Abstract
Objective
To define the performance characteristics of the LOCI® method for cardiac troponin I on the Dimension EXL system.
Designs and methods
Three different levels of commercial control (mean concentrations 0.426, 1.42, and 18.64 µg/L) were used for the imprecision study, quantifying separately within-run and between-run over 20 days. The limit of blank (LoB) and limit of detection (LoD) were assessed with 20 replicates of a sample without troponin I. Linearity was assessed by regression analysis. In addition, we studied inaccuracy, carry-over and limit of quantitation and conducted a method comparison with the Stratus CS (n=69). The reference interval was determined in 146 healthy blood donors using non-parametric method.
Results
The within-run imprecision (coefficient of variation [CV], %) obtained at each level was 2.4, 1.4% and 2.2%, while the between-run imprecision (CV,%) was 3.3%, 2.9% and 2.5%. Total imprecision was 4.06%, 3.3% and 3.4% for each control level. The limit of quantitation which corresponds to the troponin I concentration at which CV=10% was 0.05 µg/L. Method comparison with the Stratus CS assay produced the equation: Dimension EXL=−0.002698+1.0233⁎(Stratus CS) with a confidence interval from −0.01562 to 0.00626 for the intercept and (0.979 to 1.0875) for the slope. The 99th percentile obtained for the reference population was 0.047 µg/L.
Conclusions
The LOCI method for cardiac troponin I on the Dimension EXL meets all guidelines recommended criteria referring to limit of quantitation, imprecision and shows excellent transferability with the Stratus CS method.
Keywords: Cardiac troponin, LOCI, Dimension EXL
Highlights
-
•
We found an acceptable imprecision. The linearity and limit of detection meet the manufacturer׳s specifications.
-
•
The 99th percentile in the reference population was 0.047 µg/L. The 10% CV cut-off was 0.050 µg/L.
-
•
The correlation with Stratus CS method was excellent.
1. Introduction
Cardiac troponins I or T are the preferred and recommended biomarkers of coronary acute syndromes [1], [2], [3] due to their high myocardial tissue specificity and high clinical sensitivity. Detection of a rise and/or fall of the troponin measurements, coupled with clinical symptoms and/or electrocardiogram is necessary to diagnose acute myocardial infarction [4]. Blood samples for the measurement of troponin should be drawn for biomarker testing at hospital presentation followed by sampling at 6–9 h, although the new high sensitivity assays make possible the measurement on admission and 3–6 h later, shortening the turnaround time [2]. An increased cTn concentration is defined as a value exceeding the 99th percentile of a normal reference population, on condition that precision at this level is optimal [1]. An acceptable imprecision is described by coefficient of variation (CV) at 99th percentile ≤10% [5], although few assays meet this requirement. Assays with CV>20% at the 99th percentile should not be used [6]. The 99th percentile decision limit for Tn assays should be determined in each local laboratory by internal studies using the specific assay [5].
While antibodies against troponin T are produced by a single manufacturer, there are several manufacturers in the case of troponin I. In addition the cTnI molecule shows a great complexity due to phosphorylation and other post-translational modifications. So, cTnI contains two serine residues that can be phosphorylated in vivo by protein kinase A, which means that 4 forms of the protein can coexist in the cell (dephosphorylated, 2 monophosphorylated and biphosphorylated). Phosphorylation changes the conformation of troponin I and modifies its interaction with anti-TnI antibodies [7]. Other post-translational modifications include oxidation, reduction or partial digestion by proteases [8]. Monoclonal antibodies specific to several epitopes of cTnI are able to recognize all known modifications circulating in the blood. The diversity of epitopes and manufacturers makes it easy to understand the lack of standardization of cTnI assays [9].
The existence of different isoforms of troponin, the different specificity of the immunoassay antibodies, the lack of standardization of the cardiac troponin I assays and the importance of accurate measurement of troponin levels [5] make it necessary for each laboratory to establish the metrological characteristics of their assay and the 99th percentile of their reference population [5], [10].
The aim of this study was to assess the analytical performance characteristics of the luminescent oxygen channeling immunoassay (LOCI™) method for cTnI on the Dimension EXL. In addition we determined the 99th percentile of the upper reference limit in a healthy population and the 10% coefficient of variation cut-off. Finally we performed a method comparison with the Stratus CS, which is used as a backup method in our laboratory.
2. Materials and methods
The method evaluated for cTnI measurement was LOCI™. LOCI technology utilizes paired synthetic beads in a homogeneous immunoassay method. The first bead (Chemibead) is coated with an analyte-specific antibody and contains a chemiluminescent dye. The second bead (Sensibead) is coated with the protein streptavidin and contains a photosensitive dye. In addition to the beads an analyte-specific biotinylated antibody reagent is used. The antigen of interest binds the biotinylated antibody and the antibody bound to the surface of the Chemibead. The two beads form a complex based on the analyte-specific antibody binding and the strong affinity of streptavidin for the biotinylated antibody. When this bead complex is exposed to light at 680 nm, the photosensitive dye bound to the Sensibead releases singlet oxygen. This singlet oxygen rapidly diffuses to and is absorbed by the Chemibead, causing a chemiluminescent reaction which is measured by spectrophotometry. The performance characteristics of this method were previously published for the Dimension Vista analyser [11]. Our study was performed on Dimension EXL analyzer (Siemens Healthcare Diagnostics, Erlangen, Germany).
2.1. Imprecision
Imprecision was determined according to the Spanish Clinical Chemistry Society (SEQC, Sociedad Española de Química Clínica) guideline [12]. Briefly, three levels of commercial quality controls (Liquicheck Cardiac Markers Plus, Bio-Rad Laboratories, Hercules, CA, USA) with different concentrations of cTnI (0.475, 1.57 and 18.8 µg/L) were tested 20 times during a day in the same analytical run for calculating the within-run imprecision. In addition 20 aliquots per level were frozen for the between-run imprecision determination. One aliquot per level was thawed and analyzed over twenty days. Within-run and between-run imprecision were expressed as coefficient of variation (%CV). Total imprecision was calculated as the square root of the sum of the squares of the within- and between-run CVs.
2.2. Linearity
Determination of linearity was carried out as described in the CLSI guideline EP6-A [13]. Two samples, one a serum pool with increased cTnI (42.16 µg/L) and the other diluent matrix without cTnI (CTNI ADIL, Siemens Healthcare Diagnostics, Erlangen, Germany), were used to prepare seven intermediate dilutions of each specimen. A triplicate analysis in each mix was performed and the linearity was verified using regression analysis.
2.3. Limit of detection
Determination of the limits of blank (LoB) and detection (LoD) was carried out as described in the CLSI guideline EP17-A [14]. In accordance with this guideline, we measured the limit of blank defined as the highest value we expect to see in a series of results in a sample that contains no analyte, measuring the mean and standard deviation of commercial sample diluent (n=20). The limit of detection (LoD) was calculated according to the expression: LoD=LoB+1.645 SD, where SD is the standard deviation of samples at the LoD.
2.4. Limit of quantitation
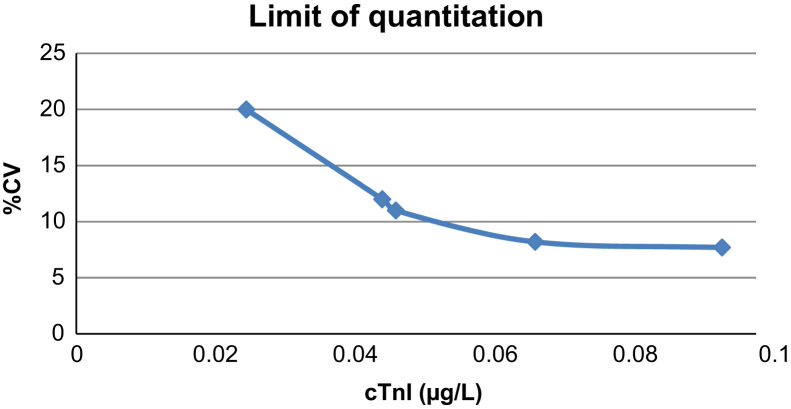
The limit of quantitation was determined for cTnI using five serum pools with mean values of 0.047, 0.0250, 0.045, 0.0675 and 0.095 µg/L. Samples were run in duplicate, once each day, for a total of 20 days. An Analysis of Variance (ANOVA) was performed to determinate the within-laboratory imprecision expressed as %CV. The mean concentration (X) for each of the serum pools was then plotted against the corresponding %CV (Y). The limit of quantitation at imprecision (CV) of 10% was found by interpolation.
2.5. Carry-over assessment
For carry-over evaluation we followed the CLSI protocol [15], using a pool with high cTnI concentrations (A) (mean concentration of 8.56 µg/L) and other one with concentration close to limit of detection (B) (0.05 µg/L). The high pool was first analyzed in duplicate followed by analysis of the low pool in triplicate, obtaining the sequence: A1, A2, B1, B2, B3, which was repeated 10 times during the same day. The mean of 10 replicate measurements of the low concentration pool was compared with the results obtained after analyzing the high pool using the appropriate test. Previously we carried out the Kolmogorov–Smirnov test to assess the normality of distribution. In addition the Broughton [16] carryover constant was found using the following equation:
where Aim, Bim are the means of the results obtained for the determinations carried out in the order i (i=1, 2, 3).
2.6. Upper reference limit (URL)
The 99th percentile was determined according to the Laboratory Medicine Practice Guidelines of the NACB and IFCC Committee for Standardization of Markers of Cardiac Damage [5]. Briefly, 146 healthy volunteers were selected from the healthy donors from our local blood bank. Donors reporting consumption of drugs of abuse, smokers and alcohol drinkers were excluded. The individuals included had no history of disease and no prescription medications documented. Blood samples were collected into lithium heparin tubes and plasma samples were obtained by centrifugation at 1700g for five minutes. The samples were processed on the Dimension® EXL™ on different days.
2.7. Method comparison
We carried out this comparison as described in the CLSI guideline EP9-A2 [17] using the Stratus CS cTnI assay (Siemens Healthcare Diagnostics, Erlangen, Germany) as the comparison procedure. We used plasma from 69 patients with cTnI concentrations distributed over the whole analytical range (0.017–40 µg/L). Seven samples were undetectable with the EXL method (<0.017 µg/L), and 6 of them gave low values (0.017–0.05 µg/L), 36 had values moderately higher than cut-off (0.05–0.5 µg/L), 9 had high values (0.5–2 µg/L) and 9 had extremely high values (>2 µg/L). Samples were analyzed consecutively on the Dimension EXL and Stratus CS over 10 days. To assess the correlation between the methods we used the Passing-Bablok regression procedure [18]. In addition we performed a Bland–Altman plot [19] to assess the agreement between two assays. Med Calc 11.3 Statistical Package (MedCalc Software, Ostend, Belgium) and Excel 2007+Analyse.it (Analyse-It Software, Leeds, UK) were used for the calculations and graphs.
3. Results
3.1. Imprecision
The results for within-run, between-run and total imprecision at the three levels of cTnI tested are shown in Table 1.
Table 1.
Imprecision of cTnI LOCI method on the Dimension EXL. SD=standard deviation, CV=coefficient of variation.
| Pool 1 | Pool 2 | Pool 3 | ||
|---|---|---|---|---|
| Within-run | Mean (µg/L) | 0.4264 | 1.419 | 18.639 |
| SD (µg/L) | 0.01 | 0.025 | 0.419 | |
| CV% | 2.39 | 1.64 | 2.251 | |
| Between-run | Mean (µg/L) | 0.409 | 1.377 | 17.969 |
| SD (µg/L) | 0.0134 | 0.04 | 0.451 | |
| CV% | 3.277 | 2.9 | 2.513 | |
| Total imprecision | CV% | 4.057 | 3.337 | 3.373 |
The total imprecision obtained at the low, medium and high levels was 4.06%, 3.3% and 3.4%, respectively, which indicate that the assay shows acceptable imprecision.
3.2. Linearity
The least-square regression after analysis of the successive dilutions of the samples with high (42.16 µg/L) and low (diluent) values of cTnI yielded the following regression equation: Y=−0.156+1.02X, where Y is the theoretical concentration and X the mean concentration for each dilution. The 95% confidence interval (CI) for the intercept (−0.155–1.24) includes zero and the CI for the slope (0.964–1.076) includes unity.
3.3. Limit of blank and limit of detection
The mean measured concentration after 25 replicate measurements of analyte-free diluent was 0.0056 µg/L. The limit of blank, defined as the upper limit in a series of results in samples without analyte, [14] was 0.009 µg/L. The limit of detection, according to EP17-A, was calculated as 0.0122 µg/L.
3.4. Limit of quantitation
The precision profile for cTnI is shown in Fig. 1. The 10% CV cut-off was determined to be 0.050 µg/L
Fig. 1.
Limit of quantitation (precision profile) of cTnI on the Dimension EXL. Testing was performed in duplicate over 20 non-consecutive test days. The coefficient of variation (CV,%) for each of the serum pools was then plotted against the mean cTnI concentration.
3.5. Upper reference limit (URL)
The 99th percentile for 146 healthy individuals, calculated using a non-parametric method was 0.047 µg/L. Use of the method proposed by Harris and Boyd [20] permitted us to conclude that there were no significant differences between men and women.
3.6. Carry-over assessment
The Gaussian distribution of results for the B1 and B3 series was confirmed by the Kolmogorov–Smirnov test. In consequence a t-test was applied for mean comparison, and showed no significant differences between the two sequences (p=0.699). Significant carry-over was thus excluded. In addition we calculated the Broughton׳s constant (k) as described above and found k=0.039%. The difference of means carryover (B1–B3) was 0.00325 μg/L.
3.7. Method comparison
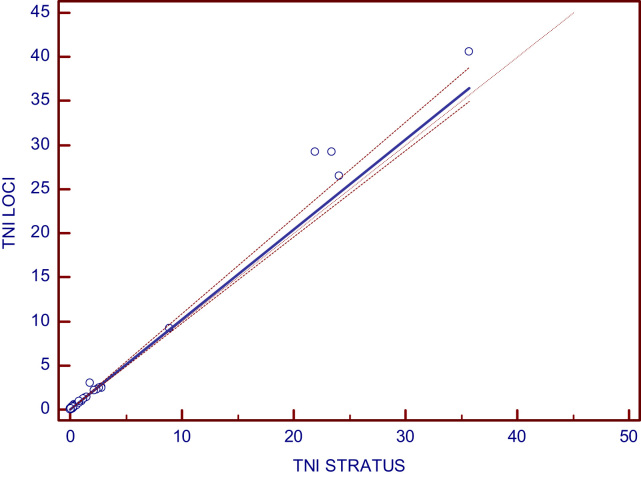
Method comparison testing for cTnI included a total of 69 plasma specimens covering the range of 0.017–40.57 µg/L. The non parametric Passing and Bablok regression yielded the following equation Dimension EXL=−0.0027+1.023x (Stratus CS) and the Pearson correlation coefficient was r=0.997. The confidence intervals for intercept, slope and r are shown in Table 2.
Table 2.
Results of Passing-Bablok regression for the comparison between the Dimension EXL (y) and the Stratus CS cTnI (x) methods. The 95% confidence intervals are shown in parentheses.
| Regression equation | y=−0.0027+1.023x |
|---|---|
| Intercept (95% CI) | −0.0027 (−0.01562 to 0.00626) |
| Slope (95% CI) | 1.023 (0.979–1.0875) |
| r(95% CI) | 0.997 (0.9948–0.9980) |
The 95% confidence intervals encompass zero for the intercept and 1 for the slope, so we can conclude that both methods produce interchangeable results.
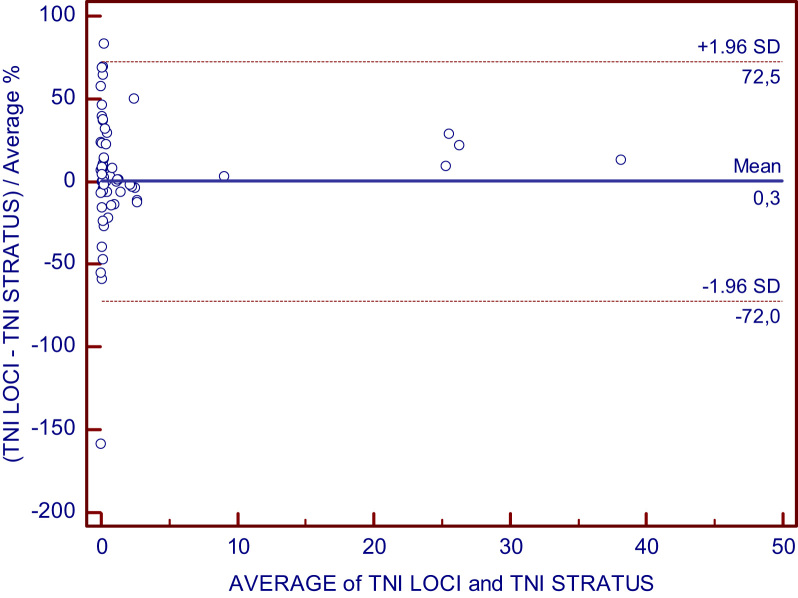
The Passing-Bablok regression graph and Bland–Altman plot are shown in Fig. 2, Fig. 3.
Fig. 2.
Passing-Bablok linear regression for the comparison between Dimension EXL and Stratus CS cTnI methods.
Fig. 3.
Bland–Altman plot for the comparison between Dimension EXL and Stratus CS for cTnI assay.
4. Discussion
Measurement of cardiac troponin is the cornerstone of diagnosis of acute coronary syndrome, which make essential to assess the fundamental characteristics of the assays used – total imprecision, 99th percentile of the reference population and functional sensitivity. In addition, lack of harmonization between different assays for cTnI requires comparisons between different methods, especially when they are used in the same institution [9].
We found that the analytical performance characteristics of LOCI™ method for cTnI on the Dimension EXL are acceptable. The imprecision is similar to the manufacturer׳s claims and the method is linear across a wide analytical range (0.017–40 µg/L). The limit of detection calculated according to the EP-17-A guideline [14] was 0.012 µg/L.
The 99th percentile in our reference population was 0.047 µg/L, lower than the cut-off claimed by the manufacturer, which highlights the need to determine cut-offs for each population [3], [5]. The limit of quantification or functional sensitivity was 0.05 µg/L. The limit of quantitation is of particular significance in the diagnosis of acute coronary syndrome. We obtained the same limit of quantification as declared by the manufacturer.
Sample carryover was not statistically significant according to t-test (p=0.699). The Broughton׳s constant (k) indicates that there is no significant contamination between samples, given that values less than 1% are permitted [16]. In addition we can conclude that there is no clinically significant carryover, because the difference of mean values between the samples most and least affected (B1–B3) was 0.00325 ug/L, which is less than the imprecision at low concentrations.
A method comparison with cTnI measurement on the Stratus CS was essential for us because this method is our laboratory׳s alternative method. The correlation was adequate (r=0.997), without proportional or constant errors as indicated by the confidence intervals for the intercept and slope.
Previous studies of Arrebola et al. [11] and Kelley et al. [21] have assessed the analytical performance for LOCI cardiac troponin I on the Dimension Vista™, but we have not found studies published for the LOCI method on the Dimension EXL™.
In conclusion the LOCI method for cardiac troponin I on the Dimension EXL meets all guidelines and recommended criteria for limit of quantitation and imprecision, and the transferability with the Stratus CS method is excellent.
Conflict of interest
The authors declare that they do not have any conflict of interest with the company who provided reagents and instrumentation.
Acknowledgments
The authors thank Siemens Healthcare Diagnostics for the gift of one reagent kit for the purposed of the study.
References
- 1.Thygesen K., Alpert J.S., White H.D. Universal definition of myocardial infarction. Circulation. 2007;116:2634–2653. doi: 10.1161/CIRCULATIONAHA.107.187397. [DOI] [PubMed] [Google Scholar]
- 2.Thygesen K., Alpert J.S., Jaffe A.S., White H.D., Simoons M.L., Chaitman B.R. Third universal definition of myocardial infarction. J Am Coll Cardiol. 2012;60:1581–1598. doi: 10.1016/j.jacc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Wu A., Apple F., Gibler B., Jesse R., Warshaw M., Valdés R. National academy of clinical biochemistry standards of laboratory practice: recommendations for the use of cardiac markers in coronary artery diseases. Clin Chem. 1999;45:1104–1121. [PubMed] [Google Scholar]
- 4.Jaffe A.S. Chasing troponin: how long can you go if you can see the rise? J Am Coll Cardiol. 2006;48:1763–1764. doi: 10.1016/j.jacc.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Apple F., Jesse R., Newby L.K., Wu A.H., Christenson R.H. National academy of clinical biochemistry and IFCC committee for standardization of markers of cardiac damage laboratory medicine practice guidelines: analytical issues for biochemical markers of acute coronary syndromes. Clin Chem. 2007;53:547–551. doi: 10.1373/clinchem.2006.084715. [DOI] [PubMed] [Google Scholar]
- 6.Thygesen K., Mair J., Katus H., Plebani M., Venge P., Collinson P. Study group on biomarkers in cardiology of the ESC working group on acute cardiac care. recommendations of the use of cardiac troponin measurement in acute cardiac care. Eur Heart J. 2010;31:2197–2204. doi: 10.1093/eurheartj/ehq251. [DOI] [PubMed] [Google Scholar]
- 7.Apple F.S., Collinson P.O. Analytical characteristics of high-sensitivity cardiac troponin assays. Clin Chem. 2012;58:54–61. doi: 10.1373/clinchem.2011.165795. [DOI] [PubMed] [Google Scholar]
- 8.Adams J.E., Bodor G.S., Davila-Roman V.G., Delmez J.A., Apple F.S., Landenson J.H. Cardiac troponin I: a marker with high specificity for cardiac injury. Circulation. 1993;88:101–106. doi: 10.1161/01.cir.88.1.101. [DOI] [PubMed] [Google Scholar]
- 9.Tate J.R., Bunk D.M., Christenson R.H., Katrukha A., Noble J.E., Porter R.A. Standardization of cardiac troponin I measurement: past and present. Pathology. 2010;42:402–408. doi: 10.3109/00313025.2010.495246. [DOI] [PubMed] [Google Scholar]
- 10.Morrow D.A., Cannon C.P., Jesse R.L., Newby L.K., Ravkidie J., Storrow A.B. National academy of clinical biochemistry laboratory medicine practice guidelines: clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Circulation. 2007;115:365–375. doi: 10.1161/CIRCULATIONAHA.107.182882. [DOI] [PubMed] [Google Scholar]
- 11.Arrebola M.M., Lillo J.A., Diez De Los Rios M.J., Rodríguez M., Dayaldasani A., Yahyaoui Pérez V. Analytical performance of a sensitive assay for cardiac troponin I with LOCI™ technology. Clin Biochem. 2010;43:998–1002. doi: 10.1016/j.clinbiochem.2010.04.073. [DOI] [PubMed] [Google Scholar]
- 12.Canalias Reverter F. Recomendaciones para el estudio de la precisión de los procedimientos de medida en el laboratorio clínico. Sociedad Española de Bioquímica Clínica y Patología Molecular. Comité Científico. Quím. Clín. 2003;22:63–65. [Google Scholar]
- 13.Clinical and Laboratory Standard Institute . NCCLS; Wayne PA: 2003. Evaluation of the linearity of quantitative measurement procedures: a statistical approach: approved guideline. NCCLS guideline EP6-A. [April] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute . NCCLS; Wayne PA: 2003. Protocols for determination of limits of detection and limits of quantitation: approved guideline. NCCLS guideline EP-17-A. [April] [Google Scholar]
- 15.National Commitee for Clinical Laboratory Standards . NCCLS; Wayne PA: 2002. Preliminary evaluation of quantitative clinical laboratory methods. EP10-A2; p. 5. [Google Scholar]
- 16.Broughton P., Gowenlock A.H., McCormack J.J., Neil D. A revised scheme for the evaluation of automatic instruments for use in clinical chemistry. Ann Clin Biochem. 1974;11:207–212. doi: 10.1177/000456327401100164. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute . NCCLS; Wayne PA: 2002. Method comparison and bias estimation using patient samples: approved guideline. NCCLS guideline EP9-A2. [September] [Google Scholar]
- 18.Passing H., Bablok W. A new biometrical procedure for testing the equality of measurements from two different analytical method comparison studies in clinical chemistry. J Clin Chem Clin Biochem. 1983;21:709–720. doi: 10.1515/cclm.1983.21.11.709. [DOI] [PubMed] [Google Scholar]
- 19.Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327:307–310. [PubMed] [Google Scholar]
- 20.Harris E., Boyd J. On dividing reference data into subgroups to produce separate reference ranges. Clin Chem. 1990;36:265–270. [PubMed] [Google Scholar]
- 21.Kelley W.E., Lockwood M., Cervelli D.R., Sterner J., Scott M.G., Duh S.H. Cardiovascular disease testing on the Dimension Vista® system.: Biomarkers of acute coronary syndromes. Clin Biochem. 2009;42:1444–1451. doi: 10.1016/j.clinbiochem.2009.05.020. [DOI] [PubMed] [Google Scholar]