Abstract
The currently favoured method for wheat (Triticum aestivum L.) transformation is inapplicable to many elite cultivars because it requires callus culture and regeneration. Here, we developed a simple, reproducible, in planta wheat transformation method using biolistic DNA delivery without callus culture or regeneration. Shoot apical meristems (SAMs) grown from dry imbibed seeds were exposed under a microscope and subjected to bombardment with different-sized gold particles coated with the GFP gene construct, introducing DNA into the L2 cell layer. Bombarded embryos were grown to mature, stably transformed T0 plants and integration of the GFP gene into the genome was determined at the fifth leaf. Use of 0.6-µm particles and 1350-psi pressure resulted in dramatically increased maximum ratios of transient GFP expression in SAMs and transgene integration in the fifth leaf. The transgene was integrated into the germ cells of 62% of transformants, and was therefore inherited in the next generation. We successfully transformed the model wheat cultivar ‘Fielder’, as well as the recalcitrant Japanese elite cultivar ‘Haruyokoi’. Our method could potentially be used to generate stable transgenic lines for a wide range of commercial wheat cultivars.
Introduction
Wheat (Triticum aestivum L.) is a major staple crop that is cultivated worldwide. Difficulty in transformation has meant that the application of genetic engineering in wheat has lagged behind that of other crops, such as rice and maize. The first successful transformation of wheat was reported using particle bombardment of embryogenic callus1. Subsequently, an Agrobacterium-mediated transformation method using immature, embryo-derived, regenerable callus was developed2–4. To improve the efficiency of culture-based transformation, several model cultivars have been modified under various experimental conditions5–8. However, the application of these methods is limited to those cultivars with characteristics suitable for culture and regeneration. Many elite commercial cultivars lack this property, and are thus difficult to transform. In addition, culture-based transformation methods are generally time-consuming and suffer from somatic variations.
To avoid the problems associated with tissue culture and regeneration steps in transformation, alternative in planta methods, which introduce transgenes directly into intact plant tissue, have been developed in several plant species. In Arabidopsis thaliana, the Agrobacterium-mediated floral-dip method is most widely used to transform immature florets9,10, targeting ovule cells10. In planta transformation methods have been reported in other plant species, such as Medicago truncatula 11, Solanum lycopersicum and some cereals12–15, but these are not used as standard protocols as they are often inefficient and irreproducible.
Here we report a novel in planta wheat transformation method using biolistic particle delivery. In planta particle bombardment (iPB) utilises the meristematic tissues of mature embryos and does not require embryogenic callus culture, regeneration, or antibiotic selection. We show that this method can produce stably transformed transgenic wheat plants, not only in an experimental cultivar (‘Fielder’), but also in a commercial elite cultivar (‘Haruyokoi’).
Results
Optimization of bombardment conditions for in planta transformation
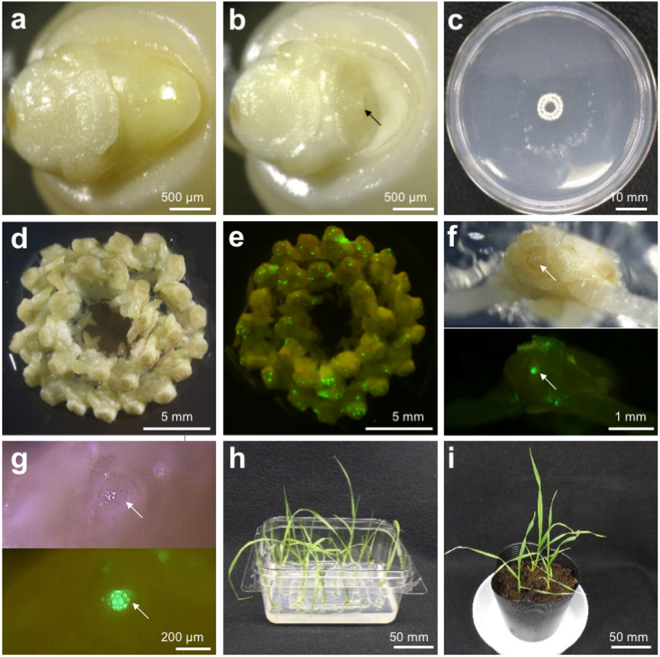
To establish microprojectile-mediated DNA transfer to the meristematic cells, the GFP gene driven by the maize ubiquitin promoter (Pubi) was used as a reporter. Coleoptiles and the first three leaf primordia were removed from imbibed ‘Fielder’ seeds to expose shoot apical meristems (SAMs) (Fig. 1a and b). SAM-exposed apical tissues were excised from seeds and arranged in circles on a plate (Fig. 1c). Gold particles of 0.6–1.6 µm in diameter were coated with the reporter plasmid and bombarded into the apical tissue under 1100 psi (7.6 MPa) or 1350 psi (9.3 MPa) helium pressure. After 12 h, the bombarded apical tissues were observed under a fluorescence microscope to check for transient GFP expression in the SAM (Fig. 1d and e). GFP signals were detected on the entire surface of the SAM in several bombarded plants; those carrying five or more signal spots were considered GFP-positive (Fig. 1f and g). On the other hand, no wound-induced GFP signal (auto-fluorescence) was observed in the SAM of apical tissues bombarded without GFP plasmid (Supplementary Fig. S1). As shown in Table 1, the percentage of GFP-positive SAMs gradually increased with decreasing gold particle size under 1350 psi (7.5, 35.0, and 74.2%). By contrast, the percentage of GFP-positive SAMs decreased when the particles were accelerated at 1100 psi (21.7, 15.8, and 13.3%). This result suggests that particle size and pressure affects delivery efficiency in a complex manner, but the highest efficiency was observed with 0.6-μm gold particles and 1,350 psi pressure (Table 1).
Figure 1.
Procedure for in planta particle bombardment (iPB) transformation of wheat. (a) Coleoptiles and leaf primordia from mature embryos were excised under a microscope, (b) and arranged on a culture plate with Murashige and Skoog’s (MS) medium before (c) transformation. (d) Bright field and (e) fluorescence-merged images of the whole apical tissues 12 h after bombardment. (f) Bright field (upper) and fluorescence (lower) images of a single apical tissue. (g) Close-up images of a SAM region of apical tissue. (h) Wheat plants grown on MS medium 3 weeks after bombardment. (i) Bombarded wheat plants 2 weeks after transfer to soil. Shoot apical meristems (SAMs) are indicated by arrows in panels (b), (f) and (g).
Table 1.
Effects of particle size and pressure on gene delivery efficiency.
| Particle size (μm) | Helium pressure (psi) | No. of bombarded plants | No. of plants carrying GFP within SAM *(%) | No. of transgenic plants** (%) |
|---|---|---|---|---|
| 1.6 | 1,100 | 120 | 26 (21.7) | 0 (0.0) |
| 1,350 | 9 (7.5) | 0 (0.0) | ||
| 1.0 | 1,100 | 120 | 19 (15.8) | 1 (0.8) |
| 1,350 | 42 (35.0) | 2 (1.7) | ||
| 0.6 | 1,100 | 120 | 16 (13.3) | 1 (0.8) |
| 1,350 | 89 (74.2) | 5 (4.2) |
*Wheat plants carrying 5 or more GFP signal spots were considered GFP-positive.
**Wheat plants were considered to be transgenic based on positive genomic PCR in the fifth leaf of T0 progeny.
Transient GFP expression in SAMs suggested that stable transformation might occur within the meristematic region. Bombarded embryos were grown on basal Murashige and Skoog (MS) medium for 2–3 weeks to allow full root and shoot development, and were then transferred to soil (Fig. 1h and i). Chromosomal integration of the GFP gene was tested in the fifth leaf by genomic polymerase chain reaction (PCR) (Table 1). The GFP gene was detected when 0.6-µm and 1.0-µm particles were used. No plants bombarded with 1.6-μm particles showed integrated GFP (Table 1). With 1350 psi pressure and 0.6-μm particles, the maximum number (5) of transgenic plants were obtained, which correlated with the efficiency of transient GFP expression in SAMs (Table 1). The transgene was detected in subsequently developed leaves (Supplementary Fig. S2). These results suggested that, within the range of conditions tested, 0.6-μm particles and 1350-psi pressure is the optimum combination for transgene delivery into the SAM and the generation of transgenic plants.
Integration and inheritance of the transgene in transgenic plants
Larger-scale screening of transgenic plants was conducted using the optimal conditions (0.6-μm particles and 1350-psi pressure). Of the 577 bombarded embryos, 379 that transiently expressed GFP in their meristematic regions were selected and grown to adulthood. Eight putative transgenic plants were identified by genomic PCR analysis of flag leaf tissue (Table 2). T1 seeds from the primary spike of these plants were subjected to further analysis. Genomic PCR analysis revealed that five (FG1–5) of the eight lines inherited the transgene to the T1 generation (Table 2, Supplementary Table S1, Fig. 2a). Existence of GFP in the genome was also confirmed by DNA gel blot analysis (Fig. 2b). The number of hybridising bands varied from two (FG3) to over ten (FG5) (Fig. 2b). Chi-square analysis revealed that the FG3 line segregated in a Mendelian fashion (measured value = 50:2, expected value = 15:1, χ 2 = 0.20, P > 0.3). These results suggest that GFP was directly introduced into a germline cell within SAMs and was inherited to the next generation.
Table 2.
Comparison of transformation efficiency between ‘Fielder’ and ‘Haruyokoi’.
| Cultivar | No. of bombarded wheats | No. of transgenic plants in T0 progeny* (%) | No. of transgenic plants in T1 progeny** (%) | No. of T1 plants expressing GFP (%) |
|---|---|---|---|---|
| Fielder | 577 | 8 (1.39) | 5 (0.87) | 1 (0.17) |
| Haruyokoi | 569 | 13 (2.28) | 4 (0.70) | 2 (0.35) |
*Wheat plants were considered to be transgenic based on positive genomic PCR in the flag leaf of T0 progeny.
**Wheat plants were considered to be transgenic based on positive genomic PCR in T1 progeny.
Figure 2.
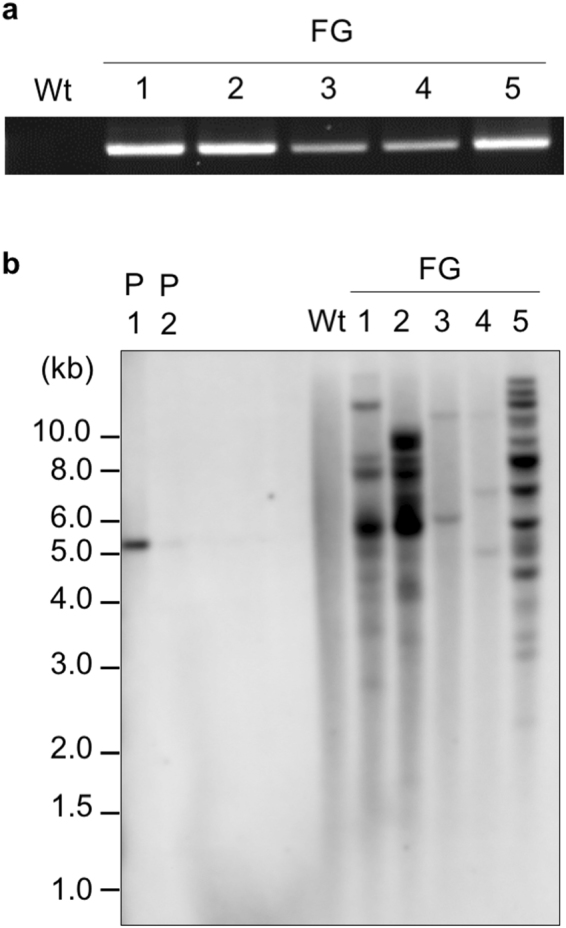
Integration and inheritance of the GFP gene in transformed wheat lines. (a) Genomic polymerase chain reaction (PCR) analysis of five independent transgenic ‘Fielder’ lines (FG1–5) and wild-type (Wt) lines. Genomic DNA was extracted from each first leaf of T1 progeny. The full-length gel image is shown in Supplementary Fig. S6. (b) DNA gel blot analysis of transgenic wheat lines (T2 progeny) and Wt lines using genomic DNA from leaves digested with HindIII. One-hundred picograms (P1) and 10 pg (P2) of linearised GFP vector (5.1 kb) were used as positive controls.
Expression of the transgene in transgenic plants
We tested GFP transgene expression in T1 plants by analysing GFP mRNA and GFP protein in the transgenic plants by reverse transcription (RT)-PCR and immunoblot analysis, respectively. Both GFP mRNA and GFP protein accumulated in FG1, but not in FG5 which shows a complicated transgene integration pattern (Figs 2b and 3a,b). In the other lines (FG2–4), less mRNA accumulated than in FG1, but protein accumulation was undetectable (Fig. 3a and b). Expression of GFP was also confirmed by fluorescence detection. In FG1, GFP fluorescence was observed predominantly in the endosperms and aleurone cells of T1 seeds, T1 seedlings, and in T1 leaf stomata (Fig. 3c and d, Supplementary Fig. S3). The particular GFP expression pattern in transgenic T1 plants was consistent with that of the Pubi-driven GFP gene previously reported16.
Figure 3.
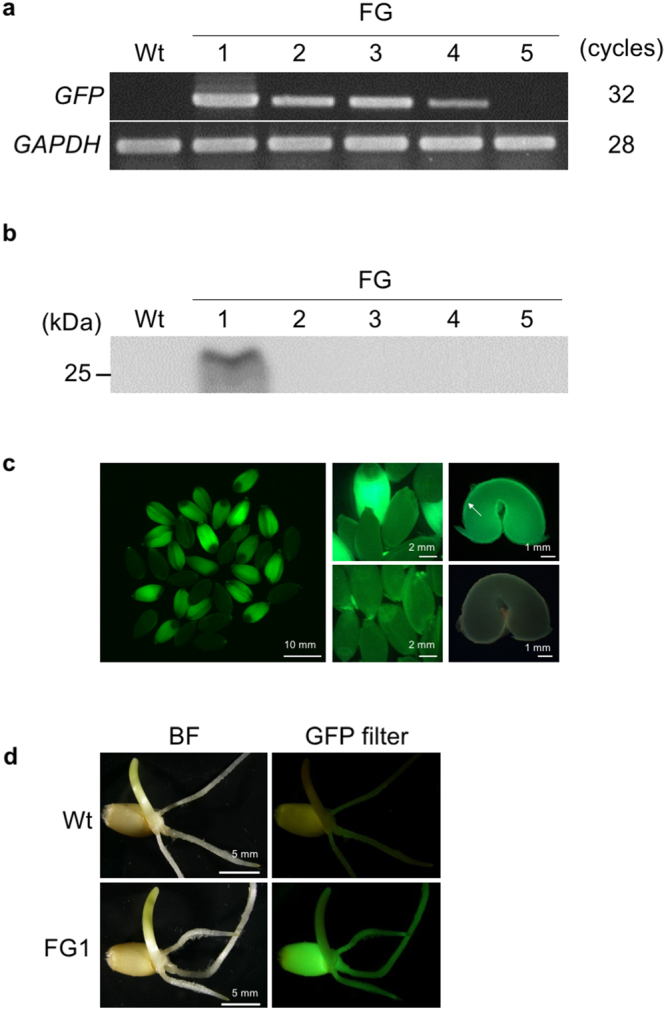
Expression of GFP in transgenic wheat plants. (a) Reverse transcription polymerase chain reaction (RT-PCR) analysis of GFP in ‘Fielder’ lines (FG1–5) and wild-type (Wt) plants. The GAPDH gene (Genbank accession number: EF592180) was used as a housekeeping control. Full-length gel images are shown in Supplementary Fig. S7. (b) Immunoblot analysis of green fluorescent protein (GFP) in transgenic plants (FG1–5). Total protein (20 µg per lane) extracted from each T1 leaf tissue was separated using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted. GFP was detected with anti-GFP antibodies. The full-length gel image is shown in Supplementary Fig. S8. (c) GFP accumulation in T1 seeds of a transgenic wheat line (FG1). Left: fluorescence image of whole T1 seeds harvested from the primary panicle of FG1. Middle: close-up views of T1 (upper) and Wt (lower) seeds. Right: fluorescence images of sections of FG1 T1 (upper) and Wt (lower) seeds. The aleurone cell layer is indicated by an arrow. (d) Bright-field (BF, left) and GFP (right) images of T1 seedlings in Wt (top) and FG1 (bottom) lines.
Genotype analysis in T1 plants
To determine whether all T1 seed derived from one transgenic T0 line had the same genotype, T1 seeds derived from spikes of the main shoot and five tillers of FG1 were germinated and their genotypes analysed by genomic PCR and DNA gel blots. Spikes from the main shoot and four of the tillers (all except the third tiller in FG1), contained mostly transgenic T1 seeds (Supplementary Table S2). In DNA gel blot analysis of the T1 seedlings, hybridisation signals from the spike of the main shoot and tillers were similar (Supplementary Fig. S4). These results suggest that T1 seeds produced by a single T0 transgenic plant have the same genotype, and one mature-embryo SAM cell is able to differentiate to produce T1 seeds on the primary and subsidiary spikes.
Stable transformation in a commercial wheat cultivar
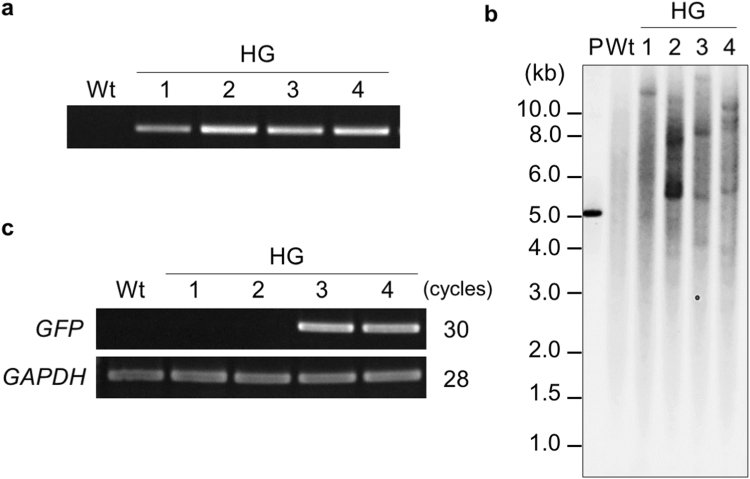
Tissue culture and Agrobacterium-mediated infection limit the applicability of transformation in many wheat commercial cultivars. Since our method involves no embryogenic callus culture or Agrobacterium infection, we applied it to a Japanese elite cultivar ‘Haruyokoi’, whose transformation had never been reported. Of the 569 bombarded embryos, 505 expressed GFP in their SAMs. Using genomic PCR analysis of the T0 progeny flag leaves, 13 of the 505 plants were selected as putative transgenic plants. Genomic PCR analysis of T1 plants showed that four (HG1–4) out of the 13 plants inherited the transgene (Table 2, Fig. 4a, Supplementary Table S1). Integration of GFP into their genomes was also confirmed by DNA gel blot analysis (Fig. 4b). Semi-quantitative RT-PCR and GFP fluorescence analyses revealed that two lines (HG3, HG4) expressed GFP in T1 plants (Fig. 4c, Supplementary Fig. S5). Overall transformation efficiency was comparable to that of the model cultivar ‘Fielder’. These results suggest that the iPB method described here can be used to create transgenic plants in wheat cultivars that are resistant to conventional tissue culture-based methods.
Figure 4.
Integration and expression of the GFP gene in transgenic ‘Haruyokoi’. (a) Genomic polymerase chain reaction (PCR) analysis of four T1 transgenic (HG1–4) lines and wild-type (Wt) plants. Genomic DNA was extracted from each first leaf. The full-length gel image is shown in Supplementary Fig. S5. (b) Reverse transcription-PCR (RT-PCR) analysis of GFP expression in HG1–4 lines and Wt plants. The GAPDH gene was used as a housekeeping control. Full-length gel images are shown in Supplementary Fig. S6. (c) DNA gel blot analysis of transgenic wheat lines (T2 progeny) and Wt lines using genomic DNA from leaves digested with HindIII. Two-hundred picograms of the linearised GFP vector (5.1 kb) used as a positive control (P).
Discussion
To develop a wheat transformation method that requires no embryogenic callus culture, we applied a biolistic delivery system to apical meristem tissue of imbibed wheat embryos. Our iPB method successfully produced transgenic wheat plants in ‘Haruyokoi’, a leading Japanese cultivar, as well as a model transformation cultivar, ‘Fielder’. Based on transient GFP expression and chromosome integration, transformation efficiency in the T0 generation significantly increased when 0.6-µm particles and 1350-psi helium pressure was used (Table 1, Supplementary Fig. S2). We further confirmed that the introduced GFP gene was inherited and stably expressed in the T1 generation (Table 2, Figs 2a,b and 3a). As demonstrated with ‘Haruyokoi’, since the iPB method requires no embryogenic callus culture or regeneration processes, it can potentially be applied to many commercial cultivars that are difficult to transform via conventional tissue culture-based methods. However, transformation efficiency in the T1 generation was approximately 0.8% (Table 2), which is lower than that of the conventional tissue culture-based methods. Further improvement and optimisation is necessary if this method is to be used as a standard protocol.
The use of particle bombardment to directly deliver genes into plant cells is a widely-used technique. However, it can sometimes cause instability in transgene expression. In this study, two patterns of instability were observed. In one of the transgenic ‘Fielder’ lines (FG5), which contains over 10 copies of the transgene, no GFP mRNA and or GFP protein was observed (Figs 2b and 3a,b). Transgenes sometimes integrate into a single plant genome locus as a multimer, which may result in gene silencing caused by homologous rearrangements17,18. In two other lines, FG3 and FG4, transgene expression was low, although the transgene copy number was also low (Figs 2b and 3a,b). Genomic PCR analysis of the promoter region of GFP (Pubi) in the FG3 and FG4 genomes revealed that a ~500-bp segment from the 5′-end of Pubi is missing in these lines (data not shown). This suggests that partial deletion of the promoter might lead to incomplete gene expression (gene repression) in these lines.
SAMs are organised in three cell layers: L1–L3. The L1 and L3 layers are destined to develop into epidermal and vascular cells, respectively. Germ cells such as pollen and egg cells are derived from L2 cells19,20. Within the L2 layer, cells that differentiate into male/female gametes are unspecified. Since we observed that at least a portion of transgenic T0 plants inherited the transgene into the T1 generation, we conclude that particle delivery successfully occurred in the L2 layer cells.
The microprojectile size and acceleration pressure are key controlling factors for particle bombardment. The ratio of SAMs transiently expressing GFP decreased with decreasing particle size in the 1100 psi pressure condition (Table 1). This suggests that a higher velocity might be required for small particles to be able to penetrate the SAM surface. Conversely, when 1,350 psi pressure was used, the ratio increased with decreasing particle size (Table 1). This is likely because the velocity of gold particles accelerated at 1350 psi was sufficient to penetrate the SAM surface and many more plasmids were delivered into SAM cells compared to the other conditions. We also observed that the number of putative T0 transgenic plants appeared to increase in line with the above-mentioned ratio (Table 1) and stably transformed transgenic wheat plants were obtained using a combination of 0.6-µm gold particles and 1350 psi helium pressure (Table 2). When 1.6-µm particles and 1350 psi pressure was used, no T0 transgenic plants were generated (Table 1). Therefore, we conclude that, within the range of tested conditions, the combination of 0.6-µm particles and 1350 psi acceleration pressure is the best to deliver transgenes into the SAM L2 layer.
In summary, the iPB method described here is able to stably transform wheat without embryogenic callus culture and can be applied to a wide range of wheat cultivars that are difficult to transform with conventional culture-based methods.
Methods
Preparation of mature embryos
Mature seeds of wheat (Triticum aestivum L.) were sterilised by soaking in sodium hypochlorite (6.0%, 20 min) with a detergent (alkyl ether sulphate), rinsed several times with sterile distilled water, and germinated overnight on moist filter paper at 22 °C. The parts of the coleoptile and leaf primordia covering the SAM were excised with a needle (ϕ 0.2 mm; TERUMO, Japan) under a stereomicroscope. Embryos were subsequently separated from endosperms and placed upright on Murashige and Skoog (MS) medium21 supplemented with maltose (30 g L−1), 2-morpholinoethanesulfonic acid (MES) monohydrate (0.98 g L−1, pH 5.8), plant preservative mixture (3%; Nacalai Tesque, Japan), and phytagel (7.0 g L−1; Sigma–Aldrich, USA). Thirty embryos per plate were placed in a circle (diameter 0.8 cm).
Expression vector
The sGFP gene22 was cloned into the plasmid pUba23 in place of the bar gene. The resulting vector (pUba–GFP) allows expression of sGFP under the control of a maize ubiquitin gene promoter (Pubi) and the nopaline synthase terminator. Wheat SAMs were transformed with pUba-GFP.
Preparation of microprojectiles and transformation
Microprojectiles were prepared as previously reported24 with slight modifications. Briefly, the plasmid (5 μg) was mixed with gold particles (Bio-Rad, USA) of appropriate diameters in glycerol (60 mg mL−1, 50%), spermidine (10 µL, 0.1 M), and CaC12 (25 µL, 2.5 M) in a 1.5-mL tube for 5 min. After incubation at room temperature (10 min), the DNA-coated gold particles were centrifuged (at 9100 × g for 2 sec) and the supernatant removed. The pellet was washed with ethanol (70 µL, 70%) and then with 99.5% ethanol. The final pellet was resuspended in ethanol (24 µL, 99.5%) and sonicated (for 1 sec) just before use. Aliquots (6 µL) were spread onto macrocarrier membranes (Bio-Rad, USA) and allowed to evaporate on a clean bench. Bombardment was conducted using a PDS-1000/He™ device (Bio-Rad, USA) with a target distance of 6.0 cm from the stopping plate. The vacuum in the chamber was 27 inches of Hg and the helium pressure was 1100 or 1350 psi. Bombardment was repeated four times.
Microscopic analyses
SAMs in the bombarded mature embryo and the sections of T1 seeds were observed with an MZFLIII microscope equipped with a GFP filter (excitation wavelength, 470/40 nm; emission wavelength, 525/50 nm). Fluorescence images of whole T1 seeds and T1 seedlings were observed with a fluoroimager LAS3000 (FUJIFILM, Japan) and Olympus SZX16 stereomicroscope equipped with a GFP filter (Ex: BP460–495, Em: BA510IF), respectively.
Plant growth conditions
Twelve hours after transformation, mature embryos expressing GFP in the SAMs were transferred into a Phytatray™ II (Sigma-Aldrich, USA) with basal MS medium and cultivated for 2–3 weeks in a growth chamber under long day conditions (16 h light/8 h darkness, 22 °C). Seedlings were planted in pots (3 seedlings/pot, ϕ 10.5 cm) and grown in a phytotron under long day conditions (16 h light/8 h darkness, 22 °C).
Polymerase chain reaction (PCR)
For PCR analysis of the transformed plants, DNA was isolated from the indicated leaf (of the T0 progeny) and the first leaf (of the T1 and T2 progeny), as described previously25. PCR amplification to select transformants was performed with Pubi and GFP gene-specific primers (Pubi-F, 5′-TTAGCCCTGCCTTCATACGC-3′; and GFP-R, 5′-ACCATGTGATCGCGCTTCT-3′). Each round of PCR was conducted in a reaction mixture (15 µL) containing dNTP and 1 × Ex Taq buffer (0.2 mM of each), primer (300 nM of each), Ex Taq HS polymerase (0.25 U; TaKaRa Bio, Japan), and genomic DNA (about 20 ng). The mixture was denatured (for 3 min at 94 °C) in a thermocycler and then subjected to 30 cycles (T1 or T2 generation) or 32 cycles (T0 generation) of amplification (94 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 30 sec). Half of the individual PCR products was resolved by agarose gel electrophoresis and visualised by staining with ethidium bromide under UV light.
RT-PCR analysis
Total RNA was isolated from leaf discs using Trizol (Invitrogen, USA) according to the manufacturer’s protocol. First-strand cDNA was synthesised from total RNA (0.5 µg) using ReverTra Ace® qPCR RT Master Mix (Toyobo, Japan). PCR amplification was conducted as follows: initial denaturation (94 °C for 1 min), followed by the indicated number of incubation cycles (94 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 30 sec), and a final extension (at 72 °C for 5 min) using GFP gene-specific primers (GFP-F, 5′-ACGGCCACAAGTTCAGCGT-3′; and GFP-R, 5′-ACCATGTGATCGCGCTTCT-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a quantitative control: GAPDH-F, 5′-CAACGCTAGCTGCACCACTAACT-3′ and GAPDH-R, 5′-GACTCCTCCTTGATAGCAGCCTT-3′. Half of the individual PCR products were resolved by agarose gel electrophoresis and visualised by staining with ethidium bromide under UV light.
DNA gel blot analysis
Genomic DNA (30 µg), extracted by the cetyl trimethylammonium bromide (CTAB) method26, was digested with HindIII, resolved by agarose gel electrophoresis, and blotted onto a Hybond-N+ membrane (GE Healthcare, UK). A GFP gene-specific probe was directly labelled with alkaline phosphatase using the AlkPhos Direct labelling kit (GE Healthcare, UK). Hybridisation, washing and chemiluminescent detection with CDP Star were performed as recommended by the supplier (GE Healthcare).
Immunoblot analysis
Total protein (20 µg) extracted from leaf tissue (1 g) was separated by 12.5% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on to a nitrocellulose membrane. The membrane was blocked overnight in 1X TBST buffer (10 mM Tris-HCl, 150 mM NaCl, 0.05% Tween-20, pH 7.5) with fat free milk (5%) at 4 °C. Blots were incubated with anti-GFP mouse IgG1κ antibody (Roche, Switzerland), and then with horseradish peroxidase (HRP)-linked anti-mouse IgG (GE Healthcare). Cross-reacting bands were detected using enhanced chemiluminescence immunoblotting detection reagents (GE Healthcare) and a chemiluminescent analyser, LAS3000 (GE Healthcare).
Progeny analysis
PCR analysis of leaves of the T2 progeny was conducted by detecting the GFP transgene. The Chi-square (χ 2) test was used to analyse whether the observed segregation ratio agreed with a Mendelian ratio in the T2 progeny. P is the probability of the observed ratios reflecting the expected segregation ratio.
Electronic supplementary material
Acknowledgements
We would like to thank Drs Kentaro Sasaki, Etsuo Shimosaka, Midori Yoshida, and Takayuki Saito for suggestions and comments. We also thank Dr. Fumitaka Abe for providing the pUba plasmid. This work was supported in part by Cabinet Office, Government of Japan, Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO).
Author Contributions
H.H., Y.N., R.M., R.I., and N.T. designed the experiments. H.H. and Q.L. performed most of the experiments. H.H. and R.I. wrote the manuscript and generated the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-017-17188-2.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-11936-0.
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has not been fixed in the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vasil V, Castillo AM, Fromm ME, Vasil IK. Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Nat. Biotechnol. 1992;10:667–674. doi: 10.1038/nbt0692-667. [DOI] [Google Scholar]
- 2.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 3.Ishida Y, et al. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 4.Cheng M, et al. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997;115:971–980. doi: 10.1104/pp.115.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng M, Hu TC, Layton J, Liu CN, Fry JE. Desiccation of plant tissues post- Agrobacterium infection enhances T-DNA delivery and increases stable transformation efficiency in wheat. In Vitro Cell Dev. Biol. Plant. 2003;39:595–604. doi: 10.1079/IVP2003471. [DOI] [Google Scholar]
- 6.Wu H, Sparks C, Amoah B, Jones HD. Factors influencing successful Agrobacterium -mediated genetic transformation of wheat. Plant Cell Rep. 2003;21:659–668. doi: 10.1007/s00299-002-0564-7. [DOI] [PubMed] [Google Scholar]
- 7.Vasil IK. Molecular genetic improvement of cereals: transgenic wheat (Triticum aestivum L.) Plant Cell Rep. 2007;26:1133–1154. doi: 10.1007/s00299-007-0338-3. [DOI] [PubMed] [Google Scholar]
- 8.Ishida Y, Tsunashima M, Hiei Y, Komari T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 2015;1223:189–198. doi: 10.1007/978-1-4939-1695-5_15. [DOI] [PubMed] [Google Scholar]
- 9.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 10.Bechtold N, et al. The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics. 2000;155:1875–87. doi: 10.1093/genetics/155.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trieu AT, et al. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000;22:531–41. doi: 10.1046/j.1365-313x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- 12.Yasmeen A, et al. In Planta Transformation of Tomato. Plant Mol. Biol. Rep. 2009;27:20–28. doi: 10.1007/s11105-008-0044-5. [DOI] [Google Scholar]
- 13.Mu G, et al. Genetic Transformation of Maize Female Inflorescence Following Floral Dip Method Mediated by Agrobacterium. Biotechnology. 2009;11:178–183. doi: 10.3923/biotech.2012.178.183. [DOI] [Google Scholar]
- 14.Rod-in W, Sujipuli K, Ratanasut K. The floral-dip method for rice (Oryza sativa) transformation. J. Agric. Technol. 2014;10:467–474. [Google Scholar]
- 15.Zale JM, Agarwal S, Loar S, Steber CM. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 2009;28:903–913. doi: 10.1007/s00299-009-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel G, et al. Analysis of T-DNA integration and generative segregation in transgenic winter triticale (x Triticosecale Wittmack) BMC Plant Biol. 2012;12:171. doi: 10.1186/1471-2229-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohli A, et al. Transgene integration, organization and interaction in plants. Plant Mol. Biol. 2003;52:247–258. doi: 10.1023/A:1023941407376. [DOI] [PubMed] [Google Scholar]
- 18.Sparks, C. A. & Jones, H. D. Transformation of wheat by biolistics. Transgenic Crops of the World—Essential Protocols (ed. Curtis, I.). 19–34 (Dordrecht: Kluwer Academic Publishers, 2004).
- 19.Satina S, Blakeslee AF, Avery AG. Demonstrations of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras. Am. J. Bot. 1940;27:895–905. doi: 10.2307/2436558. [DOI] [Google Scholar]
- 20.Stewart, R. N. Ontogeny of the primary body in chimeral forms of higher plants. In The ClonalBasis of Development (eds. Subtelny, S. & Sussex, I. M.) 131–160 (New York: Academic Press, 1978).
- 21.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–479. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 22.Chiu W, et al. Engineered GFP as a vital reporter in plants. Curr. Biol. 1996;6:325–330. doi: 10.1016/S0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 23.Toki S, et al. Expression of a maize ubiquitin gene promoter-bar chimeric gene in transgenic rice plants. Plant Physiol. 1992;100:1503–1507. doi: 10.1104/pp.100.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagio T. Optimizing the particle bombardment method for efficient genetic transformation. Japan Agric. Res. Q. 1998;32:239–247. [Google Scholar]
- 25.Zhu H, Qu F, Zhu LH. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 1993;21:5279–5280. doi: 10.1093/nar/21.22.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.