Abstract
Background
In vertebrate embryos, a "segmentation clock" times somitogenesis. Clock-linked genes, including Lunatic fringe (Lfng), exhibit cyclic expression in the presomitic mesoderm (PSM), with a period matching the rate of somite formation. The clock period varies widely across species, but the mechanisms that underlie this variability are not clear. The half-lives of clock components are proposed to influence the rate of clock oscillations, and are tightly regulated in the PSM. Interactions between Lfng and mir-125a-5p in the embryonic chicken PSM promote Lfng transcript instability, but the conservation of this mechanism in other vertebrates has not been tested. Here, we examine whether this interaction affects clock activity in a mammalian species.
Results
Mutation of mir-125 binding sites in the Lfng 3'UTR leads to persistent, non-oscillatory reporter transcript expression in the caudal-most mouse PSM, though dynamic transcript expression recovers in the central PSM. Despite this, expression of endogenous mir-125a-5p is dispensable for mouse somitogenesis.
Conclusions
These results suggest that mir-125a sites in the Lfng 3'UTR influence transcript turnover in both mouse and chicken embryos, and support the existence of position-dependent regulatory mechanisms in the PSM. They further suggest the existence of compensatory mechanisms that can rescue the loss of mir-125a-5p in mice.
Keywords: Lunatic fringe, miRNA, mir-125a-5p, post-transcriptional regulation, 3'UTR, somitogenesis, segmentation clock
Introduction
In vertebrate segmentation, somites bud from the presomitic mesoderm (PSM), and give rise to the axial skeleton and skeletal muscle, producing a segmented adult body (Maroto et al., 2012). Future intersomitic boundaries are positioned sequentially within the PSM by a "wavefront" or "determination front" that provides positional information (Dubrulle et al., 2001). Posterior to the determination front (also called region I) (Saga and Takeda, 2001), a "segmentation clock" acts to time somite formation. Anterior to the determination front (region II), clock oscillations are arrested, and pre-somites are compartmentalized into rostral and caudal domains (Saga and Takeda, 2001).
Genes linked to the segmentation clock exhibit cyclic expression in region I, with a period that generally matches the rate of somite formation in that organism (Hubaud and Pourquie, 2014, Wahi et al., 2016), although the clock period slows as cells are displaced anteriorly (Palmeirim et al., 1997, Aulehla et al., 2008, Shih et al., 2015). The rate of somite formation and the specific oscillatory genes essential for somite formation vary widely among vertebrates (Wahi et al., 2016). Mutations in clock genes can result in congenital vertebral defects such as spondylocostal dysostoses, scoliosis, or spinal kyphosis (Turnpenny et al., 2007, Eckalbar et al., 2012), highlighting the importance of normal clock function. However, the mechanisms that differentially regulate the clock in species with distinct timing requirements are poorly understood.
In all vertebrates examined to date, at least one Notch pathway component or target exhibits cyclic expression, linking Notch activation to clock function (Liao and Oates, 2016). However, the specific Notch pathway components that oscillate in the clock vary among species. For example the Lfng RNA, which encodes a glycosyltransferase that modifies Notch and some of its ligands (Takeuchi and Haltiwanger, 2014), exhibits clock-linked expression in chicken (McGrew et al., 1998) and mouse (Forsberg et al., 1998, Aulehla and Johnson, 1999) embryos, but not in zebrafish (Prince et al., 2001). In the mouse and chicken clock, Lfng acts in a negative feedback loop that requires transcriptional activation by Notch and repression by the bHLH repressor HES7 (c-hairy1 in chickens) (Hubaud and Pourquie, 2014, Wahi et al., 2016). While the existence of species-specific clock components and regulation helps explain the variations in period between different species, changes in post-transcriptional regulation may also contribute to changes in clock period among species that share similar clock networks.
Mathematical models predict that the maintenance of synchronized clock gene oscillations requires regulation at both transcriptional and post transcriptional levels (Lewis, 2003, Feng and Navaratna, 2007, Gonzalez and Kageyama, 2009). Large changes in component half-lives generally disrupt segmentation clock function (Hirata et al., 2004, Riley et al., 2013, Williams et al., 2016). However, mathematical models and biological data suggest that, within a narrow window, changes in half-lives of clock components may influence the period of oscillations, which could contribute to species-specific clock periods (e.g. Lewis, 2003, Schroter et al., 2012, Goldbeter and Pourquie, 2008, Wiedermann et al., 2015, Herrgen et al., 2010). Spatially controlled changes in half-life regulation could also contribute to mechanisms that produce more rapid oscillations in cells in the posterior PSM that slow as cells enter the anterior PSM (Palmeirim et al., 1997, Aulehla et al., 2008, Shih et al., 2015).
In the context of segmentation, work done in both chickens and mice demonstrates that the 3’ untranslated regions (3’UTR) of cyclic genes including Hes7 and Lfng promote rapid RNA turnover in the PSM (Hilgers et al., 2005, Nitanda et al., 2014, Fujimuro et al., 2014). 3'UTR sequences often influence transcript half-life by providing binding sites for regulatory proteins or small RNAs, and several studies have suggested that regulation of cyclic transcripts by miRNAs could play a critical role in maintenance of stable oscillations (Bonev et al., 2012, Tan et al., 2012, Riley et al., 2013). miRNAs are short non-coding RNAs that repress the expression of protein coding genes usually by binding to specific sites in their 3’UTR and causing mRNA degradation and/or translational repression (Ha and Kim, 2014). In the context of the segmentation clock, we previously demonstrated that the Lfng 3'UTR contains conserved target sequences for miRNAs of the mir-125 family, and that blocking interactions between mir-125a-5p and Lfng in the chicken PSM stabilizes Lfng transcripts and perturbs segmentation clock function (Riley et al., 2013).
Whether mir-125a-5p plays a role in regulating the stability of Lfng mRNA during mouse segmentation is unknown. Some computer models predict that Lfng:mir-125 interactions could act to fine tune Notch activity in the PSM and would affect the period and amplitude of clock gene oscillations (Jing et al., 2015). This interaction could be necessary in all vertebrates in which Lfng plays a role in the clock. Alternatively, since the period of clock gene oscillations varies across these vertebrates, ranging from 90 minutes in chicken (Palmeirim et al., 1997), to 120 minutes in mouse (Tam, 1981), to 4–5 hours in humans (William et al., 2007), regulation of Lfng by members of the mir-125 family may be a chicken-specific phenomenon, controlling clock period in that species only.
To test effects of the mir-125 family miRNAs on Lfng transcript stability in mouse embryos, we mutated the putative binding sites for mir-125-5p family miRNAs and examined the PSM expression of Venus reporter constructs. We find that loss of these sites specifically perturbs transcript expression in the most caudal PSM, but that mutant transcripts recover their dynamic expression pattern in the central region of the PSM. Despite this, mutations that prevent production of one specific mir-125 family member, mir-125a-5p, have no effect on mouse somitogenesis or skeletal morphology. These findings suggest that the specific 3'UTR sequences we previously identified as mir-125a-5p binding sites are necessary for normal Lfng expression specifically in the caudal-most PSM domain. However, other mechanisms may compensate for the loss of mir-125a-5p during mouse segmentation.
RESULTS
mir-125a-5p can alter transcript expression levels through binding sites in the Lfng 3'UTR
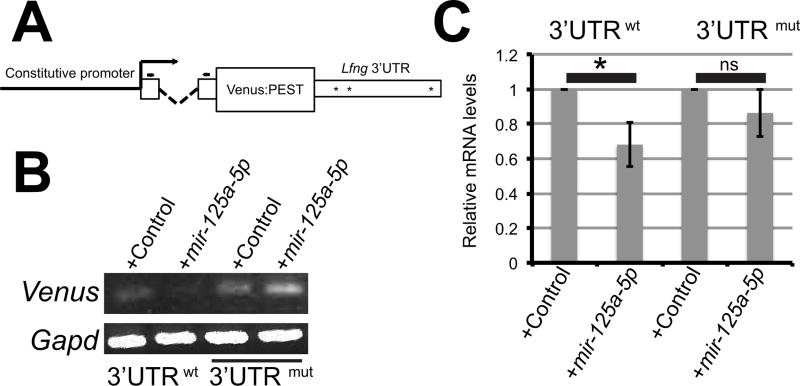
We previously demonstrated that mir-125a-5p affects protein expression from exogenous transcripts that contain the mouse Lfng 3'UTR in cell culture (Riley et al., 2013). To examine how mir-125a-5p affects expression at the transcript level, we developed Venus reporter constructs that contain mouse Lfng 3'UTR sequences (Fig 1A). Spliced Venus transcripts were detected in NIH3T3 cells. Co-transfection of pre-mir-125a-5p caused an apparent reduction in steady-state Venus transcript levels in transient transfections, and this effect was abrogated when the mir-125a-5p binding sites in the Lfng 3'UTR were mutated (Fig 1B). To extend these observations into a second cell line and to quantify this effect, stable C2C12 lines expressing Venus constructs were produced, and we observe similar results, with exogenous mir-125a-5p causing a 30% decrease in the expression level of a transcript containing the wild type Lfng 3'UTR, while mutation of the putative 125a binding sites in the 3'UTR abrogates this effect (Fig 1C). This effect is less dramatic than that observed at the protein level, where the amount of luciferase protein expressed from a transcript containing the mouse Lfng UTR was reduced by 70% by exogenous mir-125a-5p (Riley et al., 2013). However, these data support the idea that mir-125a-5p can affect transcript stability in mammalian cells via the mir-125a-5p binding sites in the mouse Lfng 3'UTR, without ruling out an additional role in regulation of translational efficiency.
Figure 1. mir-125a-5p can destabilize transcripts via mir-125 binding sites in the mouse Lfng 3’UTR in mammalian cells.
A) Destabilized Venus reporter constructs. A constitutive promoter drives the expression of a destabilized Venus protein flanked by a beta-globin intron at the 5’ end and either the wild type Lfng 3’UTR or mutated Lfng 3’UTR at the 3’ end. Asterisks indicate approximate positions of mir-125a binding sites. Arrows = primers used in PCR analyses. B) NIH3T3 cells were transiently transfected with wild type or mutant 3'UTR constructs along with control or pre-mir-125a-5p. Semi-quantitative RT-PCR demonstrates a reduction in RNA expression levels for transcripts containing wild type sequences in the presence of exogenous mir-125a-5p. C) Stable C2C12 cell lines expressing Venus constructs with a wild type or mutated Lfng 3’UTR were transfected with control or pre-mir-125a-5p. By qRT-PCR transcripts containing wild type UTR sequences are significantly reduced in the presence of mir-125a-5p, and this reduction is dependent on mir-125 binding sites. Expression levels were normalized to Gapd, and values represent the average of three independent experiments +/− SEM.
Mutation of mir-125 binding sites in the Lfng 3'UTR alters transgene expression in the caudal PSM of mouse embryos
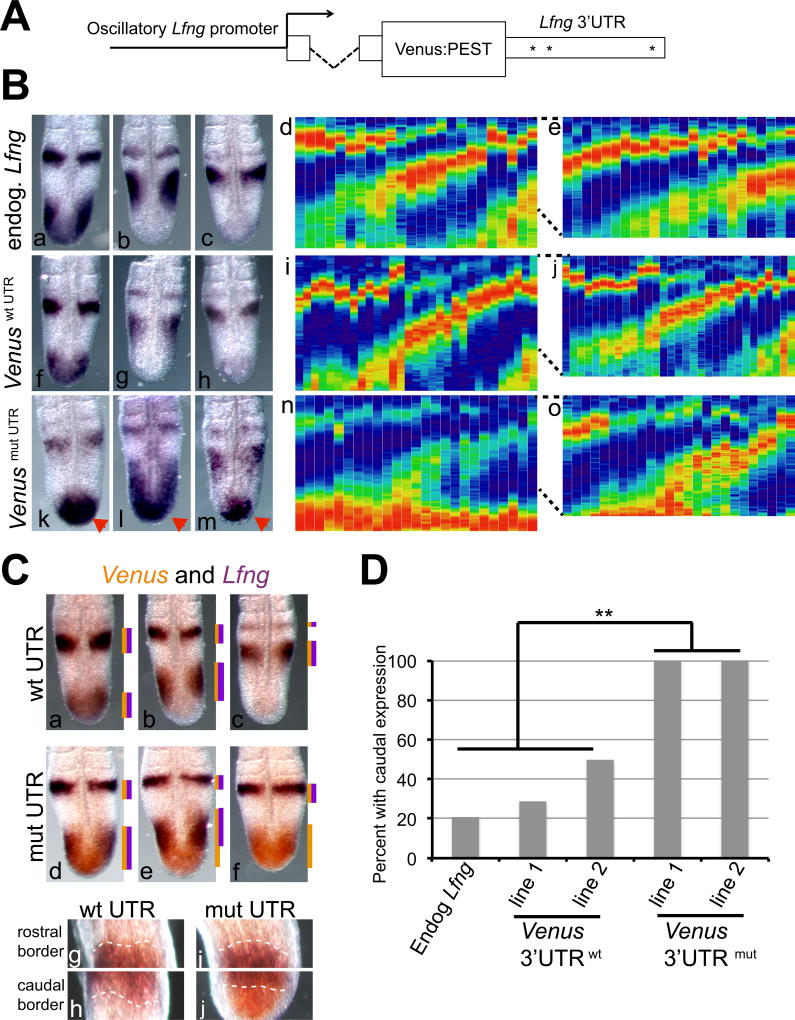
To examine the effects of mir-125 binding sites in a more physiologically relevant context, reporter transgenes were produced that express a Venus:PEST fusion protein under control of a Lfng promoter known to drive oscillatory expression in the PSM (Cole et al., 2002, Morales et al., 2002). The beta-globin intron was included at the 5' end of the transcript, allowing transcript interaction with the splicing machinery without triggering nonsense-meditated decay (Fig. 2A). Transgene constructs contained wild type Lfng 3'UTR (LvLwt) or Lfng 3'UTR sequences with mutations of the putative mir-125 binding sites (LvLmut). These constructs allow us to examine the effects of the 3'UTR on transcript expression without perturbing the feedback loops that regulate Lfng expression. We identified two independent transgenic lines that expressed detectable levels of Venus transcripts for each construct. Both LvLwt lines exhibit oscillatory Venus expression patterns in the mouse PSM, with all three previously described phases of expression (Pourquie and Tam, 2001) observed (Fig 2B). In contrast, in LvLmut embryos from both lines strong Venus expression was observed in the caudal PSM of all embryos examined (Fig 2B).
Figure 2. Mutation of the putative mir-125 binding sites perturb transgene expression patterns specifically in the caudal PSM.
A) Schematic of transgenes with a 3.8 kb fragment of the mouse Lfng 3'UTR driving expression of a Venus:PEST fusion protein, with beta-globin intron sequences 5' to the Venus coding sequence. Asterisks = approximate locations of putative mir-125 binding sites. B) In situ hybridization of Lfng (a–e) or Venus (f–o) transcripts, in 10.5 d.p.c. PSMs. Wild type embryos exhibit oscillatory expression of endogenous Lfng mRNA by RNA in situ (a–c) and by a "kymograph" expression profile (d, n=24). Venus transgenes containing a wild type Lfng 3'UTR exhibit oscillatory Venus RNA expression in the PSM by in situ analysis (f–h) and expression profile analysis (i, n=31). In transgenic embryos with mutant 3'UTR sequences, strong Venus expression is always detected in the caudal PSM (red arrowheads) by RNA in situ (k–m) and expression profile analysis (n, n=25). Expression profiles of the anterior 2/3 of each PSM reveals oscillatory Venus expression from the mutant 3'UTR containing transgene (o), similar to the wild type transgene (j) or endogenous Lfng expression pattern (e). Two independent lines for each transgene exhibited similar expression patterns, and data from one representative line are shown. C) Expression patterns of Lfng and Venus mRNA were directly compared, with Lfng expression in purple, and Venus in orange. In LvLwt embryos, expression of Venus overlaps that of the endogenous Lfng transcript (a–c). In LvLmut embryos (d–f) Venus mRNA is consistently observed in the posterior PSM, even when Lfng expression is not. Lines to right of each pattern denote extent of detection of each color. In panels g–j, increased magnification of the rostral and caudal border of the posterior stripe of an embryo in phase II (equivalent to panels b and e) are shown. At the anterior border of the stripe (g, i) there is complete overlap of Venus transcript (in orange) and Lfng transcript (in purple) in transgenic embryos with either the wild type (g) or the mutant 3'UTR (i). In contrast, at the caudal end of that stripe, there is again complete overlap of Venus transcript and Lfng transcript in transgenic embryos with the wild type 3'UTR (h) , but in embryos with the mutant 3'UTR, the orange Venus signal extends into the tailbud, past the limit of the Lfng expression domain (j).
D) Quantification reveals a significant increase in the fraction of embryos with RNA expression in the caudal PSM of embryos expressing transcripts containing the mutant 3'UTR. Data from two independent transgenic lines for each construct are quantified.
To confirm and extend these results, we directly compared transgenic Venus expression to endogenous Lfng expression in single embryos using two color in situ analysis. In embryos expressing the LvLwt transgene, we observe complete overlap of Venus and Lfng expression patterns, confirming that the LvLwt transgenes recapitulate endogenous Lfng expression (Fig 2C). However in embryos expressing LvLmut constructs, Venus transcripts are always detectable in the caudal region of the PSM, and do not always overlap with Lfng expression. This is especially apparent in embryos that are in phases II and III of oscillatory Lfng expression but display Venus expression in the tail bud where Lfng expression has been downregulated (Fig 2C, panels e,f)
To examine the oscillatory expression profiles in our transgenic lines across a larger number of embryos, we created expression profiles (frequently called "kymographs") including multiple embryos for each line. Signal intensity was converted to color information with the highest expression in each embryo defined as 100%. The oscillatory expression profile of endogenous Lfng mRNA is similar to the Venus expression profile in LvLwt lines, but again in the LvLmut lines we observe persistent, strong Venus expression in the caudal tailbud (Fig 2B compare d, i, n). To quantify this effect we compared the fraction of embryos that exhibit strong expression (defined as red or orange signal) in the caudal 1/3 of the PSM among endogenous Lfng, LvLwt, and LvLmut. We observe a statistically significant change in expression patterns in both LvLmut lines (Fig 2D), confirming that the loss of the mir-125a binding sites in the Lfng 3'UTR affects transcript expression in the caudal PSM of mouse embryos.
Venus expression in the anterior PSM of LvLmut embryos retains dynamic expression patterns
The high Venus expression in the caudal PSM of all LvLmut embryos compromises the detection of dynamic expression in the anterior PSM of these embryos, as the oscillatory expression peaks in the anterior PSM may be at lower levels than the robust caudal expression. To assess dynamic expression in the central and anterior PSM of LvLmut embryos, we repeated the kymograph expression profile analysis but focused on signal levels from the anterior 2/3 of the PSM, excluding the expression in the caudal 1/3 of each embryo. In the anterior PSM, dynamic expression was observed in both LvLmut lines, and the expression profiles were similar to those observed in LvLwt lines and for exogenous Lfng (Fig 2B compare e, j, o). These results suggest that mutation of the mir-125 binding sites in the Lfng 3'UTR disrupts mRNA regulation specifically in the caudal-most region of the PSM but not the rest of the PSM.
mir-125a-5p expression can be eliminated without disruption of nearby miRNAs
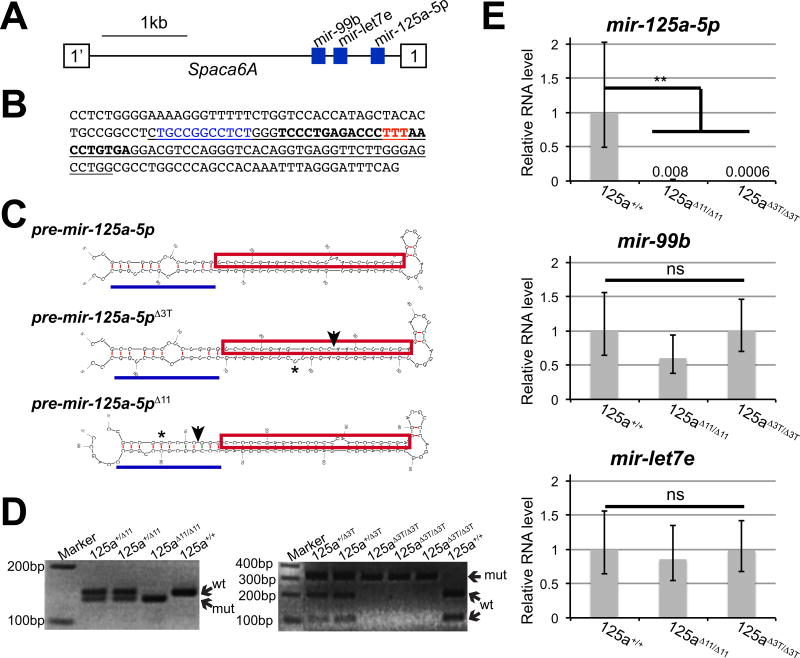
These results support the hypothesis that putative mir-125 binding sites in the mouse Lfng 3'UTR act to regulate the transcript in the segmentation clock, but do not specifically address the function of mir-125a-5p itself. To examine the potential role of mir-125a-5p in the segmentation clock we eliminated expression of the mature mRNA in vivo via CRISPR/Cas9 genome editing (Yang et al., 2013, Fujihara and Ikawa, 2014). Two independent mutant alleles were produced that affect mir-125a-5p sequences. mir-125a-5pΔ11 has an 11 bp deletion just 5' of the mature miRNA sequence (Fig. 3B). This mutation affects the pri-miRNA stem loop, and is predicted to disrupt DROSHA recognition sites and affect the production of pre-mir-125a-5p (Zeng et al., 2005, Han et al., 2006, Auyeung et al., 2013) (Fig. 3C). mir-125a-5pΔ3T has a deletion of 3 contiguous "T" nucleotides in the mature mir-125a-5p sequence, which will prevent production of mature mir-125a-5p. (Fig. 3B and C).
Figure 3. Crispr/CAS9 mutation of the endogenous mir-125a-5p locus.
A) Schematic of the mir-99b-let7e-125a cluster. The three miRNAs are located in an intron of the Spaca6A gene. B) Genomic sequences surrounding mir-125a-5p_ are shown with the pre-mir hairpin underlined and the mature mir-125a-5p in bold. The mir-125a-5pΔ11 allele has an 11 bp deletion (blue) at the base of the hairpin, while the mir-125a-5pΔ3T allele has a deletion of 3 bp (red) in the mature miRNA sequence. C) Predicted structures of mir-125a-5p precursor transcripts produced from the mir-125a-5pΔ11 and mir-125a-5pΔ3T alleles predicted by Mfold. Mature miRNA sequence is boxed in red, and the hairpin structure recognized by Drosha is highlighted with a blue line. Sites of deletions are indicated by arrows. Potentially important changes in hairpin structures are highlighted (asterisks). D) PCR genotyping identifies mouse genotypes, demonstrating that homozygous offspring are produced from intercrosses. Wild type and mutant alleles are indicated. E) By qRT-PCR, we observe that mir-125a-5p expression is lost in homozygous mir-125a-5pΔ11 and mir-125a-5pΔ3T mutant embryos compared to the wild type controls (numbers above bars represent average measured relative RNA level in each genotype). The mature miRNA expression levels of mir-99b and let-7e are not significantly altered in mutant embryos. Expression levels were normalized to sno-234 expression, with wild type set to 1. Error bars represent SEM from three independent samples assayed in triplicate. Statistical significance calculated using Student’s T test.
Homozygous mutant embryos were produced and genotyped by PCR (Fig. 3D) Quantitative PCR analysis demonstrates that homozygous mir-125a-5pΔ11 or mir-125a-5pΔ3T mutant embryos exhibit a complete loss of mir-125a-5p expression confirming that the mutations disrupt production of the mature miRNA (Fig 3E). mir-125a-5p is located in a genomic cluster in close proximity to miR-99b and let-7e (Fig. 3A), but RT-PCR demonstrates that these mutations do not affect the expression of these nearby miRNAs (Fig 3E), confirming that our mutant lines represent simple mir-125a-5p loss of function mutations.
mir-125a-5p mutant mice are viable and morphologically normal
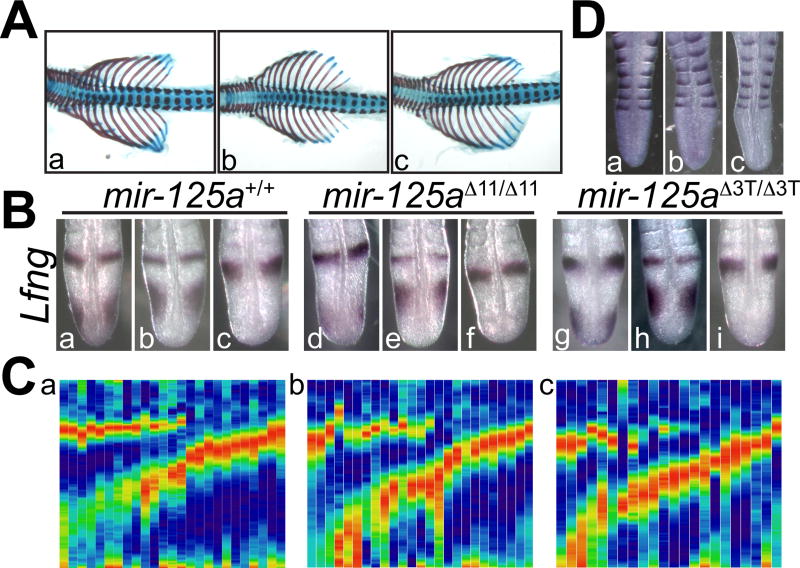
mir-125a-5pΔ11 and mir-125a-5pΔ3T founder mice were outcrossed for several generations to wild type FVB/JN mice to reduce concerns of off site mutations, and then intercrossed to produce homozygous mutant offspring. Homozygous embryos of both lines are weaned at Mendelian rates (Table 1), and appear outwardly normal, exhibiting none of the defects expected from perturbations of Lfng expression. Skeletal analysis does not reveal any changes in the number and morphology of ribs and vertebrae of mutant mice compared to wild type and heterozygous littermates (Fig. 4A), indicating that mir-125a-5p expression is dispensable for normal skeletal development. Similar findings were observed in homozygous mutant offspring of homozygous mutant females indicating that embryo development is not being rescued by maternally expressed mir-125a-5p (data not shown).
Table 1. Loss of mir-125a-5p has no effects on viability at weaning.
Mice were genotyped at weaning from intercrosses of heterozygous animals. Observed numbers of each genotype are shown, with the expected number in parentheses. χ2 analysis and associated P values indicate that homozygous mutant mice are obtained at expected rates. Data include mice from 5 individual litters for mir-125a-5pΔ3T and 8 litters for mir-125a-5pΔ11.
| Allele | total | wild type | heterozygote | homozygote | χ2 | P |
|---|---|---|---|---|---|---|
| mir-125a-5pΔ11 | 64 | 17 (16) | 34 (32) | 13 (16) | 0.75 | 0.69 |
| mir-125a-5pΔ3T | 49 | 13 (12.5) | 24 (24.5) | 12 (12.5) | 0.06 | 0.97 |
Figure 4. Lfng expression, somite patterning, and skeletal morphology are normal in mir-125a-5p mutant mice.
A) Skeletal morphology was assessed by alizarin red /alcian blue staining. Normal skeletal morphology and somite patterning are observed in wild type (a), mir-125a-5pΔ11 (b) and mir-125a-5pΔ3T (c) embryos. B). Lfng expression at 10.5 d.p.c. is observed in all three phases of oscillatory expression in wild type, mir-125a-5pΔ11 and mir-125a-5pΔ3T embryos. C) Expression profiles of multiple embryos further support the idea that Lfng expression patterns are unperturbed after the loss of mir-125a-5p. D) Somite patterning was examined using the caudal marker Uncx. Normal somite patterning is observed in wild type (a), mir-125a-5pΔ11 (b) and mir-125a-5pΔ3T (c) embryos. Quantification of somites, vertebrae, and ribs (data not shown) confirms normal somitogenesis and skeletal development in the absence of mir-125a-5p.
Lfng and Uncx expression patterns are unaffected in mir-125a-5p mutant embryos
To specifically examine whether the loss of mir-125a-5p has an effect on Lfng expression in the mouse PSM, we examined endogenous Lfng expression in mutant embryos. All three previously described phases of Lfng expression were observed in both mutant lines (Fig 4B), and expression profiles produced as described above appeared normal in both mutant lines (Fig 4C). Specifically, we did not observe dramatically increased levels of Lfng expression in the caudal 1/3 of the PSM. These results suggest that mir-125a-5p is not required for the maintenance of Lfng oscillations in the mouse PSM.
Since Lfng expression is required for normal somite patterning (Zhang et al., 2000), we examined somite compartment identity and patterning by examining the expression of Uncx, a marker for the posterior somite compartment. As suggested by the normal skeletal morphology and Lfng expression described above, we found no changes in Uncx expression in homozygous mir-125a-5p mutant embryos (Fig 4D). Further, we found no change in the number of somites between the forelimb and hindlimb (wild type: 21.8 +/− 0.9 (n=21); mir-125a-5pΔ11/Δ11: 21.6 +/− 1.4 (n=8); mir-125a-5pΔ3T/Δ3T: 21.7 +/− 0.6 (n=14)) indicating that we have not altered the period of the segmentation clock by subtly altering the half-life of the Lfng RNA. Taken together these results indicate that mir-125a-5p function is dispensable in the context of the mouse segmentation clock.
DISCUSSION
The mechanisms that contribute to species-specific clock periods are not fully understood. Similarly, although we know that clock oscillations slow as cells move anteriorly in the PSM, we know very little about how this is controlled (Oates et al., 2012). Changes in the post-transcriptional control of clock components either between species or in a spatially-specific way along the PSM could influence embryonic coordination of spatial and temporal information during segmentation. We previously found that mir-125a could regulate Lfng in the chicken PSM (Riley et al., 2013), and here we examine whether this regulatory relationship is conserved in mice.
The mouse Lfng 3'UTR contains 3 putative binding sites for mir-125 family miRNAs, and we used reporter constructs to examine whether these sequences influence transcript expression in the PSM. Perhaps surprisingly, we observe disruptions in transcript expression only in the caudal-most 1/3 of the mouse PSM, where embryos expressing the LvLmut transgene exhibit persistent Venus transcript expression, while embryos expressing LvLwt transgenes recapitulate endogenous Lfng expression throughout the PSM. We have not formally ruled out an effect on transgene transcription in the LvLmut lines, but given that the differences between the transgenes are confined to the 3'UTR it is more likely that loss of the putative mir-125a sites perturbs transcript turnover in the caudal PSM. The observation of similar expression patterns in multiple independent transgenic lines also argues against insertion site effects. However, we have been unable to successfully utilize the beta-globin intron sequences as in situ probes, and thus can not formally demonstrate that these transgenes are exhibiting oscillatory transcription in the caudal PSM, and that the persistent signal is due to increased transcript half-life.
However, when we focus on the central region of the PSM, in between the tailbud and the determination front, we observe dynamic Venus expression in LvLmut embryos that matches the endogenous Lfng expression pattern. This finding is reminiscent of the expression pattern observed when Lfng:125a interactions were perturbed in chicken embryos, where stable expression was observed in the caudal PSM, but a region without Lfng expression was consistently seen between the posterior and anterior Lfng expression domains (Riley et al., 2013). Taken together, these findings support the idea that the post-transcriptional regulation of Lfng may be position-dependent within the PSM, and that the mir-125a binding sites are more critical in the posterior PSM, where endogenous oscillations are faster. Thus, one intriguing interpretation of our findings would be interactions between mir-125a and Lfng could influence the oscillatory period of transcript expression particularly in the caudal-most region of the PSM.
If true, this finding would suggest that other regulatory factors that are enriched more anteriorly in the PSM may target the Lfng 3'UTR through other cis-acting elements to maintain oscillatory expression of the transcript in cells that have exited the tail bud. This would suggest the existence of at least three distinct regulatory regions in the PSM. In the most anterior PSM (called region II in(Saga and Takeda, 2001)), Lfng transcripts are stably expressed in the presumptive rostral domain of the presomites, and this expression pattern is controlled by distinct cis-acting enhancers from those required for robust oscillatory expression linked to the clock (Cole et al., 2002, Morales et al., 2002, Shifley et al., 2008). Our data here suggest that as cells move through the posterior 2/3 of the PSM (called region I in(Saga and Takeda, 2001)), they encounter changes in post-transcriptional regulation that may influence the slowing period of the clock. Indeed, work from other groups has suggested that additional regions of the Lfng 3'UTR may be sufficient to destabilize an exogenous RNA in some contexts (Chen et al., 2005).
Within region I of the PSM, several activity gradients have been described including Wnt (Aulehla et al., 2003, Aulehla et al., 2008), Fgf (Sawada et al., 2001, Dubrulle and Pourquie, 2004), and recently glycolysis (Bulusu et al., 2017, Oginuma et al., 2017) all of which are high in the caudal PSM and reduced approaching the determination front. It is interesting to speculate that one or more of these gradients could influence the position-specific expression of molecules or activities that influence the half-lives of clock-linked transcripts, coordinating the deceleration of the clock as cells approach the determination front.
Despite the effects of mutation of mir-125 binding sites in the context of reporter transgenes, the loss of one specific family member in mice, mir-125a-5p, has no effect somitogenesis or clock function. In previous work in chick, we focused on that specific mir-125 family member as it was enriched in the PSM compared to mature somites. Using anti-mirs and target protectors we demonstrated that interactions between mir-125 and Lfng transcripts are required for oscillatory Lfng expression in the chick PSM. However, here we find that loss of mir-125a-5p in mice does not stabilize Lfng expression in the caudal PSM and has no effect on mouse somitogenesis.
Several models could reconcile our apparently contradictory findings. One possibility is that mir-125a-5p:Lfng interactions could provide a species-specific method of regulation that is important in chicken embryos but not mice. In this case, both mir-125a-5p and the conserved sites would be dispensable for somitogenesis in mice, and the transgene effects observed here may be artifacts. However, the fact that multiple independent LvLmut transgene lines had similar defects in Venus expression in the caudal region argues against this model. A more likely explanation would be that the requirements for mir-125a-5p:Lfng interactions in the caudal PSM are conserved between mice and chickens and that the differences observed here result from the different techniques used to disrupt these interactions.
In chicken embryos, introduction of anti-mirs or target protectors produce an acute loss of mir-125 function, while in this study loss of mir-125a-5p in mice is chronic and could trigger upregulation of other pathways to counteract for the absence of mir-125a-5p. This type of compensation has been documented by a previous study where loss of mir-125a-5p in mice resulted in upregulation of another miRNA that functionally compensated for the loss of mir-125a-5p (Tatsumi et al., 2016). In addition other members of the mir-125 family may have redundant functions in the PSM. Both mir-125b and mir-351, which share a seed sequence with mir-125a-5p, are expressed in the mouse PSM, though not enriched compared to mature somites (Riley et al., 2013). It is possible that Lfng expression is regulated by the combined activity of all these family members, and that loss of only mir-125a-5p is not sufficient to disrupt normal segmentation. In this case, our previous results in chicken embryos might suggest that the anti-mir-125a-5p morpholino may inhibit the activity of other closely related miRNAs, a possibility that has been suggested by others (Bartel, 2009, Li et al., 2011). However, our work here does not formally rule out the possibility is that factors other than mir-125a-5p may bind to the sequences that we refer to as ‘mir-125a-5p binding sites’ and destabilize the transcript, which would make mir-125a-5p dispensable in the context of mouse segmentation. Experiments that mutate the "mir-125a" sites in the endogenous Lfng locus or examination of mice with loss of function mutations in multiple mir-125 family members will be necessary to distinguish among these possibilities.
In conclusion, our data from reporter transgenes support the idea that putative mir-125 binding sequences in the Lfng 3'UTR regulate transcript turnover in the caudal-most region of the mouse PSM. However, loss of mir-125a-5p has no effect on Lfng oscillations and the mouse segmentation clock. These results suggest the intriguing possibility that distinct, position-dependent modes of transcript half-life within the PSM may contribute to the slowing of clock oscillations as cells are displaced anteriorly in the PSM.
EXPERIMENTAL PROCEDURES
Plasmid production
The 5’ beta-globin intron/Venus cassette from pRARE-Venus-Ubiq-dsRed was a gift from Randall Moon. PEST sequences amplified from pdEGFP-N1(Clontech) (5’-TTGGCGCGCCAGCCATGGCTTCCCGCCG and 5’-TCTAGACTACACATTGATCCTAGCA), were inserted in frame with VENUS. The PGK promoter was amplified from the ploxp plasmid (5’-TCAGTACTTTTCCCAAGGCAGT and 5’-ATTGGCTGCAGGTCGAAAG) and the CMV promoter came from pCDNA3.1 (Invitrogen). The 3.8 kb BamHI-NotI fragment of mouse Lfng promoter was used in transgenes. Wild type mouse Lfng 3'UTR amplified from genomic DNA (5'-AATCTAGACAGTCGTGGTTGAAACTCTGT and 5'- TTATCGATTGGCTGTCCTGGAACTCACTC). For mutant 3'UTR constructs, putative mir-125 binding sites were replaced with HinDIII recognition sequences (underlined) by site-directed mutagenesis using primers: Site 1: 5'- TTCTTAAGCTTGTGGCAGGTCTGT and 5'-TGCCACAAGCTTAAGAAGACGACT; Site 2 5'-TCCACTAAGCTTTCCGTGGGTGCT and 5'-CACGGAAAGCTTAGTGGAGCCCCA; 1Site 3 5'-AAGCTCAAGCTTAATTGATGTGTT and 5'-TCAATTAAGCTTGAGCTTTTCCCT
Tissue culture
NIH3T3 (ATCC CRL-1658), mouse fibroblasts cells, were grown in DMEM + 10% FBS. For transient transfections 40,000 cells/well were transfected with 500ng of reporter plasmid along with 50 nMol of either Scrambled control (Ambion, negative control NC#1 #17110) or pre-mir-125a-5p (Ambion, pre mir #17100 PM10492) using Lipofectamine®2000 (Invitrogen). Cells were incubated for 24–48 hours before isolating RNA for RT-PCR. Stable C2C12 (ATCC CRL-1772) lines were selected with 800ug/ml Geneticin. Venus expression was confirmed by RT-PCR and visualization of VENUS protein (data not shown).
RT-PCR
RNA was purified (Zymo Quick Miniprep) and converted to cDNA (SuperscriptIII Reverse Transcriptase, Invitrogen) following manufacturer instructions. Venus transcripts (primers 5’ACACGCTGAACTTGTGGC and 5’CTCCGGATCGATCCTGAGAA) and Gapd transcripts (primers 5’GGTGCTGAGTATGTCGTGGAGT and 5’GGGCGGAGATGATGACCCTT) were amplified with SYBR Green Master Mix (Applied Biosystems) in a Step-One ABI machine: 50°C (2 mins), 95°C (10 mins) then 40 cycles: 95°C (15 s) and 60°C (1 min); then melt cycle: 95°C (15 s), 60°C (1 min), 95°C (15 s) and 60°C (15 s). Venus expression was normalized to Gapd expression, and values for Scrambled controls were set to 1. Analyses used three biologically independent samples assayed in triplicate. Statistical analysis used the Student’s T-test with p<0.05 considered significant.
For miRNA expression analysis, RNA was collected from 9.5 d.p.c. embryos. Real time PCR utilized TaqMan microRNA assays (Applied Bioscience) specific for mmu-mir-125a-5p (2198), mmu-mir-99b (436) and mmu-let-7e (2406). sno135 and sno-234 were used as reference genes, and sno234 was used for normalization. Results indicate mean +/− SEM from three biologically independent samples for each genotype assayed in triplicate, assessed using the Student’s T test with p<0.05 considered significant.
Mouse production and maintenance
Mice were maintained under Ohio State University IACUC supervision with an approved animal use protocol that complies with the NIH “Guide for Care and Use of Laboratory Animals”. Transgenic lines were produced at the Genetically Engineered Mouse Modeling Core at OSU. For standard transgenic mice, LvLwt or LvLmut plasmid inserts were injected into the male pronucleus of FVB/NTac embryos, and lines were maintained on the FVB/N background. PCR genotyping used primers 5’CGAGGAGCTGTTCACCGG and 5’CGTGCTGCTTCATGTGGTCG, which produce a transgene specific band of 200 bp. Two independent transgenic lines for LvLwt and two for LvLmut were used for further analysis.
Guide RNAs were designed at the CRISPR design tool (http://crispr.mit.edu/) (Yang et al., 2013) to target the mir-125a-5p locus. Two independent targeting sequences (O1:GTCCACCATAGCTACACTGC and O2:GGACGTCCTCACAGGTTAAA) were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid #42230) as described (Cong et al., 2013). Circular plasmids were injected into fertilized eggs of C57BL/6Tac embryos as described (Fujihara and Ikawa, 2014). Founder mice were screened by High Resolution Melt Analysis as described (Talbot and Amacher, 2014) with primers 5’GCTTAGGGTATCTGTTTCTG and 5’GAGGAGAAGATAGTGACCTT. PCR analysis confirmed that no plasmid sequences were found in the genomes of founder mice. PCR amplification and sequencing of the mir-125a locus was performed as described to characterize mutations (Talbot and Amacher, 2014). PCR genotyping of the mir-125a-5pΔ11 allele used primers 5'CCTCTGGGGAAAAGGGTTTT and 5'GAAATCCCTAAATTTGTGGCT. Analysis on 4% agarose gels distinguishes the 138 bp mutant band and 149 bp wild type band. PCR genotyping of the mir-125a-5pΔ3T allele used primers 5'GCTTAGGGTATCTGTTTCTG and 5'GAGGAGAAGATAGTGACCTT followed by digestion of the PCR product with MseI, producing a mutant band of 308 bp and wild type bands of 204 bp and 108 bp. Founder mir-125a-5p mutant mice were crossed to FVB/NJ mice, and successive backcrosses to the FVB/NJ line were used to eliminate second site mutations. Analyses used embryos starting with the N3 generation, and no changes in phenotype were observed in crosses in the N8 generation.
Whole mount in situ hybridization
Embryos were collected from timed pregnancies, (day of plug identification = 0.5 days post copulation (d.p.c.). In situ hybridization was performed as previously described (Shifley et al., 2008). For detection of Venus expression, the Venus-PEST region was amplified (5’CGAGGAGCTGTTCACCGG and 5’ TCTAGACTACACATTGATCCTAGC) for use as a probe. Expression analysis of Lfng (Johnston et al., 1997) and Uncx (Mansouri et al., 1997) used previously described probes. Oscillatory expression profiles were created by transforming in situ images of 10.5 d.p.c. embryos into heat maps using the FIJI software. Images were converted to greyscale and the plot profile of the PSM from the most recently somite boundary to the tip of the PSM was generated and pseudo-colored in FIJI using the "physics" LUT. Maximum intensity is coded as red and minimum intensity is coded as dark blue. Fischer exact test was used to assess differences in distribution of expression patterns, with p<0.05 being considered as significant.
Two color in situ analysis was performed as described using digoxigen and fluorescein-labeled probes (Shifley and Cole, 2008). Embryos were simultaneously incubated with two probes. After incubation with an AP-conjugated anti-fluorescein antibody and detection with BCIP-NBT (purple precipitate), embryos were heated to 65°C for 15 minutes and washed into 100% MeOH to inactivate the antibody. Embryos were then washed back into MABT prior to incubation with AP-conjugated anti-digoxigenin antibody and detection with BCIP-INT (orange precipitate).
Acknowledgments
We thank the Cole lab for comments. This work was supported in part by NSF grant #IOS-0919649 (SEC), and a Pelotonia Foundation Graduate Fellowship (KW). Mouse strain production (Genetically Engineered Mouse Modeling Core), and qRT-PCR analysis, and DNA sequencing (Genomics Shared Resource) were subsidized by the OSUCCC support grant (NIH P30 CA016058).
References
- Aulehla A, Johnson RL. Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev Biol. 1999;207:49–61. doi: 10.1006/dbio.1998.9164. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquie O. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Stanley P, Papalopulu N. MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulusu V, Prior N, Snaebjornsson MT, Kuehne A, Sonnen KF, Kress J, Stein F, Schultz C, Sauer U, Aulehla A. Spatiotemporal analysis of a glycolytic activity gradient linked to mouse embryo mesoderm development. Dev Cell. 2017;40:331–341. doi: 10.1016/j.devcel.2017.01.015. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kang L, Zhang N. Negative feedback loop formed by lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis. 2005;43:196–204. doi: 10.1002/gene.20171. [DOI] [PubMed] [Google Scholar]
- Cole SE, Levorse JM, Tilghman SM, Vogt TF. Clock regulatory elements control cyclic expression of lunatic fringe during somitogenesis. Dev Cell. 2002;3:75–84. doi: 10.1016/s1534-5807(02)00212-5. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquie O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal hox gene activation. Cell. 2001;106:219–32. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquie O. Fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- Eckalbar WL, Fisher RE, Rawls A, Kusumi K. Scoliosis and segmentation defects of the vertebrae. Wiley Interdiscip Rev Dev Biol. 2012;1:401–423. doi: 10.1002/wdev.34. [DOI] [PubMed] [Google Scholar]
- Feng P, Navaratna M. Modelling periodic oscillations during somitogenesis. Math Biosci Eng. 2007;4:661–673. doi: 10.3934/mbe.2007.4.661. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse lunatic fringe expression, in four-hour cycles at two- hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Fujihara Y, Ikawa M. CRISPR/Cas9-based genome editing in mice by single plasmid injection. Methods Enzymol. 2014;546:319–336. doi: 10.1016/B978-0-12-801185-0.00015-5. [DOI] [PubMed] [Google Scholar]
- Fujimuro T, Matsui T, Nitanda Y, Matta T, Sakumura Y, Saito M, Kohno K, Nakahata Y, Bessho Y. Hes7 3'UTR is required for somite segmentation function. Sci Rep. 2014;4:6462. doi: 10.1038/srep06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Pourquie O. Modeling the segmentation clock as a network of coupled oscillations in the notch, wnt and FGF signaling pathways. J Theor Biol. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Kageyama R. Hopf bifurcation in the presomitic mesoderm during the mouse segmentation. J Theor Biol. 2009;259:176–189. doi: 10.1016/j.jtbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Herrgen L, Ares S, Morelli LG, Schroter C, Julicher F, Oates AC. Intercellular coupling regulates the period of the segmentation clock. Curr Biol. 2010;20:1244–1253. doi: 10.1016/j.cub.2010.06.034. [DOI] [PubMed] [Google Scholar]
- Hilgers V, Pourquie O, Dubrulle J. In vivo analysis of mRNA stability using the tet-off system in the chicken embryo. Dev Biol. 2005;284:292–300. doi: 10.1016/j.ydbio.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Hubaud A, Pourquie O. Signalling dynamics in vertebrate segmentation. Nat Rev Mol Cell Biol. 2014;15:709–721. doi: 10.1038/nrm3891. [DOI] [PubMed] [Google Scholar]
- Jing B, Yuan J, Yin Z, Lv C, Lu S, Xiong H, Tang H, Ye G, Shi F. Dynamic properties of the segmentation clock mediated by microRNA. Int J Clin Exp Pathol. 2015;8:196–206. [PMC free article] [PubMed] [Google Scholar]
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian fringe genes implicated in boundary determination and the notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BK, Oates AC. Delta-notch signalling in segmentation. Arthropod Struct Dev. 2016 doi: 10.1016/j.asd.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Yokota Y, Wehr R, Copeland NG, Jenkins NA, Gruss P. Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev Dyn. 1997;210:53–65. doi: 10.1002/(SICI)1097-0177(199709)210:1<53::AID-AJA6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Maroto M, Bone RA, Dale JK. Somitogenesis. Development. 2012;139:2453–2456. doi: 10.1242/dev.069310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquie O. The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D. Periodic lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Nitanda Y, Matsui T, Matta T, Higami A, Kohno K, Nakahata Y, Bessho Y. 3'-UTR-dependent regulation of mRNA turnover is critical for differential distribution patterns of cyclic gene mRNAs. FEBS J. 2014;281:146–156. doi: 10.1111/febs.12582. [DOI] [PubMed] [Google Scholar]
- Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: Structure, function and dynamics of the vertebrate segmentation clock. Development. 2012;139:625–639. doi: 10.1242/dev.063735. [DOI] [PubMed] [Google Scholar]
- Oginuma M, Moncuquet P, Xiong F, Karoly E, Chal J, Guevorkian K, Pourquie O. A gradient of glycolytic activity coordinates FGF and wnt signaling during elongation of the body axis in amniote embryos. Dev Cell. 2017;40:342–353. doi: 10.1016/j.devcel.2017.02.001. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pourquie O, Tam PP. A nomenclature for prospective somites and phases of cyclic gene expression in the presomitic mesoderm. Dev Cell. 2001;1:619–620. doi: 10.1016/s1534-5807(01)00082-x. [DOI] [PubMed] [Google Scholar]
- Prince VE, Holley SA, Bally-Cuif L, Prabhakaran B, Oates AC, Ho RK, Vogt TF. Zebrafish lunatic fringe demarcates segmental boundaries. Mech Dev. 2001;105:175–80. doi: 10.1016/s0925-4773(01)00398-7. [DOI] [PubMed] [Google Scholar]
- Riley MF, Bochter MS, Wahi K, Nuovo GJ, Cole SE. Mir-125a-5p–mediated regulation of lfng is essential for the avian segmentation clock. Dev Cell. 2013;24:554–561. doi: 10.1016/j.devcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Takeda H. The making of the somite: Molecular events in vertebrate segmentation. Nat Rev Genet. 2001;2:835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, Takeda H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128:4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- Schroter C, Ares S, Morelli LG, Isakova A, Hens K, Soroldoni D, Gajewski M, Julicher F, Maerkl SJ, Deplancke B, Oates AC. Topology and dynamics of the zebrafish segmentation clock core circuit. PLoS Biol. 2012;10:e1001364. doi: 10.1371/journal.pbio.1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifley ET, Cole SE. Lunatic fringe protein processing by proprotein convertases may contribute to the short protein half-life in the segmentation clock. Biochim Biophys Acta. 2008;1783:2384–2390. doi: 10.1016/j.bbamcr.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Shifley ET, Vanhorn KM, Perez-Balaguer A, Franklin JD, Weinstein M, Cole SE. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development. 2008;135:899–908. doi: 10.1242/dev.006742. [DOI] [PubMed] [Google Scholar]
- Shih NP, Francois P, Delaune EA, Amacher SL. Dynamics of the slowing segmentation clock reveal alternating two-segment periodicity. Development. 2015;142:1785–1793. doi: 10.1242/dev.119057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Haltiwanger RS. Significance of glycosylation in notch signaling. Biochem Biophys Res Commun. 2014 doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–585. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP. The control of somitogenesis in mouse embryos. J Embryol Exp Morphol. 1981;65(Suppl):103–128. [PubMed] [Google Scholar]
- Tan SL, Ohtsuka T, Gonzalez A, Kageyama R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells. 2012;17:952–961. doi: 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- Tatsumi N, Hojo N, Yamada O, Ogawa M, Katsura Y, Kawata S, Morii E, Sakamoto H, Inaba R, Tsuda A, Fukuda I, Moriguchi N, Hasuwa H, Okabe M, Fujiki F, Nishida S, Nakajima H, Tsuboi A, Oka Y, Hosen N, Sugiyama H, Oji Y. Deficiency in WT1-targeting microRNA-125a leads to myeloid malignancies and urogenital abnormalities. Oncogene. 2016;35:1003–1014. doi: 10.1038/onc.2015.154. [DOI] [PubMed] [Google Scholar]
- Turnpenny PD, Alman B, Cornier AS, Giampietro PF, Offiah A, Tassy O, Pourquie O, Kusumi K, Dunwoodie S. Abnormal vertebral segmentation and the notch signaling pathway in man. Dev Dyn. 2007;236:1456–1474. doi: 10.1002/dvdy.21182. [DOI] [PubMed] [Google Scholar]
- Wahi K, Bochter MS, Cole SE. The many roles of notch signaling during vertebrate somitogenesis. Semin Cell Dev Biol. 2016;49:68–75. doi: 10.1016/j.semcdb.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Wiedermann G, Bone RA, Silva JC, Bjorklund M, Murray PJ, Dale JK. A balance of positive and negative regulators determines the pace of the segmentation clock. Elife. 2015;4:e05842. doi: 10.7554/eLife.05842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William DA, Saitta B, Gibson JD, Traas J, Markov V, Gonzalez DM, Sewell W, Anderson DM, Pratt SC, Rappaport EF, Kusumi K. Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev Biol. 2007;305:172–186. doi: 10.1016/j.ydbio.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Shifley ET, Braunreiter KM, Cole SE. Disruption of somitogenesis by a novel dominant allele of lfng suggests important roles for protein processing and secretion. Development. 2016;143:822–830. doi: 10.1242/dev.128538. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Martin GV, Kelley MW, Gridley T. A mutation in the lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr Biol. 2000;10:659–662. doi: 10.1016/s0960-9822(00)00522-4. [DOI] [PubMed] [Google Scholar]