The PD-associated LRRK2 G2019S gene abnormally alters marrow myelopoiesis and peripheral myeloid cell differentiation, leading to decreased Th17 cell activity.
Keywords: Crohn, myeloid cells, intestine, T cells, inflammation
Abstract
Parkinson’s disease (PD) is a neurodegenerative disease, whereas Crohn’s disease is an inflammatory bowel disease. Interestingly, polymorphisms in the LRRK2 gene have been identified as risk factors for both diseases. LRRK2 G2019S is the most prevalent mutation found in PD. To gain insights into the role of the LRRK2 G2019S gene on the development and activation of the immune system in the brain–gut axis, we investigated the effect of LRRK2 G2019S on bone marrow myeloid progenitors and myeloid cell function in the periphery. We used bacterial artificial chromosome transgenic rats harboring the human LRRK2 G2019S gene. LRRK2 G2019S transgene decreased the numbers of monocytic and granulocytic progenitors in the bone marrow. However, the numbers of peripheral, immature myeloid cells with suppressive activity were increased in the gut and blood circulation of LRRK2 G2019S compared with control rats in various acute and chronic inflammatory responses. In inflammatory conditions, Th17 cell activity was suppressed, but tissue-associated phylum Bacteroidetes was abnormally increased in the intestine of LRRK2 G2019S rats. The abnormally expanded myeloid cells because of the LRRK2 G2019S gene were highly suppressive on Th17 cell differentiation. Moreover, we found that inhibition of LRRK2 kinase affects myeloid progenitors and myeloid cell differentiation. Taken together, the results indicate that abnormal LRRK2 activity can alter bone marrow myelopoiesis, peripheral myeloid cell differentiation, and intestinal immune homeostasis. These findings may have ramifications in immune and inflammatory responses in patients with LRRK2 abnormalities.
Introduction
PD is a neurodegenerative disease primarily affecting dopaminergic neurons of the substantia nigra and other brain nuclei, whereas CD is an inflammatory bowel disease. Polymorphisms in many genes, including the leucine-rich repeat kinase 2 (LRRK2) gene, also called dardarin and Park8, are linked to PD. The G2019S mutation in the LRRK2 gene is the most common genetic cause of PD and is found in both dominantly inherited familial and sporadic PD [1–5]. Polymorphisms in the LRRK2 gene are also linked to CD [6–8]. Therefore, the LRRK2 G2019S gene presents a good experimental model to study the role of a single mutation in disease-linked genes in the pathogenesis of both PD and CD.
The LRRK2 gene produces a large protein with 286 amino acids and at least 7 domains, including the N-terminal armadillo repeats (ARM), ankyrin repeats, leucine-rich repeat, Ras of complex proteins (ROCs), C terminus of ROC, kinase, and C-terminal WD40. The ROC domain binds and hydrolyzes GTP, induces LRRK2 dimerization, and activates the kinase domain [5, 9–11]. Other domains are involved in LRRK2 interactions with many proteins, including 14-3-3 proteins, Wnt signaling pathway proteins, mitogen-activated kinase, and microtubules [12–15]. The G2019S mutation is localized to the kinase domain and increases kinase activity [16]. Because of the association of LRRK2 gene polymorphisms with PD, most research activities on LRRK2 have been focused on neuronal cells. However, LRRK2 is widely expressed in the body, including in immune cells [17–20], and LRRK2 regulates diverse biologic functions, including mitochondrial function, cellular signaling, neurite growth, cellular vesicle trafficking, and autophagy [21–25]. Much effort has been focused on identified substrates, such as Rab GTPases, which are phosphorylated by LRRK2 [22, 26, 27].
Mounting evidence suggests that LRRK2 may regulate the immune system. However, the function of LRRK2 in the innate and adaptive arms of the immune system remains largely unclear. Recent studies indicate that LRRK2 affects certain myeloid cells. LRRK2-deficient mice were highly susceptible to colitis [28], and this is associated with the function of LRRK2 in restraining NF-AT. LRRK2 regulates monocyte adhesion to endothelial cells [29], and the LRRK2 G2019S mutation increases chemotactic activity of myeloid cells [30]. Importantly, LRRK2 expression appears to be highest in circulating immune cells, such as myeloid cells and B cells, compared with other cells, including brain tissue cells [19, 20]. LRRK2 expression is increased by IFN-γ in Mϕs and in inflamed CD lesions [20, 31]. Taken together, these data suggest that it is critical to understand how alterations in LRRK2-mediated immune function may underlie both PD and CD.
We hypothesized that the LRRK2 G2019S gene affects myeloid cell differentiation and peripheral T cell phenotype, which can influence immunity and inflammatory responses in peripheral tissues, such as the intestine and other tissues. Using BAC transgenic rats harboring the human LRRK2 G2019S gene, which manifest preclinical features of PD in the absence of an end-stage phenotype [25], we determined the effect of LRRK2 G2019S gene on bone marrow myelopoiesis, peripheral myeloid differentiation, and effector T cell phenotype. Here, we report that the LRRK2 G2019S gene abnormally alters marrow myelopoiesis and peripheral myeloid cell differentiation, leading to decreased Th17 cell activity. These findings may have ramifications in our understanding of dysregulated immune responses in patients with LRRK2 polymorphisms.
MATERIALS AND METHODS
Animals and in vivo treatments
Control and G2019S hemizygote Sprague-Dawley rats on NTac:SD background were obtained from Taconic Biosciences (Hudson, NY, USA). The LRRK2 G2019S rats were developed by the laboratory of C. Li and the Michael J. Fox Foundation (New York, NY, USA). All experiments with animals were approved by the Purdue Animal Care and Use Committee. To induce an acute inflammatory response in the colon with TNBS, 7–10-mo-old control and LRRK2 G2019S male rats were fasted overnight and received an intrarectal administration of TNBS (50 mg/kg of body weight in 50% ethanol; Sigma-Aldrich, St. Louis, MO, USA) as described previously [32]. TNBS-treated animals were sacrificed 6 d after TNBS challenge. To induce chronic inflammatory response in the colon with DSS (MP Biomedicals, Solon, OH, USA), 7–10-mo-old control and LRRK2 G2019S male rats were fed drinking water containing 4–5% DSS for ∼25 d (Supplemental Fig. 1). To induce a systemic inflammatory response, 6–7-mo-old, male rats were injected once with LPS (1.2 million EU/kg, i.p; Sigma-Aldrich) on d 0 and were sacrificed on d 15. Frequencies of indicated myeloid and T cells in the colon, blood, and brain tissues were examined by flow cytometry as described below.
Cell isolation and flow cytometry
Bone marrow cells from 6–11-mo-old, male rats were prepared by flushing femurs with PBS using 27-gauge needles. Blood was collected with a rat guillotine in heparin-containing tubes following decapitation. Bone marrow and blood cells were treated with a hypotonic lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA) to lyse RBCs. To prepare colon lamina propria cells, colon tissues were cut into 1-cm tissues and treated with EDTA (10 mM; Sigma-Aldrich) to remove epithelial cells and mucus followed by digestion with collagenase at 37°C for 60 min to release lamina propria cells. For isolation of spleen CD4+ T cells, splenocytes were stained with PE-conjugated Ab to CD4 (clone OX-35) and then with anti-PE microbeads. The stained cells were selected with AutoMACS (Miltenyi Biotec, San Diego, CA, USA). Cells were stained with Abs to RP-1 (RP1) and His-48 (HIS48) for characterization of myeloid cells. For Th1 or Th17 cells, cells were stained with Abs to CD4 (OX-38; BioLegend, San Diego, CA, USA) and then activated in RPMI 1640 medium, supplemented with 10% FBS, PMA (50 ng/ml), ionomycin (1 μM), and monensin (2 mM; Sigma-Aldrich) for 4 h. Cells were then fixed in 2% paraformaldehyde, permeabilized, and stained with Abs to IL-17A (eBio17B7; Thermo Fisher Scientific, Waltham, MA, USA) and IFN-γ (DB-1). Stained cells were analyzed with a Canto II cytometer (BD Biosciences, San Diego, CA, USA). Cell sorting was performed by FACS Aria (BD Biosciences). Sorted cells were plated on slide glasses with a slide centrifuge (Cytospin-4; Thermo Fisher Scientific) and stained with a Wright-Giemsa staining solution (Sigma-Aldrich).
Cell culture
Bone marrow cells were differentiated with mM-CSF (20 ng/ml) for 7 d and examined for surface phenotype. When indicated, LRRK2 kinase inhibitors (all from Bio-Techne, Minneapolis, MN, USA), such as GSK2578215A (10 μM) and LRRK2-IN-1 (1 μM), were added to culture. Cultured bone marrow cells were activated with LPS (1 μg/ml) for 24 h and cocultured with spleen CD4+ T cells. CD4+ T cells (1 × 105) were cocultured with cultured bone marrow cells (1 × 104/well) in U-bottom, 96-well plates for 5 d in the presence of SEB (5 μg/ml) for TCR activation. For Th17 culture, hTGF-β1 (5 ng/ml), mIL-6 (20 ng/ml), mIL-1β (10 ng/ml), mIL-23 (10 ng/ml), mIL-21(10 ng/ml), mTNF-α (20 ng/ml), anti–mIL-4 (11B11, 10 µg/ml), and anti–mIFN-γ (XMG1.2, 10 µg/ml) were used. For Th1 culture, hIL-2 (100 U/ml), mIL-12 (10 ng/ml), and anti–mIL-4 (10 μg/ml) were used. For Tnp, hIL-2 (100 U/ml) was added. All cytokines were from BioLegend. To assess suppressive activity of myeloid cells on T cell proliferation, the dilution of CFSE because of cell division was assessed by flow cytometry.
Myeloid colony forming assays
Bone marrow cells (5 × 104) were plated in each plate (3 cm) containing 0.2% agar-containing RPMI-1640 medium supplemented with 10% FBS, mM-CSF (10 ng/ml), murine granulocyte (G)-CSF (10 ng/ml), murine G Mϕ-CSF (10 ng/ml), and/or rat SCF (50 ng/ml). Bone marrow cells (5 × 105) were also plated in 2.1% methylcellulose-containing RPMI-1640 medium supplemented with 30% FBS, hemin (4 mM), human erythropoietin (20 U/ml), mM-CSF (10 ng/ml), murine G-CSF (10 ng/ml), rat SCF (50 ng/ml), and/or mIL-3 (10 ng/ml). The plates were incubated for 7 d in a CO2 incubator at lowered (5%) oxygen concentration, and all myeloid colonies bigger than cell clusters (∼40 cells) were counted.
PCR analysis of mRNA expression and tissue-associated bacteria
For measuring mRNA expression, total RNA of proximal colon tissues or cultured bone marrow cells was extracted using TRIzol solution (Thermo Fisher Scientific), and converted to cDNA with SuperScript II reverse transcriptase (Thermo Fisher Scientific). SYBR green-based quantitative PCR was performed on ABI 7300 (Thermo Fisher Scientific). The signals were normalized with the mouse Actb (β-actin) signal. For measuring bacteria levels in colon tissues, we followed the method published previously [33]. Briefly, 16S rRNA gene-amplifying primers specific for each bacterial group were used for a SYBR green-based quantitative RT-PCR assay to measure bacteria in DNA isolated from colon tissues with QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA). The signals were normalized with the signal of the mouse genomic Actb gene. Primers used for this study are listed in Supplemental Table 1.
Statistical analysis
Student’s t test (1 or 2-tailed) was used to determine the significance of differences between 2 groups. Differences in body weight changes after TNBS or DSS treatment were examined by a repeated measures ANOVA (SAS, version 9.2; SAS Institute, Cary, NC, USA). P ≤ 0.05 was considered significant.
RESULTS
Decreased numbers of bone marrow myeloid progenitors in LRRK2 G2019S transgenic rats
To assess the effect of the PD-associated human LRRK2 G2019S gene on myelopoiesis, we examined numbers of bone marrow myeloid progenitors with colony-forming assays. The numbers of myelopoietic progenitors responsive to stimulation with SCF and GM-CSF or GM-CSF alone; SCF and M-CSF or M-CSF alone; and SCF and G-CSF or G-CSF alone were significantly decreased in the bone marrow of LRRK2 G2019S rats compared with control rats (Fig. 1A). The decrease of myeloid progenitors described above was confirmed with a complementary methylcellulose colony assay containing M-CSF and SCF or GM-CSF, SCF, EPO, and IL-3 (Fig. 1B).
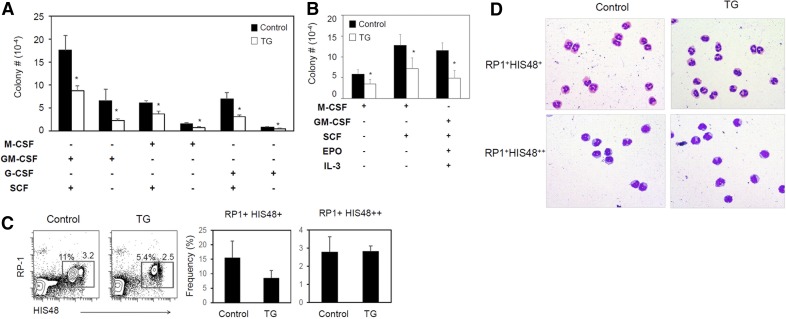
Figure 1. Decreased myelopoiesis in LRRK2 G2019S rats.
(A) Numbers of myeloid cell colonies formed in agar from the bone marrow progenitors of control and LRRK2 G2019S rats. (B) Numbers of myeloid cell colonies formed in methylcellulose from the bone marrow of control and LRRK2 G2019S rats. (C) Expression of RP-1 and HIS48 by peripheral blood myeloid cells. (D) Wright-Giemsa staining of sorted RP-1+ cell subsets. Blood RP-1+ cells were sorted into HIS48+ and HIS48++ cells. Representative dot plots and images and pooled data (n = 3–6) from 2–3 individual experiments are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
We also examined the frequency and phenotype of peripheral myeloid cells in the LRRK2 G2019S and control rats. RP-1+ cells represent peripheral myeloid cells, including primarily granulocytic myeloid cells in rats [34], and HIS48 is expressed by both granulocytic and monocytic myeloid cells [35]. RP-1+ myeloid cells are divided into HIS48+ and HIS48++ cells (Fig. 1C). The numbers of these subsets were somewhat decreased in the peripheral blood of LRRK2 G2019S rats, but the decrease was not statistically significant (Fig. 1C). The 2 subsets were sorted and stained with a Giemsa method. RP1+ HIS48+ cells were of largely granulocytic lineage whereas HIS48++ cells were largely monocytic lineage myeloid cells (Fig. 1D).
Abnormal expansion of peripheral RP-1+ myeloid cells in LRRK2 G2019S rats during inflammatory responses
Tissue inflammation is associated with both PD and CD [36, 37]. Next, we examined peripheral myeloid cell responses during steady state and inflammatory responses in control and G2019S transgenic rats (Fig. 2 and Supplemental Fig. 1). TNBS induces a T cell–mediated inflammatory response in the colon as a hapten [32]. In steady states, few RP-1+ myeloid cells were detected in colon tissues, but their numbers were increased after an acute challenge with TNBS. In TNBS-treated rats, RP-1+ myeloid cell numbers were more increased in the colon of LRRK2 G2019S than in that of control rats following the TNBS challenge. Because TNBS-induced colitis is an acute inflammatory response, we also employed a more chronic intestinal inflammation model induced with DSS during a 25-d period. In this model, DSS damages intestinal epithelial barrier, resulting in bacterial invasion and tissue inflammation. In DSS-induced colitis, the numbers of colonic RP-1+ myeloid cells were more greatly increased in LRRK2 G2019S rats compared with control rats (Fig. 2A). However, we did not find any clear differences in RP-1+ cell numbers between the brain tissues of control and LRRK2 G2019S rats (Fig. 2A). Because TNBS and DSS treatments induce inflammatory responses mainly in the intestine, we also employed a systemic inflammation model induced with LPS injection. LPS administration i.p. resulted in many more RP-1+ myeloid cells in the blood of LRRK2 G2019S than control rats (Fig. 2B). These results indicate that the LRRK2 G2019S gene causes abnormal myeloid cell expansion in inflamed but not in unaffected tissues. In all inflammation models, we did not see any clear differences in weight change between WT and LRRK2 G2019S rats (Supplemental Fig. 1).
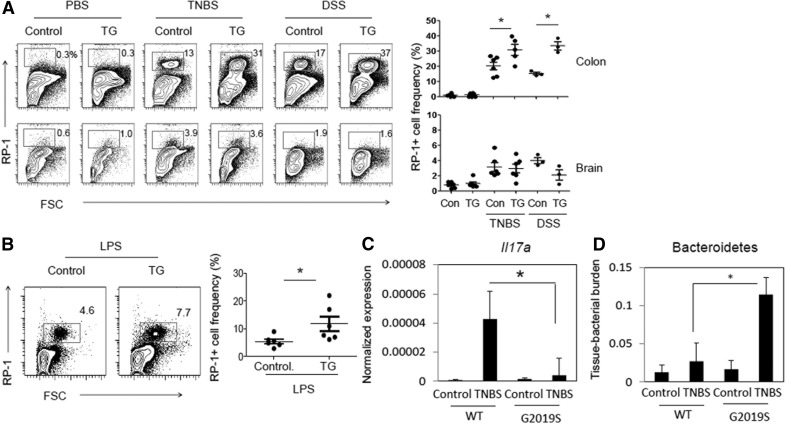
Figure 2. Increased myeloid cell expansion in the intestine and blood, but not the brain, of LRRK2 G2019S rats challenged with TNBS, DSS, or LPS.
(A) RP-1+ cell frequency was examined in the colonic lamina propria. Control and LRRK2 G2019S TG rats were administered TNBS (rectal injection, 50 mg/kg of body weight) or DSS (4∼5%) as described in Supplemental Fig. 1. Rats were sacrificed 6 (TNBS) or 25 (DSS) d later, and indicated organs and tissues were examined for RP-1+ cell frequency. (B) Control and LRRK2 G2019S TG rats were injected with LPS (1.2 million EU/kg per on d 0) and sacrificed 15 d later, and blood cells were examined for RP-1+ cell frequency. (C) Colon tissues were examined for il17a mRNA expression. Expression levels were normalized by that of β-actin. (D) Colon tissues were examined for levels of Bacteroidetes based on Bacteroidetes-specific 16S rRNA gene levels. Tissue Bacteroidetes levels were normalized by β-actin gene levels. Representative dot plots and pooled data (n = 3–6) from 2 individual experiments are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
Because IL-17 is a key cytokine that regulate microbiota in intestinal inflammation, we examined the expression of the il17a gene and levels of tissue-associated microbiota in control and TNBS-treated rats. Il17a expression was greatly induced by the TNBS treatment in control rats but this induction was greatly dampened in the colon of LRRK2 G2019S rats (Fig. 2C). Because the phylum Bacteroidetes is a dominant tissue-associated bacterial group increased in patients with Crohn’s disease [38], we assessed Bacteroidetes levels in colon tissues. Colon tissue-associated Bacteroidetes were abnormally increased in LRRK2 G2019S rats (Fig. 2D). These results indicate dysregulated intestinal immune homeostasis in LRRK2 G2019S rats.
Decreased Th17 cell response in the gut of LRRK2 G2019S transgenic rats
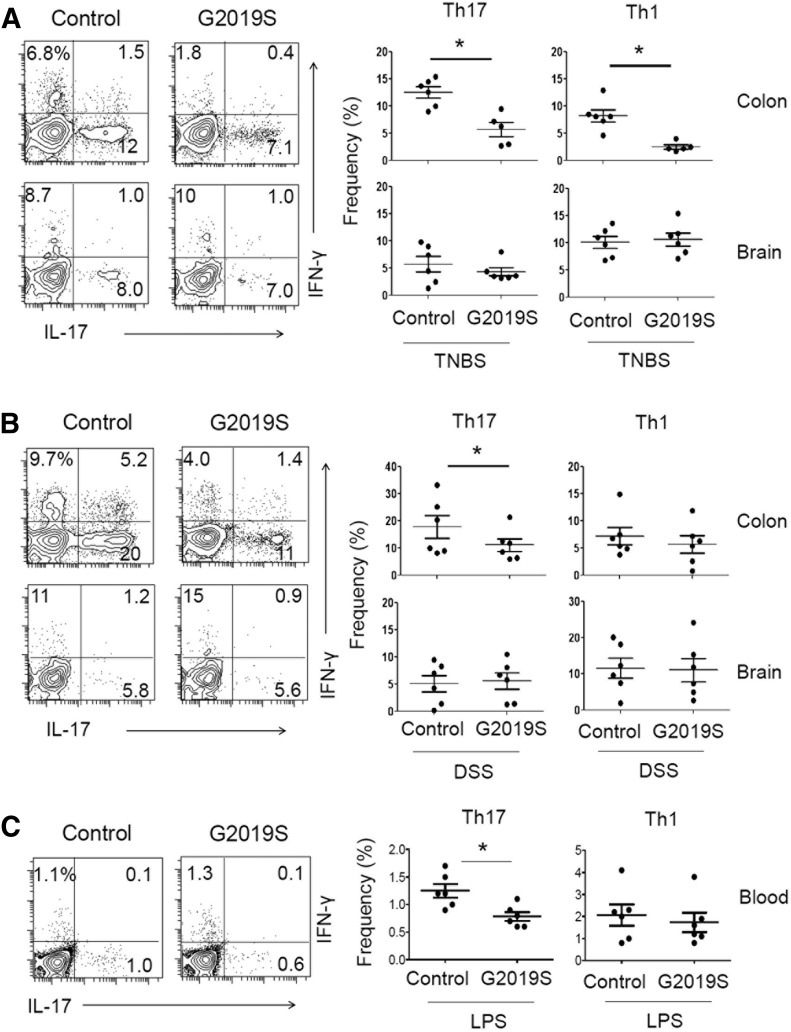
Effector T cells, such as IL-17–producing Th17 and IFN-γ–producing Th1 cells, are generated during acute and chronic inflammatory responses [39]. We found that numbers of Th17 and Th1 cells were decreased in the colon, but not brain, tissues of TNBS-treated LRRK2 G2019S rats (Fig. 3A). Numbers of Th17, but not Th1, cells were decreased also in DSS-treated LRRK2 G2019S compared with control rats (Fig. 3B). Numbers of Th17 cells were also decreased in the blood circulation of LPS-treated rats (Fig. 3C). These results indicate that effector T cell responses, particularly Th17 cell responses in affected tissues, are dampened in the LRRK2 G2019S rats during both intestinal and systemic inflammatory responses.
Figure 3. Th17 cell frequency in the colon, brain, and blood of LRRK2 G2019S rats during intestinal or systemic inflammatory responses.
Frequencies of IL-17+ Th17 cells and IFN-γ+ Th1 cells in the colon, brain, or blood cells were examined in rats challenged with TNBS (A), DSS (B), and LPS (C). Control and G2019S rats were treated with TNBS, DSS, or LPS, as described in Supplemental Fig. 1. Rats were sacrificed 6 (A), 25 (B), or 15 d (C) later, and cells were isolated from the indicated tissues were examined for intracellular cytokine expression. Representative dot plots and pooled data (n = 5–6) from 2 individual experiments are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
Myeloid cells generated from LRRK2 G2019S rats have T cell–suppressing activity in vitro
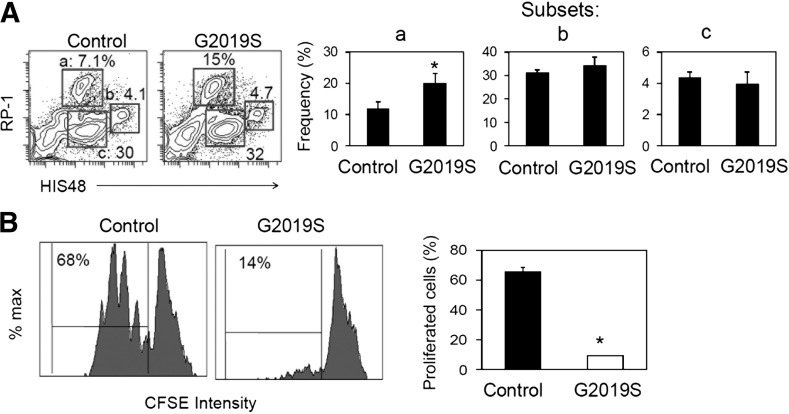
Myeloid cells can either support or suppress effector T cell responses, depending on their differentiation stages and milieu. It has been reported that RP1- or HIS48-expressing rat myeloid cells display suppressive activity on T cell proliferation [35, 40]. We cultured bone marrow cells isolated from control and LRRK2 G2019S TG rats in the presence of M-CSF. In vitro culture of bone marrow cells generated RP1/HIS48-expressing myeloid cells. The bone marrow cells of control and LRRK2 G2019S rats were different in generating RP-1high myeloid cells, but not other cells expressing HIS48 (Fig. 4A). Interestingly, the myeloid cells differentiated from LRRK2 G2019S rat marrow cells displayed greater suppressive activity on the proliferation of spleen CD4+ T cells compared with myeloid cells generated from the control rat marrow cells (Fig. 4B). These results indicate that the LRRK2 G2019S gene abnormally promotes the generation of suppressive myeloid cells in inflammatory conditions.
Figure 4. LRRK2 G2019S mutation promotes generation of suppressive myeloid cells in vitro.
(A) Rat bone marrow cells were differentiated with mM-CSF (20 ng/ml) for 7 d, and examined for RP-1 and HIS48. (B) T cell proliferation-suppression activity of myeloid cells was examined. Myeloid cells prepared as in (A) with M-CSF (20 ng/ml) for 7 d and then activated with LPS (1 μg/ml) for 24 h before coculture with isolated rat CD4+ T cells in the presence of SEB (5 μg/ml) for 3 d. Representative dot plots and pooled data (n = 3) are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
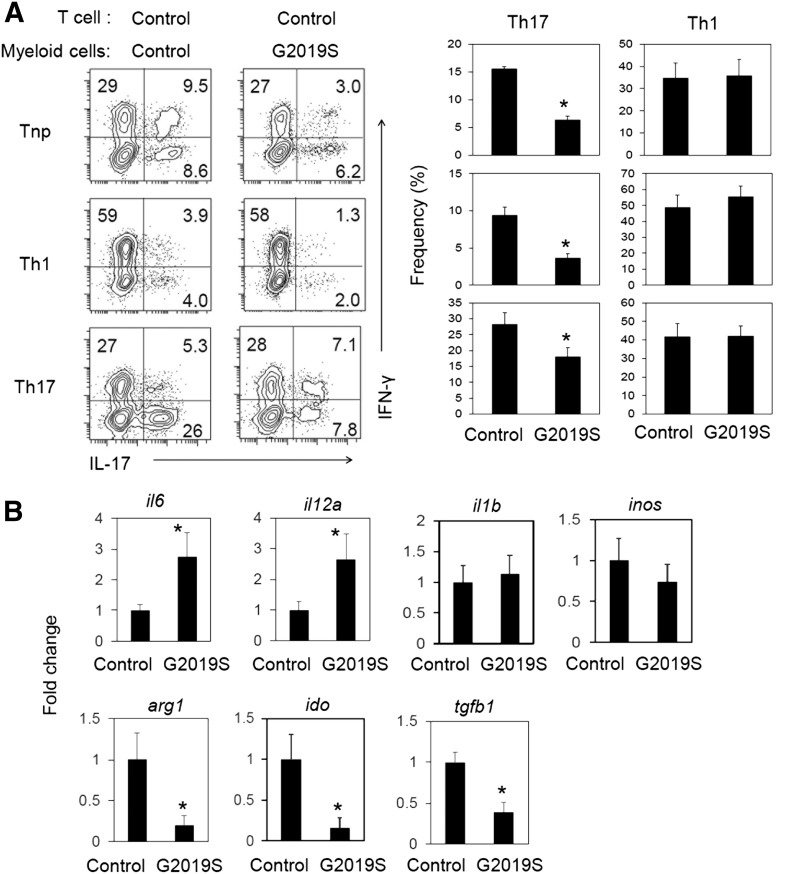
Myeloid cells generated from LRRK2 G2019S transgenic rats are defective in supporting Th17 cell differentiation in vitro
We, next, studied the effect of the myeloid cells harboring the human LRRK2 G2019S gene on effector T cell differentiation. We cocultured M-CSF–activated bone marrow–derived myeloid cells and CD4+ T cells for 5 d in the presence of the superantigen SEB in various cytokine conditions that promote nonpolarized (Tnp), Th17, and Th1 differentiation. Under all cytokine conditions, Th17 cell differentiation was suppressed by the LRRK2 G2019S myeloid cells compared with control myeloid cells (Fig. 5A). However, control and LRRK2 G2019S myeloid cells were not different in supporting Th1 cell differentiation. To gain further insights into the phenotype of myeloid cells, we examined the expression of selected genes in cultured control and LRRK2 G2019S myeloid cells (Fig. 5B). The control and transgenic myeloid cells were different in mRNA expression in that the expression of il6 (IL-6) and il12a (IL-12a) was increased, whereas that of arg1 (arginase 1), ido (indoleamine 2,3-dioxygenase), and tgfb1 (Tgfb1) was decreased. Altered expression of these genes indicates that the LRRK2 G2019S transgene profoundly affects myeloid cell differentiation and function. Moreover, LRRK2 inhibitors suppressed myeloid colony formation (Supplemental Fig. 2), indicating the importance of normal (or natural) levels of LRRK2 activity in maintaining myelopoietic activity.
Figure 5. LRRK2 G2019S gene in myeloid cells suppresses Th17 cell differentiation in vitro.
(A) Effects of in vitro–cultured myeloid cells on CD4+ T cell differentiation. LPS-activated myeloid cells were cocultured with rat spleen CD4+ T cells in the presence of SEB (5 μg/ml) for 5–6 d. Frequencies of IL-17+ Th17 cells and IFN-γ+ Th1 cells were examined after coculture. (B) Expression of selected genes in cultured myeloid cells. Rat bone marrow cells were differentiated with M-CSF (20 ng/ml) for 7 d and then activated with LPS for 24 h before quantitative RT-PCR analysis of indicated genes. Representative dot plots and pooled data (n = 5) are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
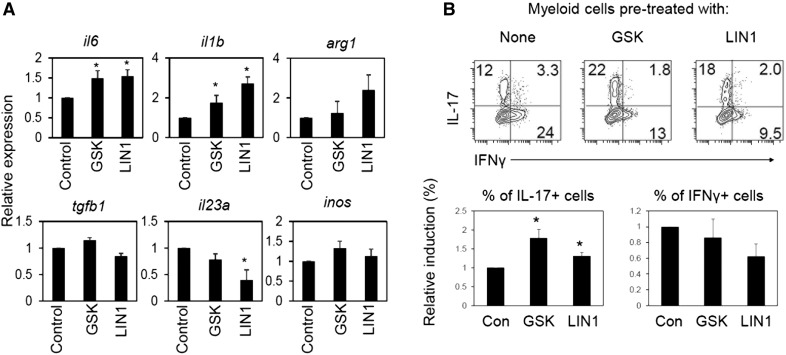
LRRK2 inhibitors altered the expression of il6, il1b, il23a, and arg1, but not Tgfb1 and inos (inducible nitric oxide synthase), in cultured myeloid cells (Fig. 6A). To gain further insights into the effect of suppressed LRRK2 activity in myeloid cells on T cell differentiation, we cocultured LRRK2 inhibitor-pretreated myeloid cells with spleen CD4+ T cells and examined T cell differentiation into Th17 cells. The myeloid cells, pretreated with LRRK2 inhibitors (GSK2578215A and LRRK2-IN-1), had somewhat greater activity in inducing Th17 cells than control myeloid cells had (Fig. 6B). Their activity to support Th1 cells was somewhat decreased, but that difference from control cells was not statistically significant. Thus, suppressed LRRK2 kinase activity can also affect the differentiation and phenotype of myeloid cells.
Figure 6. Inhibition of LRRK2 kinase activity affects myeloid cell differentiation and phenotype.
(A) Effects of LRRK2 kinase inhibitors on selected gene expression in bone marrow–derived myeloid cells. (B) Function of bone marrow–derived myeloid cells, pretreated with LRRK2 kinase inhibitors, in regulating CD4+ T cell differentiation into Th17 and Th1 cells. Rat bone marrow cells were differentiated with M-CSF (20 ng/ml) for 7 d in the presence of indicated LRRK2 inhibitors (GSK2578215A or LRRK2-IN-1) and then activated with LPS for 24 h before quantitative RT-PCR analysis of indicated genes in (A) or for coculture in (B). LPS-activated myeloid cells were cocultured with rat spleen CD4+ T cells in the presence of SEB (5 μg/ml) for 5–6 d, and frequencies of IL-17+ Th17 cells and IFN-γ+ Th1 cells were examined after coculture. Representative dot plots and pooled data (n = 5) are shown. *Significant differences (P ≤ 0.05) from control groups. Error bars indicate means ± sem.
DISCUSSION
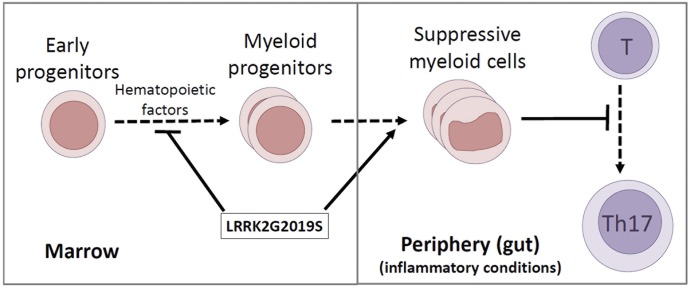
The results of this study reveal a potentially important role of LRRK2 in regulating myelopoiesis and myeloid cell functions in the periphery. The G2019S mutation lies in the kinase domain of LRRK2 protein and increases its kinase activity. Moreover, the LRRK2 G2019S BAC transgenic rats express human LRRK2 at levels 5–8-fold greater than that of natural rat LRRK2 [41], thus increasing the overall activity of LRRK2. We identified a negative role of the human LRRK2 G2019S gene in bone marrow myelopoiesis (Fig. 7). Interestingly, the mutant LRRK2 gene induces peripheral expansion of myeloid cells with a suppressive function on T cells during inflammatory responses. Myeloid cells with the LRRK2 G2019S gene were defective in supporting Th17 cell response. Given the importance of myeloid cells and Th17 cells in the immune system [39], our findings present a potential link between the LRRK2 G2019S gene and dysregulated immune and inflammatory responses.
Figure 7. LRRK2 G2019S gene affects bone marrow myelopoiesis and peripheral myeloid cell differentiation and function.
The LRRK2 G2019S gene decreases the numbers of myeloid progenitors in the bone marrow of transgenic rats but abnormally increases peripheral expansion of suppressive myeloid cells, which suppress Th17 cell differentiation. Because myeloid cells and Th17 cells have central roles in promoting immunity and mediating inflammatory responses, the LRRK2 G2019S–induced dysregulation is likely to have significant effects on the inflammatory responses in various tissues.
We found that the LRRK2 G2019S gene affects numbers of myeloid progenitors responsive to myelopoietic cytokines that stimulate granulocyte, Mϕ, and granulocyte-Mϕ progenitors. Moreover, suppressed natural rat LRRK2 activity by LRRK2 inhibitors also caused decreased myelopoietic activity, at least in vitro. These results indicate that optimal regulation of LRRK2 activity is important for homeostasis of myelopoiesis. Myelopoiesis in the bone marrow is regulated by various cell types and hematopoietic cytokines. Homeostasis of myelopoiesis is required for production of optimal numbers of myeloid lineage cells for innate and adaptive immunity. Myelopoiesis is often suppressed in certain pathologic conditions caused by mutations in key genes or after ablative therapies in patients with cancer and autoimmune diseases.
LRRK2 regulates many important intracellular signaling molecules, such as MAPK, NF-AT, and protein kinase A [28, 42, 43], which profoundly regulate myeloid cell maturation. In general, the LRRK2 G2019S gene causes increased phosphorylation of MAPK and related signaling pathways [44, 45]. LRRK2-deficient mice had microglial abnormalities, such as decreased inflammatory response to LPS ,and increased motility because up-regulated expression of CX3CR1, whereas microglial cells in LRRK2 G2019S mice had decreased motility because of defective focal adhesion kinase signaling [46, 47]. Myelopoietic growth factors, such as M-CSF and GM-CSF, activate MAPK/ERK1/2 pathways and NF-AT [48, 49], which are also known to be regulated by LRRK2 [28, 43]. Therefore, aberrant regulation of these signaling pathways because of abnormal LRRK2 activity could also change myeloid cell responses to cytokines. Further studies of these effects, in the future, may enhance our understanding.
Maturation and terminal differentiation of myeloid progenitors continue in the periphery, and this is greatly altered during inflammatory responses [50, 51]. We observed that the RP-1+ myeloid population was abnormally increased in LRRK2 G2019S rats upon inflammatory challenges. RP-1+ myeloid cells have immune suppressive functions, mimicking that of myeloid-derived suppressor cells in mice and humans [35, 40]. This response during inflammatory responses contrasts with the decreased myelopoiesis in the bone marrow of steady-state LRRK2 G2019S animals. The expansion of RP-1+ cells occurs only in active sites of inflammatory responses, such as the intestine, but not in the brain in LRRK2 G2019S rats. In this regard, we previously determined that the brain of LRRK2 G2019S rats has preclinical features but not the end-stage phenotype of PD [25]. Detailed analysis of the nigrostriatal dopamine system in the transgenic rats should be performed, and that should be correlated with intestinal inflammatory responses in the future. Interestingly, the function of the expanded myeloid cells was suppressive on T cells rather than activating, indicating that they are either immature or suppressive myeloid cells. The suppressive phenotype is often associated with immature myeloid cells or alternatively activated Mϕs, which are often found in cancer and chronic infection or inflammatory responses [52, 53]. In addition to LRRK2 G2019S model, we also examined the effect of decreased LRRK2 kinase activity on myeloid cells using LRRK2 inhibitors. M-CSF–activated myeloid cells, differentiated in the presence of LRRK2 inhibitors, had increased activity in supporting Th17 cell differentiation. This result complements the finding that the gain-of-function G2019S mutation in myeloid cells decreased Th17 differentiation. We observed altered expression of Th17-inducing cytokines, such as IL-6, IL-1β, and TGFβ1, when LRRK2 function was altered by either the G2019S gene or inhibitors, but the changes in cytokine expression do not accurately explain the changes in Th17-inducing activity of myeloid cells, warranting additional studies.
Peripheral myeloid cells affect T cell differentiation into effector T cells, such as Th17 cells, which produce IL-17 to promote neutrophil recruitment and activation [39]. Moreover, Th17 cells stimulate intestinal epithelial cells for barrier immunity [54]. We found that the myeloid cells generated from the marrow progenitors of LRRK2 G2019S animals are defective in supporting Th17 cell activity. We also observed that Bacteroidetes levels in colon tissues were abnormally increased in LRRK2 G2019S vs. control rats. We speculate that the defective Th17 response, associated with LRRK2 G2019S gene, has the potential to affect immunity and inflammatory responses in humans.
In summary, the human LRRK2 G2019S gene causes decreased bone marrow myelopoiesis and dysregulated peripheral myeloid phenotype during inflammatory responses, leading to defective Th17 generation (Fig. 7). The dysregulation of peripheral immune cells discovered in this study is likely to affect peripheral immunity and associated inflammatory responses and has the potential to affect inflammatory conditions in multiple organs, including the intestine and brain.
AUTHORSHIP
J.P. performed experiments, analyzed data, and prepared most of the figures, with experimental contributions from J.-W.L. S.C.C. and H.E.B. performed colon forming assays. C.H.K. and J.R.C. conceived the project and obtained funding. C.H.K., J.R.C., and J.P. drafted the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in part, by grants from the Michael J. Fox Foundation to C.H.K. and J.R.C., the National Multiple Sclerosis Society, and the U.S. National Institutes of Health (Grants R01AI074745, R01DK076616, R01AI080769, and R01AI121302) to C.H.K. We thank M. Kim and J. Wise (Purdue University) for aid in animal studies.
Glossary
- BAC
bacterial artificial chromosome
- CD
Crohn’s disease
- DSS
dextran sulfate sodium
- EPO
human erythropoietin
- h
human
- m
mouse
- mM-CSF
murine macrophage-CSF
- PD
Parkinson disease
- ROC
Ras of complex protein
- SCF
stem cell factor
- SEB
staphylococcal enterotoxin B
- TNBS
2,4,6-trinitrobenzenesulfonic acid
Footnotes
The online version of this paper, found at www.jleukbio.org, contains supplemental information.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Nichols W. C., Pankratz N., Hernandez D., Paisán-Ruíz C., Jain S., Halter C. A., Michaels V. E., Reed T., Rudolph A., Shults C. W., Singleton A., Foroud T.; Parkinson Study Group-PROGENI investigators (2005) Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet 365, 410–412. [DOI] [PubMed] [Google Scholar]
- 2.Di Fonzo A., Rohé C. F., Ferreira J., Chien H. F., Vacca L., Stocchi F., Guedes L., Fabrizio E., Manfredi M., Vanacore N., Goldwurm S., Breedveld G., Sampaio C., Meco G., Barbosa E., Oostra B. A., Bonifati V.; Italian Parkinson Genetics Network (2005) A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson’s disease. Lancet 365, 412–415. [DOI] [PubMed] [Google Scholar]
- 3.Gilks W. P., Abou-Sleiman P. M., Gandhi S., Jain S., Singleton A., Lees A. J., Shaw K., Bhatia K. P., Bonifati V., Quinn N. P., Lynch J., Healy D. G., Holton J. L., Revesz T., Wood N. W. (2005) A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 365, 415–416. [DOI] [PubMed] [Google Scholar]
- 4.Greggio E. (2012) Role of LRRK2 kinase activity in the pathogenesis of Parkinson’s disease. Biochem. Soc. Trans. 40, 1058–1062. [DOI] [PubMed] [Google Scholar]
- 5.Rudenko I. N., Cookson M. R. (2014) Heterogeneity of leucine-rich repeat kinase 2 mutations: genetics, mechanisms and therapeutic implications. Neurotherapeutics 11, 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett J. C., Hansoul S., Nicolae D. L., Cho J. H., Duerr R. H., Rioux J. D., Brant S. R., Silverberg M. S., Taylor K. D., Barmada M. M., Bitton A., Dassopoulos T., Datta L. W., Green T., Griffiths A. M., Kistner E. O., Murtha M. T., Regueiro M. D., Rotter J. I., Schumm L. P., Steinhart A. H., Targan S. R., Xavier R. J., NIDDK IBD Genetics Consortium, Libioulle C., Sandor C., Lathrop M., Belaiche J., Dewit O., Gut I., Heath S., Laukens D., Mni M., Rutgeerts P., Van Gossum A., Zelenika D., Franchimont D., Hugot J. P., de Vos M., Vermeire S., Louis E., Belgian-French IBD Consortium, Wellcome Trust Case Control Consortium, Cardon L. R., Anderson C. A., Drummond H., Nimmo E., Ahmad T., Prescott N. J., Onnie C. M., Fisher S. A., Marchini J., Ghori J., Bumpstead S., Gwilliam R., Tremelling M., Deloukas P., Mansfield J., Jewell D., Satsangi J., Mathew C. G., Parkes M., Georges M., Daly M. J., Parkes M., Georges M., Daly M. J. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., Anderson C. A., Bis J. C., Bumpstead S., Ellinghaus D., Festen E. M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C. G., Montgomery G. W., Prescott N. J., Raychaudhuri S., Rotter J. I., Schumm P., Sharma Y., Simms L. A., Taylor K. D., Whiteman D., Wijmenga C., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Cohen A., Colombel J. F., Cottone M., Stronati L., Denson T., De Vos M., D’Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van Gossum A., Guthery S. L., Halfvarson J., Verspaget H. W., Hugot J. P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newmvaan W., Panés J., Phillips A., Proctor D. D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A. H., Stokkers P. C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S. R., Brant S. R., Rioux J. D., D’Amato M., Weersma R. K., Kugathasan S., Griffiths A. M., Mansfield J. C., Vermeire S., Duerr R. H., Silverberg M. S., Satsangi J., Schreiber S., Cho J. H., Annese V., Hakonarson H., Daly M. J., Parkes M. (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson C. A., Boucher G., Lees C. W., Franke A., D’Amato M., Taylor K. D., Lee J. C., Goyette P., Imielinski M., Latiano A., Lagacé C., Scott R., Amininejad L., Bumpstead S., Baidoo L., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Colombel J. F., Denson L. A., De Vos M., Dubinsky M., Edwards C., Ellinghaus D., Fehrmann R. S., Floyd J. A., Florin T., Franchimont D., Franke L., Georges M., Glas J., Glazer N. L., Guthery S. L., Haritunians T., Hayward N. K., Hugot J. P., Jobin G., Laukens D., Lawrance I., Lémann M., Levine A., Libioulle C., Louis E., McGovern D. P., Milla M., Montgomery G. W., Morley K. I., Mowat C., Ng A., Newman W., Ophoff R. A., Papi L., Palmieri O., Peyrin-Biroulet L., Panés J., Phillips A., Prescott N. J., Proctor D. D., Roberts R., Russell R., Rutgeerts P., Sanderson J., Sans M., Schumm P., Seibold F., Sharma Y., Simms L. A., Seielstad M., Steinhart A. H., Targan S. R., van den Berg L. H., Vatn M., Verspaget H., Walters T., Wijmenga C., Wilson D. C., Westra H. J., Xavier R. J., Zhao Z. Z., Ponsioen C. Y., Andersen V., Torkvist L., Gazouli M., Anagnou N. P., Karlsen T. H., Kupcinskas L., Sventoraityte J., Mansfield J. C., Kugathasan S., Silverberg M. S., Halfvarson J., Rotter J. I., Mathew C. G., Griffiths A. M., Gearry R., Ahmad T., Brant S. R., Chamaillard M., Satsangi J., Cho J. H., Schreiber S., Daly M. J., Barrett J. C., Parkes M., Annese V., Hakonarson H., Radford-Smith G., Duerr R. H., Vermeire S., Weersma R. K., Rioux J. D. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J. M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., Thomas K. J., Baekelandt V., Cookson M. R. (2008) The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 283, 16906–16914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Civiero L., Vancraenenbroeck R., Belluzzi E., Beilina A., Lobbestael E., Reyniers L., Gao F., Micetic I., De Maeyer M., Bubacco L., Baekelandt V., Cookson M. R., Greggio E., Taymans J. M. (2012) Biochemical characterization of highly purified leucine-rich repeat kinases 1 and 2 demonstrates formation of homodimers. PLoS One 7, e43472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger Z., Smith K. A., Lavoie M. J. (2010) Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry 49, 5511–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandhi P. N., Wang X., Zhu X., Chen S. G., Wilson-Delfosse A. L. (2008) The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J. Neurosci. Res. 86, 1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancho R. M., Law B. M., Harvey K. (2009) Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signalling pathways. Hum. Mol. Genet. 18, 3955–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C. H., Chan D., Greggio E., Saha S., Guillily M. D., Ferree A., Raghavan K., Shen G. C., Segal L., Ryu H., Cookson M. R., Wolozin B. (2010) MKK6 binds and regulates expression of Parkinson’s disease-related protein LRRK2. J. Neurochem. 112, 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzamko N., Deak M., Hentati F., Reith A. D., Prescott A. R., Alessi D. R., Nichols R. J. (2010) Inhibition of LRRK2 kinase activity leads to dephosphorylation of Se910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J. 430, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. USA 102, 16842–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44, 595–600. [DOI] [PubMed] [Google Scholar]
- 18.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607. [DOI] [PubMed] [Google Scholar]
- 19.Hakimi M., Selvanantham T., Swinton E., Padmore R. F., Tong Y., Kabbach G., Venderova K., Girardin S. E., Bulman D. E., Scherzer C. R., LaVoie M. J., Gris D., Park D. S., Angel J. B., Shen J., Philpott D. J., Schlossmacher M. G. (2011) Parkinson’s disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J Neural Transm (Vienna) 118, 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardet A., Benita Y., Li C., Sands B. E., Ballester I., Stevens C., Korzenik J. R., Rioux J. D., Daly M. J., Xavier R. J., Podolsky D. K. (2010) LRRK2 is involved in the IFN-γ response and host response to pathogens. J. Immunol. 185, 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. (2006) The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52, 587–593. [DOI] [PubMed] [Google Scholar]
- 22.MacLeod D. A., Rhinn H., Kuwahara T., Zolin A., Di Paolo G., McCabe B. D., Marder K. S., Honig L. S., Clark L. N., Small S. A., Abeliovich A. (2013) RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. (published correction in Neuron 2013. 77, 994.) Neuron 77, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodson M. W., Leung L. K., Lone M., Lizzio M. A., Guo M. (2014) Novel ethyl methanesulfonate (EMS)-induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo. Dis. Model. Mech. 7, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arranz A. M., Delbroek L., Van Kolen K., Guimarães M. R., Mandemakers W., Daneels G., Matta S., Calafate S., Shaban H., Baatsen P., De Bock P. J., Gevaert K., Vanden Berghe P., Verstreken P., De Strooper B., Moechars D. (2015) LRRK2 functions in synaptic vesicle endocytosis through a kinase-dependent mechanism. J. Cell Sci. 128, 541–552. [DOI] [PubMed] [Google Scholar]
- 25.Lee J. W., Cannon J. R. (2015) LRRK2 mutations and neurotoxicant susceptibility. Exp. Biol. Med. (Maywood) 240, 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin N., Jeong H., Kwon J., Heo H. Y., Kwon J. J., Yun H. J., Kim C. H., Han B. S., Tong Y., Shen J., Hatano T., Hattori N., Kim K. S., Chang S., Seol W. (2008) LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 314, 2055–2065. [DOI] [PubMed] [Google Scholar]
- 27.Dodson M. W., Zhang T., Jiang C., Chen S., Guo M. (2012) Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 21, 1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M. J. (2011) The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 12, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hongge L., Kexin G., Xiaojie M., Nian X., Jinsha H. (2015) The role of LRRK2 in the regulation of monocyte adhesion to endothelial cells. J. Mol. Neurosci. 55, 233–239. [DOI] [PubMed] [Google Scholar]
- 30.Moehle M. S., Daher J. P., Hull T. D., Boddu R., Abdelmotilib H. A., Mobley J., Kannarkat G. T., Tansey M. G., West A. B. (2015) The G2019S LRRK2 mutation increases myeloid cell chemotactic responses and enhances LRRK2 binding to actin-regulatory proteins. Hum. Mol. Genet. 24, 4250–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuss M., Adamopoulou E., Kahle P. J. (2014) Interferon-γ induces leucine-rich repeat kinase LRRK2 via extracellular signal-regulated kinase ERK5 in macrophages. J. Neurochem. 129, 980–987. [DOI] [PubMed] [Google Scholar]
- 32.Wirtz S., Neufert C., Weigmann B., Neurath M. F. (2007) Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546. [DOI] [PubMed] [Google Scholar]
- 33.Kim M. H., Kang S. G., Park J. H., Yanagisawa M., Kim C. H. (2013) Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406.e1-10. [DOI] [PubMed] [Google Scholar]
- 34.Gotoh S., Itoh M., Fujii Y., Arai S., Sendo F. (1986) Enhancement of the expression of a rat neutrophil-specific cell surface antigen by activation with phorbol myristate acetate and concanavalin A. J. Immunol. 137, 643–650. [PubMed] [Google Scholar]
- 35.Dolen Y., Gunaydin G., Esendagli G., Guc D. (2015) Granulocytic subset of myeloid derived suppressor cells in rats with mammary carcinoma. Cell. Immunol. 295, 29–35. [DOI] [PubMed] [Google Scholar]
- 36.Torres J., Mehandru S., Colombel J. F., Peyrin-Biroulet L. (2016) Crohn’s disease. Lancet.389, 1741–1755. [DOI] [PubMed] [Google Scholar]
- 37.De Virgilio A., Greco A., Fabbrini G., Inghilleri M., Rizzo M. I., Gallo A., Conte M., Rosato C., Ciniglio Appiani M., de Vincentiis M. (2016) Parkinson’s disease: autoimmunity and neuroinflammation. Autoimmun. Rev. 15, 1005–1011. [DOI] [PubMed] [Google Scholar]
- 38.Gophna U., Sommerfeld K., Gophna S., Doolittle W. F., Veldhuyzen van Zanten S. J. (2006) Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim C. H. (2009) Migration and function of Th17 cells. Inflamm. Allergy Drug Targets 8, 221–228. [DOI] [PubMed] [Google Scholar]
- 40.Jia W., Jackson-Cook C., Graf M. R. (2010) Tumor-infiltrating, myeloid-derived suppressor cells inhibit T cell activity by nitric oxide production in an intracranial rat glioma + vaccination model. J. Neuroimmunol. 223, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J. W., Tapias V., Di Maio R., Greenamyre J. T., Cannon J. R. (2015) Behavioral, neurochemical, and pathologic alterations in bacterial artificial chromosome transgenic G2019S leucine-rich repeated kinase 2 rats. Neurobiol. Aging 36, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muda K., Bertinetti D., Gesellchen F., Hermann J. S., von Zweydorf F., Geerlof A., Jacob A., Ueffing M., Gloeckner C. J., Herberg F. W. (2014) Parkinson-related LRRK2 mutation R1441C/G/H impairs PKA phosphorylation of LRRK2 and disrupts its interaction with 14-3-3. Proc. Natl. Acad. Sci. USA 111, E34–E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liou A. K., Leak R. K., Li L., Zigmond M. J. (2008) Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiol. Dis. 32, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White L. R., Toft M., Kvam S. N., Farrer M. J., Aasly J. O. (2007) MAPK-pathway activity, Lrrk2 G2019S, and Parkinson’s disease. J. Neurosci. Res. 85, 1288–1294. [DOI] [PubMed] [Google Scholar]
- 45.Chen C. Y., Weng Y. H., Chien K. Y., Lin K. J., Yeh T. H., Cheng Y. P., Lu C. S., Wang H. L. (2012) (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ. 19, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma B., Xu L., Pan X., Sun L., Ding J., Xie C., Koliatsos V. E., Cai H. (2016) LRRK2 modulates microglial activity through regulation of chemokine (C-X3-C) receptor 1 -mediated signalling pathways. Hum. Mol. Genet. 25, 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi I., Kim B., Byun J. W., Baik S. H., Huh Y. H., Kim J. H., Mook-Jung I., Song W. K., Shin J. H., Seo H., Suh Y. H., Jou I., Park S. M., Kang H. C., Joe E. H. (2015) LRRK2 G2019S mutation attenuates microglial motility by inhibiting focal adhesion kinase. Nat. Commun. 6, 8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbosa C. M., Bincoletto C., Barros C. C., Ferreira A. T., Paredes-Gamero E. J. (2014) PLCγ2 and PKC are important to myeloid lineage commitment triggered by M-SCF and G-CSF. J. Cell. Biochem. 115, 42–51. [DOI] [PubMed] [Google Scholar]
- 49.Gomez-Cambronero J., Colasanto J. M., Huang C. K., Sha’afi R. I. (1993) Direct stimulation by tyrosine phosphorylation of microtubule-associated protein (MAP) kinase activity by granulocyte-macrophage colony-stimulating factor in human neutrophils. Biochem. J. 291, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cannistra S. A., Griffin J. D. (1988) Regulation of the production and function of granulocytes and monocytes. Semin. Hematol. 25, 173–188. [PubMed] [Google Scholar]
- 51.Silva M. T. (2010) Neutrophils and macrophages work in concert as inducers and effectors of adaptive immunity against extracellular and intracellular microbial pathogens. J. Leukoc. Biol. 87, 805–813. [DOI] [PubMed] [Google Scholar]
- 52.Netherby C. S., Abrams S. I. (2017) Mechanisms overseeing myeloid-derived suppressor cell production in neoplastic disease. Cancer Immunol. Immunother. doi: 10.1007/s00262-017-1963-5 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dorhoi A., Kaufmann S. H. (2015) Versatile myeloid cell subsets contribute to tuberculosis-associated inflammation. Eur. J. Immunol. 45, 2191–2202. [DOI] [PubMed] [Google Scholar]
- 54.Blaschitz C., Raffatellu M. (2010) Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 30, 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.