aguilera MO, Beron W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8:1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, Ardura M, Chung W, Smith E, Wise C, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Omran T, Fahiminiya S, Sorfazlian N, Almuriekhi M, Nawaz Z, Nadaf J, Khadija KA, Zaineddin S, Kamel H, Majewski J, Tropepe V. Nonsense mutation in the WDR73 gene is associated with Galloway-Mowat syndrome. J Med Genet. 2015;52:381–390. doi: 10.1136/jmedgenet-2014-102707. [DOI] [PubMed] [Google Scholar]
- Blom M, Reis K, Nehru V, Blom H, Gad AK, Aspenstrom P. RhoD is a Golgi component with a role in anterograde protein transport from the ER to the plasma membrane. Exp Cell Res. 2015;333:208–219. doi: 10.1016/j.yexcr.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Burgess T, Brown NJ, Stark Z, Bruno DL, Oertel R, Chong B, Calabro V, Kornberg A, Sanderson C, Kelly J, et al. Characterization of core clinical phenotypes associated with recurrent proximal 15q25.2 microdeletions. Am J Med Genet A. 2014;164A:77–86. doi: 10.1002/ajmg.a.36203. [DOI] [PubMed] [Google Scholar]
- Byrne S, Jansen L, U.K.-I. JM, Siddiqui A, Lidov HG, Bodi I, Smith L, Mein R, Cullup T, Dionisi-Vici C, et al. EPG5-related Vici syndrome: a paradigm of neurodevelopmental disorders with defective autophagy. Brain. 2016;139:765–781. doi: 10.1093/brain/awv393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Siripala AD, Leong JM, Welch MD. Membrane-deforming proteins play distinct roles in actin pedestal biogenesis by enterohemorrhagic Escherichia coli. J Biol Chem. 2012;287:20613–20624. doi: 10.1074/jbc.M112.363473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci. 2015;128:193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Colin E, Huynh Cong E, Mollet G, Guichet A, Gribouval O, Arrondel C, Boyer O, Daniel L, Gubler MC, Ekinci Z, et al. Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway-Mowat syndrome. Am J Hum Genet. 2014;95:637–648. doi: 10.1016/j.ajhg.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB. Actin nucleation by WH2 domains at the autophagosome. Nat Commun. 2015;6:7888. doi: 10.1038/ncomms8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- Doelken SC, Seeger K, Hundsdoerfer P, Weber-Ferro W, Klopocki E, Graul-Neumann L. Proximal and distal 15q25.2 microdeletions-genotype-phenotype delineation of two neurodevelopmental susceptibility loci. Am J Med Genet A. 2013;161A:218–224. doi: 10.1002/ajmg.a.35695. [DOI] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM, Burckstummer T. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad AK, Nehru V, Ruusala A, Aspenstrom P. RhoD regulates cytoskeletal dynamics via the actin nucleation-promoting factor WASp homologue associated with actin Golgi membranes and microtubules. Mol Biol Cell. 2012;23:4807–4819. doi: 10.1091/mbc.E12-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, Artal-Martinez de Narvajas A, Koenig AO, Billadeau DD. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell. 2012;23:3215–3228. doi: 10.1091/mbc.E12-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks RN, Puffenberger EG, Baple E, Harding B, Crino P, Fogo AB, Wenger O, Xin B, Koehler AE, McGlincy MH, et al. Recessive nephrocerebellar syndrome on the Galloway-Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73. Brain. 2015;138:2173–2190. doi: 10.1093/brain/awv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Zajac AL, Holzbaur EL, Ostap EM, Dominguez R. WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr Biol. 2015;25:1791–1797. doi: 10.1016/j.cub.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016;5:e12245. doi: 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi N, Chen Y, Wang S, Chen M, Zhao M, Yang G, Ma M, Su Q, Luo S, Shi J, et al. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol. 2015;17:1112–1123. doi: 10.1038/ncb3215. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulding DA, Record J, Malinova D, Thrasher AJ. Actin cytoskeletal defects in immunodeficiency. Immunol Rev. 2013;256:282–299. doi: 10.1111/imr.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Ktistakis NT. Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem. 2013;55:17–27. doi: 10.1042/bse0550017. [DOI] [PubMed] [Google Scholar]
- Rosti RO, Dikoglu E, Zaki MS, Abdel-Salam G, Makhseed N, Sese JC, Musaev D, Rosti B, Harbert MJ, Jones MC, et al. Extending the mutation spectrum for Galloway-Mowat syndrome to include homozygous missense mutations in the WDR73 gene. Am J Med Genet A. 2016;170A:992–998. doi: 10.1002/ajmg.a.37533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti RO, Sotak BN, Bielas SL, Bhat G, Silhavy JL, Aslanger AD, Altunoglu U, Bilge I, Tasdemir M, Yzaguirrem AD, et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J Med Genet. 2017;54:399–403. doi: 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Russo AJ, Mathiowetz AJ, Hong S, Welch MD, Campellone KG. Rab1 recruits WHAMM during membrane remodeling but limits actin nucleation. Mol Biol Cell. 2016;27:967–978. doi: 10.1091/mbc.E15-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter K, Waschbusch D, Anft M, Hugging D, Kind S, Hanisch J, Lakisic G, Gautreau A, Barnekow A, Stradal TE. JMY is involved in anterograde vesicle trafficking from the trans-Golgi network. Eur J Cell Biol. 2014;93:194–204. doi: 10.1016/j.ejcb.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Shen QT, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang HW. Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J Cell Biol. 2012;199:111–124. doi: 10.1083/jcb.201204010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- Vodopiutz J, Seidl R, Prayer D, Khan MI, Mayr JA, Streubel B, Steiss JO, Hahn A, Csaicsich D, Castro C, et al. WDR73 mutations cause infantile neurodegeneration and variable glomerular kidney disease. Hum Mutat. 2015;36:1021–1028. doi: 10.1002/humu.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbe M, Kozyrev SV, Farias FH, Bremer HD, Hedlund A, Pielberg GR, Seppala EH, Gustafson U, Lohi H, Carlborg O, et al. Multiple Changes of gene expression and function reveal genomic and phenotypic complexity in SLE-like disease. PLoS Genet. 2015;11:e1005248. doi: 10.1371/journal.pgen.1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kenny SJ, Ge L, Xu K, Schekman R. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife. 2015;4:e11205. doi: 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.