Abstract
Plant growth-promoting Bacillus amyloliquefaciens FZB42 induces systemic salt tolerance in Arabidopsis and enhances the fresh and dry weight. However, the underlying molecular mechanism that allows plants to respond to FZB42 and exhibit salt tolerance is largely unknown. Therefore, we performed large-scale transcriptome sequencing of Arabidopsis shoot tissues grown under salt stress with or without FZB42 inoculation by using Illumina sequencing to identify the key genes and pathways with important roles during this interaction. In total, 1461 genes were differentially expressed (FZB42-inoculated versus non-inoculated samples) at 0 mM NaCl, of which 953 were upregulated and 508 downregulated, while 1288 genes were differentially expressed at 100 mM NaCl, of which 1024 were upregulated and 264 were downregulated. Transcripts associated with photosynthesis, auxin-related, SOS scavenging, Na+ translocation, and osmoprotectant synthesis, such as trehalose and proline, were differentially expressed by FZB42 inoculation, which reduced the susceptibility to salt and facilitated salt adaptation. Meanwhile, etr1-3, eto1, jar1-1, and abi4-102 hormone-related mutants demonstrated that FZB42 might induce plant salt tolerance via activating plants ET/JA signaling but not ABA-dependent pathway. The results here characterize the plant transcriptome under salt stress with plant growth-promoting bacteria inoculation, thereby providing insights into the molecular mechanisms responsible for induced salt tolerance.
Introduction
Soil salinity is a major issue that affects agriculture and approximately 20% of agricultural land is salt-stressed at present1, 2. Indeed, salt has become one of the main abiotic stress factors that limit agricultural productivity. Mechanistically, apart from causing plant ion imbalance and osmotic stress, excess salinity inhibits metabolism, including photosynthesis, protein and lipid synthesis, thereby limiting the crop growth and yield, and it may even lead to plant death3, 4.
In order to reduce the severe effects of salt stress on plants, many approaches have been developed. Many studies have addressed this issue by focusing on genetic engineering, but this approach is time-consuming and may cause possible environmental risks, so genetically engineered plants are rarely used in the field5, 6. Recently, the use of microbes has become a new alternative for improving stress tolerance in plants7, 8. Plant-growth-promoting rhizobacteria (PGPR) comprise a diverse group of rhizosphere-colonizing bacteria that promote plant growth via direct or indirect mechanisms9, 10, which may be correlated with the ability to resist various pathogens, the production of phytohormones, the release of volatiles, and the production of phytase and siderophores to enhance the availability of minerals in the soil11–14. In addition to their growth-promoting activity, some PGPR are also known to alleviate salt stress15 and the use of PGPR for salt tolerance enhancement has significant advantages compared with other approaches.
Many PGPRs have been studied and applied as useful and efficient agents for inducing salt tolerance in plants due to their ability to promote plant growth and to aggressively colonize plant roots. Recently, it has been shown that the tolerance of salt stress in many agricultural crops can be enhanced by various PGPR genera, such as Azospirillum, Pseudomonas, Burkholderia, Bacillus, Arthrobacter, Azotobacter, and Enterobacter 16. For example, inoculating plants such as wheat, radish, Arabidopsis thaliana, and maize with Bacillus sp. in saline conditions can prevent poor growth and improve the performance of plants in adverse conditions17, 18. The interaction between plants and PGPRs is a complex and reciprocal process. Thus, when PGPRs induce salt tolerance in plants, the expression patterns of plant genes will exhibit a corresponding response to their stimulation. However, most previous studies have focused mainly on the physiological aspects of plants during this interactive process and there have been no reports of the transcriptome profiling of genes involved in the induction of systemic salt tolerance conferred by PGPR in plants.
Bacillus amyloliquefaciens FZB42 was isolated from the rhizosphere soil of lettuce and it has been used widely in commercial applications to support the production of a broad range of economically important plants19. In recent years, many studies have been performed with FZB42 to determine its plant growth-promoting characteristics and biocontrol activities14, 20, 21. The molecular mechanism of FZB42 confers resistance to salt stress in plants is still unknown.
In the present study, we evaluated the effects of FZB42 inoculation on the growth of A. thaliana under salt stress (100 mM NaCl) and identified plant genes with important roles in response to this bacterium by transcriptome profiling in A. thaliana shoot tissues using Illumina sequencing. Our study is to characterize the plant transcriptome under salt stress after colonization by PGPR, thereby providing insights into the molecular mechanism that allows PGPR to induce salt tolerance in plants.
Results
Bacillus amyloliquefaciens FZB42 promoted Arabidopsis growth under non-salt stress and salt stress conditions
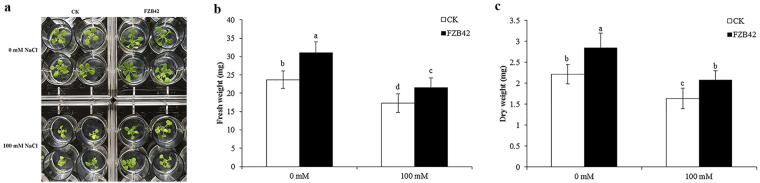
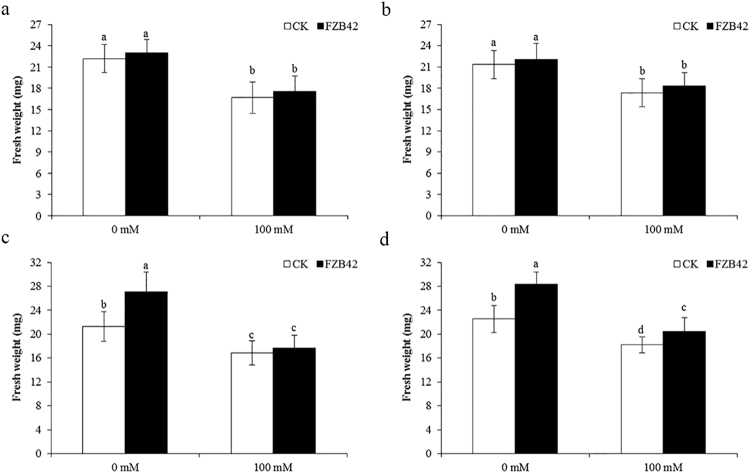
To confirm that FZB42 alleviated plant salt stress, plant growth was monitored in the presence or absence of 100 mM NaCl after inoculation with FZB42. After salt stress treatment for 10 days, the biomass of Arabidopsis seedlings inoculated with FZB42 was higher based on measurements of the fresh and dry weight of the shoot parts of plants (Fig. 1). Compared with non-inoculated seedlings, FZB42-inoculated Arabidopsis exhibited 31.2% and 24.7% increases in the plant fresh weight at 0 and 100 mM NaCl, respectively (Fig. 1b). Similarly, differences were also observed in the dry biomass accumulation, where FZB42-inoculated Arabidopsis exhibited 28.3% and 27.2% increases in the plant dry weight at 0 and 100 mM NaCl compared with non-inoculated seedlings, respectively (Fig. 1c). These results suggest that FZB42 promotes plant growth under non-stress and saline conditions.
Figure 1.
Effects of FZB42 on growth and salt tolerance in Arabidopsis thaliana. Plant seedlings were grown for 10 days in half-strength MS medium, before transplanting into 12-well microtiter plates. After treatment for 3 days with FZB42, NaCl was added to obtain a final concentration of 100 mM. At 10 days after bacterial treatment, the shoot parts were removed and measured to determine the fresh weight. The dry weight of plants was measured after in a drying oven at 75 °C for 2 days. (a) Photographs were taken of the plants after exposure to salt stress with or without FZB42. (b) FZB42 increased the fresh weight. (c) FZB42 increased the dry weight. White and black bars represent FZB42-inoculated and non-inoculated treatments, respectively. Error bars mean standard deviation. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range tests, P < 0.05).
Characterization of the sequenced Illumina libraries
To identify genes related to FZB42-induced plant salt tolerance, twelve Col-0 Illumina libraries including three biological replicates for treatments were constructed from shoot tissues treated or untreated with FZB42 and grown at 0 and 100 mM NaCl. The FPKM (fragments per kilobase of exon per million fragments mapped) expression data was tested by correlation analysis to evaluate sampling between three biological replicates. All the correlation coefficiencies between biological pairs were over 0.98 (Supplementary Fig. S1). An average raw reads about 27,673,695 with paired-ends of 150 bp were obtained from twelve samples, which comprised 24,612,344, 23,716,667, 33,409,535, and 24,784,139 clean reads (paired-ends), respectively (Supplementary Table S1). After mapping the clean reads obtained from the twelve libraries onto the Arabidopsis genome, at least 83% of the clean reads could be mapped onto the reference database. The proportion of uniquely mapped reads was over 76.8%, whereas multiple mapped reads were found mainly in the rRNA or intergenic regions. The mapped reads from the four samples were distributed mainly in exon regions, followed by intergenic and intron regions (Supplementary Fig. S2). Sequencing saturation was analyzed to assess whether the sequencing depth was sufficient for transcriptome coverage. The results showed that the number of detected genes was saturated when the total reads ≥2 million (Supplementary Fig. S3), thereby satisfying the requirements for further analysis.
KEGG pathways
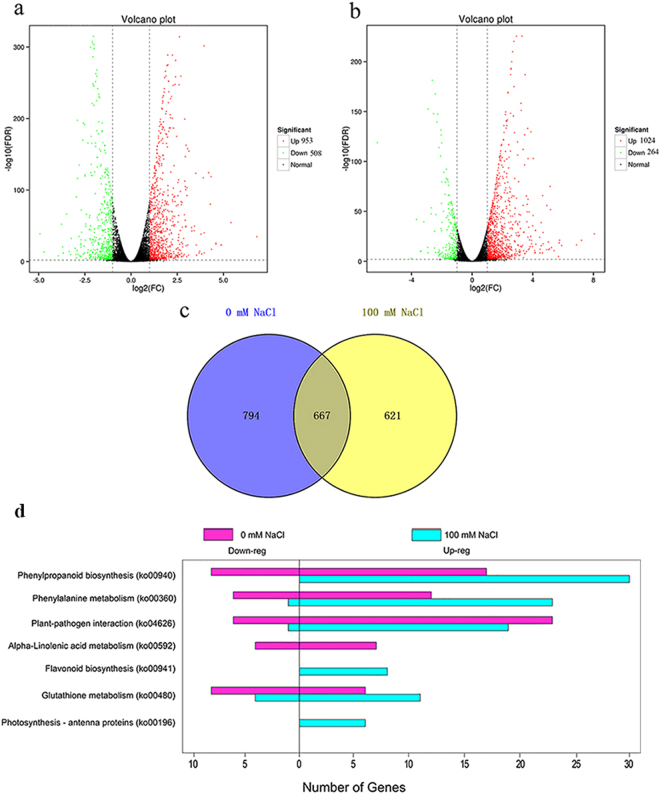
To elucidate the molecular mechanisms associated with the plant response to FZB42 under salt stress conditions, we identified the differentially expressed genes (DEGs) in A. thaliana under the four experimental treatments. Compared with non-inoculated plants, there were 1461 and 1288 DEGs at 0 and 100 mM NaCl, respectively, where 953 were upregulated and 508 were downregulated at 0 mM NaCl, and 1024 were upregulated and 264 were downregulated at 100 mM NaCl (Fig. 2a,b). In total, 794 genes were specifically regulated at 0 mM NaCl and 621 DEGs were identified only under salt stress condition (Fig. 2c) (Supplementary Table S2).
Figure 2.
Differentially expressed genes in FZB42-inoculated versus non-inoculated plants at 0 and 100 mM NaCl. The volcano plot shows the differentially expressed genes (DEGs) with 0 mM NaCl (a) and 100 mM NaCl (b) for FZB42-inoculated versus non-inoculated plants. DEGs were identified using a threshold of FDR ≤ 0.01 and |log2 (fold change)| ≥ 1. The red dots represent upregulated DEGs and the green dots indicate downregulated DEGs. The black dots show genes without obvious changes between two corresponding samples. The Venn diagram shows the number of specific and common DEGs in FZB42-inoculated versus non-inoculated plants at 0 and 100 mM NaCl (c) (see Supplementary Table S2). Pathway enrichment analysis for DEGs. The gene numbers with significant enrichment are shown for upregulated and downregulated genes at 0 and 100 mM NaCl together (Corrected_P-value < 0.05) (d) (see Supplementary Table S3).
To further understand the biological functions of the DEGs, all of the annotated genes were mapped onto terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to search for significantly enriched genes involved in specific metabolic pathways. At 0 mM NaCl, 402 of 1461 DEGs mapped onto the KEGG database for Arabidopsis (Supplementary Fig. S4a) and 5 terms were significantly enriched (Corrected P-value < 0.05). Similarly, at 100 mM NaCl, 353 of 1288 DEGs mapped onto the KEGG database for Arabidopsis (Supplementary Fig. S4b) and six terms were significantly enriched (Corrected P-value < 0.05) (Fig. 2d, Supplementary Table S3). Four terms comprising phenylpropanoid biosynthesis, phenylalanine metabolism, glutathione metabolism, and plant-pathogen interaction were significantly enriched at both 0 and 100 mM NaCl. Interestingly, one term related to plant photosynthesis process, photosynthesis-antenna proteins, was only significantly enriched at 100 mM NaCl. Six genes in the photosynthesis-antenna proteins pathway were all upregulated, which suggested that FZB42 might enhance plant photosynthesis under salt stress compared with non-stress condition.
Effects of FZB42 on the expression patterns of DEGs
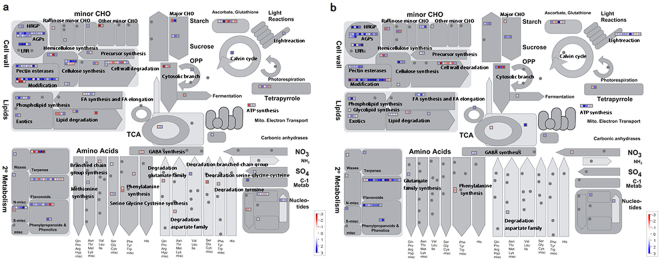
MapMan was used to integrate and visualize the DEGs according to their functions in metabolic pathways (Figs 3 and 4, Supplementary Tables S4 and S5). According to the Arabidopsis transcriptome database TAIR 10, two image annotator modules for overview and biological process were used to map BINs/subBINS data. This allowed us to explore the genes that are induced by FZB42-inoculation by focusing on genes related to energy metabolism, major and minor carbohydrate metabolism, hormone metabolism, redox, transcription factors (TFs), and stress responses22.
Figure 3.
Schematic overview of differentially expressed genes (DEGs) related to different metabolic processes in the Mapman ontology. Upregulated and downregulated genes in FZB42-inoculated versus non-inoculated plants at 0 and 100 mM NaCl are shown in (a,b). Blue squares indicate upregulated DEGs and the red squares indicate downregulated DEGs. The scale bar is shown as log2.
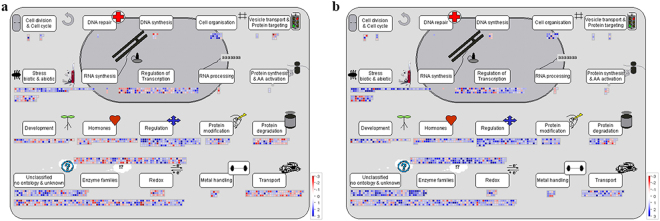
Figure 4.
Overview of cell functions related to differentially expressed genes (DEGs) in FZB42-inoculated versus non-inoculated plants at 0 mM (a) and 100 mM NaCl (b) according to Mapman. Blue squares indicate upregulated DEGs and red squares indicate downregulated DEGs.
Energy metabolism
An overview of the DEGs modulated by FZB42 according to their corresponding metabolic pathways was generated using MapMan (Fig. 3a,b). In total, Mapman mapped out of 1461 and 1288 DEGs, 203 and 180 were differentially mapped at 0 and 100 mM NaCl, respectively. FZB42 modified the expression pattern of some genes related to major and minor CHO metabolism. Compared with 0 mM NaCl, the numbers of upregulated genes related to the light reaction in photosynthesis and ATP synthesis were higher at 100 mM NaCl, suggesting that FZB42 might promote plants produce more energy under salt stress condition. These results were consistent with those shown in Fig. 2. Furthermore, several genes related to photosynthesis like the ones encoding for the photosynthetic pigments (tetrapyrroles) also showed differential expression patterns under the two conditions. However, one gene (ribulose-5-phosphate isomerase) encoding the enzyme of calvin cycle was only significantly induced to 3.8-fold at 0 mM NaCl (Supplementary Tables S4 and S6).
Several pathways related to the biosynthesis and degradation of amino acids and trehalose metabolism were also modulated by FZB42. Among them, we observed that delta1-pyrroline-5-carboxylate synthase 1 (P5CS1), a key enzyme in proline production, was only upregulated in FZB42-inoculated plants in the presence of salt (Fig. 3b). As an important osmoprotectant, proline was usually accumulated in plant cells when plants were in response to abiotic stresses. The positive regulation of P5CS1 was conducive to the accumulation of proline, which helped plants to survive unfavourable salt conditions. For trehalose metabolism, at 0 mM NaCl, one gene (trehalose-6-phosphatase synthase S8, ATTPS8) was upregulated, whereas two genes were detected under salt stress condition, of which haloacid dehalogenase-like hydrolase (HAD) superfamily protein (TPPH) showed positive and trehalose-6-phosphate phosphatase (ATTPPB) showed negative regulation (Supplementary Tables S4 and S6).
Genes associated with growth and development
At 100 mM NaCl, 4/11 genes associated with genetic material replication and synthesis were upregulated, including ribonuclease, DNA synthesis, RNA synthesis, and purine permease (Fig. 4b), whereas 8/15 genes were upregulated at 0 mM NaCl (Fig. 4a). Furthermore, two DNA repair related genes (DNA repair protein and methyladenine glycosylase family protein) and one DNA repair related gene (methyladenine glycosylase family protein) were all upregulated at 0 and 100 mM NaCl, respectively (Supplementary Tables S5 and S6).
Similarly, the transcript levels of several genes involved in plant growth-related processes also increased in FZB42-inoculated seedlings at 0 and 100 mM NaCl, including extensins (i.e., proline-rich extensin-like family protein and expansin-like proteins), cell wall-modification enzymes (i.e., pectinesterase and polygalacturonase). Polygalacturonase is an enzyme that catalyzes carbohydrate synthesis, which is an important component of the pectin network that comprises plant cell walls. In addition, genes encoding other cell wall modification enzymes (xyloglucan endotransglucosylase/hydrolase) that participate in cell wall loosening were also modified by FZB42. At 100 mM NaCl, ten transcripts encoding xyloglucan endotransglucosylase/hydrolase were all upregulated, whereas only 11/14 genes were upregulated at 0 mM NaCl. The upregulation of these genes might benefit the plant cell wall extension (Supplementary Tables S5 and S6).
In addition, two genes involved in the cell cycle (crooked neck protein and CYCD4;1) and one gene involved in cell division (mitogen-activated protein kinase kinase kinase 16) were upregulated by FZB42 at 100 mM NaCl (Fig. 4b). Four genes involved in the cell cycle and cell division were upregulated at 0 mM NaCl (Fig. 4a, Supplementary Tables S5 and S6). Changes in the transcription levels of cell wall modification and cell division-related genes are beneficial for cell development and division, thereby allowing plants grow stronger.
Stress-response genes
We analyzed DEGs modified by abiotic and biotic stresses in plants inoculated with FZB42 at 0 and 100 mM NaCl (Fig. 4a,b, Supplementary Tables S5 and S6). Interestingly, compared with salt stress, more genes involved in biotic and abiotic stresses were differentially expressed by FZB42 at 0 mM NaCl.
At 0 mM NaCl, 34 abiotic stress-related genes and 67 biotic stress-related genes were detected, of which 21 and 59 genes were upregulated, respectively. Similarly, at 100 mM NaCl, 32 abiotic stress-related genes and 52 biotic stress-related genes were identified, of which 19 and 51 genes were significantly upregulated, respectively. Interestingly, 20 genes encoding PR-proteins were all upregulated at 100 mM NaCl whereas 36 PR-proteins genes were induced by FZB42 0 mM NaCl, of which 32 genes were upregulated. In addition, five genes responding to cold stress were only detected at 100 mM NaCl and all of them were upregulated by FZB42. While three genes responding to drought and salt were upregulated at 0 mM NaCl, two genes responding to drought and salt were identified at 100 mM NaCl and AT5G04060 showed negative regulation and AT4G30650 showed positive regulation by FZB42.
Heat-shock proteins (Hsps) are molecular chaperones and a ubiquitous feature of cells, which prevent the stress-induced denaturation of other proteins23. In this study, 18 and seventeen Hsps were differentially expressed in FZB42-inoculated plants at 0 and 100 mM NaCl. Interestingly, a lower number of genes (35.3%) was upregulated at 100 mM NaCl compared with 0 mM NaCl (50%).
Many transcripts encoding for antioxidases that function as reactive oxygen species (ROS) scavenger were also modified by FZB42. Our results showed genes involved in antioxidant responses such as glutathione-S-transferase (11 genes 54.5% of which were upregulated), peroxidases (10 genes 90% were upregulated), and redox (21 genes 38.1% of which were upregulated) were differentially regulated by FZB42 at 0 mM NaCl (Fig. 4a). An increase in the number of upregulated antioxidant-related genes was found at 100 mM NaCl compared with 0 mM NaCl. At 100 mM NaCl, genes involved in antioxidant responses such as glutathione-S-transferase (13 genes 84.6% of which were upregulated), peroxidases (18 genes 100% of which were upregulated, fold change ranged between 2.1 and 118), and redox (14 genes 57.1% of which were upregulated) were differentially regulated by FZB42 (Fig. 4b). In particular, the upregulated of the redox.thioredoxin was particularly found at 100 mM NaCl (Fig. 4b, Supplementary Tables S5 and S6).
Hormone-related genes
The expression patterns of genes involved in jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), auxin, salicylic acid (SA), and brassinosteroid, were differentially regulated by FZB42 at 0 and 100 mM NaCl (Fig. 4, Supplementary Tables S5 and S6).
Jasmonic acid and Ethylene metabolism
Both JA and ET are considered to be stress-responsive phytohormones and they are capable of eliciting defence responses. Compared with 100 mM NaCl, the treatment with 0 mM NaCl produced a much higher number of DEGs related to JA and ET metabolism after FZB42-inoculation.
In terms of JA metabolism, three genes involved in JA synthesis were found to be upregulated by FZB42, including two lipoxygenases (AT1G17420 and AT1G72520) and one allene oxide cyclase (AT3G25780), downstream of the lipoxygenases in JA synthesis, which had a fold change of 2.3, 2.8 and 2.6, respectively, compared with non-FZB42 inoculation under salt stress condition. In addition, three JA-responsive defense-related genes (AT2G26010, AT5G44420, and At1g75830), were also upregulated under salt stress condition. Four genes involved in JA synthesis were also found to be upregulated by FZB42 under non-stress condition, whereas jasmonic acid carboxyl methyltransferase (JMT), a key enzyme for jasmonate-regulated plant responses, and jacalin lectin family protein (AT1G52100) were downregulated (Fig. 4a,b, Supplementary Table S6).
Similar to JA, RNAseq data also suggestes that FZB42 induces ethylene biosynthesis under salt stress condition. Four ACC synthases (ACS7, ACS2, ACS8, and ACS11), the key enzymes in ethylene synthesis, were upregulated 4.4, 5, 5.1 and 3.3-fold by FZB42 at 100 mM NaCl. Likewise, 1-aminocyclopropane-1-carboxylate oxidase (AT1G77330) that plays an important role in converting the ACC into ethylene was upregulated 3.3-fold at 100 mM NaCl. Under salt stress condition, FZB42 also affected 5 ethylene signal transduction genes, all of which were upregulated except HLS1 (AT4G37580). At 0 mM NaCl, two genes involved in ethylene biosynthesis were detected by FZB42, only one of which was upregulated. The transcriptional levels of seven ethylene signal transduction genes (71.4% upregulated) were also affected by FZB42 under non-stress condition (Fig. 4a,b, Supplementary Tables S5 and S6).
Abscisic acid metabolism
9-cis epoxycarotenoid dioxygenase (NCED) genes encode key enzymes for abscisic acid biosynthesis24. In our study, one NCED gene (NCED3, AT3G14440) showed decreased expression level at both 0 and 100 mM NaCl. NCED4 (AT4G19170) was only significantly downregulated at 100 mM NaCl. Specifically, two genes involved in abscisic acid signal transduction were only detected at 100 mM NaCl, both of which ABI1 (AT4G26080) and MARD1 (AT3G63210) showed negative regulation (Fig. 4a,b, Supplementary Tables S5 and S6), suggesting FZB42 conferred plant salt tolerance in ABA-independent pathway.
Auxin metabolism
Two genes involved in auxin biosynthesis were identified in FZB42-inoculated plant at 0 mM NaCl, of which one (TGG2) was upregulated and one (UGT1) was downregulated. Similar to this, two genes involved in auxin biosynthesis were also detected in FZB42-inoculated plants at 100 mM NaCl, including ILL5 (upregulated 54.4-fold) and UGT1 (downregulated 2.3-fold). 16 and nineteen auxin induced-regulated-responsive-activated genes were identified in FZB42-inoculated plant at 0 mM and 100 mM NaCl, respectively, of which 10 and fifteen were differentially upregulated (Fig. 4a,b, Supplementary Tables S5 and S6).
Salicylic acid metabolism
FZB42 affected the expression of 6 transcripts involved in SA synthesis, all of which were upregulated except UGT74E2 (AT1G05680) and GAMT2 (AT5G56300) at 0 mM NaCl. Intriguingly, 4 transcripts involved in SA synthesis were all upregulated at 100 mM NaCl (Fig. 4a,b, Supplementary Tables S5 and S6).
Bassinosteroid metabolism
As plant steroidal, brassinosteroids (BRs) regulate plant growth and development. Overexpression of CHI2/SHK1/SOB7 (CYP72C1, At1g17060) could cause BR deficiency25, 26. The transcriptional level of CHI2/SHK1/SOB7 was downregulated by FZB42 at 0 mM and 100 mM NaCl, respectively. This indicated that BR might accumulate in FZB42-inoculated plants compared with non-inoculated plants. Furthermore, one gene EXL5 (AT2G17230) involved bassinosteroid signal transduction was identified, which was differentially upregulated at both 0 and 100 mM NaCl. (Fig. 4a,b, Supplementary Tables S5 and S6).
Transcription factors
In total, 58 (32 upregulated) and 49 (37 upregulated) differentially expressed TFs were identified in FZB42-inoculated plants at 0 mM and 100 mM NaCl, respectively (Fig. 4a,b, Supplementary Tables S5 and S6). Several TF families with pivotal roles in response to abiotic stress were differentially regulated, including MYB, WRKY, bZIP, AP2/EREBP, and bHLH. These included seven common DEGs from the MYB family comprising MYB6, MYB77, MYB10, MYB3, MYBL2, MYBL, and RVE2, as well as specific 16 at 0 mM NaCl and 8 at 100 mM NaCl. MYB3, MYB6, MYB15, MYB74, MYB77, and two MYB-related gene family CCA1-like genes (CCA1 and LHY) except MYB3 and one MYB-related gene family CCA1-like gene (RVE2) were all upregulated by FZB42 under salt stress condition; interestingly, all of these transcripts responded to ethylene and JA27. Furthermore, within the WRKY transcription factor family, 4 (100% upregulated) and twelve (91.7% upregulated) WRKY genes were obsered at 0 and 100 mM NaCl, respectively. Similarly, three WRKY transcriptional factors (TFs) (WRKY 30, 46 and 48) identified in this study under salt stress were also known to respond to ethylene28, 29, further pointing to a role of ethylene and JA in inducing salt tolerance conferred by FZB42. FZB42 affected the expression of 15 and 15 (AP2/EREBPs) at 0 mM and 100 mM NaCl, 26.7% and 60% of which were upregulated, respectively. All the five bZIPs except HYH and one bZIP (AT5G04840) were all upregulated at 0 and 100 mM NaCl, respectively. In addition, we also identified 11 (81.8% upregulated) and six (83.3% upregulated) bHLHs at 0 and 100 mM NaCl, respectively.
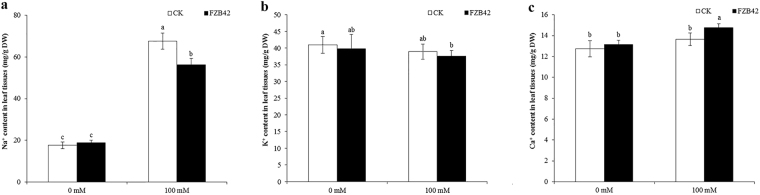
FZB42 affected the Na+, K+, and Ca2+ balance under saline conditions
We measured the ion contents of Na+, K+, and Ca2+, which are involved in photosynthesis and energy transfer. We found that all three of these ions exhibited greater changes in FZB42-inoculated plants compared with non-inoculated plants at 0 mM and 100 mM NaCl. And similar results were obtained by Ghaffari et al.22 studying root-colonized fungus Piriformospora indica induced barley salt stress tolerance. At 100 mM NaCl, the Na+ content of shoot tissues from FZB42-inoculated plants reduced by 16.4% compared with that from non-inoculated plants, which suggested that FZB42 decreased Na+ accumulation under salt stress condition (Fig. 5a). This result was consistent with the RNA-seq data, which detected three transcripts encoding Na+/H+ antiporter (NHX1, CHX16, and CHX17) and one transcript encoding sodium transporter (HKT1), and all of these four genes showed positive regulation. However, FZB42 had no effects on K+ accumulation, although there was only a slight decrease in the K+ content in FZB42-inoculated plants at both 0 and 100 mM NaCl (Fig. 5b). The K+/Na+ ratio in the shoot tissues was increased by FZB42 inoculation under salt stress condition, which was beneficial for salt adaptation by plants. The Ca2+ level was only significantly increased in FZB42-inoculated plants under salt stress condition (P < 0.05), despite the fact that a slightly high Ca2+ level was also detected in FZB42-inoculated plants under non-stress condition, but no significant difference was detected (Fig. 5c). Similar to the K+/Na+ ratio, there was a higher Ca2+/Na+ ratio in FZB42-inoculated plants compared with non-inoculated plants under salt stress condition.
Figure 5.
Effects of FZB42 on the ion contents of shoot tissues under salt stress. (a) Na+ content. (b) K+ content. (c) Ca2+ content. White and black bars represent FZB42-inoculated and non-inoculated treatments, respectively. Error bars mean standard deviation. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range tests, P < 0.05).
Inducible proline accumulation
The accumulation of osmoprotectants, including glycine betaine, trehalose, and proline, is a common stress avoidance mechanism in plants such as Arabidopsis. To investigate whether proline accumulation was facilitated by FZB42 under salt stress, the proline contents of Arabidopsis shoots were measured in the four treatments. In agreement with its transcriptional regulation, the highly upregulated expression of the key proline synthesis enzyme P5CS1 showed that FZB42 could significantly enhance the accumulation of proline in plants treated with 100 mM NaCl. Under salt stress condition, the proline accumulation increased by over 32% when inoculated with FZB42 (79.3 ± 2.9 μg/g shoot) compared with non-inoculated seedlings (60.1 ± 2.7 μg/g shoot). The induced accumulation of proline by FZB42 helped Arabidopsis to resist the osmotic stress produced by high salinity. In addition, there was no significant difference in the amount of proline accumulation between FZB42-inoculated and non-inoculated plants under non-stress condition.
Mutant analysis confirms the role of ET, JA and ABA in salt tolerance
In this study, we found 15 ethylene-responsive transcriptional factors were differentially regulated in response to FZB42 inoculation under salt stress. To further confirm the role of ethylene in FZB42 inducing plants salt tolerance, thus, Arabidopsis mutants etr1-3 (defective in ethylene signaling) and eto1 (ethylene overproducing mutant) were further analyzed for salt tolerance. It appeared that FZB42 did not promote etr1-3 mutants growth at both 0 and 100 mM NaCl. Although the shoot weights of etr1-3 mutants were slightly increased in FZB42-inoculated mutants than those of non-inoculated mutants, no significant differences were detected (Fig. 6a). Interestingly, similar results were also obtained in the ethylene-overproducing mutants eto1, FZB42 did not promote growth in eto1 mutants under non-stress and stress conditions (Fig. 6b). The results of etr1-3 and eto1 mutants indicated that ethylene signaling might be required for FZB42-plant growth promotion and inducing salt tolerance but a fine regulation of ethylene homeostasis was also necessary in this process. Likewise, JA insensitive mutants jar1-1 treated with FZB42 also showed no significant tolerance to salt than non-inoculated mutants under stress condition, whereas significant shoot weight was detected in FZB42-inoculated mutants compared with non-inoculated plants at 0 mM NaCl (Fig. 6c). Lastly, abi4-102 mutants (abscisic acid insensitive) inoculated by FZB42 exhibited better growth, as judged from shoot weight. There were significant differences in shoot weight between FZB42-inoculated and non-inoculated abi4-102 mutants under both non-stress and stress conditions (Fig. 6d). These results were in agreement with our transcriptome data, genes related to ethylene and jasmonic acid synthesis were all upregulated by FZB42, whereas genes related to abscisic acid synthesis were all downregulated by FZB42 at 100 m M NaCl (Fig. 4b, Supplementary Tables 5 and S6). Taken together, the mutant analysis confirms that FZB42 might induce systemic salt tolerance via activating plant ET/JA signaling but not ABA-dependent pathway.
Figure 6.
Effects of FZB42 on growth and salt tolerance in different Arabidopsis mutants. Four mutants were grown for 10 days in half-strength MS medium, before transplanting into 12-well microtiter plates. After treatment for 3 days with FZB42, NaCl was added to obtain a final concentration of 100 mM. At 10 days after bacterial treatment, the shoot parts were removed and measured to determine fresh weights. (a) etr1-3 mutants. (b) eto1 mutants. (c) jar1-1 mutants. (d) abi4-102 mutants. White and black bars represent FZB42-inoculated and non-inoculated treatments, respectively. Error bars mean standard deviation. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range tests, P < 0.05).
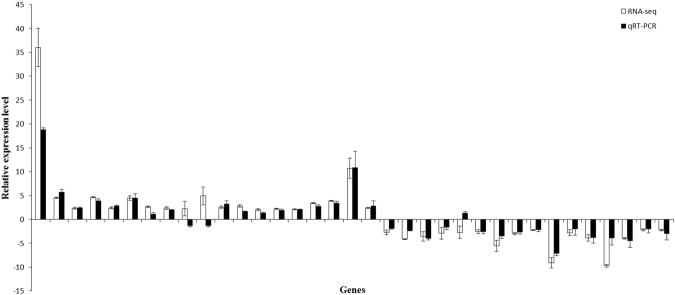
Validation of DGEs by qRT-PCR
To validate the RNA-seq results, 35 selected DEGs were analyzed by qRT-PCR using specific primers (Supplementary Table S7). Among the 35 selected DEGs, nineteen were upregulated (fold change ranged between 2 and 36) and sixteen were downregulated (fold change ranged between −9.7 and −2.2). Most of the qRT-PCR results obtained from the 35 selected genes were in general agreement with their corresponding changes in transcript abundance according to RNA-seq (Fig. 7), thereby confirming the reliability of the Illumina results.
Figure 7.
Comparisons of the relative expression levels determined by qRT-PCR (black bars) and RNA-seq (white bars) for 35 selected DEGs. The y-axis shows the relative expression levels of 35 selected genes. The x-axis indicates the 35 selected genes (from left to right: AT5G44420, AT3G23250, AT4g26200, AT1g17420, AT4G39210, AT4G23700, AT1g29910, AT2G05070, ATCG00120, ATCG00580, AT2G39800, AT2G05100, AT2G34420, AT3G27690, AT5G54270, AT4G39770, AT1G64170, AT1G14540, AT1G70290, AT3g14440, AT4G13250, AT1G78090, AT4G36110, AT5G23370, AT3G24500, AT3G03480, AT4G08390, AT1G05560, AT5G51720, AT4G32810, AT3G12580, AT3G04800, AT1G64900, AT3G44990, and AT1G10070). Bars represent the mean ± SD based on three replicates.
Discussion
Previous studies have shown that PGPRs can elicit induced systemic resistance (ISR) to reduce the susceptibility of plants to diseases caused by biotic stresses, such as pathogenic fungi, bacteria, viruses, and nematodes30. In addition, some PGPRs also confer plants induced systemic tolerance to abiotic stresses such as salinity and drought31. The present study demonstrated that FZB42 alleviated salt stress in Arabidopsis and improved its growth under hydroponic conditions (Fig. 1), as shown in previous studies3, 18, 32, 33. Illumina sequencing was used to identify genes induced by FZB42 in Arabidopsis shoot tissues in response to salt stress.
Salinity usually causes ion toxicity due to ionic imbalance with a higher Na+ content in plant tissues. Thus, reducing the Na+ content is important for improving salt tolerance. Once Na+ influxes into plant cells, Na+/H+ antiporters are usually induced for Na+ recirculation and sequestration. It has been shown that Arabidopsis NHX1 encodes a tonoplast Na+/H+ antiporter that is responsible for Na+ sequestration and it’s overexpression enhances Arabidopsis salt tolerance34, 35. In this study, NHX1 was upregualted by FZB42 at 100 mM NaCl, indicating that FZB42 could alleviate alleviates salt stress in Arabidopsis by increasing vacuolar Na+ compartmentalization and then minimizing toxic ion accumulation (Fig. 5a). The expressions of NHX1 in maize and Arabidopsis were also significantly induced by inoculation with Bacillus amyloliquefaciens SQR9, a close relative of FZB42, under salt stress condition33, 36. However, this result was contrary to the observed repression of NHX1 in rice inoculated with Bacillus amyloliquefaciens NBRISN13 and grown under salt stress37. That was probably due to the differences in plant species and sample collection time. The HKT1 gene, a high-affinity K+ transporter, is known to be differentially induced in plant roots and shoots by volatiles emitted by Bacillus subtilis GB03, thereby reducing accumulation of Na+ throughout plant tissues18. Interestingly, HKT1 was also significantly induced by FZB42 and SQR9 through root colonization under salt stress33, showing that Bacillus amyloliquefaciens could alleviate Na+ toxicity through regulating HKT1 expression. Moreover, many studies showed that the exopolysaccharides (EPSs) produced by PGPRs could bind Na+ and subsequently decreased the amount of Na+ available for plant uptake38–40, and many EPS-producing PGPRs have been used to help plants alleviating salt stress41, 42. We also found that FZB42 adhered aggressively to Arabidopsis roots via biofilm formation (Supplementary Fig. S5). Thus, EPSs in the biofilm on Arabidopsis roots might also play important roles in binding Na+ to restrict the uptake of Na+ and ultimately decrease the plant Na+ content.
Salinity usually causes an increased level of ethylene in plants that ultimately inhibit plants growth43. Some PGPRs containing ACC deaminase that can sequester ACC (the immediate precursor of ethylene) have been documented to enhance plants salt tolerance via a reduction in levels of ethylene and consequently facilitate the growth of many crops such as rice, tomato, cotton, and maize under salinity condition44–47. And there are growing evidences showing that certain PGPRs without ACCD also induce plant salt tolerance; for example, although Bacillus pumilus WP8 and Pseudomonas putida RBP1 had no ACCD, they did successfully increase tomato salt tolerance48. SQR9 that failed to secrete ACCD also enhanced maize, Zea mays and Arabidopsis salt tolerance33. These findings implied that these PGPRs were not via reducing ethylene production to confer plant salt tolerance, supporting the view that reducing ethylene production is not the sole approach accounting for salt tolerace in all PGPRs. In contrast, low concentrations of ethylene appeared to enhance plant growth49. And several previous studies showed ethylene signaling was required for certain PGPRs-growth promotion. Chen et al.50 showed that Variovorax paradoxus 5C-2 promoted growth of ethylene overproducing mutant (eto1-1), but growth promotion was not found in ethylene-insensitive mutants (etr1-1 and ein2-1). Poupin et al.51 found that Burkholderia phytofirmans PsJN-growth promotion was also related to ethylene signaling because it failed to stimulate the development of Arabidopsis mutants with an impaired ethylene (ein2-1). In this study, genes related to ethylene synthesis were differentially modified by FZB42 under non-stress and stress conditions. It was noted that five genes including (ACS2, ACS7, ACS8, ACS11, and ACO5) were all upregulated by FZB42 at 100 mM NaCl (Fig. 4b). On the other hand, mutants data showed FZB42 did not promote growth in etr1-1 and eto1 mutants (Fig. 6a,b) under non-stress and stress conditions. All of these results indicated that ethylene signaling might play important roles not only in FZB42-growth promotion but also in inducing plants salt tolerance but depending a fine ethylene homeostasis.
Jasmonic acid is potent signal compounds, and it accumulates rapidly when plants are under abiotic stress52. Many studies showed that JA is in response to salt stress, acting as positive regulators of salt tolerance53, 54; for example, salt tolerant tomato showed increased levels of jasmonic acid and JA content changed in response to salt-stress55. In this study, all of the genes involved in jasmonic acid synthesis were differentially upregulated by FZB42 at 0 and 100 mM NaCl. On the other hand, similar to ethylene mutants, FZB42 was also unable to induce salt tolerance in jar1-1 mutant line of Arabidopsis (Fig. 6c) at 100 mM NaCl. The results of this study collectively suggested that FZB42 might use the ethylene and jasmonic acid transduction pathway to induce Arabidopsis salt systemic tolerance. This result was in agreement with the previous study done by Cho et al.56, who showed that that jasmonic acid and ethylene-regulated defense genes might play important roles in Pseudomonas chlororaphis O6 mediating systemic tolerance against abiotic and biotic stresses.
As important phytohormones, both JA and ET are considered to be stress-responsive hormones57, which can activate the plant defence network by regulating the expression of many TFs. Indeed, several TFs were identified in our transcriptomic analysis (Fig. 4a,b), including ERF, MYB, and WRKY. And most of these TFs played critical roles in biotic and abiotic stress responses in plants58. ERF TFs are located downstream of genes in the ET signaling pathway. The overexpression of many of these ERF genes conferred tolerance to various abiotic stresses in plants59, 60. Several MYB and WRKY TFs that respond to ET and JA were also upregulated by FZB42 under salt stress, further suggesting that JA/ET signaling might be involved in plant mechanism for salt stress adaptation conferred by FZB42.
The accumulation of osmoprotectants, including glycine betaine, trehalose, and proline, is a common stress avoidance mechanism in plants such as Arabidopsis. TSS and proline contents were highly enhanced in wheat treated with Bacillus subtilis SU47 and Arthrobacter sp. SU18 under salt stress17. The expression of proline biosynthetic gene P5CS1 was highly upregulated in Enterobacter sp. EJ01 infected seedlings under salt condition3. Similar to the results of Kim et al.3, P5CS1 was significantly upregulated by FZB42 under salt stress condition (Fig. 4b). Moreover, a highly proline content was also found in FZB42 treated plants. The increased amount of proline may function as an osmolyte or reactive oxygen species (ROS) scavenger, and its induced production by FZB42 under salt stress agreed with similar previous findings in cotton, rice, tomato, and Arabidopsis 3, 30, 61.
Abiotic stresses such as salt and drought can induce oxidative stress by generating ROS, which are scavenged by plants antioxidases such as catalase, superoxide dismutase, peroxidase, glutathione-S-transferase, and redox enzymes62. Researches with application of PGPR showed that PGPRs could significantly increase antioxidase activities of plants. Tomato treated with Enterobacter sp. EJ01 exhibited higher APX (ascorbate peroxidase) activities under salt condition than did EJ01-free seedlings3. The activities of POD and CAT was highly enhanced in B. amyloliquefaciens SQR9 treated maize than that of SQR9-free maize under salt stress33. In the current study, the enhanced expression levels of genes related to peroxidases, glutathione-S-transferases, and redox in FZB42-inoculated plants compared with non-inoculated plants showed that FZB42 could enhance the capacity of plants for scavenging ROS.
In conclusion, this study demonstrated that FZB42 can improve plant growth and enhance salt tolerance. In order to identify genes with important roles in the response to FZB42, we investigated the transcriptome profiles of Arabidopsis shoot tissues under salt stress using Illumina sequencing while the root samples were also collected for further analysis. The RNA-seq data indicated that FZB42 induced the upregulation of genes related to photosynthesis, auxin, ROS scavenging, Na+ translocation, osmoprotectants such as trehalose and proline as well as ethylene and jasmonic acid signaling under salt stress condition, thereby suggesting that these pathways might have roles in plant salt tolerance induced by FZB42 (Fig. 8). The results of this study provide a useful resource that may facilitate plant functional genomic studies, including many candidate genes as potential markers of tolerance to salt stress.
Figure 8.
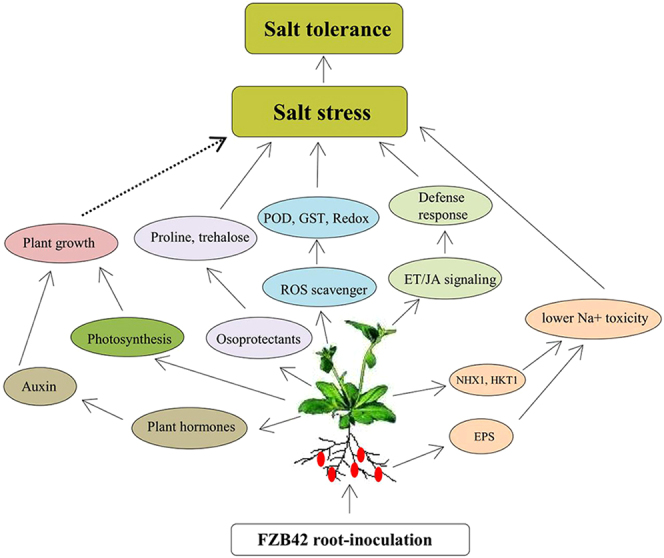
Pathways of FZB42 induced plants salt tolerance. Solid arrows indicated plant compounds or genes affected by FZB42, which had direct effects in enhancing plants salt tolerance. Broken arrows indicated plant growth affected by FZB42, which had indirect effects in enhancing plants salt tolerance. FZB42 root-inoculation enhanced plants photosynthesis and altered the expression levels of genes related to auxin, which promoted plants growing stronger and then enhanced plants tolerance to salt.
Methods
Plant materials and growth conditions
Arabidopsis thaliana (Col-0) seeds were surface sterilized with 2% NaClO for 3–4 min, washed five times with sterile water, and then planted on Petri dishes containing half-strength Murashige and Skoog (MS) solid medium, 0.8% (wt/vol) agar, and 1.5% (wt/vol) sucrose (pH 5.8) for germination. The growth conditions comprised 22 °C with a 16/8 h light/dark photoperiod.
Culture conditions for B. amyloliquefaciens FZB42
B. amyloliquefaciens FZB42 (deposited as strain 10A6 in the culture collection at the Bacillus Genetic Stock Center) was grown in Luria-Bertani liquid medium at 37 °C with shaking at 200 rpm. After incubating overnight, cells were obtained by centrifugation at 10,000 × g for 6 min, washed once, and re-suspended in sterile water to 5.0 × 107 colony-forming units/mL for use as an inoculum.
Hydroponic culture and salt stress
Ten-day-old seedlings at appropriate growth states were transplanted singly into the wells of 12-well microtiter plates containing half-strength MS medium without sugar, and the medium was replaced every 3 days. Three days after transplantation, a half of 12-well microtiter plates was treated with FZB42 with a final concentration of 106 CFUs ml−1 (B: only FZB42 inoculation); and the other half was treated with an equal amount of water as the non-inoculated control (CK: only water). At 3 days after bacterial inoculation, a half of 12-well microtiter plates (FZB42-inoculated and non-inoculated) was supplemented with 100 mM NaCl (B + S: 100 mM NaCl + FZB42 inoculation; S: only 100 mM NaCl). After salt treatment for 7 days, the shoot parts of seedlings were harvested to obtain physiological measurements and then frozen in liquid nitrogen, before storing at −80 °C until RNA extraction. This process was repeated three times as three biological replicates and each replicate included 72 plants.
RNA isolation and Illumina sequencing
Total RNA was extracted from shoot tissues of 30 plants of three independent biological replicates using an RNeasy Plant Mini Kit according to the manufacturer’s instructions. The quality of RNA samples was assessed by UV-absorbance spectrophotometry and 1% agarose gel electrophoresis. After using OligodT magnetic beads to enrich poly(A) + mRNA, random hexamer primers were employed to synthesize cDNA. Finally, sequencing adaptors were ligated to the short fragments and suitable cDNA fragments were selected and purified. Subsequently, the final cDNA library templates from the four treatments were obtained by PCR. Sequencing was performed by Beijing Biomarker Technologies Corporation (Beijing, China) using a HiSeq. 2000 system (Illumina).
Analysis of Illumina sequencing results
After removing adaptors, the unknown nucleotides and those of low quality were filtered from the raw reads to obtain clean reads, which were then mapped onto the Arabidopsis thaliana reference genome version TAIR10. Gene expression levels were calculated using FPKM values (fragments per kilobase of exon per million fragments mapped) by the Cufflinks software63. Genes with a false discovery rate (FDR) < 0.01 and |log2(fold change)| ≥ 1 were defined as differentially expressed genes (DEGs). DEGs were aligned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) using BLASTX. Furthermore, the Mapman system was used to classify the regulated transcripts at 0 and 100 mM NaCl.
Shoot tissues proline quantification
The shoot tissues proline content was quantified according to a previously described method64 with some slight modifications, as follows. First, 100 mg of shoot tissues were homogenized in 10 mL of 3% aqueous sulfosalicylic acid and filtered through filter paper. Next, 2 mL of the filtrate was reacted with 2 mL acid-ninhydrin and 2 mL glacial acetic acid in a 15 mL centrifuge tube for 1 h at 100 °C. The reaction mixture was then extracted with 4 mL of toluene for 15–20 s. The absorbance of the chromophore containing toluene aspirated from the aqueous phase was read at 520 nm using a spectrophotometer with toluene as the blank.
Determination of Na+, K+, and Ca2+
Na+, K+, and Ca2+ were determined by inductively coupled plasma optical emission spectrometry (ICP-OES, PerkinElmer Optima 8000, USA). Shoot tissues were digested using HNO3 according to a previously described method18.
Mutant analysis
Four kinds of Arabidopsis mutants etr1-3 (ethylene insensitive mutant), eto1 (ethylene overproducing mutant), jar1-1 (jasmonate insensitive mutant) and abi4-102 (abscisic acid insensitive mutant) donated by Clay were further studied for salt tolerance conferred by FZB42 by measuring the fresh weight of seedlings treated as described above.
qRT-PCR analysis
qRT-PCR analysis was performed for 35 DGEs using ACT2 (GenBank: AT3G18780) and L2 (GenBank: AT2G44065) as genes to normalize the results. The primers used in this study are shown in Supplementary Table S7. First-strand cDNA synthesis was performed using a PrimeScriptTM RT reagent Kit with gDNA Eraser (perfect Real Time), which could remove genomic DNA. qRT-PCR was performed with a Mx3000 P system (Applied Biosystems) using SYBR® Premix Ex Taq™ II (TliRNaseH Plus) (TaKaRa) according to the manufacturer’s instructions. qRT-PCR employed the following cycling conditions: 95 °C for 30 s, and 40 cycles at 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s, followed by extension. Each PCR analysis was repeated at least three times. The expression levels of genes were calculated using the threshold cycle2−ΔΔCt method65.
Statistical analysis
The data were analyzed by SAS 9.0 software. One-way ANOVA and Duncan multiple-range tests were performed to determine significant differences in four treatments.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China No. 31370447, the Chinese Academy of Sciences under the “One Hundred Talents” program No. Y127L41 and Progect Group of Ningxia Agricultural Comprehensive Development (PGNACD).
Author Contributions
S.F.L., R.Y.W. designed the experiments. S.F.L. performed the experiment. S.F.L. analysed the data and wrote the paper. R.Y.W., Y.W. revised the manuscript. Contributed reagents/materials/analysis tools: H.T.H., X.L., X.Z., Y.B.Z., Z.K.X.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11308-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Flowers TJ, Yeo AR. Breeding for salinity resistance in crop plants: where next? Funct Plant Biol. 1995;22:875–884. [Google Scholar]
- 2.Zhu JK. Plant salt tolerance. Trends Plant Science. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 3.Kim K, et al. Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol Cells. 2014;37:109–117. doi: 10.14348/molcells.2014.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meloni DA, Oliva MA, Martinez CA, Cambraia J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ Exp Bot. 2003;49:69–76. doi: 10.1016/S0098-8472(02)00058-8. [DOI] [Google Scholar]
- 5.Hu, S., Tao, H., Qian, Q. & Guo, L. Genetics and molecular breeding for salt-tolerance in rice. Rice Genomics and Genetics. 3 (2012).
- 6.Sergeeva E, Shah S, Glick BR. Tolerance of transgenic canola expressing a bacterial ACC deaminase gene to high concentrations of salt. World J Microbiol Biotechnol. 2006;22:277–282. doi: 10.1007/s11274-005-9032-1. [DOI] [Google Scholar]
- 7.Mayak S, Tirosh T, Glick BR. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Bioch. 2004;42:565–572. doi: 10.1016/j.plaphy.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Gamalero E, Berta G, Massa N, Glick BR, Lingua G. Interactions between Pseudomonas putida UW4 and Gigasporarosea BEG9 and their consequences for the growth of cucumber under salt-stress conditions. J Appl Microbiol. 2010;108:236–245. doi: 10.1111/j.1365-2672.2009.04414.x. [DOI] [PubMed] [Google Scholar]
- 9.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 10.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255:571–586. doi: 10.1023/A:1026037216893. [DOI] [Google Scholar]
- 11.Van Loon LC. Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol. 2007;119:243–254. doi: 10.1007/s10658-007-9165-1. [DOI] [Google Scholar]
- 12.Lucy M, Reed E, Glick BR. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek. 2004;86:1–25. doi: 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- 13.Ryu CM, et al. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idris EE, Iglesias DJ, Talon M, Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe In. 2007;20:619–626. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- 15.Bacilio M, Rodriguez H, Moreno M, Hernandez JP. Mitigation of salt stress in wheat seedling by a gfp-tagged Azospirillum lipoferum. Biol Fert Soils. 2004;40:188–193. doi: 10.1007/s00374-004-0757-z. [DOI] [Google Scholar]
- 16.Paul D, Lade H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: a review. Agrono Sustain Dev. 2014;34:737–752. doi: 10.1007/s13593-014-0233-6. [DOI] [Google Scholar]
- 17.Upadhyay SK, Singh JS, Saxena AK, Singh DP. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biology. 2012;14:605–611. doi: 10.1111/j.1438-8677.2011.00533.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, et al. Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol Plant Microbe In. 2008;21:737–744. doi: 10.1094/MPMI-21-6-0737. [DOI] [PubMed] [Google Scholar]
- 19.Fan B, et al. Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J Biotechnol. 2011;151:303–311. doi: 10.1016/j.jbiotec.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Koumoutsi A, et al. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186:1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen XH, Koumoutsi A, Scholz R, Borriss R. More than anticipated–production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J Mol Microb Biotech. 2008;16:14–24. doi: 10.1159/000142891. [DOI] [PubMed] [Google Scholar]
- 22.Ghaffari MR, et al. Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant mol biol. 2016;90:699–717. doi: 10.1007/s11103-016-0461-z. [DOI] [PubMed] [Google Scholar]
- 23.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Leng P, Zhang G, Li X. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J Plant Physiol. 2009;166:1241–1252. doi: 10.1016/j.jplph.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi N, et al. shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. The Plant J. 2005;42:13–22. doi: 10.1111/j.1365-313X.2005.02357.x. [DOI] [PubMed] [Google Scholar]
- 26.Turk EM, et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. The Plant J. 2005;42:23–34. doi: 10.1111/j.1365-313X.2005.02358.x. [DOI] [PubMed] [Google Scholar]
- 27.Yanhui C, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant mol biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell. 2006;18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koyama T, et al. A regulatory cascade involving class II Ethylene Response Factor transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 2013;162:991–1005. doi: 10.1104/pp.113.218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloepper JW, Ryu CM, Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004;94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Siddikee MA, Glick BR, Chauhan PS, Yim WJ, Sa T. Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol Bioch. 2011;49:427–434. doi: 10.1016/j.plaphy.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, et al. Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol Plantarum. 2016;158:34. doi: 10.1111/ppl.12441. [DOI] [PubMed] [Google Scholar]
- 34.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 35.Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19:765–8. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, et al. Beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 induces plant salt tolerance through spermidine production. Mol Plant Microbe In. 2017;30:423–432. doi: 10.1094/MPMI-02-17-0027-R. [DOI] [PubMed] [Google Scholar]
- 37.Nautiyal CS, et al. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol Bioch. 2013;66:1–9. doi: 10.1016/j.plaphy.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Geddie JL, Sutherland IW. Uptake of metals by bacterial polysaccharides. J Appl Biomater. 1993;74:467–472. [Google Scholar]
- 39.Ashraf M, Hasnain S, Berge O, Mahmood T. Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol Fert Soils. 2004;40:157–162. [Google Scholar]
- 40.Upadhyay SK, Singh JS, Singh DP. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere. 2011;21:214–222. doi: 10.1016/S1002-0160(11)60120-3. [DOI] [Google Scholar]
- 41.Xu Z, Shao J, Li B, Yan X, Shen Q. Contribution of bacillomycin D In Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl Environ Microb. 2013;79:808–815. doi: 10.1128/AEM.02645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashraf M, Hasnain S, Berge O. Effect of exo-polysaccharides producing bacterial inoculation on growth of roots of wheat (Triticum aestivum L.) plants grown in a salt-affected soil. Int J Environ Sci Technol. 2006;3:43–51. doi: 10.1007/BF03325906. [DOI] [Google Scholar]
- 43.Botella M, et al. Polyamine, ethylene and other physico-chemical parameters in tomato (Lycopersicon esculentum) fruits as affected by salinity. Physiol Plant. 2000;109:428–34. doi: 10.1034/j.1399-3054.2000.100409.x. [DOI] [Google Scholar]
- 44.Bal HB, Nayak L, Das S, Adhya TK. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil. 2013;366:93–105. doi: 10.1007/s11104-012-1402-5. [DOI] [Google Scholar]
- 45.Kausar, R., Shahzad, S. M., Arshad, M. & Anjum, M. A. Screening and evaluation of rhizobacteria containing ACC-deaminase for growth promotion of wheat (Triticum aestivum L.) under salinity stress. J Agric Sci. 47 (2009).
- 46.Wu Z, Yue H, Lu J, Li C. Characterization of rhizobacterial strain Rs-2 with ACC deaminase activity and its performance in promoting cotton growth under salinity stress. World J Microb Biot. 2012;28:2383–2393. doi: 10.1007/s11274-012-1047-9. [DOI] [PubMed] [Google Scholar]
- 47.Nadeem SM, Zahir ZA, Naveed M, Arshad M. Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can J Microbiol. 2007;53:1141–1149. doi: 10.1139/W07-081. [DOI] [PubMed] [Google Scholar]
- 48.Shen M, et al. Effect of plant growth-promoting rhizobacteria (PGPRs) on plant growth, yield, and quality of tomato (Lycopersicon esculentum Mill.) under simulated seawater irrigation. J Gen Appl Microbiol. 2012;58:253–262. doi: 10.2323/jgam.58.253. [DOI] [PubMed] [Google Scholar]
- 49.Ma JH, Yao JL, Cohen D, Morris B. Ethylene inhibitors enhance in vitro root formation from apple shoot cultures. Plant Cell Rep. 1998;17:211–214. doi: 10.1007/s002990050380. [DOI] [PubMed] [Google Scholar]
- 50.Chen, L., Dodd, I. C., Theobald, J. C., Belimov, A. A. & Davies, W. J. The rhizobacterium Variovorax paradoxus 5C-2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J Exp Bot. ert031 (2013). [DOI] [PMC free article] [PubMed]
- 51.Poupin, M. J., Greve, M., Carmona, V. & Pinedo, I. A complex molecular interplay of auxin and ethylene signaling pathways is involved in Arabidopsis growth promotion by Burkholderia phytofirmans PsJN. Front Plant Sci. 7 (2016). [DOI] [PMC free article] [PubMed]
- 52.Xu YI, et al. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. The Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong W, et al. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol. 2013;161:1217–1228. doi: 10.1104/pp.112.211854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, et al. Awheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014;164:1068–1076. doi: 10.1104/pp.113.227595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedranzani H, et al. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003;41:149–158. doi: 10.1023/A:1027311319940. [DOI] [Google Scholar]
- 56.Cho SM, et al. Identification and transcriptional analysis of priming genes in Arabidopsis thaliana induced by root colonization with Pseudomonas chlororaphis O6[J] Plant Pathol J. 2011;27:272–279. doi: 10.5423/PPJ.2011.27.3.272. [DOI] [Google Scholar]
- 57.Bari R, Jones JDG. Role of plant hormones in plant defenceresponses. Plant mol biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 58.Lindemose S, O’Shea C, Jensen MK, Skriver K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci. 2013;14:5842–5878. doi: 10.3390/ijms14035842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 60.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medeiros FHV, et al. Transcriptional profiling in cotton associated with Bacillus subtilis (UFLA285) induced biotic-stress tolerance. Plant Soil. 2011;347:327–337. doi: 10.1007/s11104-011-0852-5. [DOI] [Google Scholar]
- 62.Ramanjulu S, Bartels D. Drought and desication-induced modulation of gene expression in plants. Plant Cell Environ. 2002;25:141–151. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- 63.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.