Abstract
Background
Trichorhinophalangeal syndrome (TRPS) patients tend to have alopecia that appears to be androgenetic, and this genetic model might give clues to the pathogenesis of hair loss or hair morphogenesis.
Objective
This study was conducted to identify additional genetic evidence of TRPS and hair morphogenesis from a TRPS patient.
Methods
From one TRPS type I patient, we extracted RNA and profiled whole transcriptome in non-balding and balding scalp areas using high-throughput RNA sequencing.
Results
We found a total of 26,320 genes, which comprised 14,892 known genes with new isoforms and 4,883 novel genes from the non-balding and balding areas. Among these, a total of 1,242 genes showed different expression in the two scalp areas (p<0.05 and log2 fold-change >0). Several genes related to the skin and hair, alopecia, and the TRPS1 gene were validated by qRT-PCR. Twelve of 15 genes (KRT6C, KRTAP3-1, MKI67, GPRC5D, TYRP1, DSC1, PMEL, WIF1, SOX21, TINAG, PTGDS, and TRPS1) were down-regulated (10 genes: p<0.01; SOX21 and PTGDS: p>0.05), and the three other genes (HBA2, GAL, and DES) were up-regulated (p<0.01) in the balding scalp. Many genes related to keratin and hair development were down-regulated in the balding scalp of the TRPS type I patient. In particular, the TRPS1 gene might be related to androgen metabolism and hair morphogenesis.
Conclusion
Our result could suggest a novel perspective and evidence to support further study of TRPS and hair morphogenesis.
Keywords: Androgenetic alopecia, Differentially expressed gene, Transcriptome, Trichorhinophalangeal syndrome, TRPS1
INTRODUCTION
Type I trichorhinophalangeal syndrome (TRPS) presents with craniofacial dysmorphism, skeletal abnormality, and sparse scalp hairs1. TRPS patients tend to have alopecia that appears to be androgenetic, and thus, this genetic model might give clues to the pathogenesis of hair loss or hair morphogenesis, as has been found in previous studies2. Fantauzzo and Christiano1 reported that the target genes of Trps1, Wif1, Sox18, and Sox21 played an important role in vibrissa follicle morphogenesis by analyzing the gene expression profiles between wild-type and Trps1Δgt/Δgt mutant mouse embryos to understand hair morphogenesis. This is very interesting because sparse scalp hair is a common feature of TRPS. Herein, we analyzed whole transcriptome from non-balding and balding scalp areas from the TRPS patient using high-throughput sequencing and attempted to identify important genetic information about TRPS symptoms and hair morphogenesis.
MATERIALS AND METHODS
Information of patient with TRPS type I
A 15-year-old boy visited with sparse and slowly growing scalp hairs that had been that way since his childhood. Especially, his fronto-temporal hair line regressed to the vertex and his vertex hair density and thickness decreased compared to the occiput hairs. He had the typical TRPS phenotypes, including a bulbous nose, a long philtrum, and abnormally short fingers and toes. We took tissue from the non-balding (occiput area) and balding portions (vertex area) of his scalp for genetic analysis (Supplementary Fig. 1). This study was approved by the institutional review board of Dankook University Hospital (IRB no. DKUH 2014-08-005).
RNA sequencing
We extracted total RNA from the tissues using trizol reagent, and then enriched mRNA by oligo-dT and synthesized to cDNA. We subjected the cDNA to end-repair and poly-A addition and connected it with 5′ and 3′adaptors on both ends3. By separating on a BluePippin 2% agarose gel (Sage Science, Beverly, MA, USA), we selected and amplified suitable fragments. The final library sizes and qualities were evaluated with an Agilent High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, USA). Subsequently, we performed high-throughput RNA sequencing using an Illumina Hiseq2500 sequencer (Illumina, San Diego, CA, USA). Among total output reads, we mapped high-quality reads to the human reference genome (Ensembl release 72)4.
Differentially expressed genes and gene ontology analysis
We calculated the gene expression level based on fragments per kilobase of exon per million mapped reads (FPKM) using Cufflinks v2.1.13 from Ensembl release 72. We generated gene-level count data using HTSeq-count v0.5.4p35. Based on this, we analyzed differentially expressed genes (DEGs) using the gene TCC6. We calculated normalization factors using iterative DEGES/edgeR. We filtered DEGs based on p-value <0.05 and log2 fold change >0. To characterize their molecular function, we analyzed gene ontology (GO) (www.geneontology.org). p-value <0.001 was considered statistically significant.
Quantitative real-time polymerase chain reaction
We synthesized a total of 500 ng of RNA to cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) and an RNase inhibitor (Promega). We designed a primer pair for target genes using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (Supplementary Table 1). We amplified 15 genes and a GAPDH gene as a control to normalize expression using the Eco Real-Time PCR System (Illumina). We confirmed the absence of any non-specific amplified products through melting curve analysis at 55℃~95℃. All reactions were performed in triplicate and analyzed by delta-delta Ct method.
RESULTS
Dataset from RNA sequencing
We processed a total of ten billion raw reads in the filtering step and mapped 94.9% and 94.8% of the clean reads on the human reference genome (Table 1). Based on these data, we found a total of 26,320 genes, which comprised 14,892 known genes with new isoforms and 4,883 novel genes. At the transcript level, we found a total of 218,609 transcripts expressed (FPKM >0) in either the non-balding and balding scalps.
Table 1. Summary of RNA-sequencing.
| Sample | Raw reads | Clean reads (%) | Mapped reads (%) | Properly paired (%) | Gene | Transcript | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum | Known | Known (+new isoforms) | Novel | Sum | Known | Novel | |||||
| Nonbalding | 53,351,054 | 50,338,798 (94.4) | 47,780,868 (94.9) | 36,570,528 (72.6) | 26,320 | 3,426 | 14,892 | 4,883 | 218,609 | 150,194 | 68,415 |
| Balding | 54,289,736 | 51,192,244 (94.3) | 48,539,550 (94.8) | 36,789,924 (71.9) | |||||||
Identifying differentially expressed genes
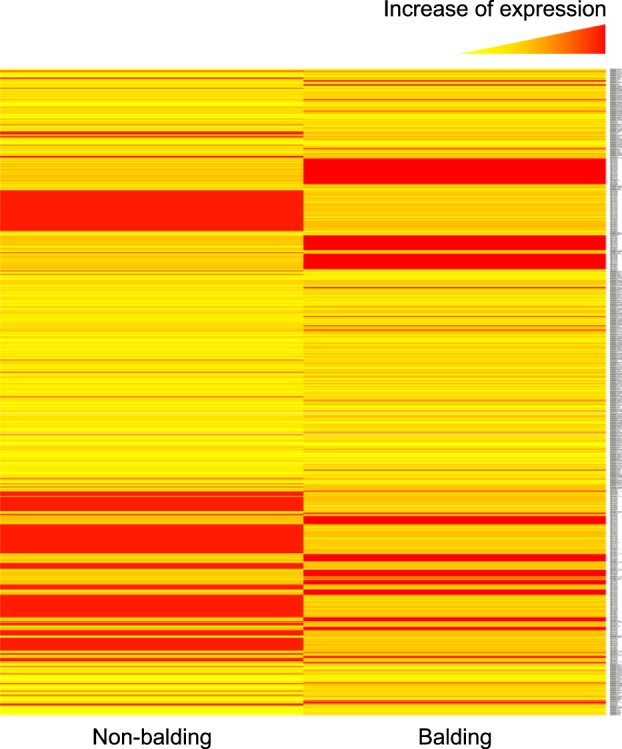
Based on FPKM value, we analyzed gene expression levels and identified DEGs between the non-balding and balding scalp samples. The total number of DEGs was 1,242, comprising transcripts expressed in both samples and in either sample (with p-value <0.05 and log2 fold-change >0) (Fig. 1). Compared to non-balding sample, up- and down-regulated genes were 636 and 606 in balding scalp; specifically, 557 genes showed sample-specific expression.
Fig. 1. Heat map of differently expressed genes in the non-balding and balding scalp samples. A total of 1,242 differentially expressed genes (DEGs) were identified through RNA sequencing (p-value <0.01 and log2 fold-change >0). The left and right columns display, respectively, the results for the non-balding and balding scalp areas. Up-regulated to down-regulated genes are indicated by red and yellow, respectively.
Gene ontology analysis
To characterize the DEGs, we conducted GO analysis. A total of 1,242 DEGs were associated with 3,108 GO terms; at the cut-off of p-value <0.001, 45, 17 and 11 GO terms were associated with biological processes, molecular function, and cellular components, respectively (Supplementary Table 2, 3). The hair-related terms were follicle morphogenesis and development, the hair cycle, hair cell differentiation, and keratinization and were associated with the down-regulated SOX21 gene.
Verifying DEGs using quantitative real-time polymerase chain reaction
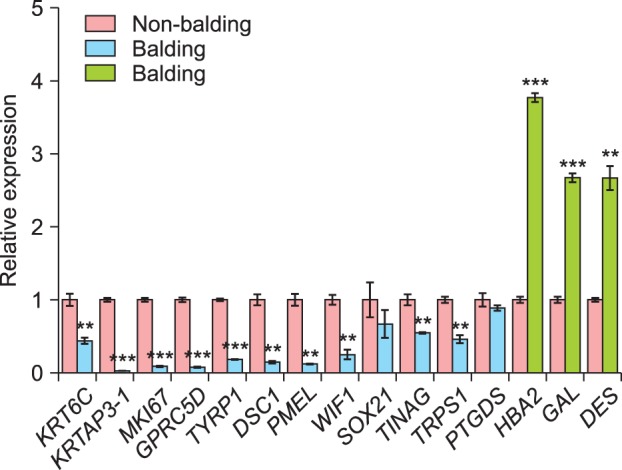
We validated 15 DEGs in the non-balding and balding scalps samples using quantitative real-time polymerase chain reaction (Supplementary Table 1), preferentially selecting skin, hair, and alopecia-related genes; in addition, we validated the WIF1, SOX21, TINAG and TRPS1 genes studied in TRPS1Δgt/Δgt mutant mouse1. Additionally, we validated HBA2, GAL, DES (the up-regulated genes with the lowest p-values) and PTGDS, and this result was consistent with the RNA sequencing (Fig. 2). Thirteen of the 15 genes were statistically significant (p<0.01), including six genes with p-value <0.001. The SOX21 and PTGDS genes were down-regulated but not significantly (p>0.05).
Fig. 2. Validation of 15 differentially expressed genes by quantitative real-time polymerase chain reaction. Three genes were up-regulated (green bar) and 12 were down-regulated (blue bar) in the non-balding scalp. Thirteen genes showed statistical significance (p-value<0.05); the exceptions were SOX21 and PTGDS. (**p<0.01, and ***p<0.001).
DISCUSSION
Interestingly, the sparse hairs of TRPS patients are thin and miniaturized just as in androgenic alopecia. Therefore, we intended to find a genetic difference the between non-balding and balding scalp of a TRPS type I patient and identify a candidate genes related to hair loss or morphogenesis. Among 1,242 of DEGs, we could find lots of keratin and keratin associated genes which might be due to sampling from scalps. Two keratin-related genes (KRT6C and KRTAP3-1) were down-regulated in balding scalp. The MKI67 down-regulated in balding scalp is involved in active proliferation of cells and are reported low expression in hair follicle stem cells7. Down-regulation of MKI67 in balding scalp of TRPS type I patient in our study seemed to suggest degenerated or abnormal hair cell cycle.
A key factor in TRPS pathogenesis, the TRPS1 gene was down-regulated in the balding area. Originally, TRPS1 is a transcription factor to repress its target genes via binding to GATA motif of the promoter region1. However, a recent study has revealed that TRPS1 activated the expression of target gene. Fantauzzo and Christiano1 showed Trps1 activated Wnt inhibitors and other transcription factor essential for follicle morphogenesis in mouse. Study of a TRPS mutant mouse suggests that TRPS1 might be necessary for hair follicular formation and shows that the Wnt inhibitor and extracellular matrix protein were regulated by TRPS1 during early hair morphogenesis8. Decreased TRPS1 protein can disrupt endochondral cartilage differentiation and cell interactions in hair follicle development9. In addition, TRPS1 protein expression is down-regulated by androgens in human prostate cancer, and thus the TRPS1 gene might play a role in androgen metabolism in prostate cancer10. Though the correlation of TRPS1 gene and androgen metabolism has not yet been studied in the alopecia, we could expect the further study about this correlation because the male pattern baldness is associated with androgen metabolism.
WIF1 and SOX21, the target genes of TRPS1, were down-regulated in a TRPS1Δgt/Δgt mutant mouse and in the balding scalp of a TRPS type I patient1. WIF1 is a Wnt inhibitor and is expressed in dermal papilla, like TRPS1 gene. In a previous study, Wnt-related genes including WNT11 and WIF1 were up-regulated in a 120-day-old goat embryo in which secondary hair follicles and mature primary hair follicles were present, which indicates that Wnt signaling is involved in early hair follicle formation11. The SOX21 gene was shown to regulate the layered differentiation of hair follicles12. Its disruption showed the human alopecia-like phenotype in a mouse with progressive hair loss. Interestingly, target gene expression in TRPS was not inversely proportional to that in Fantauzzo's TRPS1 Δgt/Δgt mutant mouse1.
We compared the gene expression patterns with those of androgenetic alopecia by Garza et al.13. KRT6C and GPRC5D were down-regulated and the HBA2 gene was up-regulated in balding scalp in both studies. However, PTGDS expression was not significant, unlike in a previous study13. The GPRC5D gene was dramatically up-regulated in hair follicle keratinization and differentiation in the skin of an old embryo (120-day) in which secondary hair follicles had developed and primary hair follicles had matured, indicating its role in keratinization and hair follicle morphogenesis11. PTGDS might be involved in androgenetic alopecia13, but it is not related to hair loss in TRPS.
In conclusion, we expect our results to suggest novel perspectives and support further study to understand TRPS and hair morphogenesis.
ACKNOWLEDGMENT
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2015R1D1A1A02059462) and the research fund of Dankook University in 2016.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-29-597-s001.pdf.
Photograph of the trichorhinophalangeal syndrome patient. He has sparse and slowly growing scalp hairs (up), bulbous nose, long philtrum, and thin upper lip (bottom). A red circle indicates vertex area where tissue was obtained but, occiput is not appeared in this photograph.
Information of primer pairs used in quantitative real-time polymerase chain reaction
DEGs and GO
Go terms
References
- 1.Fantauzzo KA, Christiano AM. Trps1 activates a network of secreted Wnt inhibitors and transcription factors crucial to vibrissa follicle morphogenesis. Development. 2012;139:203–214. doi: 10.1242/dev.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, et al. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature. 2010;464:1043–1047. doi: 10.1038/nature08875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 8.Malik TH, Von Stechow D, Bronson RT, Shivdasani RA. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol. 2002;22:8592–8600. doi: 10.1128/MCB.22.24.8592-8600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gai Z, Gui T, Muragaki Y. The function of TRPS1 in the development and differentiation of bone, kidney, and hair follicles. Histol Histopathol. 2011;26:915–921. doi: 10.14670/HH-26.915. [DOI] [PubMed] [Google Scholar]
- 10.Chang GT, Jhamai M, van Weerden WM, Jenster G, Brinkmann AO. The TRPS1 transcription factor: androgenic regulation in prostate cancer and high expression in breast cancer. Endocr Relat Cancer. 2004;11:815–822. doi: 10.1677/erc.1.00853. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Wang X, Yan H, Zeng J, Ma S, Niu Y, et al. Comparative transcriptome analysis of fetal skin reveals key genes related to hair follicle morphogenesis in cashmere goats. PLoS One. 2016;11:e0151118. doi: 10.1371/journal.pone.0151118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiso M, Tanaka S, Saba R, Matsuda S, Shimizu A, Ohyama M, et al. The disruption of Sox21-mediated hair shaft cuticle differentiation causes cyclic alopecia in mice. Proc Natl Acad Sci U S A. 2009;106:9292–9297. doi: 10.1073/pnas.0808324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza LA, Liu Y, Yang Z, Alagesan B, Lawson JA, Norberg SM, et al. Prostaglandin D2 inhibits hair growth and is elevated in bald scalp of men with androgenetic alopecia. Sci Transl Med. 2012;4:126ra34. doi: 10.1126/scitranslmed.3003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photograph of the trichorhinophalangeal syndrome patient. He has sparse and slowly growing scalp hairs (up), bulbous nose, long philtrum, and thin upper lip (bottom). A red circle indicates vertex area where tissue was obtained but, occiput is not appeared in this photograph.
Information of primer pairs used in quantitative real-time polymerase chain reaction
DEGs and GO
Go terms


