Abstract
Personality traits have been suggested as potential endophenotypes for Bipolar Disorder (BP), as they can be quantitatively measured and show correlations with BP. The present study utilized data from 2745 individuals from 686 extended pedigrees originally ascertained for having multiplex cases of BP (963 cases of BPI or schizoaffective BP). Subjects were assessed with the NEO Personality Inventory, Revised (NEO PI-R) and genotyped using the Illumina HumanLinkage-24 Bead Chip, with an average genetic coverage of 0.67 cM. Two point linkage scores were calculated for each trait as a quantitative variable using SOLAR (Sequential Oligogenic Linkage Analysis Routines). Suggestive evidence for linkage was found for neuroticism at 1q32.1 (LOD=2.52), 6q23.3 (2.32), 16p12 (2.79), extraversion at 4p15.3 (2.33), agreeableness at 4q31.1 (2.37), 5q34 (2.80), 7q31.1 (2.56), 16q22 (2.52), and conscientiousness at 4q31.1 (2.50). Each of the above traits have been shown to be correlated with the broad BP phenotype in this same sample. In addition, for the trait of openness, we found significant evidence of linkage to chromosome 3p24.3 (rs336610, LOD=4.75) and suggestive evidence at 1q43 (2.74), 3p26 (2.71), 5q35.1 (3.03), 11q14.3 (2.61), 11q21 (2.30), and 19q13.1 (2.52). These findings support previous linkage findings of the openness trait to chromosome 19q13 and the agreeableness trait to 4q31 and identify a number of new loci for personality endophenotypes related to bipolar disorder.
Keywords: genome-wide, linkage studies, Latino, NEO, personality, bipolar disorder
INTRODUCTION
Bipolar disorder is an often devastating mental illnesses, with high emotional, social and economic impact on the lives of patients and family members [Jin and McCrone, 2015; Miller et al., 2014]. In recent decades, there has been significant progress in developing diagnostic methods for reliably diagnosing severe bipolar disorder (bipolar disorder type I) and some related bipolar “spectrum” disorders (bipolar type II disorder), and there has also been recent progress in identifying some of the genetic loci (through linkage and association analyses) which contribute to severe bipolar disorder. Controversy remains, however, about the diagnostic borders of bipolar disorder, particularly in relation to schizophrenia and schizoaffective disorder, other types of mood disorder, and to personality disorders [Doherty and Owen, 2014; Bezerra-Filho et al., 2015]. Genetic studies have provided one approach to better identify the biologic underpinnings of bipolar disorder and to study the overlap of bipolar disorder with other psychiatric conditions and psychologic measures.
The majority of genetic studies for bipolar disorder to date have focused on attempts to find linkage or association to categorical diagnoses such as BPI. Although these have shown some success in identifying chromosomal regions [Escamilla and Zavala, 2008] and associated genotypes [Sklar et al., 2011] that contain or are near putative genetic variants that predispose to bipolar disorder, a large portion of the genetic basis of bipolar disorder remains unaccounted for [Gershon et al., 2011]. More recently, alternative quantitative phenotypes have been proposed as means of overcoming some of the obstacles the psychiatric field has had to identifying the genetic factors contributing to the bipolar phenotype. Although likely sharing, with bipolar disorder, complex causes (both genetic and environmental) contributing to the phenotype, personality dimensions, as measured with scales such as the NEO (Costa and McRae 1992), are one set of quantitative traits which may serve as promising endophenotypes for bipolar disorder [Greenwood et al., 2013]. Aside from genetic studies, it has previously been noted that there are associations between some of the personality traits (which are quantitative in nature and applicable to all human beings) and some categorical psychiatric conditions like depression and schizophrenia [Koorevaar et al., 2013; Guerra et al., 2000]. Identification of the genetic components of personality traits, at the same time as studies are underway to identify genetic components of bipolar disorder and other psychiatric conditions, offer an opportunity to better understand the interactions and components of the biological components that shape psychological experience and psychiatric illness.
The Genetics of Bipolar Disorder in Latino Populations (GBDLP) study employed both categorical diagnoses (DSM-IV) and quantitative measures (including measures of personality) in our assessments of a large number of individuals from extended pedigrees with at least one case of bipolar disorder, to allow for the opportunity to identify genes involved in both categorical illnesses and quantitative traits that might show some overlap with bipolar disorder. We have previously reported on the heritability of factors in the Five Factor personality model (NEO) and their correlation with bipolar disorder in this sample of of pedigrees [Hare et al., 2011]. In those studies we reported NEO factor heritability scores as follows: 0.25 for agreeableness, 0.24 for conscientiousness, 0.24 for extraversion, 0.23 for neuroticism, and 0.32 for openness to experience. This confirmed, in these families, that the five NEO factors are indeed heritable and therefore partly due to genetic inheritance, as previously shown in other research studies of personality conducted using twin-based designs [Bergeman et al., 1993; Jang and Livesley, 1996; Jang et al., 1998; Viken et al., 1994; Pincombe et al., 2007; Rettew et al., 2008; Wray et al., 2007]. In addition, in Hare et al. (2011) we also showed that categorical diagnoses for bipolar disorder were heritable in the GBDLP families (heritabilities from 0.49 to 0.43 for narrow and broad phenotype models). Finally, we were able to show, within the GBDLP families, that four of the NEO personality factors showed significant correlations with the broad phenotype of BP (BPI, BPII, schizoaffective BP, recurrent unipolar depression). Neuroticism correlated positively with the broad BP phenotype, while conscientiousness, extraversion, and agreeableness correlated negatively with the broad BP phenotype.
As a first step in finding genes of relevance for bipolar disorder in the Latino population, we completed a genome-wide linkage scan for bipolar disorder in the GBDLP families, identifying loci for BP at 8q24 and 14q32 [Gonzalez et al., 2014]. Here, we now report on a quantitative trait linkage analysis for the five NEO factors in this same set of families. Our goal was to identify any genetic loci showing linkage to one or more of the five NEO personality factors, which to our knowledge is the first time this has been done in families of Latino ancestry. In addition, we were interested to see if there was any overlap of personality factor linkage sites with the sites previously shown to be in linkage with the bipolar disorder phenotype in these families, or if the personality factors indicated linkage to areas not directly linked to the categorical phenotype of bipolar disorder.
MATERIALS AND METHODS
Study Population
The present study utilized data from the Genetics of Bipolar in Latino Populations study, a multicenter study designed to investigate genetic determinants of bipolar disorder. This sample consists of 686 extended pedigrees (all of Mexican or Central American ancestry) originally ascertained for having multiplex cases of Bipolar Type I disorder. Samples were recruited from the southwest United States (Los Angeles, CA; Albuquerque, NM; El Paso and San Antonio, TX), Mexico (Mexico City and Monterrey), Guatemala, and Costa Rica. The average pedigree size consisted of 5.48 individuals, ranging in size from 2 to 21 subjects. All participating subjects gave informed consent using a process that was approved by each participating site’s Institutional Review Board and by the National Institute of Mental Health. The study was conducted in accordance with the principles of the Helsinki Declaration of 1975 and in compliance with the Code of Ethics of the World Medical Association.
Phenotypic Assessment
The majority of subjects (including all available family members of BP subjects) completed assessments using the short form of the NEO Personality Inventory, Revised (NEO PI-R) [Costa and McRae, 1992]. 2768 subjects completed the assessments administered by certified study personnel in their preferred language (English or Spanish). For Spanish speaking subjects, a validated Spanish translation of the NEO short form [Abella et al., 2011] was used. In reviewing the data question # 26 of the NEO was felt to be poorly translated in the Spanish version, so this item was treated as missing data across all study samples. Additional missing data from the NEO PI-R was addressed using recommended guidelines. Twenty participants were removed due to missing data points in more than 10 of the possible 60 items. Three additional participants were removed because of missing data points in more than 3 items within a subsection. Additional missing values (which constituted less than .17 percent of the total NEO data) were imputed using a neutral response of “2”. Factor scores for neuroticism, extraversion, openness to experience, agreeableness, and conscientiousness were calculated as described previously [Hare et al., 2011]. A total of 2745 participants were retained for further analysis. Lifetime ratings for depression and mania were scored by best estimators for each subject using the Lifetime Dimensions of Psychosis Scale [Levinson et al., 2002]. Categorical phenotypes, although not utilized in the present analysis, were designated using a consensus best estimate process, as previously described in Escamilla et al. (2007).
Genotyping
For each subject, a total of 5574 single nucleotide polymorphisms (SNOs) were genotyped at the Center for Inherited Disease Research using the Illumina HumanLinkage-24 Bead Chip, with an average genetic coverage of 0.67 cM and a total of 5574 SNPs. Pedigrees were examined for consistency of familial relationships using PREST [McPeek and Sun, 2000] and genotype quality was tested using PLINK (Purcell et al., 2007; http://pngu.mgh.harvard.edu/purcell/plink/) and MERLIN software programs as described previously [Gonzalez et al., 2014].
Statistical Analysis
The SOLAR (Sequential Oligogenic Linkage Analysis Routines) computer package [Almasy and Blangero, 1998] was used to estimate heritability and linkage to chromosomal locations affecting variation in personality scores based on the NEO PI-R. A maximum likelihood-based variance decomposition approach was used to estimate heritability for each of the factor scores for neuroticism, extraversion, openness to experience, agreeableness, and conscientiousness.
To correct for intermarker linkage disequilibrium, we selected a subset of markers to use in linkage analysis, eliminating markers that were in strong disequilibrium with each other. PLINK (Purcell et al., 2007; http://pngu.mgh.harvard.edu/purcell/plink/) version 1.07 was utilized. We performed pruning at a threshold of r2=0.2, which is below the level of r2=0.3 recommended by Sobota et al. (2015). After pruning, the remaining subset of markers numbered 3523, and only these were used in subsequent linkage analyses.
Two-point linkage analyses was performed to locate areas of the genome that harbor QTLs that influence each respective personality trait based on identity-by-descent (IBD) allele sharing among family members. Distributions of phenotypes were examined prior to analysis to ensure normalization of data. Age, sex, age-sex interaction, language of assessment, and lifetime ratings of mania and depression were tested as covariates in all analyses and retained if a likelihood ratio test was significant at p<0.10. Thresholds of significance were set at LOD scores > 3.6 and suggestive at LOD scores > 2.2, based on Lander and Kruglyak’s (1995) suggested guidelines for the interpretation of linkage studies when the mode of inheritance is uncertain. We did calculate empirical p values by permuting phenotypic data and performing 1000 permuted genome wide linkage analyses for each of the five phenotypes. These suggested an empirical genome-wide level of 2.20 for significant linkage (i.e. this was the highest individual LOD score out of 1000 permutations across the genome for each of the 5 NEO phenotypes) and a suggestive level of 1.34 for suggestive linkage. Since these empirically suggested values were so low, we chose to use the more stringent criteria of Lander and Kruglyak. Based on the empirically calculated levels, all the marker linkages reported here would be considered evidence of significant linkage.
To test whether the any of the five NEO traits were correlated with each other, we performed a correlation analysis between each possible two traits, with age, gender, and site included as co-variates. For these correlation analyses, we randomly chose one subject per family.
RESULTS
Two-point QTL Analyses
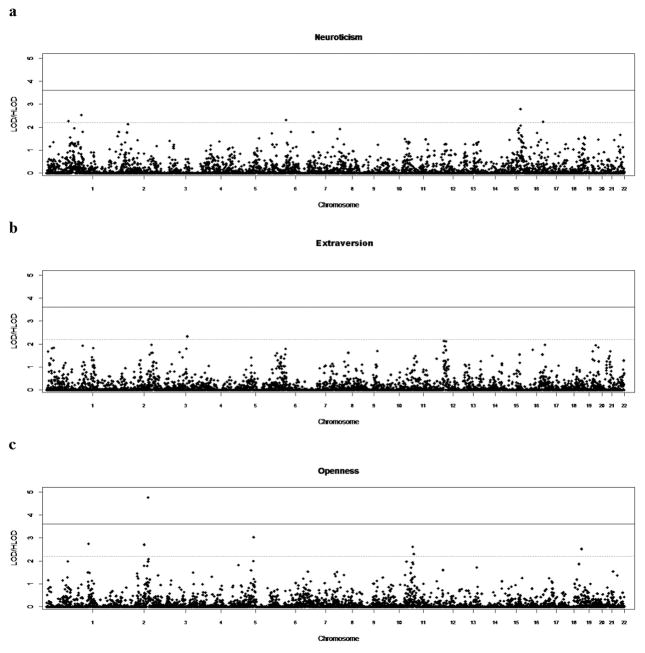
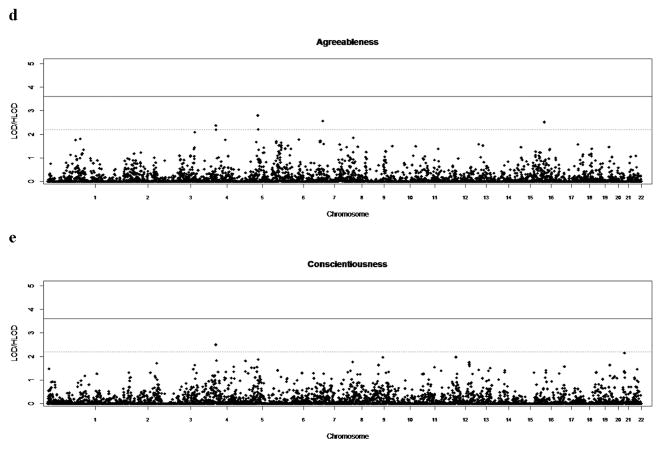
Linkage plots for each of the NEO traits are shown in figure 1, and markers which gave two-point LOD scores in the suggestive to significant range are described in table 1. Suggestive evidence for linkage was found for neuroticism at five loci (1p22 (rs508020, LOD=2.2598), 1q32.1 (rs1052238, LOD=2.5230), 6q23.3 (rs1041480, LOD=2.3152), 16p12 (rs936347, LOD=2.7873) and 17q11.2 (rs1472081, LOD=2.2374)), extraversion at one locus 4p15.3 (rs6845573, LOD=2.3334), agreeableness at four loci (4q31.1 (rs1425566, LOD=2.3735; rs1389331, LOD=2.1980), 5q34 (rs2069347, LOD=2.7996; rs878953, LOD=2.2085), 7q31.1 (rs2028030, LOD=2.5635), and 16q22 (rs722579, LOD=2.5185)), and conscientiousness at one locus 4q31.1 (rs1425566, LOD=2.5001). Each of the above traits have been shown to be correlated with the broad BP phenotype in this same sample (Hare et al. 2011). In addition, for the trait of openness, we found significant evidence of linkage to chromosome 3p24.3 (rs336610, LOD=4.7535) and suggestive evidence at 1q43 (rs7520974, LOD=2.7434), 3p26 (rs1850264, LOD=2.7087), 5q35.1 (rs791346, LOD=3.0312), 11q14.3 (rs160195, LOD=2.6129), 11q21 (rs1255182, LOD=2.3043), and 19q13.1 (rs4239577, LOD=2.5204).
Figure 1.
Genome-wide two-point results from non-parametric linkage (NPL) analysis. The X axis depicts the whole autosomal genome divided into 22 chromosomes. The Y axis shows the log of odds (LOD) scores from NPL analysis. (a) Neuroticism, (b) Extraversion, (c) Openness, (d) Agreeableness, (e) Conscientiousness. The solid horizontal line to indicate the empirical genome-wide significant cutoff (LOD scores > 3.6) and another dashed horizontal line to indicate the empirical genome-wide suggestive cutoff (LOD scores > 2.2) as determined for each trait are represented.
Table 1.
Two-point linkage results at loci with significant (LOD>3.6) or suggestive (LOD>2.2) LOD scores in a genome-wide scan of NEO personality factors in Latino pedigrees
| Trait | Locus | Marker | Position (in bp) | LOD | Nearest Gene (location of gene in bp) |
|---|---|---|---|---|---|
| Neuroticism | |||||
| 1q32.1 | rs1052238 | 198665496 | 2.5230 | PTPRC (198638968–198757476) | |
| 6q23.3 | rs1041480 | 135051938 | 2.3152 | HBS1L (134960378–135054898) | |
| 16p12 | rs936347 | 17176938 | 2.7873 | XYLT1 (17102324–17470881) | |
| Extraversion | |||||
| 4p15.3 | rs6845573 | 8061774 | 2.3334 | ABLIM2 (7965310–8158832) | |
| Openness | |||||
| 1q43 | rs7520974 | 239903960 | 2.7434 | CHRM3 (239386565–239911462) | |
| 3p24.3 | rs336610 | 18608523 | 4.7535 | LOC101927777 (18445237–18530114) KCNH8(19147093–19535643) |
|
| 5q35.1 | rs791346 | 173765741 | 3.0312 | LOC101928136 (173707599–173746209) LOC101928154 (173786790–173790942) |
|
| 11q14.3 | rs160195 | 88559805 | 2.6129 | GRM5 (88504576–89065961) | |
| 11q21 | rs1255182 | 95739227 | 2.3043 | LOC100129203(95230186–95234404) FGFR3P2 (95756954–95757444) |
|
| 19q13.1 | rs4239577 | 33803962 | 2.5204 | KCTD15 (33796846–33815761) | |
| Agreeableness | |||||
| 4q31.1 | rs1425566 | 133361842 | 2.3735 | PCDH10(133149290–133208606) PABPC4L(134196333–134201748) |
|
| 5q34 | rs2069347 | 163440189 | 2.7996 | CCNG1 (163437270–163445016) | |
| 5q34 | rs878953 | 164499486 | 2.2085 | LOC102546299 (164470279–164542984) | |
| 7q31.1 | rs2028030 | 107719560 | 2.5635 | SLC26A4(107660635–107717809) LOC101927974 (107742751–107744051) |
|
| 16q22 | rs722579 | 70494321 | 2.5185 | COG4 (70480569–70523554) | |
| Conscientiousness | |||||
| 4q31.1 | rs1425566 | 133361842 | 2.5001 | PCDH10 (133149290–133208606) PABPC4L(134196333–134201748) |
Candidate Gene Identification
Of the single nucleotide polymorphisms which gave suggestive or significant linkage signals, table 1 also lists the gene within which these SNPs fell or (for those SNPs falling in regions outside of genes) the genes closest (proximally and distally) to the SNP. Genes nearest to the peak linkage scores for neuroticism include CDC14A, PTPRC, HBS1L, XYLT1, PIPOX/MYO18A. The candidate gene near the peak score for extraversion was ABLIM2. Genes closest to the peak scores for openness included LOC101927777/KCNH8, LOC101927174/CHL1, LOC101928136/LOC101928154, GRM5, LOC100129203/FGFR3P2, and KCTD15. For agreeableness, the genes nearest the peaks included PCDH10/PABPC4L, CCNG1, LOC102546299, SLC26A4/LOC101927974, COG4, and the peak for conscientiousness was nearest to the PCDH10/PABPC4L genes.
Correlation Analyses of the Five NEO traits
Of the possible correlations tested between each two traits, we identified two significant correlations. The NEO raw score for Neuroticism correlated negatively with the raw score for Agreeableness (r= −0.509, p< 0.001). The NEO raw scores for Agreeableness and Conscientiousness showed a moderate positive correlation (r= 0.497, p< 0.001).
DISCUSSION
A novel significant locus for the openness to experience trait
The current study offers additional evidence to the literature regarding the location of genes which contribute to the personality traits of neuroticism, extraversion, openness, agreeableness and conscientiousness, and is the first such study that we are aware of to identify these loci in a Latin American population sample. Only one of these loci showed evidence at the level of genome-wide significance (using the standard suggested by Lander and Kruglyak), and this was for the trait of openness to experience, on chromosome 3, at the locus 3p24.3 (rs336610, LOD = 4.75). Previous linkage screens for openness, summarized in Amin et al. (2012), conducted in samples from the Netherlands and Australia, and comprising from 1096 to 2244 subjects, did not find significant evidence of linkage for the openness trait, and to our knowledge, this is the first significant linkage finding for this trait in any population studied thus far. In these previous studies, the 3p24.3 locus was also not indicated as showing suggestive evidence of linkage, and thus this is a novel locus for this trait. The exact location of peak linkage signals do not necessarily correlate with the genetic variants contributing to the signal. Nevertheless, it is of interest to note the gene nearest the SNP giving the strongest two-point LOD score for this trait. rs336610 is near the KCNH8 gene. KCNH8 codes for a potassium channel protein that is widely distributed in the brain and is involved in neuronal excitability [Zou et al., 2003; Wisniewska et al., 2012].
A previous meta-analysis which included the four Danish and Australian study samples, using a binning strategy, did find evidence of a locus at 11q24 showing linkage to the openness trait. In the current study, we found suggestive evidence of linkage for the openness trait at 11q14 and 11q21, which may indicate overlap with this previous meta-analysis. Of potential interest, the SNP giving the suggestive linkage score for 11q21 was closest to the fibroblast growth factor receptor 3 pseudogene 2 (FGFR3P2), which has been associated with smoking cessation. We also found suggestive evidence for this trait at loci on chromosome 1, 5, and 19 (table), none of which have been highlighted as suggestive in the previous linkage studies mentioned above. Of potential interest, the linkage signal on chromosome 5 was provided by a SNP closest to the Glutamate receptor gene GRM5, which has been highlighted as a promising neuroreceptor for several neuropsychiatric phenotypes including smoking addiction, attention deficit/hyperactivity disorder [Gregory et al., 2014; Hindorff et al., 2014; Hulka et al., 2014; Kandratavicius et al., 2013; Tsamis et al., 2013; Vallano et al., 2013; Welter et al., 2014] and even bipolar disorder [Fatemi et al., 2013; Dhingra et al., 2012; Nakanishi et al., 1998].
We should emphasize, however, that openness to experience as a personality trait did not show association to the bipolar disorder phenotype in our current study sample [Hare et al., 2011]. Therefore, although these findings for the openness trait may be of importance in identifying the genetic contribution to openness, in our sample, it may have limited relevance with regard to identifying genetic sites contributing in some way to the bipolar phenotype.
Linkage regions for traits associated with bipolar disorder in this sample
Although the current study did not find significant linkage signals for any of the other NEO traits, we did find suggestive linkage evidence for the traits of neuroticism, extraversion, agreeableness, and conscientiousness, all of which, in our current sample, have been shown to be associated with the phenotype of bipolar disorder. As such, these loci may be indicating regions harboring not just genes for these traits, but genes of special relevance for bipolar disorder in the Latino population.
Neuroticism
Suggestive loci for neuroticism were found at two loci (at 6q3.3, and 16p12). Of note, neither of these loci overlap with significant or suggestive LOD scores for the neuroticism trait from the previous studies conducted in the Netherlands and Australia [Amin et al., 2013] or from GWAS analyses for this trait conducted in the Korean population [Kim et al., 2015]. This suggests that the gene loci contributing to this trait in the Latino population differ from genetic loci contributing to this trait in European or Asian ancestry populations studied to date. Alternatively, the loci contributing to this trait in the current sample may be different due to the fact that these families have a high loading for bipolar disorder, which may alter how psychological phenotypes are expressed in these families.
Extraversion
There was only one locus in the current study which showed suggestive linkage to the trait of extraversion (which is negatively correlated with the bipolar phenotype in our sample). This locus, at 4p15.3 extraversion (rs6845573, LOD=2.3334) has not previously been highlighted as linked to or associated with extraversion in the previous studies conducted in the Netherlands, Australia and Korea [Amin et al., 2013; Kim et al., 2015].
Agreeableness
In the current sample, the trait for agreeableness showed evidence of suggestive linkage to 4q31.1, 5q34, 7q31.1, and 16q22. The first of these loci, at 4q31.1, is very close to a locus for agreeableness at 4q32, which had a LOD score of 2.20 in the Erasmus Rucphen family study [Amin et al., 2012]. This same locus showed suggestive evidence of linkage to the trait for conscientiousness. The fact that we show this same region linked to the traits of agreeableness and conscientiousness may be related to our finding that these two traits show moderate correlation between each other and are therefore not completely independent.
Conscientiousness
Finally, the current study gave suggestive evidence of linkage to the trait for conscientiousness (negatively correlated to bipolar disorder in our sample) at 4q31.1. This locus appears to be a novel one for this trait and has not previously been reported in the Danish, Australian or Korean studies mentioned above. The SNP giving evidence of linkage to this locus (rs1425566) is near the protocadherin 10 gene (PCDH10). The encoded protein, a cadherin-related neuronal receptor, is involved in cell-cell connections in the brain. Cadherins have been shown to regulate neuronal migration, gray matter differentiation, neural circuit formation and synapse formation and remodeling. In addition several neuropsychiatric disorders, including autism and bipolar disorder have been associated with cadherin genes [Redies et al., 2012]. As mentioned above, this same SNP showed suggestive evidence of linkage to the agreeableness trait in the current paper, suggesting this SNP might influence both of these traits or, given that these traits are correlated with each other, this SNP may be associated with a common endophenotype that is captured by both the measures for agreeableness and conscientiousness.
With regard to efforts to identify the genes which underlie the genetic factors of personality, the current study is the first individual study to identify a significant linkage site for the openness trait, as defined in the NEO Five Factor Scale. The study also gives support to previous studies which have found evidence of linkage for openness in the 11q21–24 region and for agreeableness in the 4q31–32 region. Additional loci giving suggestive loci for neuroticism, extraversion, openness, and conscientiousness found in this study appear unique thus far for this Latino population. It is not unusual, when mapping the genes for complex disorders, to have different linkage results, based on the particular set of families included in any analysis. Further studies will be needed to determine if there are indeed unique loci contributing to the genetic component of these personality traits in the Latino population or whether these findings were due to stochastic issues unrelated to particular ethnic origins in the study samples. It is also quite possible that one or more of the findings here might be a false positive, given that only one is at the level of genome-wide significance. Future studies which might incorporate quantitative association analyses may also contribute to identifying the genes involved in these personality factors, and linkage studies such as those presented here may help investigators to narrow in on specific regions of interest for these traits.
In terms of identifying genes which contribute in some manner to the phenotype of bipolar disorder, it is interesting to note that in a categorical linkage analysis for bipolar disorder, done with these same families, only three loci reached the level of evidence for suggestive or significant linkage. Those regions, at 2q13–14, 8q24, and 14q32, do not overlap with any of the 19 regions here which met suggestive or significant evidence of linkage to the five personality traits measured in these same subjects. Twelve of these regions (all those showing linkage to neuroticism, extraversion, agreeableness, and conscientiousness), given that they are in linkage with traits known to correlate with bipolar disorder in this sample, therefore offer the possibility to (through fine mapping) identify additional gene variants which contribute to bipolar disorder.
Although there are a number of interesting genes highlighted in this discussion section as being in close proximity with the linkage peaks mentioned, we must urge caution in extrapolating from linkage findings to identifying the genes within a linkage region which might carry variations which directly contribute to the traits of personality and bipolar disorder. Further studies, including fine mapping and sequencing of the genetic material in the region of these peaks, will be necessary to identify which genes and variants actually contribute to these behavioral phenotypes. Association studies, in subjects drawn from the same population, may be another way to narrow in on the genetic loci and genes involved in these interesting and complex phenotypes.
Acknowledgments
This project was funded by the NIMH Genetics Initiative study: Genetics of Bipolar Disorder in Latino Populations (5RO1MH069856, Principal Investigator: Dr. Escamilla). The authors have no conflicts of interest to report. We thank the many families and family members who participated in these studies. We also thank Dr. John Blangero for consultation regarding the use of SOLAR in these analyses.
References
- Abella V, Panksepp J, Manga D, Bárcena C, Iglesias JA. Spanish validation of the Affective Neuroscience Personality Scales. Span J Psychol. 2011;14(2):926–935. doi: 10.5209/rev_sjop.2011.v14.n2.38. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N, Schuur M, Gusareva ES, Isaacs A, Aulchenko YS, Kirichenko AV, et al. A genome-wide linkage study of individuals with high scores on NEO personality traits. Mol Psychiatry. 2012;17:1031–1041. doi: 10.1038/mp.2011.97. [DOI] [PubMed] [Google Scholar]
- Amin N, Hottenga JJ, Hansell NK, Janssens AC, de Moor MH, Madden PA, et al. Refining genome-wide linkage intervals using a meta-analysis of genome-wide association studies identifies loci influencing personality dimensions. Eur J Hum Genet. 2013;21(8):876–882. doi: 10.1038/ejhg.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeman CS, Chipuer HM, Plomin R, Pedersen NL, McClearn GE, Nesselroade JR, et al. Genetic and environmental effects on openness to experience, agreeableness, and conscientiousness: an adoption/twin study. J Personality. 1993;61:159–182. doi: 10.1111/j.1467-6494.1993.tb01030.x. [DOI] [PubMed] [Google Scholar]
- Bezerra-Filho S, Almeida AG, Studart P, Rocha MV, Lopes FL, Miranda-Scippa Â. Personality disorders in euthymic bipolar patients: a systematic review. Rev Bras Psiquiatr. 2015 May 1; doi: 10.1590/1516-4446-2014-1459. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Costa PT, McCrae RR. NEO Personality Inventory-Revised (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) Professional Manual. Psychological Assessment Resources; Odessa, FL: 1992. [Google Scholar]
- Dhingra A, Ramakrishnan H, Neinstein A, Fina ME, Xu Y, Li J, et al. Gβ3 is required for normal light ON responses and synaptic maintenance. J Neurosci. 2012;32(33):11343–11355. doi: 10.1523/JNEUROSCI.1436-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6(4):29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla MA, Ontiveros A, Nicolini H, Raventos H, Mendoza R, Medina R, et al. A genome-wide scan for schizophrenia and psychosis susceptibility loci in families of Mexican and Central American ancestry. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(2):193–199. doi: 10.1002/ajmg.b.30411. [DOI] [PubMed] [Google Scholar]
- Escamilla MA, Zavala JM. Genetics of bipolar disorder. Dialogues Clin Neurosci. 2008;10(2):141–152. doi: 10.31887/DCNS.2008.10.2/maescamilla. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Rooney RJ, Thuras PD. mRNA and protein expression for novel GABAA receptors θ and ρ2 are altered in schizophrenia and mood disorders; relevance to FMRP-mGluR5 signaling pathway. Transl Psychiatry. 2013;18:3, e271. doi: 10.1038/tp.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, Alliey-Rodriquez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry. 2011;168(3):253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Camarillo C, Rodriguez M, Ramirez M, Zavala J, Armas R, et al. A genome-wide linkage scan of bipolar disorder in Latino families identifies susceptibility loci at 8q24 and 14q32. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(6):479–491. doi: 10.1002/ajmg.b.32251. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Badner JA, Byerley W, Keck PE, McElroy SL, Remick RA, et al. Heritability and linkage analysis of personality in bipolar disorder. J Affet Disord. 2013;151(2):748–755. doi: 10.1016/j.jad.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Nguyen ED, Malosh C, Mendenhall JL, Zic JZ, Bates BS, et al. Identification of specific ligand-receptor interactions that govern binding and cooperativity of diverse modulators to a common metabotropic glutamate receptor 5 allosteric site. ACS Chem Neurosci. 2014;5(4):282–295. doi: 10.1021/cn400225x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra RJ, Nestor PG, O’Donnell BF. Personality traits in schizophrenia: a comparison with a community sample. J Nerv Ment Dis. 2000;188(1):31–35. doi: 10.1097/00005053-200001000-00006. [DOI] [PubMed] [Google Scholar]
- Hare E, Contreras J, Raventos H, Flores D, Jerez A, Nicolini H, et al. Genetic structure of personality factors and bipolar disorder in families segregating bipolar disorder. J Affect Disord. 2012;136(3):1027–1033. doi: 10.1016/j.jad.2011.04.057. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, et al. European Bioinformatics Institute, European Bioinformatics Institute. [Accessed [date of access]];A Catalog of Published Genome-Wide Association Studies. 2014 Available at: www.genome.gov/gwastudies.
- Hulka LM, Treyer V, Scheidegger M, Preller KH, Vonmoos M, Baumgartner MR, et al. Smoking but not cocaine use is associated with lower cerebral metabotropic glutamate receptor 5 density in humans. Mol Psychiatry. 2014;19(5):625–632. doi: 10.1038/mp.2013.51. [DOI] [PubMed] [Google Scholar]
- Jang KL, Livesley J. Heritability of the Big Five personality dimensions and their facets: a twin study. J Pers. 1996;64:577–592. doi: 10.1111/j.1467-6494.1996.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Jang KL, McCrae RR, Angleitner A, Riemann R, Livesley WJ. Heritability of facet-level traits in a cross-cultural twin sample: support for a hierarchical model of personality. J Pers Soc Psychol. 1998;74:1556–1565. doi: 10.1037//0022-3514.74.6.1556. [DOI] [PubMed] [Google Scholar]
- Jin H, McCrone P. Cost-of-illness studies for bipolar disorder: systematic review of international studies. Pharmacoeconomics. 2015;33(4):341–353. doi: 10.1007/s40273-014-0250-y. [DOI] [PubMed] [Google Scholar]
- Kandratavicius L, Rosa-Neto P, Monteiro MR, Guiot MC, Assirati JA, Jr, Carlotti CG, Jr, et al. Distinct increased metabotropic glutamate receptor type 5 (mGluR5) in temporal lobe epilepsy with and without hippocampal sclerosis. Hippocampus. 2013;23(12):1212–1230. doi: 10.1002/hipo.22160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Kim HN, Roh SJ, Lee MK, Yang S, Lee SK, et al. GWA meta-analysis of personality in Korean cohorts. J Hum Genet. 2015 doi: 10.1038/jhg.2015.52. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Koorevaar AM, Comijs HC, Dhondt AD, van MArwijk HW, van der Mast RC, Naarding P, et al. Big Five personality and depression diagnosis, severity and age of onset in older adults. J Affect Disord. 2013;151(1):178–185. doi: 10.1016/j.jad.2013.05.075. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mowry BJ, Escamilla MA, Faraone SV. Psychosis scale (LDPS): description and interrater reliability. Schizophr Bull. 2002;28(4):683–695. doi: 10.1093/oxfordjournals.schbul.a006972. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Dell’Osso B, Ketter TA. The prevalence and burden of bipolar depression. J Affect Disord. 2014;169(Suppl 1):S3–11. doi: 10.1016/S0165-0327(14)70003-5. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Nakajima Y, Masu M, Ueda Y, Nakahara K, Watanabe D, et al. Glutamate receptors: brain function and signal transduction. Brain Res Brain Res Rev. 1998;26(2–3):230–235. doi: 10.1016/s0165-0173(97)00033-7. [DOI] [PubMed] [Google Scholar]
- Pincombe JL, Luciano M, Martin NG, Wright MJ. Heritability of NEO-PI-R extraversion facets and their relationship with IQ. Twin Res Hum Genet. 2007;10:462–469. doi: 10.1375/twin.10.3.462. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am Jour Hum Genet. 2007:81. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Rettew DC, Rebollo-Mesa I, Hudziak JJ, Willemsen G, Boomsma DI. Non-additive and additive genetic effects on extraversion in 3314 Dutch adolescent twins and their parents. Behav Genet. 2008;38:223–233. doi: 10.1007/s10519-008-9192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota RS, Shriner D, Kodaman N, Goodloe R, Zheng W, Yu-Tang Gao, Edwards TL, Amos CI, Williams SM. Addressing population-specific multiple testing burderns in genetic association studies. Ann Hum Genet. 2015;79(2):136–147. doi: 10.1111/ahg.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamis KI, Mytilinaios DG, Njau SN, Baloyannis SJ. Glutamate receptors in human caudate nucleus in normal aging and Alzheimer’s disease. Curr Alzheimer Res. 2013;10(5):469–475. doi: 10.2174/1567205011310050002. [DOI] [PubMed] [Google Scholar]
- Vallano A, Fernandez-Duenas V, Garcia-Negredo G, Quijada MA, Simon CP, Cuffí ML, et al. Targeting striatal metabotropic glutamate receptor type 5 in Parkinson’s disease: bridging molecular studies and clinical trials. CNS Neurol Disord Drug Targets. 2013;12(8):1128–1142. [PubMed] [Google Scholar]
- Viken RJ, Rose RJ, Kaprio J, Koskenvuo M. A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol. 1994;66:722–730. doi: 10.1037//0022-3514.66.4.722. [DOI] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42(Database issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska MB, Nagalski A, Dabrowski M, Misztal K, Kuznicki J. Novel β-catenin target genes identified in thalamic neurons encode modulators of neuronal excitability. BMC Genomics. 2012;17(13):635. doi: 10.1186/1471-2164-13-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Birley AJ, Sullivan PF, Visscher PM, Martin MG. Genetic and phenotypic stability of measures of neuroticism over 22 years. Twin Res Hum Genet. 2007;10:695–702. doi: 10.1375/twin.10.5.695. [DOI] [PubMed] [Google Scholar]
- Zou A, Lin Z, Humble M, Creech CD, Wagoner PK, Krafte D, et al. Distribution and functional properties of human KCNH8 (Elk1) potassium channels. Am J Physiol Cell Physiol. 2003;285(6):C1356–1366. doi: 10.1152/ajpcell.00179.2003. [DOI] [PubMed] [Google Scholar]