Abstract
Porcine stool-associated RNA virus (posavirus), and Human stool-associated RNA virus (husavirus) are viruses in the order Picornavirales recently described in porcine and human fecal samples. The tentative group (Posa and Posa-like viruses: PPLVs) also includes fish stool-associated RNA virus (fisavirus) as well as members detected in insects (Drosophila subobscura and Anopheles sinensis) and parasites (Ascaris suum). As part of an agnostic deep sequencing survey of animal and human viruses in Vietnam, we detected three husaviruses in human fecal samples, two of which share 97–98% amino acid identity to Dutch husavirus strains and one highly divergent husavirus with only 25% amino acid identity to known husaviruses. In addition, the current study found forty-seven complete posavirus genomes from pigs, ten novel rat stool-associated RNA virus genomes (tentatively named rasavirus), and sixteen novel bat stool-associated RNA virus genomes (tentatively named basavirus). The five expected Picornavirales protein domains (helicase, 3C-protease, RNA-dependent RNA polymerase, and two Picornavirus capsid domain) were found to be encoded by all PPLV genomes. In addition, a nucleotide composition analysis revealed that the PPLVs shared compositional properties with arthropod viruses and predicted non-mammalian hosts for all PPLV lineages. The study adds seventy-six genomes to the twenty-nine PPLV genomes currently available and greatly extends our sequence knowledge of this group of viruses within the Picornavirales order.
Keywords: virus discovery, next generation sequencing, husavirus, posavirus, rasavirus, basavirus, Picornavirales
1. Introduction
The order Picornavirales includes a wide range of viruses that infect a variety of hosts. According to the latest International Committee on Taxonomy of Viruses (ICTV) classification (ICTV 2017), the order comprises five families: the families Dicistroviridae and Iflaviridae contain members which infect insects (e.g. cripavirus and deformed wing virus), the family Secoviridae members which infect plants (e.g. turnip ringspot virus), the family Picornaviridae members infecting vertebrates (e.g. enteroviruses) and the family Marnaviridae. The latter contains only Heterosigma akashiwo virus for which algae is the natural host (Le Gall et al. 2008).
Although members of the Picornavirales are highly diverse, they share a number of common features, including a single stranded positive-sense RNA genome and co-linear genes encoding a helicase, protease, and RNA-dependent RNA polymerase (RdRP) replication block (Le Gall et al. 2008). The genome lengths for Picornavirales range from 7.2 to 9.8 kb. Typically, the encoded polyprotein is cleaved by virus-encoded proteases (Blom et al. 1996). Generally, members of the Picornavirales are monopartite, although some members of the Secoviridae have genomes with two segments (Le Gall et al. 2008).
Increasing improvements in next-generation sequencing (NGS) has identified a number of divergent members of the order Picornavirales. Porcine stool-associated RNA viruses (posaviruses) were found in the feces of healthy pigs and water collected from swine farms (Shan et al. 2011, Hause et al. 2015, 2016), fish stool-associated RNA virus (fisavirus) was identified in the intestinal content of a healthy carp (Reuter et al. 2015), and human stool-associated RNA virus (husavirus) was identified in the feces of predominantly healthy humans (Oude Munnink et al. 2015). Although structually closely related (based on the genome organisation), these viruses display broad genetic diversity with often less than 40% amino acid identity in specific coding regions thereby suggesting a deep evolutionary history of the virus family.
Although posaviruses can be detected at high frequency in pig fecal samples (21%), a recent study using immunoprecipitation coupled with PCR detection assay showed that posavirus antibodies were infrequently detected (Hause et al. 2016). The possibility that posaviruses may not infect pigs but rather infect gut commensal organisms or have a dietary or environmental origin is supported by blast analysis of posavirus sequences that showed that some posavirus strains have greatest sequence similarity to an RNA sequence from the parasite Ascaris suum (Shan et al. 2011; Wang et al. 2011). Furthermore, a mRNA sequence from the mosquito Anopheles sinensis and a virus recently identified in the fruit fly Drosophila subobscura have been described showing some sequence identity to posaviruses (Webster et al. 2016). Although the viruses have been identifed in samples from different hosts, the true infection hosts for fisavirus, posavirus, and husavirus remain to be determined.
As part of a study to define patterns of viral zoonosis in Vietnam (Rabaa et al. 2015), we performed detailed agnostic (random-primed) whole-genome deep sequencing (Cotten et al. 2014) on fecal samples from bats, humans, pigs, and rats and rectal swabs from humans and pigs. We have analyzed these sequence data for the presence of PPLVs and we describe here a large set of novel virus genomes from human, rat, pig, and bat samples that share homology and protein domain architecture with the previous described posaviruses.
2. Results
For simplicity, we will use the term Posa and Posa-like viruses (PPLVs) throughout the manuscript. The PPLV category comprises virus and virus sequences that show >30% amino acid homology to the existing posavirus and husavirus genome sequences, do not cluster within the five established Picornavirales families and show a Picornavirales genome organization with the expected five Picornavirales protein domains (see below for further details). A search for PPLV genomes in sequences was performed as follows: short read data (3–4 million 250 nt paired end reads per sample) were de novo assembled into longer sequence contigs and a protein sequence based USEARCH analysis (Edgar 2010) was performed against a database containing all Picornavirales protein entries in GenBank, including all known posaviruses sequences. This search identified three husaviruses, forty-seven posaviruses, ten novel rasaviruses, and sixteen novel basaviruses genomes. The genome lengths of the newly identified PPLV genome sequences varied from 8,262 to 11,318 nucleotides and for all viruses the read coverage across the genome and G + C content was determined. The results of these analyses and the available demographical data for these samples are summarized in Table 1.
Table 1.
Overview of the PPLVs identified in this study.
| Illumina ID | Accession number | ENA number lane 1 | ENA number lane 2 | Lineage | Sample source | Sample type | Source age | Date of sample | Genome length | G + C content (%) | Median depth of coveragea |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Basavirus_16715_47 | KX673228 | ERR1301485 | ERR1301574 | Bv_7 | Scotophilus kuhlii | Fecal | Unknown | 10 Jun 2014 | 9,591 | 36.7 | 116 |
| Basavirus_16715_47_2 | KX673229 | ERR1301485 | ERR1301574 | Bv_3 | Scotophilus kuhlii | Fecal | Unknown | 17 Jun 2014 | 9,585 | 32.1 | 42 |
| Basavirus_16715_5 | KX673230 | ERR1301446 | ERR1301535 | Bv_6 | Scotophilus kuhlii | Fecal | Unknown | 12 Nov 2014 | 8,776 | 40.3 | 13 |
| Basavirus_16715_61 | KX673234 | ERR1301499 | ERR1301588 | Bv_7 | Scotophilus kuhlii | Fecal | Unknown | 11 Jun 2014 | 9,566 | 36.6 | 129 |
| Basavirus_16715_61_2 | KX673235 | ERR1301499 | ERR1301588 | Bv_6 | Scotophilus kuhlii | Fecal | Unknown | 11 Jun 2014 | 9,065 | 40.3 | 208 |
| Basavirus_16715_69 | KX673237 | ERR1301507 | ERR1301596 | Bv_3 | Scotophilus kuhlii | Fecal | Unknown | 11 Jun 2014 | 9,528 | 32.2 | 21 |
| Basavirus_16715_71 | KX673238 | ERR1301509 | ERR1301598 | Bv_7 | Scotophilus kuhlii | Fecal | Unknown | 11 Jun 2014 | 9,585 | 36.9 | 132 |
| Basavirus_16715_71_2 | KX673239 | ERR1301509 | ERR1301598 | Bv_3 | Scotophilus kuhlii | Fecal | Unknown | 11 Jun 2014 | 9,483 | 32.1 | 29 |
| Basavirus_16715_77 | KX673240 | ERR1301514 | ERR1301603 | Bv_1 | Scotophilus kuhlii | Fecal | Unknown | 10 Jun 2014 | 8,829 | 38.6 | 28 |
| Basavirus_16715_84 | KX673241 | ERR1301519 | ERR1301608 | Bv_7 | Scotophilus kuhlii | Fecal | Unknown | 17 Jun 2014 | 9,391 | 36.9 | 48 |
| Basavirus_16715_86 | KX673242 | ERR1301521 | ERR1301610 | Bv_3 | Scotophilus kuhlii | Fecal | Unknown | 17 Jun 2014 | 9,530 | 32.2 | 94 |
| Basavirus_16845_64 | KX673243 | ERR1301829 | ERR1301914 | Bv_3 | Scotophilus kuhlii | Fecal | Unknown | 18 Sep 2014 | 9,537 | 32.2 | 160 |
| Basavirus_16845_79 | KX673244 | ERR1301842 | ERR1301927 | Bv_2 | Scotophilus kuhlii | Fecal | Unknown | 18 Sep 2014 | 9,147 | 35.9 | 55 |
| Basavirus_16845_94 | KX673245 | ERR1301855 | ERR1301940 | Bv_4 | Scotophilus kuhlii | Fecal | Unknown | 17 Sep 2014 | 8,262 | 48.2 | 27 |
| Basavirus_17819_2 | KX673289 | ERR1302928 | ERR1303004 | Bv_5 | Scotophilus kuhlii | Fecal | Unknown | 12 Nov 2014 | 9,271 | 46.9 | 171 |
| Basavirus_17819_8 | KX673290 | ERR1302934 | ERR1303010 | Bv_5 | Scotophilus kuhlii | Fecal | Unknown | 12 Nov 2014 | 9,240 | 47.1 | 274 |
| Husavirus_16370_59 | KX673221 | ERR1301365 | ERR1301415 | Hv_1 | Homo sapiens | Fecal | 7 years | 2 Apr 2013 | 8,856 | 52.8 | 10 |
| Husavirus_16915_89 | KX673248 | ERR1302009 | ERR1302077 | Hv_2 | Homo sapiens | Rectal swab | 51 years | 24 Mar 2013 | 8,576 | 50.8 | 17 |
| Husavirus_19344_29 | KX673274 | ERS1725523 | ERS1725523 | Hv_1 | Homo sapiens | Fecal | 59 years | 2 Oct 2014 | 9,027 | 53 | 97 |
| Posavirus_12087_40 | KX673215 | ERR473400 | NA | Pv_6 | Sus domesticus | Fecal | 1 month | 12 Mar 2012 | 11,036 | 43.8 | 32 |
| Posavirus_12087_42 | KX673216 | ERR473402 | NA | Pv_6 | Sus domesticus | Fecal | 1.5 months | 14 Mar 2012 | 11,318 | 44.1 | 24 |
| Posavirus_12144_61 | KX673217 | ERR477293 | NA | Pv_1 | Sus domesticus | Fecal | 20 months | 10 Apr 2012 | 9,729 | 35.8 | 135 |
| Posavirus_14226_38 | KX673218 | ERR775480 | NA | Pv_3 | Sus domesticus | Fecal | 15 months | 6 Mar 2012 | 9,190 | 46.5 | 34 |
| Posavirus_14226_39 | KX673219 | ERR775481 | NA | Pv_3 | Sus domesticus | Fecal | 15 months | 6 Mar 2012 | 9,071 | 46.4 | 25 |
| Posavirus_14250_11 | KX673220 | ERR779984 | NA | Pv_5 | Sus domesticus | Fecal | 2 months | 21 Mar 2012 | 9,680 | 33.2 | 55 |
| Posavirus_16915_3 | KX673246 | ERR1301944 | ERR1302014 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 19 Mar 2013 | 9,296 | 46.8 | 45 |
| Posavirus_16915_5 | KX673247 | ERR1301946 | ERR1302016 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 19 Mar 2013 | 9,204 | 46.7 | 51 |
| Posavirus_17489_2 | KX673249 | ERR1302404 | ERR1302484 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 27 Mar 2013 | 9,835 | 36.5 | 127 |
| Posavirus_17489_26 | KX673250 | ERR1302422 | ERR1302502 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,827 | 36.4 | 111 |
| Posavirus_17489_27 | KX673251 | ERR1302423 | ERR1302503 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,773 | 36.5 | 16 |
| Posavirus_17489_28 | KX673252 | ERR1302424 | ERR1302504 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,824 | 36.4 | 65 |
| Posavirus_17489_30 | KX673253 | ERR1302426 | ERR1302506 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,672 | 36.4 | 193 |
| Posavirus_17489_34 | KX673254 | ERR1302428 | ERR1302508 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 8,970 | 31.5 | 2184 |
| Posavirus_17489_35 | KX673255 | ERR1302429 | ERR1302509 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,733 | 33.5 | 146 |
| Posavirus_17489_36 | KX673256 | ERR1302430 | ERR1302510 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,729 | 33.5 | 842 |
| Posavirus_17489_39 | KX673257 | ERS1725815 | ERS1725815 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,640 | 33.5 | 20 |
| Posavirus_17489_4 | KX673258 | ERR1302405 | ERR1302485 | Pv_4 | Sus domesticus | Rectal swab | 5 months | 27 Mar 2013 | 8,970 | 31.5 | 29 |
| Posavirus_17489_4_2 | KX673259 | ERR1302405 | ERR1302485 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 27 Mar 2013 | 9,647 | 36.4 | 5 |
| Posavirus_17489_40 | KX673260 | ERR1302433 | ERR1302513 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 3 Apr 2013 | 9,650 | 33.5 | 70 |
| Posavirus_17489_45 | KX673261 | ERR1302438 | ERR1302518 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 4 Apr 2013 | 9,710 | 33.2 | 18 |
| Posavirus_17489_47 | KX673262 | ERR1301975 | ERR1302045 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 4 Apr 2013 | 9,783 | 33.2 | 24 |
| Posavirus_17489_5 | KX673263 | ERR1302406 | ERR1302486 | Pv_4 | Sus domesticus | Rectal swab | 5 months | 27 Mar 2013 | 9,222 | 31.2 | 101 |
| Posavirus_17489_50 | KX673264 | ERR1302440 | ERR1302520 | Pv_6 | Sus domesticus | Rectal swab | 5 months | 4 Apr 2013 | 11,094 | 44.6 | 45 |
| Posavirus_17489_51 | KX673265 | ERR1302441 | ERR1302521 | Pv_4 | Sus domesticus | Rectal swab | 2 years | 4 Apr 2013 | 9,286 | 31.4 | 309 |
| Posavirus_17489_60 | KX673266 | ERR1302450 | ERR1302530 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 4 Apr 2013 | 9,859 | 33.4 | 39 |
| Posavirus_17489_7 | KX673267 | ERR1302407 | ERR1302487 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 27 Mar 2013 | 9,434 | 36.3 | 12 |
| Posavirus_17489_86 | KX673268 | ERR1302473 | ERR1302553 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 9,283 | 46.7 | 237 |
| Posavirus_17489_87 | KX673269 | ERR1302474 | ERR1302554 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 9,242 | 46.7 | 14 |
| Posavirus_17489_90 | KX673270 | ERR1302477 | ERR1302557 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 8,469 | 46.6 | 14 |
| Posavirus_17489_91 | KX673271 | ERR1302478 | ERR1302558 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 9,272 | 46.9 | 66 |
| Posavirus_17489_95 | KX673272 | ERR1302481 | ERR1302561 | Pv_4 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 9,216 | 30 | 153 |
| Posavirus_17489_95_2 | KX673273 | ERR1302481 | ERR1302561 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 18 Apr 2013 | 9,570 | 32.8 | 47 |
| Posavirus_17668_10 | KX673276 | ERR1302758 | ERR1302847 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,309 | 46.6 | 154 |
| Posavirus_17668_11 | KX673277 | ERR1302759 | ERR1302848 | Pv_1 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,855 | 36.5 | 200 |
| Posavirus_17668_11_2 | KX673278 | ERR1302759 | ERR1302848 | Pv_4 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,066 | 31.2 | 18 |
| Posavirus_17668_12 | KX673279 | ERR1302760 | ERR1302849 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,196 | 46 | 38 |
| Posavirus_17668_13 | KX673280 | ERR1302761 | ERR1302850 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,666 | 33.4 | 31 |
| Posavirus_17668_13_2 | KX673281 | ERR1302761 | ERR1302850 | Pv_3 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,111 | 46.1 | 52 |
| Posavirus_17668_33 | KX673282 | ERR1302780 | ERR1302869 | Pv_6 | Sus domesticus | Rectal swab | 6 months | 16 Oct 2013 | 11,171 | 43.5 | 78 |
| Posavirus_17668_33_2 | KX673283 | ERR1302780 | ERR1302869 | Pv_3 | Sus domesticus | Rectal swab | 6 months | 16 Oct 2013 | 9,289 | 46.3 | 26 |
| Posavirus_17668_33_3 | KX673284 | ERR1302780 | ERR1302869 | Pv_9 | Sus domesticus | Rectal swab | 6 months | 16 Oct 2013 | 9,498 | 47.1 | 77 |
| Posavirus_17668_4 | KX673275 | ERR1302752 | ERR1302841 | Pv_5 | Sus domesticus | Rectal swab | 5 months | 4 May 2013 | 9,651 | 33.5 | 30 |
| Posavirus_17668_47 | KX673285 | ERR1302794 | ERR1302883 | Pv_6 | Sus domesticus | Rectal swab | 5 months | 16 Oct 2013 | 11,295 | 45.1 | 99 |
| Posavirus_17668_71 | KX673286 | ERR1302816 | ERR1302905 | Pv_7 | Sus domesticus | Rectal swab | 6 months | 17 Oct 2013 | 9,404 | 46.3 | 47 |
| Posavirus_17668_83 | KX673287 | ERR1302827 | ERR1302916 | Pv_7 | Sus domesticus | Rectal swab | 5 months | 17 Oct 2013 | 9,366 | 46.9 | 46 |
| Posavirus_17668_86 | KX673288 | ERR1302830 | ERR1302919 | Pv_7 | Sus domesticus | Rectal swab | 5 months | 17 Oct 2013 | 9,377 | 46.8 | 42 |
| Rasavirus_16715_12 | KX673222 | ERR1301453 | ERR1301542 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 12 Nov 2014 | 9,601 | 40.2 | 251 |
| Rasavirus_16715_28 | KX673223 | ERR1301467 | ERR1301556 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 14 Nov 2014 | 9,606 | 40.4 | 823 |
| Rasavirus_16715_3 | KX673224 | ERR1301444 | ERR1301533 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 12 Nov 2014 | 9,580 | 40.5 | 17 |
| Rasavirus_16715_36 | KX673225 | ERR1301474 | ERR1301563 | Rv_1 | Rattus argentiventer | Fecal | Unknown | 14 Nov 2014 | 9,911 | 44.3 | 16058 |
| Rasavirus_16715_4 | KX673226 | ERR1301445 | ERR1301534 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 12 Nov 2014 | 9,619 | 40.3 | 14 |
| Rasavirus_16715_43 | KX673227 | ERR1301481 | ERR1301570 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 14 Nov 2014 | 9,562 | 40.4 | 24 |
| Rasavirus_16715_52 | KX673231 | ERR1301490 | ERR1301579 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 10 Jun 2014 | 8,678 | 40.8 | 27 |
| Rasavirus_16715_52_2 | KX673232 | ERR1301490 | ERR1301579 | Rv_1 | Rattus argentiventer | Fecal | Unknown | 10 Jun 2014 | 8,497 | 44.7 | 50 |
| Rasavirus_16715_57 | KX673233 | ERR1301495 | ERR1301584 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 10 Jun 2014 | 9,599 | 40.3 | 9097 |
| Rasavirus_16715_67 | KX673236 | ERR1301505 | ERR1301594 | Rv_2 | Rattus argentiventer | Fecal | Unknown | 11 Jun 2014 | 9,584 | 40.3 | 966 |
Median depth of coverage was determined by mapping all quality controlled reads to the final genome using Bowtie2 with –very-sensitive-local settings. Coverage was calculated as the number of reads mapped per genome/length of genome/divided by 129.
In two human fecal samples, husaviruses (KX673274 and KX673221) showed high level of amino acid sequence identity to the previously described husaviruses KT215901, KT215902, and KT215903 (97–98% amino acid identity). In contrast, an additional husavirus detected in a human rectal swab (KX673248) showed only 25% amino acid identity over the entire polyprotein with other husaviruses.
Posavirus sequences could be detected in thirty-three (of 189) pig rectal swabs (17% frequency) and in eight (of 146) pig fecal samples (5% frequency). In each of four pig rectal swabs (sample IDs 17189_4, 17819_95, 17668_11_2, and 17668_13), two distinct strains of posaviruses were identified, while in one sample (17668_33) three distinct posaviruses were identified. The posavirus sequences identified in this study have the closest sequence identity to variants detected in farmed pigs in the USA (Shan et al. 2011, Hause et al. 2015, 2016). Moreover, novel posa-like genome sequences were found in nine (of 45) rat fecal samples [provisionally named rat stool-associated RNA viruses (rasaviruses)], and in thirteen (of 135) bat fecal samples [provisionally named bat stool-associated RNA viruses (basaviruses)]. In one rat (16715_47) and in three bat fecal samples (16715_52, 16715_61, and 16715_71) two distinct rasa/basaviruses were identified.
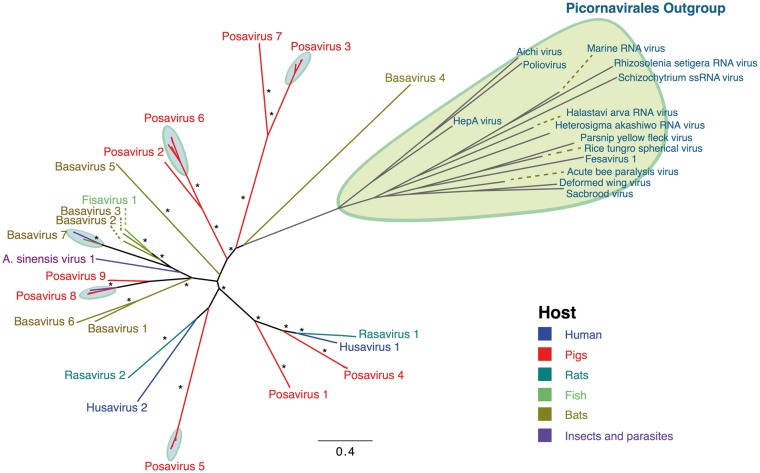
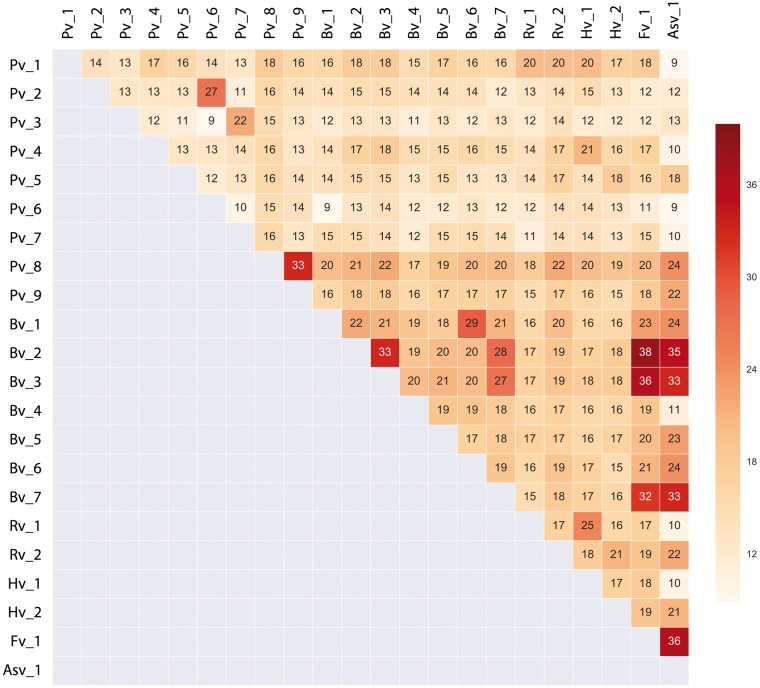
The low level of shared nucleotide identity between these novel viruses made it difficult to perform phylogenetic analyses at the whole genome level. Therefore, the protein sequence encoding the most conserved region, a provisional RdRP protein, was identified and used for phylogenetic analysis. This analysis supported a conclusion that two husaviruses (KX673221 and KX673274) belonged to a lineage that includes the previously described husaviruses (KT215901, KT215902, and KT215903), while husavirus KX673248 was distant (Fig. 1). Based on this phylogenetic analysis and using a pairwise amino acid identity cutoff of 40%, twenty-two lineages could be identified. While most lineages were found in only a single source type of sample (e.g. all porcine), the Bv_7 lineage comprised basaviruses and a virus isolated from a fruit fly Drosophila subobscura and the two lineages Pv_8 and Pv_9 comprised posaviruses and viral sequences derived from a parasite (Ascaris suum ; Shan et al. 2011) (Fig. 1). For each lineage, a representative virus genome (based on the most complete sequence of the twenty-nine sequences present in the GenBank database and the newly identified sequences) was selected and characterized in more detail (Supplementary Table S1). The length of these genomic sequences ranged from 8,576 nt to 11,318 nt with a G + C content of 31.0–53.0%. The twenty-two lineages share, on average, only 9–38% amino acid identity across the entire polyprotein (Fig. 2).
Figure 1.
Maximum-likelihood phylogenetic tree of the predicted RdRP protein domain. The maximum-likelihood phylogenetic tree was constructed in IQtree under the LG + G+I amino acid substitution model as the best-fitted model with 500 pseudo-replicates. The tree was visualized in FigTree1.4.2. Branches were colored according to the enteric samples from the hosts in which viruses were identified (blue: human, red: pig feces, dark green: rat feces, light green: fish intestinal content, brown: bat feces, and purple: insects and parasites). Significant bootstrap values (>80) are indicated with an *.
Figure 2.
Average percentage of amino acid identity across PPLVs lineages. The amino acid sequences of the complete polyprotein of different lineages were individually aligned against each other using the ClustalW in Geneious. The numbers on the x- and y-axes represent the different lineages (for more detail see Supplementary Table S1).
Phylogenetic analysis was also performed on amino acid sequences encoding the conserved helicase, 3C protease, capsid I and capsid II domains. Due to the high sequence diversity, each set of sequences was trimmed to the most conserved region of each identified conserved domain.
Neighbor-joining (NJ) and maximum-likelihood (ML) trees were constructed individually for each of the conserved domain set of sequences. The NJ tree topology of the RdRp (left panel Supplementary Fig. S1A) was relatively consistent with the ML tree topology of RdRP (Fig. 1). However, this relatively consistency was not observed in the NJ versus ML trees in other domains (left panel compared right panel, Supplementary Fig. 1), probably due to the great sequencing divergence hence challenging proper ML tree inference.
The prevalence of husaviruses among stools samples from Vietnamese individuals was 1.4% (1/71) in healthy human rectal swabs and 0.3% (2/573) in human diarrheal feces. Rasaviruses and basaviruses were detected in 22 and 9% of the rat and bat fecal samples, respectively. Pigs also commonly carry these viruses, with posavirus being found in 17% of the rectal swabs and in 5% of the fecal samples examined in this study. The frequency of virus detection was significantly higher in rectal swabs compared to fecal samples for posaviruses (P value = 0.002; Chi-squared test). The frequency of husavirus positive samples was too low to draw conclusions about the prevalence in rectal swabs compared to fecal samples (P value = 0.59; Fishers’ Exact test).
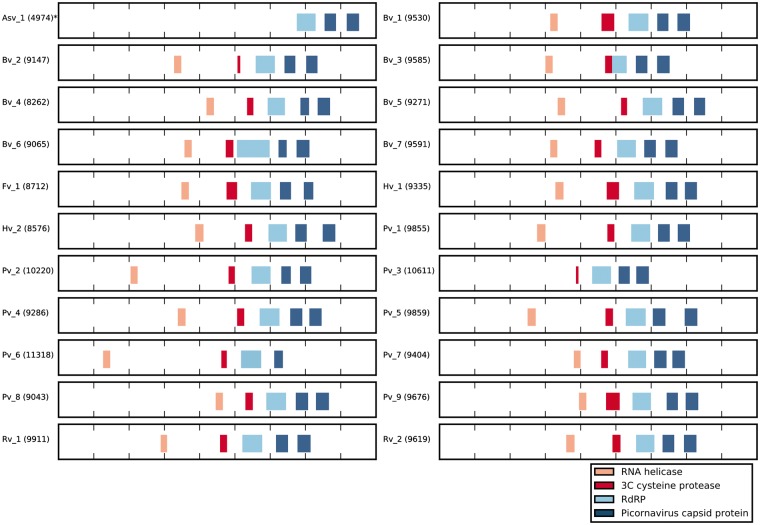
While members of the Picornavirales typically contain a Hel-Pro-Pol replication block (Le Gall et al. 2008), some of the recently identified posaviruses initially appeared to not encode a recognizable conserved protease domain (Shan et al. 2011; Hause et al. 2015, 2016). A local HMMER search (Eddy 2011) using the complete PFAM library (Marchler-Bauer et al. 2015) failed to identify a recognizable picornavirus 3C protease domain in the majority of genomes. It was unlikely that these viruses completely lacked the protease and we suspected that the failure to detect the protease domain could be due to sequence diversity in the protease domain of RNA viruses (Koonin and Dolja 1993). Accordingly, a refined 3C protease HMM profile was constructed including all newly identified protease domains in posaviruses. A search using this refined protease domain profile identifed a putative protease domain in all of the posaviruses (Supplementary Table S1). In addition, all genomes were found to encode an RNA helicase domain, an RdRP domain and two picornavirus capsid domains [with the exception of Asv_1 since this GenBank entry is only partial and posavirus_3 where no conserved RNA helicase domain could be identified (Fig. 3)].
Figure 3.
Genomic organization of different identified PPLVs lineages. The putative conserved protein domains, as determined by a conserved domain search (see ‘Methods’), and their relative position in the viral genome are illustrated. The number next to the lineage name indicates the length of each genome, the Asv1 genome (marked with an *) was only partially (<5,000 nt) sequenced. Peach blocks indicate the presence and position of an RNA helicase domain (superfamily cl21455), red blocks indicate the 3C cysteine protease domain (superfamily cl02893), light blue blocks indicate an RNA-dependent RNA polymerase (RdRP) domain (superfamily cl02808) and dark blue blocks indicate picornavirus capsid domain (superfamily cl13999). All identified conserved domains are drawn in scale related to their genome size.
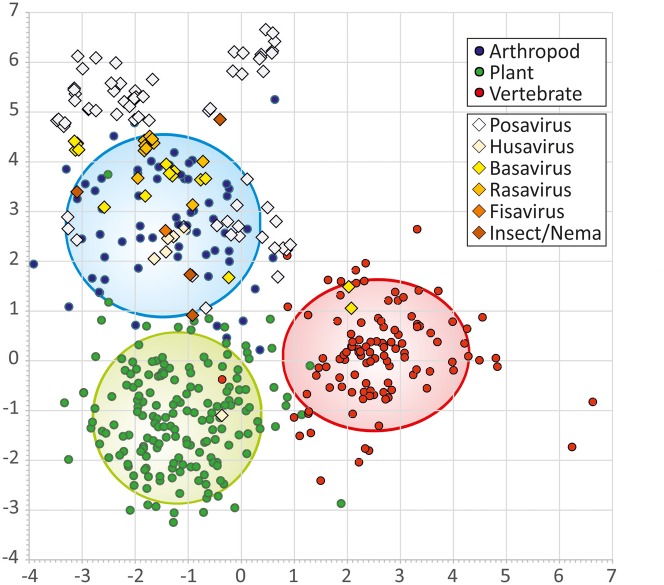
The G + C contents for all PPLV sequences were determined but no specific G + C content pattern was observed in virus sequences from different hosts. The husaviruses showed the highest G + C content (50.5–53.0%), followed by posaviruses (30.9–51.2%), rasaviruses (40.5–44.0%), and basaviruses (32.2–48.2%) (Table 1 and Supplementary Table S1). As previously described (Kapoor et al. 2010), nucleotide composition analysis (NCA) can be used to predict the host range of members of Supergroup 1 RNA virus, that includes the Picornavirales. Sequences from 105 PPLV genomes obtained in the current study and from published sources were analyzed using a pre-trained dataset of reference genomes from three categories of hosts (arthropod, plant, and vertebrate; Fig. 4). The analysis revealed that almost all posaviruses as well as all husa-, basa-, rasa-, fisa-, insect-, and nematoda viruses clustered within the arthropod group (Fig. 4). Two basaviruses (lineage Bv_2) cluster within the vertebrate group of the Picornaviridae and one husavirus (lineage Hv_2) clustered within the plant virus group of the Picornaviridae. These observations fell within the 5% error range of the analysis (95% prediction accuracy of the controls, Supplementary Table S2).
Figure 4.
Discriminant analysis of the dinucleotide bias in PPLVs compared to members of the Picornaviridae for which the infectious host is known. Viruses infecting arthropods are indicated with blue circles, viruses infecting plants in green circles and viruses infecting vertebrates in red circles. The lines indicate the 95% interval. Posaviruses are plotted in white diagonals, husavirus in light yellow, basavirus in yellow, rasavirus in dark yellow, fisavirus in orange and insect/nematode infecting viruses in dark orange.
3. Discussion
Here we report the identification of new Picornavirales members related to sequences previously identified in pig stool (posavirus) and human stools and/or rectal swabs (husavirus). In addition, we describe newly identified bat stool-associated viruses (basaviruses) and rat stool-associated viruses (rasaviruses) which have a similar genomic organization compared to posaviruses. Posaviruses are known to be widely distributed geographically with examples found in the USA (Shan et al. 2011) and in China (Zhang et al. 2014), however, this is the first detection of husavirus in human stools outside the Netherlands. These Posa and Posa-like virus genomes are collectively referred as PPLVs.
The PPLV genomes were identified based on identity to previously identified posaviruses and their lack of close protein homology to any of the Picornavirales families and the presence of a set of five protein functional domains. Using standard phylogenetic analyses, the PPLVs formed lineages which are distinct from the five established Picornavirales families (Dicistroviridae, Iflaviridae, Marnaviridae, Picornaviridae and Secoviridae) and the unassigned Picornavirales genome sequences. However, there is as much diversity between the PPLVs as there is between the PPLVs and the established Picornavirales families.
We used the USEARCH clustering algorithm in an attempt to determine how close the PPLV genomes are to existing Picornavirales genomes. All 108 PPLV genome sequences were combined with all available Picornavirales full genome sequences from GenBank (5,766 genomes, excluding those with stretches if Ns greater than 20). At various levels of homology (ranging from 60% to 90% nucleotide identities), the PPLV sequence clusters were distinct from the clusters formed from the Picornavirales genomes, i.e. there were no clusters containing both PPLV sequences and genomes classified in one of the Picornavirales families (results not shown). Thus we think it is valid to conclude that none of the PPLVs belong to established Picornavirales families. The PPLV group is however too diverse to be classified as a single virus family. Given the pace at which new virus sequences are becoming available, we believe the best approach is to deposit these sequences with a tentative identification as PPLV and as more detailed sequence data become available a better organization of these virus sequences into well-supported family or families can be made.
Members of PPLV group have now been identified in pigs, humans, fish, rats, bats, insects (Anopheles sinensis and Drosophila subobscura), and parasites (Ascaris suum). Based on phylogenetic analysis of the RdRP domain and pairwise comparisons of the entire polyprotein, we propose that the PPLVs comprise twenty-two phylogenetic lineages. These PPLVs could also be grouped in twenty-two lineages based on the NJ trees constructed from amino acid sequences encoding other conserved domains (putative helicase, protease, capsid I, and II, Supplementary Figs S1A–D).
Consistent with other members of the Picornavirales, most of the newly described PPLV sequences encoded a Hel-Pro-Pol replication block. However, in some of the genome sequences, no recognizable protease domain could be identified using conventional methods with an existing pre-made PFAM domain based on a limited number of picornavirus protease domains. However, a more detailed protease domain database based on a broader set of Picornavirales proteases, including the novel putative posavirus protease domains, revealed the presence of a protease domain across the entire range of PPLV genomes (Fig. 3).
In an attempt to infer a putative cellular host for the PPLVs, a nucleotide composition analysis (NCA) was performed. NCA incorporates composition measures of dinucleotide frequencies and has been used to predict the infectious hosts of members of RNA virus supergroup I (Kapoor et al. 2010). In a set of sequences for which the infectious host was known, the analysis was able to accurately classify viruses as either being of vertebrate, plant, or arthropod origin in around 95% of the cases (Koonin et al. 2008). Using this analysis method, the PPLV genome sequences were found to cluster with viruses from the arthropod group (Fig. 4). The two outliers of the discriminant analysis (lineage HV_2 and Bv_2) fall within the 95% confidence interval, but given their substantial sequence divergence from other PPLVs it is possible that these viruses infect another hosts.
The prevalence of husaviruses in fecal samples (0.3%) and rectal swabs (1.4%) in Vietnam was lower than the 3.5% prevalence observed in a cohort of predominantly healthy HIV-1 positive and negative individuals (Oude Munnink et al. 2015). Prevalence differences may be due to differences in RT-PCR detection versus genome assembly from next generation sequencing, the small sample numbers and/or true differences between the cohorts. Of interest, posavirus could be detected significantly more often in pig rectal swabs compared to pig fecal samples (P = 0.002), suggesting that the viruses are enriched on the rectal epidermis. This enrichment and the clustering of posaviruses with the arthropod viruses may be consistent. It is known that intestinal parasites can be found perirectally and can be detected using the scotch tape test (Enterobius_Vermicularis_Diagnostic_Test). An interesting follow-up analysis would be to determine the scotch tape virome and our prediction would be that members of the PPLV group can be found in these samples.
In summary, this study provides a large set of seventy-six new PPLV genomes, quadrupling the available genomic data for this broad group viruses. A novel Vietnamese husavirus genetically distant from the previously described husaviruses was identified and PPLV members were also detected in rat and bat feces. In addition, we were able to clarify two additional features of posavirus virology: a putative protease domain was detected in all PPLV genomes and NCA revealed that members of the PPLV group share a conserved nucleotide composition with viruses infecting members of the arthropod phylum.
4. Methods
4.1 Samples
Fecal material was collected from 135 bats (Scotophilus kuhlii), 573 humans (Homo sapiens), 146 pigs (Sus domesticus), and 45 rats (Rattus argentiventer). In addition, rectal swabs were collected from seventy-one humans and 189 pigs. These samples were collected from a 150 square kilometer area of Dong Thap province, a southern region within the Mekong Delta River in Vietnam. All fecal samples from human enrollees were diarrheal patients admitted to Dong Thap Provincial hospital, while human rectal swabs were taken from healthy farmers and family members. Pig fecal samples and rectal swabs were collected from individual pigs from breeding farms. Rat fecal samples were collected from rats, which were purchased on the market or collected from rice-field traps. The disease state of these animals is unknown. Bat fecal samples were collected from beneath roosting sites.
Ethical approval for the study was obtained from the Oxford Tropical Research Ethics Committee (OxTREC Approval No. 15-12) (Oxford, United Kingdom), the institutional ethical review board of Dong Thap Provincial Hospital (DTPH) and the Sub-Department of Animal Health Dong Thap province (Dong Thap, Vietnam).
4.2 Illumina sequencing
Fecal samples (n = 899) or rectal swabs (n = 260) were centrifuged for 10 min at 10,000 × g after which the samples were DNase treated at 37 °C for 30 min (20 U of TURBO DNase, Thermo Fisher per 100 µl of sample). Nucleic acids were extracted, transcribed into cDNA and subjected to second strand synthesis (de Vries et al. 2011, 2012). The resulting dsDNA from each sample was sheared and fractionated to 400–500 bp in length after which Illumina adapters with a unique barcode were ligated to the fragments. Resulting libraries were sequenced with the Illumina MiSeq or HiSeq platforms to generate 1–2 million 150 nt (MiSeq) or 3–4 million 250 nt (HiSeq) paired-end reads per sample.
4.3 De novo assembly and complete genome characterization
Adaptor sequences were removed and sequence reads that passed quality control were de novo assembled using SPAdes version 3.5.0 (Bankevich et al. 2012) followed by improve_assembly (Page 2012). The resulting contigs were subjected to a modified protein blast search using USEARCH (Edgar 2010) to identify novel members of the Picornavirales. To minimize the effects of Illumina cross-talk, all preliminary contigs were examined and contigs within a sample with low median coverage (greater than 10-fold lower than the major contig in the sample) were excluded from the analysis. For all PPLVs reported here, the complete or nearly complete (>8,000 nt) genome was obtained and for all viruses the genome coverage was determined by mapping all quality controlled sequence reads to the final genome. The G + C content was determined using Geneious (Kearse et al. 2012). To determine the average percentage amino acid identity across the PLLV lineages, amino acid sequences were aligned using the ClustalW in Geneious (Kearse et al. 2012).
To identify conserved protein domains encoded by the new genomes an RPS-BLAST search (Marchler-Bauer et al. 2015) against the Conserved Domain Database (CDD) was performed. The initial screen identified the helicase, RdRP, and picornavirus capsid (I and II) domains across almost all genomes. However, the 3C protease domain was identifed in only a subset of genomes, suggesting either a true absence or a mis-identification due to great sequence divergence. A modified 3C protease domain profile was generated from a protein sequence alignment of the conserved domain (pfam00548) from the CDD and used to identify the 3C protease-like regions in the new PPLV genomes. An updated alignment containing all the putative protease domains used to create a new HMM index file. A local hmmsearch analysis with this updated 3C protease profile was then performed to identify divergent putative protease domains in the PPLV genome sequences.
4.4 Discriminant analysis of the dinucleotide bias
Nucleotide composition analysis (NCA) was performed as previously described (Kapoor et al. 2010) using sequences of members of RNA virus supergroup 1 (Koonin et al. 2008) infecting vertebrates (n = 113), arthropods (n = 66), and plants (n = 172) for classification. The frequencies of each mononucleotide and dinucleotide were used for discriminant analysis to maximize discrimination between control sequences; these canonical factors were then used to infer the host origin of the RNA virus sequences obtained in the current study.
4.5 Phylogenetic analysis
All PPLV sequences identified in this study combined with all complete PPLV genomes present in the GenBank database (retrieved on 16 July 2016) were aligned using muscle (Edgar 2004). Amino acids sequences were trimmed to the region encoding for the conserved domains and alignments were manually inspected and trimmed to the most conserved part. Phylogenetic analyses were performed on the conserved putative conserved domains using IQtree (Nguyen et al. 2015), under the best-fitted amino acid model with 500 pseudo-replicates. The resulting trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
4.6 Statistical analysis
Statistical analysis was performed using the two by two table from Open Epi (Sullivan et al. 2009). As a measure of association, the Chi-squared test or the Fishers’s exact test was used.
4.7 GenBank accession numbers
All PPLV genome sequences generated in this study were deposited into the GenBank database under the accession numbers KX673215–KX673290.
Supplementary Material
Acknowledgements
The VIZIONS Consortium members (alphabetical order by surname) from the Oxford University Clinical Research Unit are: Bach Tuan Kiet, Stephen Baker, Alessandra Berto, Maciej F. Boni, Juliet E. Bryant, Bui Duc Phu, James I. Campbell, Juan Carrique-Mas, Dang Manh Hung, Dang Thao Huong, Dang Tram Oanh, Jeremy N. Day, Dinh Van Tan, H. Rogier van Doorn, Duong An Han, Jeremy J. Farrar, Hau Thi Thu Trang, Ho Dang Trung Nghia, Hoang Bao Long, Hoang Van Duong, Huynh Thi Kim Thu, Lam Chi Cuong, Le Manh Hung, Le Thanh Phuong, Le Thi Phuc, Le Thi Phuong, Le Xuan Luat, Luu Thi Thu Ha, Ly Van Chuong, Mai Thi Phuoc Loan, Behzad Nadjm, Ngo Thanh Bao, Ngo Thi Hoa, Ngo Tri Tue, Nguyen Canh Tu, Nguyen Dac Thuan, Nguyen Dong, Nguyen Khac Chuyen, Nguyen Ngoc An, Nguyen Ngoc Vinh, Nguyen Quoc Hung, Nguyen Thanh Dung, Nguyen Thanh Minh, Nguyen Thi Binh, Nguyen Thi Hong Tham, Nguyen Thi Hong Tien, Nguyen Thi Kim Chuc, Nguyen Thi Le Ngoc, Nguyen Thi Lien Ha, Nguyen Thi Nam Lien, Nguyen Thi Ngoc Diep, Nguyen Thi Nhung, Nguyen Thi Song Chau, Nguyen Thi Yen Chi, Nguyen Thieu Trinh, Nguyen Thu Van, Nguyen Van Cuong, Nguyen Van Hung, Nguyen Van Kinh, Nguyen Van Minh Hoang, Nguyen Van My, Nguyen Van Thang, Nguyen Van Thanh, Nguyen Van Vinh Chau, Nguyen Van Xang, Pham Ha My, Pham Hong Anh, Pham Thi Minh Khoa, Pham Thi Thanh Tam, Pham Van Lao, Pham Van Minh, Phan Van Be Bay, Phan Vu Tra My, Maia A Rabaa, Motiur Rahman, Corinne Thompson, Guy Thwaites, Ta Thi Dieu Ngan, Tran Do Hoang Nhu, Tran Hoang Minh Chau, Tran Khanh Toan, Tran My Phuc, Tran Thi Kim Hong, Tran Thi Ngoc Dung, Tran Thi Thanh Thanh, Tran Thi Thuy Minh, Tran Thua Nguyen, Tran Tinh Hien, Trinh Quang Tri, Vo Be Hien, Vo Nhut Tai, Vo Quoc Cuong, Voong Vinh Phat, Vu Thi Lan Huong, Vu Thi Ty Hang, Heiman Wertheim; from the Centre for Immunity, Infection and Evolution, University of Edinburgh (alphabetic order by surname): Carlijn Bogaardt, Margo Chase-Topping, Al Ivens, Lu Lu, Dung Nyugen, Andrew Rambaut, Peter Simmonds, Mark Woolhouse; from The Wellcome Trust Sanger Institute (alphabetic order by surname): Matthew Cotten, Bas Oude Munnink, Paul Kellam, My Vu Tra Phan; from the Laboratory of Experimental Virology, Department of Medical Microbiology, Center for Infection and Immunity Amsterdam (CINIMA), Academic Medical Center of the University of Amsterdam, Amsterdam, the Netherlands. (alphabetic order by surname): Martin Deijs, Lia van der Hoek, Maarten F. Jebbink, and Seyed Mohammad Jazaeri Farsani; from the Metabiota Inc. (alphabetic order by surname): Kimberly Dodd, Jason Euren, Ashley Lucas, Nancy Ortiz, Len Pennacchio, Edward Rubin, Karen E. Saylors, Tran Minh Hai, Nathan D. Wolfe.
Funding
This work was supported by the Wellcome Trust of the UK through the VIZIONS strategic award (WT/093724), by the European Union’s Horizon 2020 research and innovation programme under grant agreements No 643476 (COMPARE) and No 634650 (Virogenesis).
Conflict of interest: None declared.
Supplementary data
Supplementary data are available at Virus Evolution online.
References
- Bankevich A. et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’, Journal of Computational Biology, 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N. et al. (1996) ‘Cleavage Site Analysis in Picornaviral Polyproteins: Discovering Cellular Targets by Neural Networks’, Protein Science, 5: 2203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M. et al. (2014) ‘Full Genome Virus Detection in Fecal Samples Using Sensitive Nucleic Acid Preparation, Deep Sequencing, and a Novel Iterative Sequence Classification Algorithm’, PLoS One, 9: e93269.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries M. et al. (2011) ‘A Sensitive Assay for Virus Discovery in Respiratory Clinical Samples’, PLoS One, 6: e16118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries M. et al. (2012) ‘Performance of VIDISCA-454 in Feces-Suspensions and Serum’, Viruses, 4: 1328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2011) ‘Accelerated Profile HMM Searches’, PLoS Computational Biology, 7: e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment With High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010) ‘Search and Clustering Orders of Magnitude Faster Than BLAST’, Bioinformatics, 26: 2460–1. [DOI] [PubMed] [Google Scholar]
- Enterobius_Vermicularis_Diagnostic_Test. Available: <https://web.stanford.edu/class/humbio103/ParaSites2006/Enterobius/diagnostic%20tests.htm> accessed 22 Mar 2017.
- Hause B. M., Hesse R. A., Anderson G. A. (2015) ‘Identification of a Novel Picornavirales Virus Distantly Related to Posavirus in Swine Feces’, Virus Genes, 51: 144–7. [DOI] [PubMed] [Google Scholar]
- Hause B. M. et al. (2016). Highly diverse posaviruses in swine feces are aquatic in origin. Journal of General Virology, 97: 1362–7. [DOI] [PubMed] [Google Scholar]
- ICTV. (2017). Virus Taxonomy: The Classification and Nomenclature of Viruses. The Online (10th) Report of the ICTV [Online] <https://talk.ictvonline.org/ictv-reports/ictv_online_report/> accessed 21 Mar 2017.
- Kapoor A. et al. (2010) ‘Use of Nucleotide Composition Analysis to Infer Hosts for Three Novel Picorna-Like Viruses’, Journal of Virology, 84: 10322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. (2012) ‘Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data’, Bioinformatics, 28: 1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V. (1993) ‘Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences’, Critical Reviews in Biochemistry and Molecular Biology, 28: 375–430. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. et al. (2008) ‘The Big Bang of Picorna-Like Virus Evolution Antedates the Radiation of Eukaryotic Supergroups’, Nature Reviews Microbiology, 6: 925–39. [DOI] [PubMed] [Google Scholar]
- Le Gall O. et al. (2008) ‘Picornavirales, a Proposed Order of Positive-Sense Single-Stranded RNA Viruses with a Pseudo-T = 3 Virion Architecture’, Archives of Virology, 153: 715–27. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. (2015) ‘CDD: NCBI's Conserved Domain Database’, Nucleic Acids Research, 43: 222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B. B. et al. (2015) ‘A Novel Genus in the Order Picornavirales Detected in Human Stool’, Journal of General Virology, 96: 3440–3. [DOI] [PubMed] [Google Scholar]
- Page A. J. (2012). improve_assembly [Online]. <https://metacpan.org/pod/distribution/Bio_AssemblyImprovement/bin/improve_assembly> accessed 19 Feb 2017.
- Rabaa M. A., Tue N. T., Phuc T. M., Carrique-Mas J., Saylors K., Cotten M., Bryant J. E., Nghia H. D., Cuong N. V., Pham H. A.. et al. (2015) ‘The Vietnam Initiative on Zoonotic Infections (VIZIONS): A Strategic Approach to Studying Emerging Zoonotic Infectious Diseases’, Ecohealth, 12: 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G. et al. (2015) ‘A Novel Posavirus-Related Single-Stranded RNA Virus from Fish (Cyprinus carpio)’, Archives of Virology, 160: 565–8. [DOI] [PubMed] [Google Scholar]
- Shan T. et al. (2011) ‘The Fecal Virome of Pigs on a High-Density Farm’, Journal of Virology, 85: 11697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Dean A., Soe M. M. (2009) ‘OpenEpi: A Web-Based Epidemiologic and Statistical Calculator for Public Health’, Public Health Reports, 124: 471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. (2011) ‘Deep Small RNA Sequencing from the Nematode Ascaris Reveals Conservation, Functional Diversification, and Novel Developmental Profiles’, Genome Research, 21: 1462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. L. et al. (2016). Twenty Five New Viruses Associated with the Drosophilidae (Diptera). Biorxiv. [DOI] [PMC free article] [PubMed]
- Zhang B. et al. (2014) ‘Viral Metagenomics Analysis Demonstrates the Diversity of Viral Flora in Piglet Diarrhoeic Faeces in China’, Journal of General Virology, 95: 1603–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.