Abstract
Aims
Breast myxoid fibroadenomas (MFAs) are characterized by a distinctive hypocellular myxoid stroma, and occur sporadically or in the context of Carney Complex, an inheritable condition caused by PRKAR1A inactivating germline mutations. Conventional fibroadenomas (FAs) are underpinned by recurrent MED12 mutations in the stromal components of the lesions. We sought to investigate the genomic landscape of MFAs and compare it to that of conventional FAs.
Methods and Results
Eleven MFAs from patients without clinical and/or genetic evidence of Carney Complex were retrieved. DNA samples of tumor and matching normal tissue were subjected to massively parallel sequencing using MSK-IMPACT, an assay targeting 410 cancer genes. Genetic alterations detected by MSK-IMPACT were tested in samples where the stromal and epithelial components were separately laser capture microdissected. Sequencing revealed no germline PRKAR1A mutations and non-synonymous mutations detected in six MFAs. Interestingly, in three of the MFAs where stromal and epithelial components were separately microdissected, the mutations were found to be restricted to the epithelial rather than the stromal component. The sole exception was a lesion harboring a somatic truncating PRKAR1A mutation. Upon histologic re-review, this case was reclassified as a breast myxoma, consistent with the spectrum of tumors observed in Carney Complex patients. In this case, the PRKAR1A somatic mutation was restricted to the stromal component.
Conclusion
MFAs lack MED12 mutations and their stromal component seems not to harbor mutations in the 410 cancer genes tested. Whole-exome and/or whole-genome analyses of MFAs are required to elucidate their genetic drivers.
Keywords: fibroadenoma, MSK-IMPACT, MED12, PRKAR1A, breast myxoma
INTRODUCTION
Breast fibroadenomas (FAs) are lesions characterized by a neoplastic stroma and epithelial elements with intracanalicular or pericanalicular growth patterns.1 Myxoid FAs (MFAs) constitute a histologic subtype of breast FAs characterized by a distinctive hypocellular stromal component with abundant myxoid matrix. Less common than conventional FAs, MFAs are known to develop sporadically, however these lesions have also been associated with Carney Complex.2, 3 Carney Complex is an inheritable, autosomal dominant condition caused by inactivating germline mutations of the PRKAR1A gene in about two thirds of cases.3–6 This syndrome is characterized by spotty skin pigmentation, endocrine overactivity, and an increased risk of tumor development. The spectrum of tumors associated with Carney Complex consists mainly of myxomas, ranging from cutaneous myxomas to cardiac myxomas; psammomatous melanotic schwannomas and pituitary adenomas are also common in this condition.4, 7 MFAs occur in approximately 40% of female Carney Complex patients,3, 7 however, the actual prevalence of MFAs arising in patients with Carney Complex has yet to be defined.
Recent studies have elucidated the genomic landscape of conventional FAs.8–12 These lesions have been shown to harbor a low mutation burden and highly recurrent mutations of MED12 gene, which are found in approximately 70% of cases.8–12 Additional genes mutated in conventional FAs include RARA, FLNA and ROS1.11 These studies have also revealed that the mesenchymal but not the epithelial component of FAs harbored the somatic mutations initially identified by tumor bulk sequencing, suggesting that conventional FAs likely constitute mesenchymal rather than biphasic tumors.8 The genomic landscape of MFAs, however, has yet to be described.
Due to the clinical association between MFAs and Carney Complex and the fact that a majority of Carney Complex patients harbor PRKAR1A germline mutations, we hypothesized that sporadic MFAs might differ from conventional FAs at the genetic level and be underpinned by somatic mutations in PRKAR1A. To address this hypothesis, we performed massively parallel sequencing analysis of 11 MFAs using a targeted capture assay including PRKAR1A, and investigated whether the somatic mutations identified in MFAs would be present in the stromal or epithelial components of these lesions.
MATERIALS AND METHODS
Cases
Eleven cases of MFAs were retrieved from the pathology archives of the Department of Pathology of Memorial Sloan Kettering Cancer Center (MSKCC). Three breast pathologists reviewed the slides of the cases and confirmed the diagnosis (AM, MPM, EB). All samples were anonymized prior to the analysis and approval by the local ethics committee was obtained.
Microdissection and DNA extraction
Representative sections from 11 MFAs were subjected to microdissection to ensure >80% tumor content, as previously described,12–14 and DNA was extracted from microdissected tissue (Supplementary methods). In selected cases (MFA2, MFA6, MF7 and MFA10), the epithelial and stromal components were separately laser capture microdissected as previously described12 (Supplementary Methods).
Targeted massively parallel sequencing
DNA samples extracted from eleven MFAs and their matching normal tissue were subjected to targeted capture massively parallel sequencing at the MSKCC Integrated Genomics Operation using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay, which targets all exons of 410 genes and non-coding regions of selected genes (Supplementary Methods).12, 13
massively parallel sequencing data analysis was performed as described previously (Supplementary Methods).12, 13 Somatic genetic alterations and their pathogenicity were detected using state-of-the-art algorithms.6, 15–23 Cancer genes were defined as those described in any of three cancer gene lists.24–26 FACETS15, optimized for MSK-IMPACT sequencing assays, was employed to define allele-specific copy number alterations (CNAs) as previously described.12, 13
Sanger sequencing
Hotspot TERT promoter mutations and selected mutations identified by MSK-IMPACT were investigated in the entire cohort of MFAs and/or in the laser capture microdissected epithelial and stromal components, respectively, by Sanger sequencing, as previously described9, 27 (Supplementary Methods).
Immunohistochemistry
Expression of p53 was investigated in one case (MFA2, Supplementary Methods).
RESULTS
Cases
The MFAs included in this study (Table 1) occurred in female patients, with a median age at diagnosis of 42 years (range 33–58). Tumor size ranged from 0.5 cm to 0.9 cm. All cases showed low stromal cellularity, and intra-canalicular or mixed intra- and peri-canalicular growth patterns (Figure 1). Nine patients presented with concurrent carcinoma (Supplementary Table S1). No MFA occurred in a patient known to have Carney Complex, nor in a carrier of germline PRKAR1A mutation. Two patients were carriers of germline mutations in BRCA1 (MFA4) and CDH1 (MFA7, Supplementary Table S1).
Table 1.
Clinicopathologic features of cases presented in this study.
| Sample | Diagnosis | Age | Laterality | Size (cm) | Growth pattern | Cellularity | Epithelial to Stromal ratio | Associated features |
|---|---|---|---|---|---|---|---|---|
| MFA1 | MFA | 39 | Left | 0.6 | Mixed | Low | High | Sclerosing adenosis |
| MFA2 | MFA | 46 | Right | 0.5 | Mixed | Low | Low | - |
| MFA3 | MFA | 33 | Left | 0.9 | Intracanalicular | Low | Low | Sclerosing adenosis |
| MFA4 | MFA | 37 | Right | 0.7 | Intracanalicular | Low | Low | - |
| MFA6 | Myxoma | 42 | Left | 0.6 | N/A | Low | N/A | - |
| MFA7 | MFA | 42 | Right | 0.6 | Intracanalicular | Low | Low | - |
| MFA8 | MFA | 58 | Left | 0.6 | Mixed | Low | Low | - |
| MFA9 | MFA | 49 | Left | 0.6 | Mixed | Low | Low | - |
| MFA10 | MFA | 41 | Left | 0.5 | Intracanalicular | Low | Low | - |
| MFA11 | MFA | 51 | Left | 0.5 | Intracanalicular | Low | Low | - |
| MFA12 | MFA | 39 | Left | 0.6 | Intracanalicular | Low | Low | - |
MFA: myxoid fibroadenoma; N/A: not applicable.
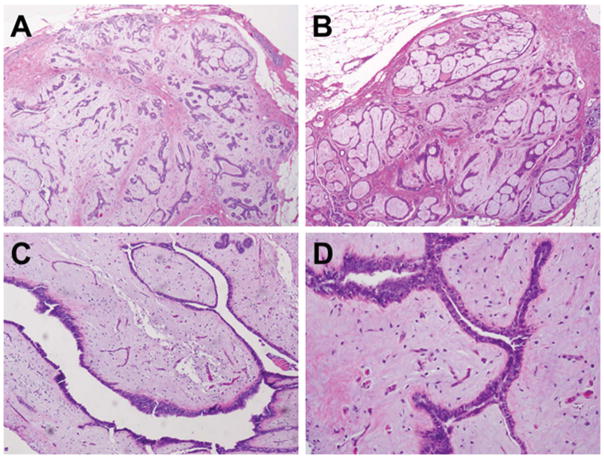
Figure 1. Morphologic features of myxoid fibroadenomas.
Representative micrographs of myxoid fibroadenomas included in this study. A) Low-power magnification (20X) of an MFA of mixed intra-canalicular and peri-canalicular architecture (MFA1). B) Low-power magnification (20X) of an MFA of intra-canalicular architecture (MFA10). C) Medium magnification (40X) of an MFA of intra-canalicular architecture (MFA3). D) High-power (200X) magnification of characteristic cleft-like architecture of intra-canalicular fibroadenomas (MFA3).
MFAs are heterogeneous at the genetic level
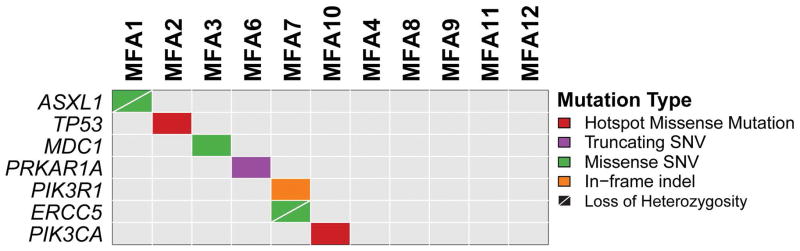
MSK-IMPACT assay was employed to identify the repertoire of somatic genetic alterations of MFAs. Sequencing was performed to a median coverage of 560× (range 269–745x, Supplementary Table S2) and revealed a median of one (range 0–3) somatic mutation per MFA (Figure 2, Table 2, Supplementary Table S3). Whilst eight cases harbored at least one synonymous (n=2) or non-synonymous somatic mutation (n=6), three cases lacked somatic mutations. Each case harboring non-synonymous somatic mutations displayed a unique mutational repertoire (Figure 2, Table 2, Supplementary Table S3). Neither recurrent mutations nor any mutation affecting MED12 were identified in the 11 MFAs analyzed. MFA6 displayed a truncating somatic single nucleotide variant in PRKAR1A. Single cases harbored pathogenic mutations in known cancer genes, including a hotspot missense TP53 mutation (R248W in MFA2), a PIK3CA mutation (H1047L in MFA10) and a disruptive deletion in PIK3R1 (L347del in MFA7). Distinct cases harbored missense mutations in ASXL1 (R244H, in MFA1) and ERCC5 (P714T in MFA7; Figure 2, Table 2; Table S3).
Figure 2. Repertoire of non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in myxoid fibroadenomas.
Heatmap indicating the non-synonymous somatic mutations identified in the MFAs analyzed. Each column represents one sample; mutated genes are reported in rows. The types of somatic mutations identified are color-coded according to the legend. Loss of heterozygosity (LOH) is identified by a diagonal bar. SNV, single nucleotide variant.
Table 2.
List of non-synonymous somatic mutations identified in this study.
| Case | Gene | cDNA Change | Protein Change | Mutant Allele Frequency | Sequencing Depth |
|---|---|---|---|---|---|
| MFA1 | ASXL1 | c.1154G>A | R244H | 6.12% | 392 |
| MFA2 | TP53 | c.932G>A | R248W | 4.11% | 632 |
| MFA3 | MDC1 | c.877G>A | H75Y | 3.83% | 287 |
| MFA6 | PRKAR1A | c.1048C>G | S307* | 4.35% | 138 |
| MFA7 | PIK3R1 | c.1655_1657del3 | L347del | 13.46% | 156 |
| MFA7 | ERCC5 | c.3563C>A | P714T | 5.75% | 348 |
| MFA10 | PIK3CA | c.3297A>T | H1047L | 6.38% | 376 |
Given that TERT promoter hotspot mutations have been described in phyllodes tumors12, 13, 28, we assessed by Sanger sequencing the presence of TERT promoter hotspot mutations in all MFAs included in this study. Confirming the findings obtained with MSK-IMPACT, which showed no TERT genetic alterations in MFAs here analyzed, Sanger sequencing revealed a lack of TERT promoter mutations (data not shown).
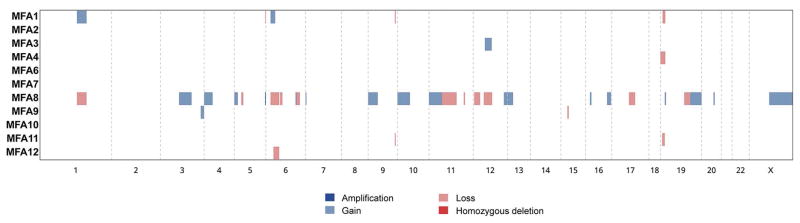
Copy number analysis revealed that the majority of MFAs displayed a flat copy number profile (Figure 3), consistent with the literature on conventional FAs.29, 30 One case (MFA8), however, displayed multiple gains and losses with a focal amplification in FLT4 (Supplementary Figure S1). No significant histologic differences were observed between MFA8 and the remaining cases.
Figure 3. Copy number alterations detected in myxoid fibroadenomas.
Heatmap depicting copy number alterations (CNAs) identified in myxoid fibroadenomas. Samples are represented on the y-axis; CNAs are represented along the x-axis according to their respective genomic location. CNAs are color-coded according to the legend.
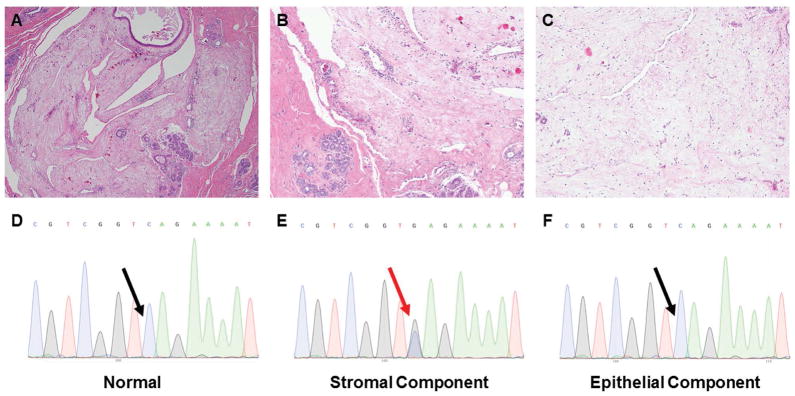
MFA6 was found to harbor a somatic loss-of-function, truncating mutation in the PRKAR1A gene (S307*). This observation would be consistent with the hypothesis that a subset of MFAs occurring in patients without a clinical history of Carney Complex may be driven by somatic rather than germline inactivation of the PRKAR1A gene. Retrospective histologic analysis, however, revealed that MFA6 differed histologically from the PRKAR1A-wild-type MFAs. Whilst all cases but MFA6 displayed a well-defined interface between the lesions and surrounding stroma and the overt biphasic architecture characteristic of breast fibroepithelial lesions (Figure 1), MFA6 was a roundish lesion composed of round to stellate-shaped cells embedded in abundant hypocellular myxoid stroma. The borders of MFA6, which lacked a well-defined interface with the adjacent breast tissue, were focally irregular, with the myxoid matrix dissecting the surrounding stroma. The few distorted ductal structures within MFA6 likely represented entrapped ducts rather than constitutive components of the lesion (Figure 4A–C). Taken together, these morphologic features led us to reclassify MFA6 as a breast myxoma, a tumor type that is more frequently observed in Carney Complex patients.3, 4, 7
Figure 4. Breast myxoma harboring a somatic truncating PRKAR1A mutation.
Representative micrographs of MFA6, which was the single case found to harbor a somatic inactivating mutation in PRKAR1A (S307*), which was reclassified as a breast myxoma upon histologic re-review. A) Low-power magnification (20X). B) Low-power magnification displaying undefined borders and dissection of surrounding stroma by the myxoid matrix (40X). C) Intermediate magnification showing highly myxoid and hypocellular stroma (100X). D) Sanger sequence electropherogram of normal sample. E) Sanger sequence electropherogram of stromal component. F) Sanger sequence electropherogram of epithelial component. Red and black arrows are representative of the presence or absence, respectively, of the S307* PRKAR1A mutation.
To validate the PRKAR1A somatic mutation identified by MSK-IMPACT in MFA6 and investigate whether the mutation would be present in the stromal cells but absent in the epithelial cells of the likely-entrapped ducts, we performed laser capture microdissection and extracted DNA of each component of MFA6 separately. Sanger sequencing confirmed that the PRKAR1A mutation was indeed present in the stromal cells, but absent in the epithelial cells and in the DNA extracted from normal tissue (Figure 4D–F). These findings confirm that the PRKAR1A mutation was somatic and provide evidence to suggest that the stromal but not the epithelial cells were the neoplastic component of this lesion.
Somatic mutations in known cancer genes are of low mutant allele fraction and restricted to the epithelial components of MFAs
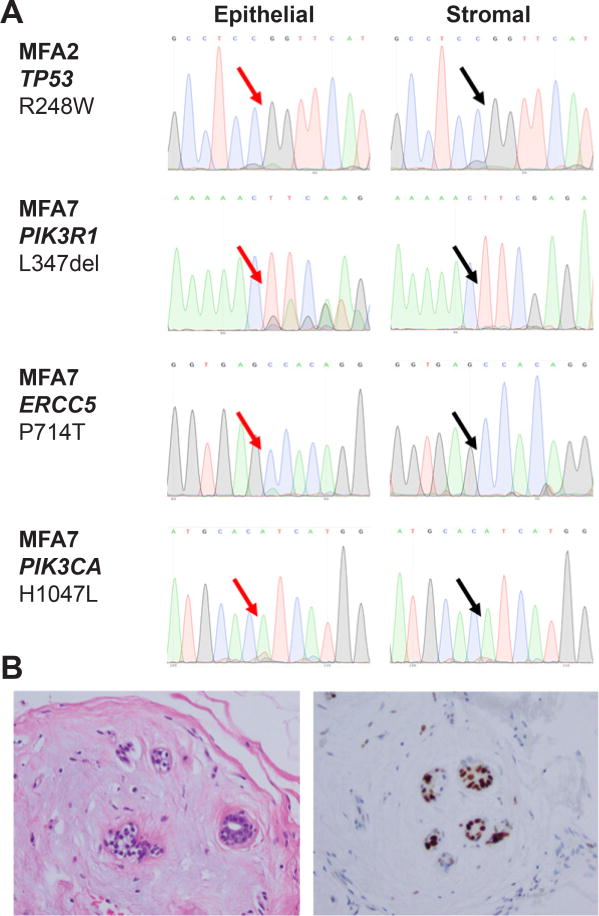
The somatic mutations identified in seven MFAs (excluding MFA6) were found to be present at low mutant allele fractions, consistent with the notion that these mutations would be subclonal. To define whether selected mutations identified by MSK-IMPACT (Table 2) would be present in the stromal or epithelial cells of MFAs, three cases were subjected to laser capture microdissection. Sanger sequencing performed on the DNA samples extracted from each component separately revealed that mutations affecting TP53 (MFA2), PIK3R1 and ERCC5 (MFA7), and PIK3CA (MFA10) were present in the epithelial components, but absent in the stromal cells of the respective cases (Figure 5A). Consistent with these results, immunohistochemistry revealed that 30% of epithelial cells, but not the stromal cells of MFA2 expressed high levels of p53 (Figure 5B).
Figure 5. Pathogenic mutations in cancer genes confined to the epithelial component of myxoid fibroadenomas.
A) TP53, PIK3R1, ERCC5 and PIK3CA Sanger sequence electropherograms of the epithelial and stromal components of MFA2, MFA7 and MFA10, respectively. Note the height of the mutant peaks is consistent with a subset of the microdissected epithelial cells harboring the mutation. Red and black arrows are representative of the presence or absence, respectively, of the respective mutations. B) Representative micrographs of MFA2; hematoxilin-eosin stain on the left and p53 immunostain on the right. Note that the epithelial cells strongly express p53, consistent with a missense hotspot mutation in TP53.
Taken together, these findings have led us to hypothesize that these mutations were likely subclonal alterations found in the epithelial components of MFAs and that the stromal components of MFAs lack somatic mutations affecting MED12 and any of the other 409 cancer genes tested.
DISCUSSION
Here we demonstrate that unlike conventional FAs, of which >70% harbor MED12 mutations in the stromal (i.e. mesenchymal) components,8–11, 31 non-Carney Complex-related MFAs lack MED12 mutations and no somatic mutations affecting the 410 cancer genes included in our sequencing assay was found to be present at high mutant allele fraction. Moreover, apart from the PRKAR1A mutation in MFA6, selected pathogenic mutations affecting known cancer genes were found to be restricted to the epithelial components of these lesions. Mutations in cancers genes, including TP5332 and RB18, have been previously detected in individual FAs. However, whether these mutations affected the epithelial or stromal cells was not investigated.
Several hypotheses can be advanced to explain our findings, including the notion that the stromal component of MFAs is not neoplastic and potentially reactive to the neoplastic epithelial component. This hypothesis, however, is unlikely to be correct, given that one case displayed gene copy number alterations, consistent with a clonal lesion, and the low mutant allele fractions of the mutations identified in the epithelial components. Arguably the most parsimonious explanation for our observations is that the genetic driver of MFAs was not encompassed by the list of genes surveyed by MSK-IMPACT, and that the somatic mutations identified in the epithelial component of the lesions analyzed in this study were subclonal. Consistent with this hypothesis, Sanger sequencing analysis of selected mutations identified by MSK-IMPACT using laser capture microdissected samples where the stromal and epithelial components were separately microdissected revealed that the mutations were restricted to the latter and the Sanger sequencing peaks in the electropherograms support the notion that these mutations were present in a subset of the epithelial cells analyzed, consistent with previous descriptions of somatic mutations in non-neoplastic breast tissue.32–34 In addition, immunohistochemical analysis of case MFA2, which harbored a TP53 missense hotspot somatic mutation (R248W) of low variant allele fraction (4.11%), demonstrated strong p53 expression in 30% of epithelial cells.
A somatic, loss-of-function mutation in PRKAR1A was detected in and restricted to the stromal cells of MFA6, which was reclassified as a breast myxoma upon histologic re-review. Although MFAs have been noted in patients with Carney Complex,3, 7 the development of myxomas is more consistent with the spectrum of tumors associated with the disease and, therefore, with the somatic inactivation of PRKAR1A.35, 36 In fact, other terms such as “breast myxomatosis” have been used to describe breast tumors arising in Carney Complex patients.3 Somatic PRKAR1A mutations may contribute to approximately 30% of tumors of the Carney Complex spectrum occurring in patients without other features of Carney Complex and without a germline inactivating mutation in PRKAR1A.6, 37 For instance, recurrent somatic PRKAR1A mutations have been described in cardiac myxomas38 and melanotic schwannomas,39 tumor types observed in Carney Complex patients.7, 39, 40
PRKAR1A encodes for cAMP-dependent protein kinase type I-α regulatory subunit and its inactivation induces protein kinase A activation and tumorigenesis.6, 7, 41 As a tumor suppressor gene, bi-allelic inactivation of PRKAR1A has been documented in tumors of the Carney Complex.39 It has been suggested, however, that PRKAR1A may be haploinsufficient, given that no LOH was found in an eyelid myxoma occurring in a patient known to be heterozygous for a common PRKAR1A germline mutation.42 Our findings of a PRKAR1A mutation with a low mutant allele fraction (Supplementary Table S2) in a breast myxoma are consistent with a heterozygous mutation, and the lack of coupled LOH further supports the notion that loss of one copy of PRKAR1A may be sufficient for myxomagenesis.
This study has several limitations. First, given the lower frequency of MFAs as compared to that of conventional FAs, the number of cases analyzed here is relatively small. Despite the small sample size, we were able to rule out the presence of MED12 in these lesions, in contrast with their presence in approximately 70% of FAs. Second, no somatic mutation was identified and validated in the stromal components of MFAs. The results of this study are limited to the 410 key cancer genes included in the MSK-IMPACT sequencing assay, hence we cannot exclude that genetic alterations in genes not included in this panel, as well as fusion-genes or epigenetic changes may play a role in the development of MFAs. Although MSK-IMPACT includes MED12, RARA and ROS1, genes mutated in FAs, we were unable to sequence FLNA owing to the limited amount of residual DNA of the MFAs included in this study and the large size of the gene. Whole-exome sequencing and/or whole-genome sequencing analyses are warranted to define whether MFAs are driven by somatic mutations affecting a gene other than those included in MSK-IMPACT. Third, our initial hypothesis that MFAs occurring in patients without Carney complex would be underpinned by PRKAR1A mutations was not confirmed. Rather, we demonstrated that only a breast myxoma, initially diagnosed as breast MFA, harbored a PRKAR1A somatic mutation. Together with the known association between breast myxomatosis and Carney Complex,3 our findings suggest that functional inactivation of PRKAR1A in breast stromal cells by germline or somatic genetic events may result in the development of breast myxomas. Moreover, our findings support the neoplastic nature of breast myxomas.
In conclusion, MFAs were found to lack MED12 somatic mutations and, therefore, differ from conventional FAs at the genetic level. Moreover, cancer gene mutations detected in MFAs were confined to the epithelium, but even in the epithelium, these mutations appear to be restricted to a subset of the cells. Therefore, we posit that the stromal component of MFAs may be driven by genetic alterations affecting a gene other than those surveyed by MSK-IMPACT, or that epigenetic alterations in genes not covered in our analyses may contribute to the pathogenesis of MFAs. Further sequencing analyses are warranted to define the genomic landscape of MFAs. A somatic inactivating mutation in the PRKAR1A gene was noteworthy, and detected in a case reclassified as a breast myxoma, a condition part of the spectrum of tumors observed in Carney Complex. Additional analyses are warranted to assess the prevalence of PRKAR1A mutations in breast myxomas and whether myxoid breast lesions occurring in Carney Complex patients are actually best considered breast myxomas rather than MFAs.
Supplementary Material
Acknowledgments
JSR-F is funded in part by a grant from the Breast Cancer Research Foundation. Research reported in this paper was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
AUTHORS’ CONTRIBUTIONS
E.B., B.W, J.S.R.-F and F.C.G conceived the study. E.B. and M.M. provided tissue samples and clinical data. E.B., A.M., M.P.M. and F.C.G. performed pathology review. J.R.L, F.C.G., F.P., J.K. and R.G-M performed experiments. K.A.B. and R.S.L. performed bioinformatics analyses. J.R.L., J.S.R-F. and F.C.G. wrote the first manuscript, which was reviewed by all co-authors.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
References
- 1.Tan BY, Acs G, Apple SK, et al. Phyllodes tumours of the breast: A consensus review. Histopathology. 2016;68:5–21. doi: 10.1111/his.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koerner FC. In diagnostic problems in breast pathology. Philadelphia: W.B. Saunders; 2009. Myxoid fibroadenoma; pp. 321–327. [Google Scholar]
- 3.Carney JA, Toorkey BC. Myxoid fibroadenoma and allied conditions (myxomatosis) of the breast. A heritable disorder with special associations including cardiac and cutaneous myxomas. Am J Surg Pathol. 1991;15:713–721. doi: 10.1097/00000478-199108000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Bertherat J. Carney complex (cnc) Orphanet J Rare Dis. 2006;1:21. doi: 10.1186/1750-1172-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang YS, Moon SD, Kim JH, Lee IS, Lee JM, Kim HS. A novel prkar1a mutation resulting in a splicing variant in a case of carney complex. Korean J Intern Med. 2015;30:730–734. doi: 10.3904/kjim.2015.30.5.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertherat J, Horvath A, Groussin L, et al. Mutations in regulatory subunit type 1a of cyclic adenosine 5′-monophosphate-dependent protein kinase (prkar1a): Phenotype analysis in 353 patients and 80 different genotypes. J Clin Endocrinol Metab. 2009;94:2085–2091. doi: 10.1210/jc.2008-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcoutsakis NA, Tatsi C, Patronas NJ, Lee CCR, Prassopoulos PK, Stratakis CA. The complex of myxomas, spotty skin pigmentation and endocrine overactivity (carney complex): Imaging findings with clinical and pathological correlation. Insights Imaging. 2013;4:119–133. doi: 10.1007/s13244-012-0208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim WK, Ong CK, Tan J, et al. Exome sequencing identifies highly recurrent med12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46:877–880. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 9.Piscuoglio S, Murray M, Fusco N, et al. Med12 somatic mutations in fibroadenomas and phyllodes tumours of the breast. Histopathology. 2015;67:719–729. doi: 10.1111/his.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfarr N, Kriegsmann M, Sinn P, et al. Distribution of med12 mutations in fibroadenomas and phyllodes tumors of the breast--implications for tumor biology and pathological diagnosis. Genes Chromosomes Cancer. 2015;54:444–452. doi: 10.1002/gcc.22256. [DOI] [PubMed] [Google Scholar]
- 11.Tan J, Ong CK, Lim WK, et al. Genomic landscapes of breast fibroepithelial tumors. Nat Genet. 2015;47:1341–1345. doi: 10.1038/ng.3409. [DOI] [PubMed] [Google Scholar]
- 12.Piscuoglio S, Ng CK, Murray M, et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and tert promoter hotspot mutations and tert gene amplification as likely drivers of progression. J Pathol. 2016;238:508–518. doi: 10.1002/path.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piscuoglio S, Geyer FC, Murray MP, et al. Massively parallel sequencing analysis of synchronous fibroepithelial lesions supports the concept of progression from fibroadenoma to phyllodes tumor. NPJ Breast Cancer. 2016 doi: 10.1038/npjbcancer.2016.35. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piscuoglio S, Burke KA, Ng CK, et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol. 2016;238:381–388. doi: 10.1002/path.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen R, Seshan VE. Facets: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic acids research. 2016 doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 17.Koboldt DC, Zhang Q, Larson DE, et al. Varscan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. Mutationtaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 21.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: Computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating prkd1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S-Y, Joseph NM, Ravindranathan A, et al. Genomic profiling of malignant phyllodes tumors reveals aberrations in fgfr1 and pi-3 kinase/ras signaling pathways and provides insights into intratumoral heterogeneity. Mod Pathol. 2016;29:1012–1027. doi: 10.1038/modpathol.2016.97. [DOI] [PubMed] [Google Scholar]
- 29.Cavalli LR, Cornelio DA, Lima RS, et al. Lack of DNA copy number alterations revealed with comparative genomic hybridization in fibroadenomas of the breast. Cancer Genet Cytogenet. 2004;153:173–176. doi: 10.1016/j.cancergencyto.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Kuijper A, Snijders AM, Berns E, et al. Genomic profiling by array comparative genomic hybridization reveals novel DNA copy number changes in breast phyllodes tumours. Cellu Oncol. 2009;31:31–39. doi: 10.3233/CLO-2009-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishima C, Kagara N, Tanei T, et al. Mutational analysis of med12 in fibroadenomas and phyllodes tumors of the breast by means of targeted next-generation sequencing. Breast Cancer Res Treat. 2015;152:305–312. doi: 10.1007/s10549-015-3469-1. [DOI] [PubMed] [Google Scholar]
- 32.Millikan R, Hulka B, Thor A, et al. P53 mutations in benign breast tissue. J Clin Oncol. 1995;13:2293–2300. doi: 10.1200/JCO.1995.13.9.2293. [DOI] [PubMed] [Google Scholar]
- 33.Yadav VK, DeGregori J, De S. The landscape of somatic mutations in protein coding genes in apparently benign human tissues carries signatures of relaxed purifying selection. Nucleic Acids Res. 2016;44:2075–2084. doi: 10.1093/nar/gkw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behjati S, Huch M, van Boxtel R, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correa R, Salpea P, Stratakis CA. Carney complex: An update. Eur J Endocrinol. 2015;173:M85–97. doi: 10.1530/EJE-15-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stratakis CA, Kirschner LS, Carney JA. Clinical and molecular features of the carney complex: Diagnostic criteria and recommendations for patient evaluation. J Clin Endocrinol Metab. 2001;86:4041–4046. doi: 10.1210/jcem.86.9.7903. [DOI] [PubMed] [Google Scholar]
- 37.Rojo Alvaro J, Martinez de Esteban JP, Pineda Arribas JJ, Ollero Garcia-Agullo MD, Munarriz Alcuaz P. Acromegaly in a patient with carney’s complex. Endocrinol Nutr. 2013;60:277–278. doi: 10.1016/j.endonu.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Maleszewski JJ, Larsen BT, Kip NS, et al. Prkar1a in the development of cardiac myxoma: A study of 110 cases including isolated and syndromic tumors. Am J Surg Pathol. 2014;38:1079–1087. doi: 10.1097/PAS.0000000000000202. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Zehir A, Sadowska J, et al. Consistent copy number changes and recurrent prkar1a mutations distinguish melanotic schwannomas from melanomas: Snp-array and next generation sequencing analysis. Genes Chromosomes Cancer. 2015 doi: 10.1002/gcc.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim MJ, Choi J, Khang SK, Kim JS, Lee JS, Cho KJ. Primary intraosseous melanotic schwannoma of the fibula associated with the carney complex. Pathol Int. 2006;56:538–542. doi: 10.1111/j.1440-1827.2006.02002.x. [DOI] [PubMed] [Google Scholar]
- 41.Horvath A, Bertherat J, Groussin L, et al. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase a (prkar1a): An update. Hum Mutat. 2010;31:369–379. doi: 10.1002/humu.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsilou ET, Chan CC, Sandrini F, et al. Eyelid myxoma in carney complex without prkar1a allelic loss. Am J Med Genet A. 2004;130A:395–397. doi: 10.1002/ajmg.a.30279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.