Abstract
Background
Phoebe (Lauraceae) comprises of evergreen trees or shrubs with approximately 100 species, distributed in tropical and subtropical Asia and Neotropical America. A total of 34 species and three varieties occur in China. Despite of economic and ecological value, only limited genomic resources are available for this genus.
Results
We sequenced the two complete chloroplast (cp) genomes of Phoebe chekiangensis and P. bournei using Illumina sequencing technology via a combined strategy of de novo and reference-guided assembly. We also performed comparative analyses with the cp genomes of P. sheareri and P. sheareri var. oineiensis previously reported. The chloroplast genomes of P. chekiangensis and P. bournei identically contain 112 genes consisting of 78 protein coding genes, 30 tRNA genes, and 4 rRNA genes, with the size of 152,849 and 152,853 bp, respectively. From the two chloroplast genomes, 131 SSRs were identified and 12 different SSRs located in five protein coding genes. The analysis showed the extremely conserved structure of chloroplast genomes with surprisingly little variations at the LSC/IR and SSC/IR boundaries. Moreover, the mean nucleotide diversity was found to be 0.162% for 77 regions, suggesting an extraordinarily low level of sequence divergence. Four highest divergent regions (trnH-psbA, rps14-trnT, petA-psbJ, ccsA-ndhD) with the percentage of nucleotide diversity higher than 0.50% were identified, which had potential use for species identification and phylogenetic studies.
Conclusion
This study will facilitate our understanding of population genetics, phylogenetic relationship and plant evolution of Phoebe species.
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-017-0192-8) contains supplementary material, which is available to authorized users.
Keywords: Phoebe chekiangensis, Phoebe bournei, Chloroplast genomes, Repeat analysis, SSRs, Divergent regions
Background
Phoebe is a genus of evergreen trees or shrubs belonging to family, Lauraceae. Phoebe comprises of approximately 100 species, distributed in tropical and subtropical Asia and neotropical America. A total of 34 species and three varieties are endemic to China (Wu et al. 2008). Phoebe species, with their high-quality wood, were widely used to make column during palace construction in the Ming and Qing dynasties and built high valuable furniture which stood for the power and status of the noble (Ding et al. 2015). The most famous and valuable Phoebe wood are called ‘wood with golden wire’, which comes from several specific and sporadically distributed rare Phoebe species, endemics to China, including Phoebe chekiangensis, Phoebe bournei, Phoebe sheareri, Phoebe zhennan, and Phoebe lichuanensis. Our target species, P. chekiangensis is distributed in Zhejiang and its adjacent areas (including Fujian, Jiangxi, and Anhui Province), whereas P. bournei is distributed in Yangtze River Basin and its south region in China. The Phoebe species have a very high economic and ecological value but with extremely limited studies on biochemical compound and population diversity (Hegde et al. 1997; Zhang et al. 2012a, b).
The chloroplast (cp) is an important organelle that plays a key role in plant photosynthesis providing energy to green plants and carbon fixation (Douglas 1990). With the rapid development of next-generation sequencing, it is now cheaper and faster to obtain genomes than by traditional Sanger sequencing. Therefore, cp genome-scale data have been increasingly used to infer phylogenetic relationships at high taxonomical levels, and even in lower taxa (Moore et al. 2007; Parks et al. 2009; Huang et al. 2014; Yang et al. 2017; Zeng et al. 2017). Although the cp genome is more conserved than the nuclear genome in plants, many mutation events in the chloroplast DNA sequence have been identified, including indels, substitutions, and inversions (Ingvarsson et al. 2003). Most angiosperm cp genomes have a quadripartite circular structure ranging from 115 to 165 kp in length, and are composed of two copies of inverted repeat (IR) regions that are separated by a large single copy (LSC) region and a small single copy (SSC) region (Wicke et al. 2011; Wang et al. 2015). Massive information for genetics, taxonomy, and phylogeny could be mined from cp genomes, because of their relatively conserved gene structure and sequence divergence between species and individuals (Parks et al. 2009; Huang et al. 2014). Furthermore, cp genomes can provide effective genetic markers for evolutionary studies from population level. In addition, analyzing the composition and structure of the cp genomes for such an important genus Phoebe can explore further genetic variations, which could improve quantitative and quality traits.
In this study, we reported the complete and annotated DNA sequences for the cp genomes of P. chekiangensis and P. bournei using next-generation sequencing platform, which is also the first comprehensive analysis on cp genomes for Phoebe combining the cp genomes of P. sheareri and P. sheareri var. oineiensis previously reported (Song et al. 2016). The specific aims of the present study were to: (1) present the complete chloroplast genome sequences and investigate global structural patterns of P. chekiangensis and P. bournei; (2) examine variations of repeat sequences and simple sequence repeats (SSRs) among the two Phoebe chloroplast genomes; (3) screen sequence divergence hotspot regions in the four Phoebe chloroplast genomes.
Methods
Plant material and DNA extraction
Young leaves of P. chekiangensis and P. bournei were sampled from single seedlings growing in the nursery located at 30.21N, 120.02E, of Zhejiang Academy of Forestry. The provenance was Hangzhou, Zhejiang Province and Mingxi, Fujian Province, PR China, which was the primary distribution area of P. chekiangensis and P. bournei, respectively. Total genomic DNA per species was extracted from 30 mg of the silica-dried leaf using the modified CTAB method (Porebski et al. 1997). The quality and concentration of the genomic DNA were assessed using agarose gel electrophoresis and an Agilent BioAnalyzer 2100 (Agilent Technologies).
Chloroplast genome illumina sequencing, assembly and annotation
Genomic DNA was used to generate short-insert (500 bp) paired-end sequencing libraries according to the Illumina standard protocol. Genomic DNA from each species was sequenced using a HiSeq™ 2000 analyzer (Illumina, San Diego, California, USA) at Beijing Genomics Institute (BGI, Shenzhen, China).The raw reads (20,777,674 and 20,787,108 bp for P. chekiangensis and P. bournei) were generated with 125 bp length and assembled into whole chloroplast genomes in a multi-step approach employing a modified pipeline that involved a combination of both reference guided and de novo assembly approaches. First, paired-end sequence reads were trimmed to remove low-quality bases (Q <20, 0.01 probability error) and adapter sequences using CLC-quality trim tool (quality_trim software included in CLC ASSEMBLY CELL package, http://www.clcbio.com/products/clc-assembly-cell/) before undertaking sequence assembly. Second, the contigs were assembled using CLC de novo assembler with the following optimized parameters: bubble size of 98, minimum contig length of 200, mismatch cost of two, deletion and insertion costs of three, length fraction of 0.9, and similarity fraction of 0.8. Third, all the contigs were aligned to the reference chloroplast genome of Machilus yunnanensis (NC028073) using BLAST (http://blast.ncbi.nlm.nih.gov/), and aligned contigs (≥90% similarity and query coverage) were ordered according to the reference chloroplast genome. Then, contigs were aligned with the reference genome to construct the draft chloroplast genome of each species in Geneious 9.0.5 software (http://www.geneious.com). Finally, clean reads were re-mapped to the draft cp genomes of two Phoebe species, and the mapping ratio were 3.41% for P. chekiangensis and 2.15% for P. bournei, and average coverage depth were 578.2 and 367.1 for P. chekiangensis and P. bournei, respectively. Using the Dual Organellar Genome Annotator (DOGMA) program (Wyman et al. 2004), the two Phoebe chloroplast genomes were annotated. Protein-coding genes were identified by using the plastid/bacterial genetic code. Intron/exon boundaries were further determined using MAFFT v7 with those of the M. yunnanensis chloroplast genomes as a reference (Katoh and Standley 2013). Using the program tRNAscan-SEwith default settings (Schattner et al. 2005), tRNA boundaries were verified. The circular chloroplast genome maps of the Phoebe were drawn using the Organellar Genome DRAW (OGDRAW) software, with subsequent manual editing (Lohse et al. 2007).
Characterization of repeat sequences and SSRs
REPuter was used to visualize both forward, palindrome, reverse and complement repeats, with a minimum repeat size of 30 bp and a sequence identity greater than 90% with hamming distance equal to 3 in P. chekiangensis and P. bournei (Kurtz and Schleiermacher 1999). The SSRs, which usually have a higher mutation rate, are easily genotyped using PCR, used as markers for phylogenetic analysis and in marker-assisted breeding when located on nuclear chromosomes. Microsatellites, or simple sequence repeats (SSRs) were detected using MISA perl script with thresholds of ten repeat units for mononucleotide SSRs, five repeat units for dinucleotide SSRs, four repeat units for trinucleotide SSRs, and three repeat units for tetra-, penta-, and hexanucleotide SSRs (Thiel et al. 2003).
Divergence hotspot identification
The four complete chloroplast genome sequences (P. chekiangensis, P. bournei, P. sheareri, and P. sheareri var. oineiensis) were aligned,using the chloroplast genome of M. yunnanensis as a reference by mVISTA program (Frazer et al. 2004). Default parameters were utilized to align the chloroplast genomes in Shuffle-LAGAN mode and a sequence conservation profile was visualized in an mVISTA plot. To discover the divergence hotspot regions in Phoebe, protein coding gene, intron, and intergenic spacer region were evaluated with DnaSP 5.10 (Librado and Rozas 2009). All the regions were sequentially extracted under the following criteria: (1) total number of mutation (Eta) >0; (2) an aligned length >200 bp. Any large structural events, such as gene order rearrangements and IR expansions/contractions, were recorded.
Results
Chloroplast genomes organization of P. chekiangensis and P. bournei
For these two Phoebe species, just three contigs which were found to be significantly homologous to the reference genome were combined to generate each chloroplast genome, with no gaps or missing nucleotides (Ns) found. The total genome size of P. chekiangensis, with a length of 152,849 bp (deposited in GenBank, Accession No. KY346511), was in close proximity to those of other Phoebe species and only 4, 6, and 27 bp smaller than that of P. bournei (152,853 bp, deposited in GenBank, Accession No. KY346512), P. sheareri var. oineiensis (152,855 bp, GenBank Accession No. KX437772), and P. sheareri (152,876 bp, GenBank Accession No. KX437773), respectively. The size of complete chloroplast genome of the four Phoebe species was within the range of angiosperms (Yang et al. 2010). Similar to the vast majority of angiosperms, showing a typical quadripartite structure, the two Phoebe chloroplast genomes consist of a pair of inverted repeats (IRs) of 18,927 bp in P. chekiangensis and 18,928 bp in P.bournei, a large single copy (LSC) region of 93,772 bp in P. chekiangensis and 93,777 bp in P. bournei and a small single copy (SSC) region of 20,775 bp in P. chekiangensis and 20,774 bp in P. bournei (Fig. 1; Table 1). Protein-coding regions accounted for 46.23% of the whole genome, while tRNA and rRNA regions accounted for 1.79 and 5.91%, respectively, and the remaining 46.07% was non-coding regions (Table 1). The overall GC content was 39.1%, whereas the GC content in the LSC, SSC and IR regions were 38.0, 34.4, and 44.4%, respectively, indicating identical levels among the two Phoebe chloroplast genomes.
Fig. 1.
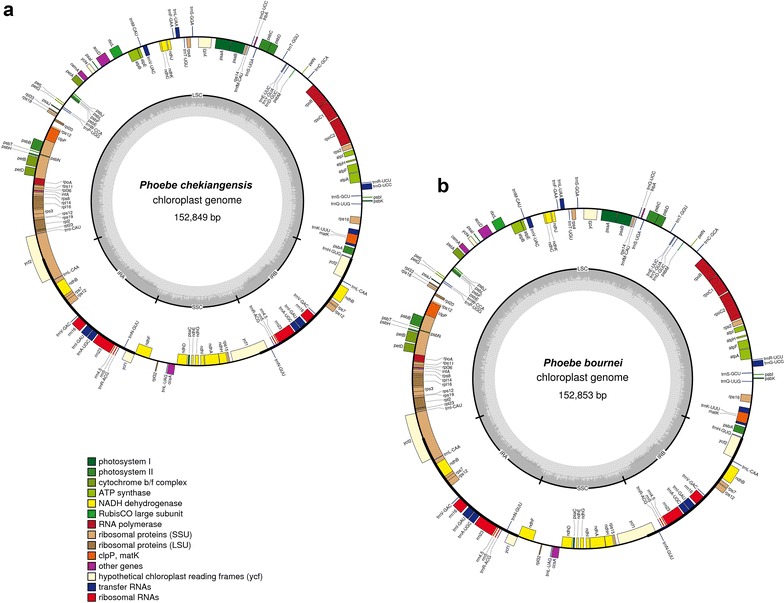
Chloroplast genome of Phoebe chekiangensis and Phoebe bournei. a Phoebe chekiangensis, b Phoebe bournei. Genes shown on the outside of the circle are transcribed clockwise, and genes inside are transcribed counter-clockwise. Large single copy (LSC), small single copy (SSC), and inverted repeats (IRa, IRb) are indicated. Genes belonging to different functional groups are color-coded. The darker gray in the inner corresponds to GC content, and the lighter gray corresponds to AT content
Table 1.
The basic characteristics of the sequencing data for P. chekiangensis and P. bournei
| Characteristics | P. chekiangensis | P. bournei |
|---|---|---|
| Clean reads | 20,777,674 | 20,787,108 |
| Average read length | 125 | 125 |
| Total cpDNA size (bp) | 152,849 | 152,853 |
| Length of large single copy (LSC) region | 93,772 | 93,777 |
| Length of inverted repeat (IR) region | 18,927 | 18,928 |
| Length of small single copy (SSC) region | 20,075 | 20,074 |
| Total CDS length | 70,668 | 70,668 |
| Total tRNA length | 2740 | 2740 |
| Total rRNA length | 9038 | 9038 |
| Total GC content (%) | 39.1 | 39.1 |
| LSC | 38.0 | 38.0 |
| IR | 44.4 | 44.4 |
| SSC | 34.4 | 34.4 |
| Total number of genes | 113 | 113 |
| Protein-coding genes | 79 | 79 |
| rRNAs genes | 4 | 4 |
| tRNAs genes | 30 | 30 |
SSR and repeat sequences analysis
Repeat sequences played a vital role in phylogenetic analysis and genome rearrangement (Nie et al. 2012). We used REPuter to analyze the repeat sequence of Phoebe cp genomes and found forward repeats, palindrome repeats and reverse repeats of at least 30 bp per repeat unit with a sequence identity of ≥90%. P. chekiangensis contained 36 repeats comprising of 13 forward repeats, 15 palindromic repeats, 6 reverse repeats and 2 complement repeats. The only quantity difference in repeat type between P. chekiangensis and P. bournei was that the latter contained 14 palindromic repeats, one less than the former (Fig. 2a; Additional file 1). The quantity of repeats with 30–40 bp length was 33 and 32 for P. chekiangensis and P. bournei, respectively. In addition, P. chekiangensis and P. bournei both contained one repeat with 41, 42, and 48 bp length (Fig. 2b; Additional file 1).
Fig. 2.
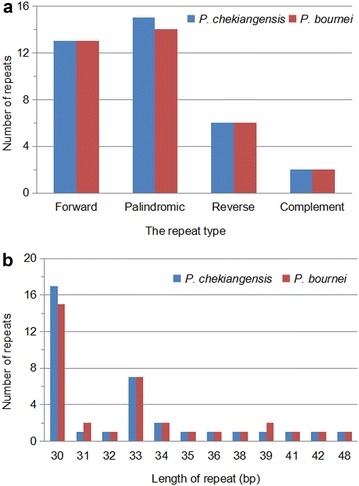
Analysis of repeated sequences in P. chekiangensis and P. bournei chloroplast genomes. a Frequency of repeats by length; b frequency of repeat types
With MISA analysis, Phoebe chloroplast genome was found to contain 66 (P. chekiangensis) and 65 (P. bournei) SSRs longer than 10 bp, of which 49 SSRs were the same for the two chloroplast genomes (similar repeat units located in similar genomic regions) (Fig. 3a; Additional file 2). Among the total 131 SSRs, most loci were located in intergenic spacer (IGS) regions (61.07%), followed by introns (23.37%) and CDS (17.56%) (Fig. 3b). We observed that 12 different SSRs were located in five protein-coding genes [ycf1 (×5), cemA(×2), rpoC2 (×2), ycf2 (×2), and matK] of the two Phoebe chloroplast genomes.
Fig. 3.
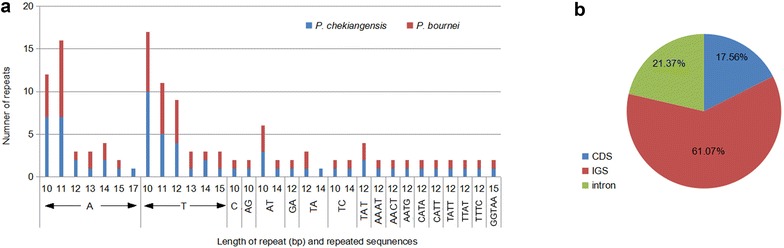
Simple sequence repeats (SSRs) in the two Pheobe chloroplast genomes. a Numbers of SSRs by length; b distribution of SSR loci. IGS intergenic spacer region
Inverted repeats (IRs) contraction and expansion of four Phoebe species
The IR region expanded into the ycf2 gene, creating a pseudogene fragment ψycf2 at the IRa/LSC border with length of 3,161 bp (P. bournei and P. sheareri var. oineiensis) and 3,162 bp (P. chekiangensi and P. sheareri). The ycf1 gene crossed the SSC/IRa region and the pseudogene fragment ψycf1 was located at the IRb region with 1381 and 1399 bp. The trnH was the unique gene with large difference among Phoebe species. The trnH genes of P. sheareri and P. sheareri var. oineiensis were separated by 21 bp at the IRa/LSC border, whereas those of P. bournei and P. chekiangensis were separated by 43 bp (Fig. 4).
Fig. 4.
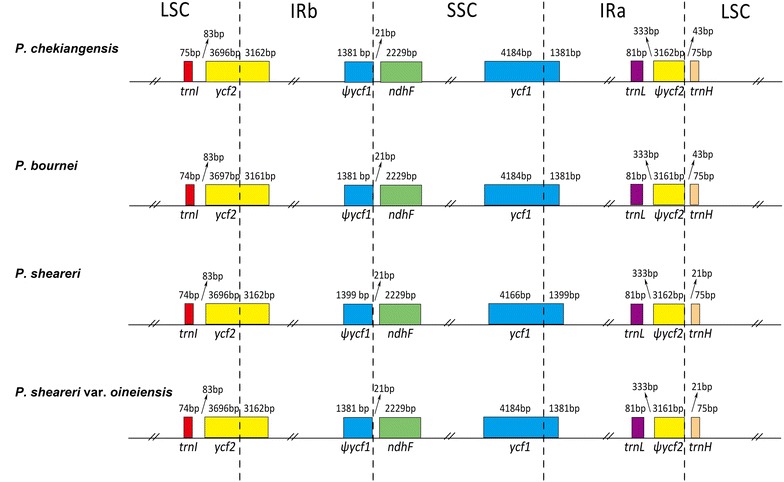
Comparison of LSC, IR, and SSC junction positions among chloroplast genomes of P. chekiangensis, P. bournei, P. sheareri, and P. shearerivar. oineiensis
Divergence sequence hotspots in Phoebe species
The overall sequence identity of the four Phoebe chloroplast genomes was compared and plotted using the mVISTA program (Frazer et al. 2004), with the annotation of M. yunnanensis as a reference to elucidate the level of sequence divergence (Fig. 5). Being largely consistent with recent studies (Yao et al. 2015; Zhang et al. 2016), most of the sequence variations were found to be located in the LSC and SSC regions, while the IR regions exhibited comparatively lower sequence diversity. The lower sequence divergence observed in the IRs rather than LSC and SSC regions for Phoebe species is likely due to copy correction between IR sequences by gene conversion (Khakhlova and Bock 2006).
Fig. 5.
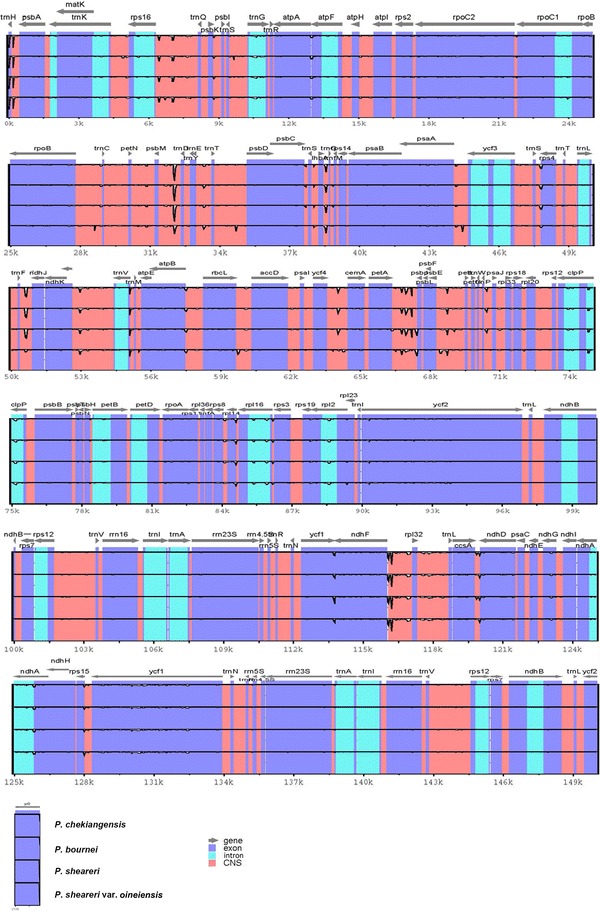
Sequence identity plots among chloroplast genomes of P. chekiangensis, P. bournei, P. sheareri, and P. sheareri var. oineiensis, with Machilus yunnanensis as a reference. Annotated genes are displayed along the top. The vertical scale represents the percent identity between 50 and 100%. Genome regions are color coded as exon, intron, and conserved non-coding sequences (CDS)
Seventy-seven regions (30 coding regions, 38 intergenic spacers, eight introns, and one rRNA) with more than 200 bp in length were eventually identified. Of these 77 regions, nucleotide diversity (Pi) ranged from 0.00018 (rrn23) to 0.01389 (ccsA-ndhD) among four Phoebe species (Fig. 6; Additional file 3). As found in most angiosperms (Choi et al. 2016), sequence divergence in intergenic regions was higher than that in genic regions of these four Phoebe chloroplast genomes. The mean value of Pi in non-coding regions was 0.221%, which was nearly more than twice as much as that (0.123% on average) in the coding regions. Intergenic regions with a percentage of Pi exceeding 0.5% were trnH-psbA (0.506%), rps4-trnT (0.716%), petA-psbJ (0.887%), and ccsA-ndhD (1.389%). However, the highest proportion of variability in genic regions was 0.251% (rps8) (Fig. 6; Additional file 3).
Fig. 6.
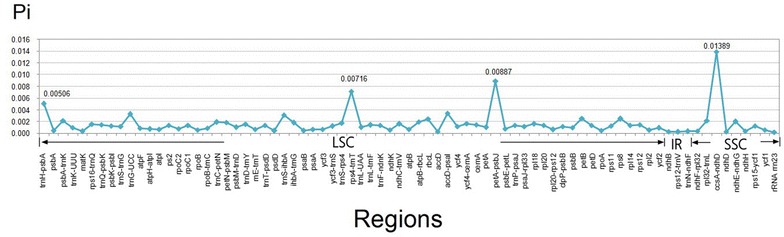
The nucleotide diversity (Pi) values were compared among four Phoebe species
Discussion
Results from this study showed the GC content of P. chekiangensis and P. bournei chloroplast genomes is close to that reported in other Lauraceae chloroplast genomes (Song et al. 2015, 2016). Although the GC percentage in the IR regions (44.4%) of the two Pheobe was higher than that of Nicotiana otophora (43%) (Asaf et al. 2016), the presence of rRNA in Pheobe (four) was lower than that of N. otophora (eight). The results in this study were incompatible to previous report which suggested that a high GC percentage in the IR regions could be due to the presence of rRNA (Qian et al. 2013). Similarly, 112 different genes, including 78 protein-coding genes (47 genes encoding photosynthesis-related proteins, four DNA dependent RNA polymerases, 20 ribosomal proteins, one translation initiation factor, four genes encoding other proteins, and two genes of unknown function), 30 tRNA genes, and four rRNA genes were annotated in P. chekiangensis and P. bournei chloroplast genomes (Fig. 1; Additional file 4). The pattern of protein coding genes was similar to that of Persea americana (Song et al. 2016). Among all the protein-coding genes, ten genes possessed a single intron, two genes (ycf3 and clpP) contain two introns, whereas six tRNA genes contain a single intron (Additional file 4).
Overall, a total of 71 repeats, including 36.6% forward repeats (26), 40.8% palindromic repeats (29), 16.9% reverse repeats (12), and 5.7% complement repeats (4), were detected in P. chekiangensis and P. bournei chloroplast genomes. About 61.07% of these repeats were distributed in intergenic spacer. The result was comparable to chloroplast genomes of most angiosperm plant (Uthaipaisanwong et al. 2012; Yao et al. 2015). Previous studies suggested that the presence of these repeats indicates that the region is a crucial hotspot for genome reconfiguration (Gao et al. 2009). One 30 bp forward repeat occurred in the ndhC-trnV intergenic spacer and one 30 bp palindromic repeat in Ψycf1 were unique to P. chekiangensis. In contrast, one 31 bp forward repeat occurred in the ndhC-trnV intergenic spacer were only contained by P. bournei (Additional file 1). Apart from the above three repeats, the others were shared between two Phoebe species. SSRs in the chloroplast with high polymorphism in copy numbers have been recognized as one of the main sources of molecular markers, and extensively used for population genetics and phylogenetic investigation (Pauwels et al. 2012; Zhang et al. 2012a, b; Zhao et al. 2015).
Among 131 SSRs longer than 10 bp, almost all mononucleotide was composed of A/T (97.7%), and majority of dinucleotides was composed of AT (60.0%). The AT richness in SSRs of the two Pheobe genome was similar to previous reports suggesting that SSRs found in the chloroplast genome were generally composed of polythymine (T) or polyadenine (A) repeats, and infrequently contained tandem cytosine (C) and guanine (G) repeats (Kuang et al. 2011; Qian et al. 2013; Chen et al. 2015). The SSRs identified in the two Phoebe species might be used in future population genetic studies as well as similar studies of other species, such as Panax ginseng (Kim and Lee 2004), Cucumis sativus (Kim et al. 2006), Vigna radiate (Tangphatsornruang et al. 2010), and Pyrus pyrifolia (Terakami et al. 2012).
Based on the analysis of inverted repeats contraction and expansion, a conclusion could be drawn that chloroplast genomes of four Phoebe species exhibited comparatively little difference at the IR/LSC and IR/SSC boundary regions, which was similar in Veroniceae (Choi et al. 2016). The genetic divergence within Lauraceaeis surprisingly low (Rohwer 2000; Song et al. 2016). The mean nucleotide diversity of the complete chloroplast genome of the four Phoebe species was only 0.162%, lower than that of two Panax species (0.40%) (Dong et al. 2014), three Veroniceae species (0.40%) (Choi et al. 2016), and nine Gossypium species (0.62%) (Xu et al. 2012), and extremely lower than that of five Epimedium species (3.97%) (Zhang et al. 2016). Molecular markers with nucleotide diversity over 1.5% have been reported before as highly variable regions and could be used to promote the further phylogenetic analysis and species identification in other seed plants (Särkinen and George 2013; Korotkova et al. 2014; Huang et al. 2014). However, due to the relative conservative of chloroplast genomes of Phoebe species, the quantity of the regions with nucleotide diversity exceeding 0.5% was only four, whereas in Nicotiana (Asaf et al. 2016), the quantity of the regions with nucleotide diversity over 0.7% was 15, and in Machilus (Song et al. 2015), the quantity of the regions with nucleotide diversity over 0.8% was 7. In this study, the highest nucleotide diversity was 1.389% (ccsA-ndhD), followed by 0.887% (petA-psbJ), 0.716% (rps4-trnT), and 0.506% (trnH-psbA). Therefore, only the above four regions with nucleotide diversity higher than 0.5% probably had the potential use for phylogeographic analyses and plant identification of Phoebe species.
Conclusions
The 77 repeat sequences were identified in the Phoebe chloroplast genomes. The analysis showed the extremely conserved structure of chloroplast genomes, the mean nucleotide diversity was found to be 0.162% for 77 regions, suggesting an extraordinarily low level of sequence divergence. Four highest divergent regions (trnH-psbA, rps14-trnT, petA-psbJ, ccsA-ndhD) were also identified, which might be useful in species identification and phylogenetic studies. Overall, this study will facilitate our understanding of population genetics, phylogenetic relationship and plant evolution of Phoebe species.
Additional files
Additional file 1: Table S1. Analyses of repeat sequences in the two Phoebe chloroplast genomes.
Additional file 2: Table S2. The simple sequence repeats in P. chekiangensis and P. bournei.
Additional file 3: Table S3. Nucleotide diversity (Pi) values and total number of mutation (Eta) in Phoebe.
Additional file 4: Table S4. Genes, separated by category, encoded by P. chekiangensis and P. bourneiplastomes.
Authors’ contributions
LYG, XWQ and LXH conceived and designed the experiment; ZWT and JDY contributed to the sampling; XWQ and LYG performed the experiment and analyzed the data. The manuscript was written by LYG. All authors read and approved the final manuscript.
Acknowledgements
We thank Yingxiong Qiu, Ruisen Lu, Shanshan Zhu of Zhejiang University for providing experiment design and laboratory support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data are deposited in NCBI.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the Major Science and Technology Program (No. 2010C12009), Key Research & Development Program (No. 2017C02028) of Zhejiang Province, and National Key Research & Development Program (No. 2016YFD0600603) of China.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-017-0192-8) contains supplementary material, which is available to authorized users.
Contributor Information
Yingang Li, Email: hzliyg@126.com.
Wuqin Xu, Email: 37224253@qq.com.
Wentao Zou, Email: 149099380@qq.com.
Dongyue Jiang, Email: jdyzjforestry@163.com.
Xinhong Liu, Email: lkyliuxh@163.com.
References
- Asaf S, Khan AL, Khan AR, Waqas M, Kang SM, Khan MA, Lee SM, Lee IJ. Complete chloroplast genome of Nicotianaotophora and its comparison with related species. Front Plant Sci. 2016;7:843–854. doi: 10.3389/fpls.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Hao ZD, Xu HB, Yang LM, Liu GX, Sheng Y, Zheng C, Zheng WW, Chen TL, Shi JS. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front Plant Sci. 2015;6:447. doi: 10.3389/fpls.2015.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Chung MG, Park SJ. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Front Plant Sci. 2016;7:342–355. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YJ, Zhang JH, Lu YF, Lin EP, Lou LH, Tong ZK. Development of EST-SSR markers and analysis of genetic diversity in natural populations of endemic and endangered plant Phoebe chekiangensis. Biochem Syst Ecol. 2015;63:183–189. doi: 10.1016/j.bse.2015.10.008. [DOI] [Google Scholar]
- Dong WP, Liu H, Xu C, Zuo YJ, Chen Z, Zhou SL. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: a case study on ginsengs. BMC Genet. 2014;15:138–145. doi: 10.1186/s12863-014-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE. Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev. 1990;8:655–661. doi: 10.1016/S0959-437X(98)80033-6. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Yi X, Yang YX, Su YJ, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol. 2009;9:130–142. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde VR, Dai P, Ladislaw C, Patel MG, Puar MS, Pachter JA. D4 dopamine receptor-selective compounds from the Chinese plant Phoebe chekiangensis. Bioorg Med Chem Lett. 1997;9:1207–1212. doi: 10.1016/S0960-894X(97)00194-7. [DOI] [Google Scholar]
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 2014;14:151–167. doi: 10.1186/1471-2148-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK, Ribstein S, Taylor DR. Molecular evolution of insertions and deletion in the chloroplast genome of Silene. Mol Biol Evol. 2003;20:1737–1740. doi: 10.1093/molbev/msg163. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR. Complete sequence and organization of the cucumber (Cucumis sativus L cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 2006;25:334–340. doi: 10.1007/s00299-005-0097-y. [DOI] [PubMed] [Google Scholar]
- Korotkova N, Nauheimer L, Ter-Voskanyan H, Allgaier M, Borsch T. Variability among the most rapidly evolving plastid genomic regions is lineage-specific: implications of pairwise genome comparisons in Pyrus (Rosaceae) and other angiosperms for marker choice. PLoS ONE. 2014;11:e112998. doi: 10.1371/journal.pone.0112998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L, Bonen L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie XJ, Lv SZ, Zhang YX. Complete chloroplast genome sequence of a major invasive species Crofton Weed (Ageratina adenophora) PLoS ONE. 2012;7:e36869. doi: 10.1371/journal.pone.0036869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84–100. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M, Vekemans X, Gode C, Frerot H, Castric V, Saumitou-Laprade P. Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance Arabidopsis halleri (Brassicaceae) New Phytol. 2012;193:916–928. doi: 10.1111/j.1469-8137.2011.04003.x. [DOI] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- Qian J, Song JY, Gao HH. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer JG. Toward a phylogenetic classification of the Lauraceae: evidence from matK sequences. Syst Bot. 2000;25:60–71. doi: 10.2307/2666673. [DOI] [Google Scholar]
- Särkinen T, George M. Predicting plastid marker variation: can complete plastid genomes from closely related species help? PLoS ONE. 2013;11:e82266. doi: 10.1371/journal.pone.0082266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Dong WP, Liu B, Xu C, Yao X, Gao J, Corlett RT. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front Plant Sci. 2015;6:662–669. doi: 10.3389/fpls.2015.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yao X, Tan YH, Gan Y, Richard TC. Complete chloroplast genome sequence of the avocado: gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can J Forest Res. 2016;46:1293–1301. doi: 10.1139/cjfr-2016-0199. [DOI] [Google Scholar]
- Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes. 2012;8:1–14. doi: 10.1007/s11295-012-0469-8. [DOI] [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Uthaipaisanwong P, Chanprasert J, Shearman J. Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq) Gene. 2012;500:172–180. doi: 10.1016/j.gene.2012.03.061. [DOI] [PubMed] [Google Scholar]
- Wang MX, Cui LC, Feng KW, Deng PC, Du XH, Wan FH, Song WN, Nie XJ. Comparative analysis of Asteraceae chloroplast genomes: structural organization, RNA editing and evolution. Plant Mol Biol Rep. 2015;33:1526–1538. doi: 10.1007/s11105-015-0853-2. [DOI] [Google Scholar]
- Wicke S, Schneeweiss GM, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZY, Peter HR, Hong DY. Flora of China. Beijing: Science press; 2008. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xu Q, Xiong G, Li P, He F, Huang Y, Wang K, Li ZH, Hua JP. Analysis of complete nucleotide sequences of 12 gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS ONE. 2012;8:e37128. doi: 10.1371/journal.pone.0037128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Zhang XW, Liu GM, Yin YX, Chen KF, Yun QZ, Zhao DJ, Al-Mssallem IS, Yu J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L) PLoS ONE. 2010;5:e12762. doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yue M, Niu C, Ma XF, Li ZH. Comparative analysis of the complete chloroplast genome of four endangered herbals of Notopterygium. Genes. 2017;8:124. doi: 10.3390/genes8040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Tang P, Li Z, Li D, Liu Y, Huang H. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS ONE. 2015;10:e0129347. doi: 10.1371/journal.pone.0129347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng SY, Zhou T, Han K, Yang YC, Zhao JH, Liu ZL. The complete chloroplast genome sequences of six Rehmannia species. Genes. 2017;8:103. doi: 10.3390/genes8030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li J, Zhao Y, Korba SS, Han Y. Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Mol Biol Rep. 2012;30:539–546. doi: 10.1007/s11105-011-0366-6. [DOI] [Google Scholar]
- Zhang R, Zhou ZC, Jin GQ, Wang SH, Wang XH. Genetic diversity and differentiation within three species of the family Lauraceae in southeast China. Biochem Syst Ecol. 2012;44:317–324. doi: 10.1016/j.bse.2012.06.007. [DOI] [Google Scholar]
- Zhang YJ, Du LW, Liu A, Chen JJ, Wu L, Hu WM, Zhang W, Kim KH, Lee SC, Yang TJ, Wang Y. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 2016;7:306–317. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yin J, Guo H. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 2015;5:696–707. doi: 10.3389/fpls.2014.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Analyses of repeat sequences in the two Phoebe chloroplast genomes.
Additional file 2: Table S2. The simple sequence repeats in P. chekiangensis and P. bournei.
Additional file 3: Table S3. Nucleotide diversity (Pi) values and total number of mutation (Eta) in Phoebe.
Additional file 4: Table S4. Genes, separated by category, encoded by P. chekiangensis and P. bourneiplastomes.
Data Availability Statement
All data are deposited in NCBI.


