Abstract
Developmental and immune-mediated disease has been linked to genetic mutation of key signaling components involved in NF-κB activation that leads to impaired activation or regulation of the canonical IKK complex. We identify patients with suspected or known defects of the NF-κB signaling pathway through clinical phenotyping and genetic sequencing. To help understand how mutations cause disease, we quantitate the kinetics and dose-response of NF-κB activation signaling events in their cells. Following activation of the canonical IKK complex, phosphorylation of the inhibitor of NF-κB proteins (IκB) leads to their degradation and the subsequent translocation of NF-κB family members from the cell cytoplasm to the nucleus. Here, we provide a method to obtain patient-derived dermal fibroblasts and quantitatively assess the integrity of the signal transduction pathway from receptor activation to nuclear p65 translocation.
Keywords: NF-κB, p65, Nuclear translocation, Fibroblasts, IL-1R, TNFR, TLR, RLR, Microscopy
1 Introduction
Stimulation of pattern recognition receptors such as the Toll-like receptors, NOD-like receptors, and RIG-I-like receptors and of certain cytokine receptors such as the TNF receptor or IL-1 receptor activates NF-κB during the immune response and in the setting of inflammatory disease [1–4]. Recruitment of signaling components to these various receptors leads to activation of the canonical inhibitor of IkBa kinase (IKK) complex and NF-κB nuclear translocation (Fig. 1). The primary substrate of the activated IKK complex is the inhibitor of NF-κB proteins (IκB) whose phosphorylation and subsequent degradation permit the nuclear translocation of NF-κB family members which otherwise remain largely in the cytosolic fraction [5]. Activated NF-κB family members once in the nucleus are further posttranslationally modified to permit DNA binding that enhances or, in some cases, represses gene expression due to specific activity at promoter and enhancer elements within the genome [5, 6].
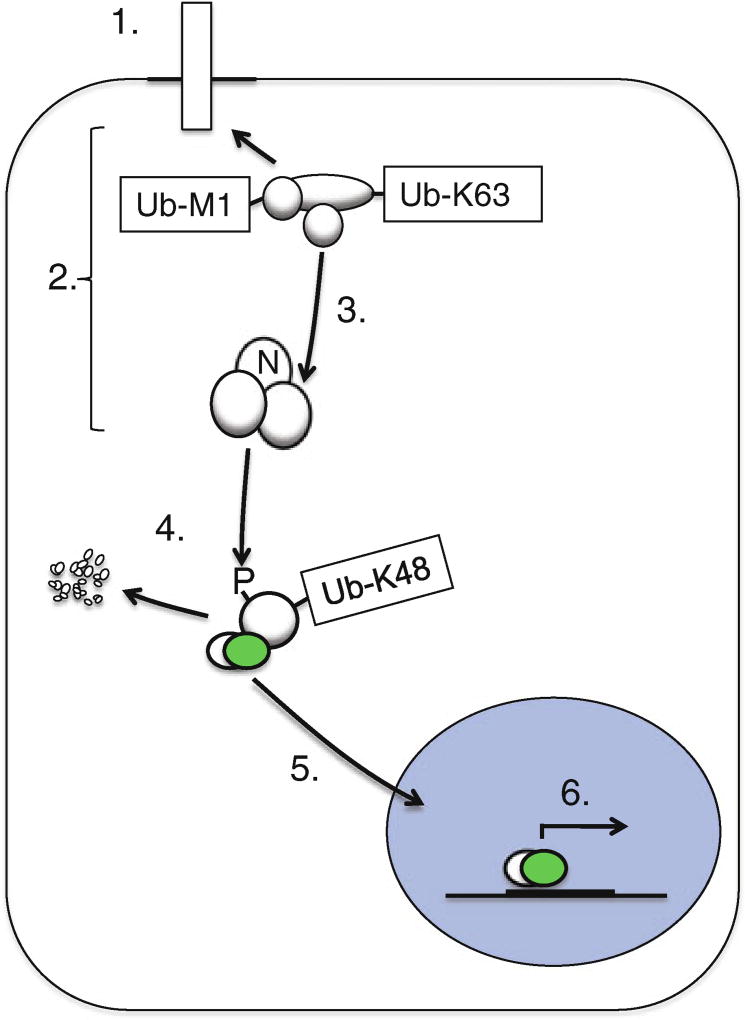
Fig. 1.
Receptor-induced signal transduction leads to NF-κB activation. Endogenous Toll-like receptors, NOD-like receptors, TNF receptor superfamily members, and cytosolic nucleic acid sensors are among the many inducers of NF-κB activation that can be studied in primary dermal fibroblasts. Receptor stimulation (1) leads to recruitment of multiple signaling proteins, termed the signalosome, (2) through posttranslational modifications such as polyubiquitination. This leads to recruitment and activation (3) of the canonical IKK complex, which is comprised of NEMO (IKKγ), IKKβ, and IKKα. The IKK complex phosphorylates IκBα proteins on specific serine residues which then lead to K48-linked polyubiquitination of IkBa and its degradation by the 26S proteasome (4). IκBα degradation permits access to nuclear localization sequences on NF-κB members which translocate to the nucleus (5), bind DNA elements, and mediate gene expression (6). Quantitative measurement of p65 nuclear translocation following receptor activation permits evaluation of the integrity of steps 1–5, above
Immunodeficiency, inflammatory disease, and/or developmental defects arise due to altered NF-κB activation [1–4, 7]. To understand how genetic mutation causes disease, we perform cellular phenotyping that includes analysis of signal transduction events in patient cells in vitro. Often, patients with Mendelian disorders in these pathways are young and severely affected, and so obtaining adequate materials to perform experiments is challenging. Dermal fibroblasts cultured from a single biopsy can be expanded greatly and cryopreserved, permitting abundant material for study. The effects of hypomorphic or hypermorphic mutant proteins on signal transduction events may be modest, yet relatively small differences “upstream” may result in large “downstream” effects on gene expression or cell survival, due to multiple successive stages of signal amplification. To enhance the sensitivity of detecting differences between samples, it is important to develop robust assays that utilize quantitation and statistical analysis. For these, time-course and dose-response experiments are of great utility which require larger numbers of test conditions. The method we describe is performed in a 96-well format, permitting multiple technical replicates to be performed for each condition, enhancing the reproducibility of experimental results. We provide a method for measuring canonical IKK induction of nuclear translocation of p65 following stimulation of primary dermal fibroblasts. The method describes a procedure to obtain cells using a simple skin biopsy; however, it can be adapted to study many different types of adherent cells such as HeLa, monocyte-derived macrophages, or mesenchymal stem cells.
2 Materials
2.1 Skin Biopsy and Plating
27-gauge needle.
5-cc Luer-Lok disposable syringe.
1 % lidocaine (10 mg/mL, Hospira, Inc, Lake Forest, IL, USA).
Betadine or chlorhexidine swab.
Disposable scalpel blade.
T75 tissue culture flask.
Dulbecco’s Modified Eagle’s Medium (DMEM).
Collagenase type IV solution (Gibco, Carlsbad, CA, USA). Make 1 mg/mL solution in DMEM.
Fibroblast culture medium: DMEM containing 15 % fetal calf serum (FCS) supplemented with penicillin (final 100 U/mL), streptomycin (final 100 µg/mL), MEM nonessential amino acids (Gibco) (stock concentration is 100x; use 10 mL per liter of medium), HEPES (final 10 mM), and sodium pyruvate 1 µM final. Combine components and then filter using a disposable tissue culture filter unit.
2.2 Cell Culture and Cell Stimulation
Lab benchtop centrifuge.
Fibroblast culture medium.
96-well flat-bottom tissue culture-treated plate.
Phosphate buffered saline (PBS).
Primary dermal fibroblasts (see Note 1).
Recombinant human TNF-alpha (R&D Systems, Minneapolis, MN, USA) (see Note 2).
Trypsin-EDTA 0.25 %.
37 °C, 5 % CO2 incubator.
50-mL conical tubes.
2.3 Immunofluorescent Staining and Image Acquisition
Permeabilization buffer: PBS containing 0.1 % Triton X-100 and 0.1 % saponin.
Alexa Fluor 488 goat anti-rabbit IgG antibody (Molecular Probes, Carlsbad, CA, USA).
Rabbit antihuman p65 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
37 % formaldehyde.
Goat serum.
PBS.
ProLong Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA, USA) (see Note 3).
IncuCyte ZOOM (see Note 4) (Essen Biosciences, Ann Arbor, MI, USA).
2.4 Image Analysis
ImageJ or FIJI software.
Excel or Prism software.
3 Methods
To define the molecular mechanisms that lead to NF-κB activation, it is useful to compare the kinetics of receptor stimulation-induced signaling between cell lines that express mutant forms of known or suspected key signaling components of this pathway. Oftentimes to achieve this, cell lines are generated that transiently or stably overexpress the mutant proteins found in individuals with immune-mediated disease. However, there is great utility in being able to study signaling characteristics from primary cells, as overexpression of certain molecules that function in signal transduction may paradoxically impair signaling, leading to spurious interpretations of their true function [8].
The shave skin biopsy is a straightforward procedure requiring minimal preparation that can be learned and performed by medically trained individuals with minimal experience (see Note 5). Prior to performing the procedure, informed consent from patients and control subjects must be obtained in accordance with institutional IRBs. In this procedure, a small piece of skin tissue is obtained which is then enzymatically treated to permit fibroblast growth on tissue culture dishes. Dermal fibroblasts undergo a finite number of divisions before reaching replicative senescence [9], and therefore it is important to plan cell preservation and storage to maximize utilization of the primary material and to ensure that experiments are performed using cells of similar passage number (see Note 6). Cells are cultured in medium and passaged by trypsinization and replating at a 1:3 ratio (the cells comprising one confluent flask are transferred onto three new flasks). For analysis of stimulation-induced p65 translocation, these cells are replated onto 96-well flat-bottom plates and stimulated with various different cytokines and recombinant pathogen-associated molecular patterns in the presence or absence of inhibitors or other controls (see Note 7). Following stimulation, cells are fixed and stained with specific antibodies to the NF-κB subunit p65. The kinetics and dose-response of NF-κB nuclear translocation from cells obtained from normal healthy controls are compared to that from individuals with primary immunodeficiency or Mendelian inflammatory disease in order to assess the integrity of signaling up to and including transcription factor nuclear translocation.
3.1 Skin Biopsy and Plating
Review procedure with the individual; obtain informed consent.
Swab the skin on the medial aspect of the forearm with antiseptic.
Fill a 5-cc syringe with 1 % lidocaine, and fit with a 27-gauge 1-in. needle.
Introduce the needle under the epidermis, injecting several microliters of lidocaine under the skin as the needle penetrates the skin. Eventually infuse approximately 50 µL of lidocaine under the skin, keeping the needle firmly in place with one hand (see Note 8).
Lift gently upward holding the syringe with the attached needle to create a tent of the skin (Fig. 2).
While the skin is under tension, slice horizontally immediately adjacent to the tip of the needle using a scalpel held in the opposite hand. A small chunk of the skin which contains cells from the epidermis and dermis will remain stuck to the needle as the skin is completely cut away.
Place the skin sample in a 15-mL conical tube containing 10 mL of DMEM (see Note 9).
Aspirate the DMEM, leaving the skin at the bottom of the conical tube. Add 1 mL of 1 % collagenase to the skin, and incubate at 37 °C 5 % CO2 for 12 h.
Vortex the sample, add 7 mL of culture medium, and plate the entire contents onto a T75 flask (see Note 10).
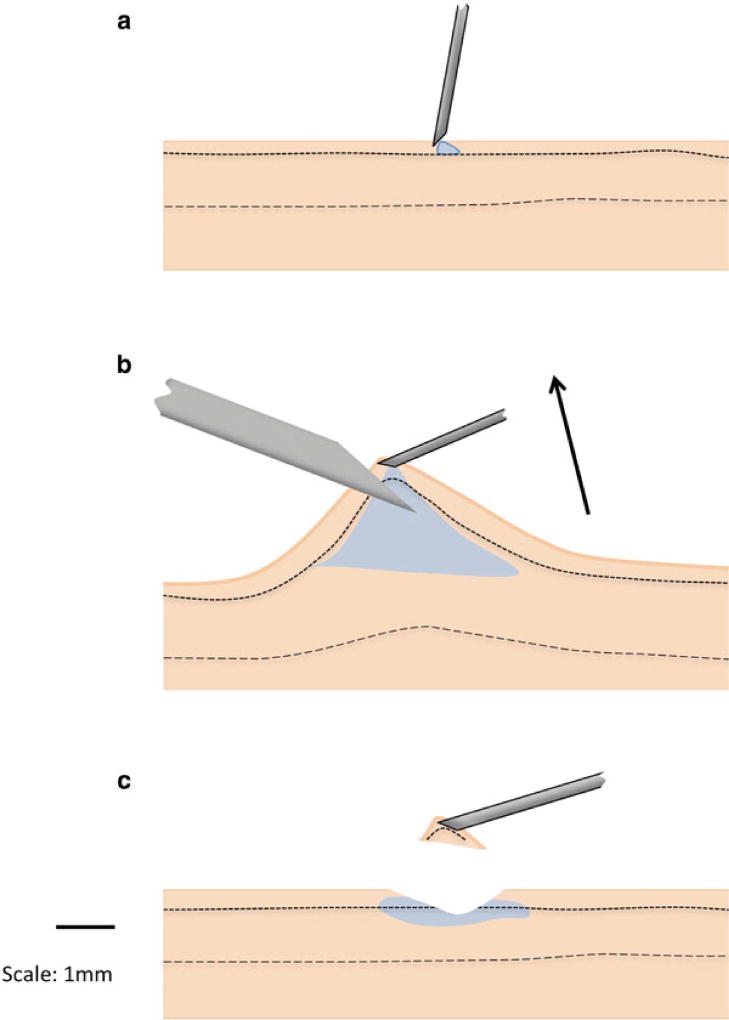
Fig. 2.
The shave skin biopsy procedure. (a) The site to be biopsied is cleaned with topical antiseptic followed by injection of 1 % lidocaine subcutaneously as a sterile fine gauge needle is introduced into the dermal layer. (b) After injecting a small “blister” of lidocaine, the needle is angled parallel to the surface of the skin, while a gentle upward force is applied to create a “tented” area of the skin, the tip of which is sliced off. (c) The skin tissue sample containing dermal and epidermal layers is transferred from the tip of the needle to a tube containing the medium
3.2 Cell Culture and Cell Stimulation
Allow cells to proliferate. Cells will reach confluence in 1–3 weeks depending on the starting amount of cells in the initial sample.
Once cells have reached confluence, passage cells by trypsinization, splitting them 1:3 in early passages (1–5).
Aspirate medium from the T75 flask of confluent fibroblasts and wash once with PBS.
Add trypsin to the T75 flask of confluent fibroblasts and incubate at 37 °C for 5 min until all the cells have lifted from the bottom of the flask.
Add 20 mL of the medium and transfer to a 50-mL conical tube.
Centrifuge for 5 min at 600 × g to pellet the cells.
Aspirate the medium and resuspend in fresh medium. Count the cells.
Seed 3 × 103 fibroblasts per well of a 96-well tissue culture plate in 200 µL of medium and culture at 37 °C, 5 % CO2 until the layer reaches 80–90 % confluence (see Note 11).
Aspirate the culture medium from wells and replace with 200 µL of warm medium.
Stimulate cells with TNF at a final concentration of 10 ng/mL in DMEM (see Note 12).
Incubate the fibroblasts at 37 °C, 5 % CO2 for 15–60 min.
3.3 Immunofluorescent Staining and Cell Imaging
Add 200 µL of pre-warmed 2 % formaldehyde solution to each well of fibroblasts and incubate at 37 °C for 10 min.
Aspirate the medium and formaldehyde from each well and wash once with 500 µL of PBS.
Permeabilize the cells by adding 200 µL of permeabilization buffer to each well and incubate at room temperature for 40 min.
Aspirate each well and wash five times with 500 µL of PBS.
Add anti-p65 antibody diluted 1:200 in permeabilization buffer for 1 h at 4 °C (see Note 13).
Aspirate the primary antibody solution and wash five times with 500 µL of PBS.
Add 200 µL of 5 % goat serum in permeabilization buffer and incubate at room temperature for 20 min.
Aspirate each well and wash five times with 500 µL of PBS.
Add anti-rabbit antibody conjugated to Alexa Fluor 488, diluted 1:750 in P/W buffer. Mix by gentle rotation and incubate at room temperature for 1 h, protected from light.
Aspirate the secondary antibody and wash five times with 500 µL of PBS.
Using the IncuCyte ZOOM fluorescence microscope (see Note 14), load the 96-well plate onto the imaging platform.
Image each well using the phase contrast, green, and red channels to observe p65 signal (Figs. 3 and 4).
Save all images as .tif files.
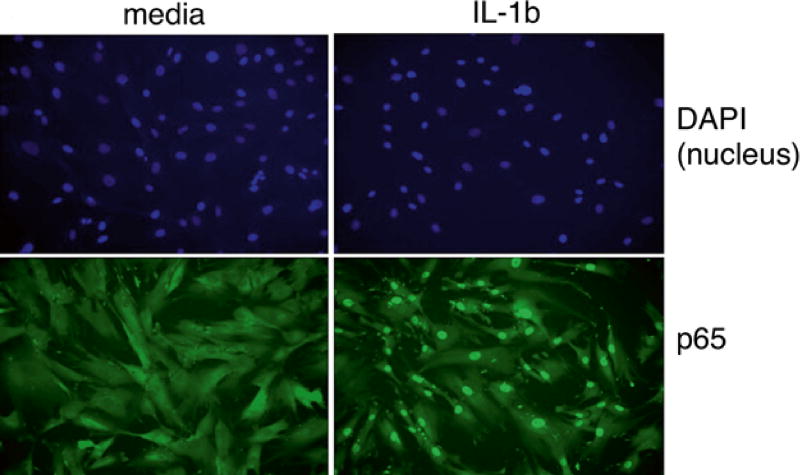
Fig. 3.
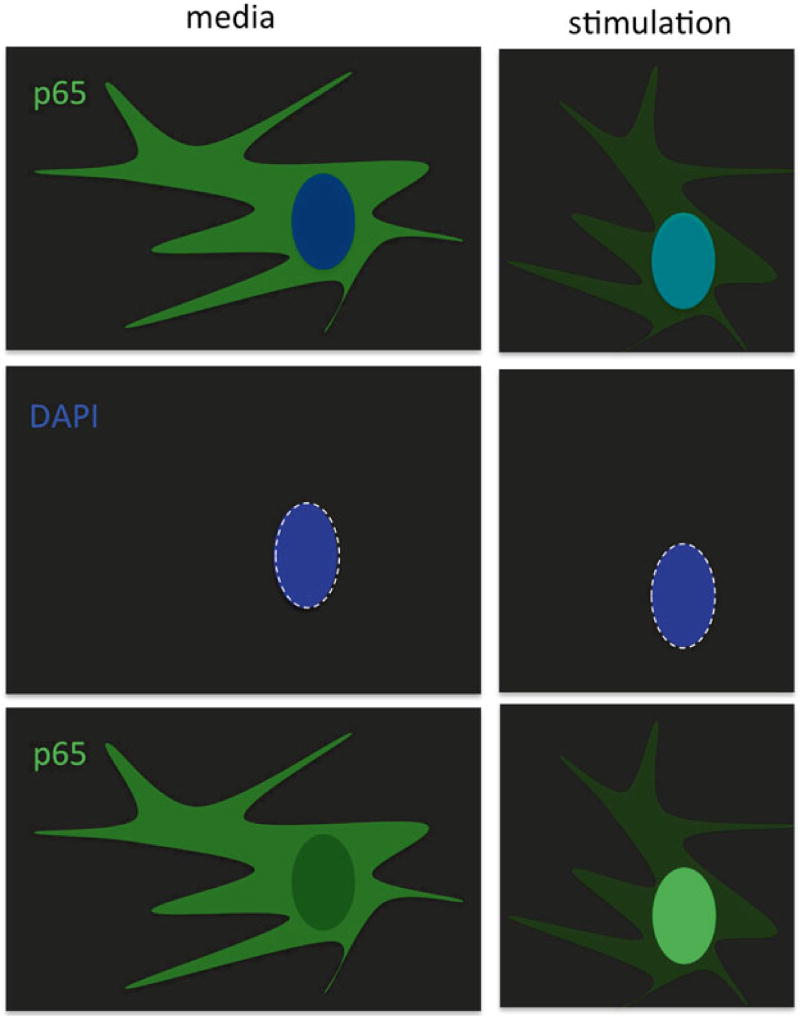
Fluorescence microscopy of primary dermal fibroblasts cultured in the medium (left panels) and stimulated for 30 min with IL-1beta (right panels). Following stimulation, cells are fixed, permeabilized, and stained for NF-κB NF-κ p65 (green) and the cell nucleus (blue)
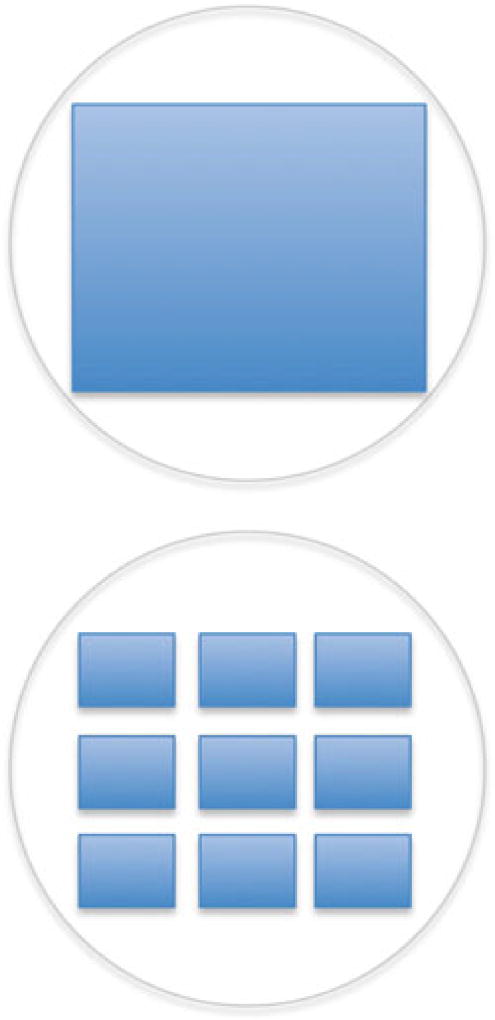
Fig. 4.
Image acquisition. Images from a 96-well plate can be acquired using a 4× objective lens (top) or four or nine regularly spaced images per well are obtained using 10× or 20× (bottom)
3.4 Quantitation of Nuclear p65
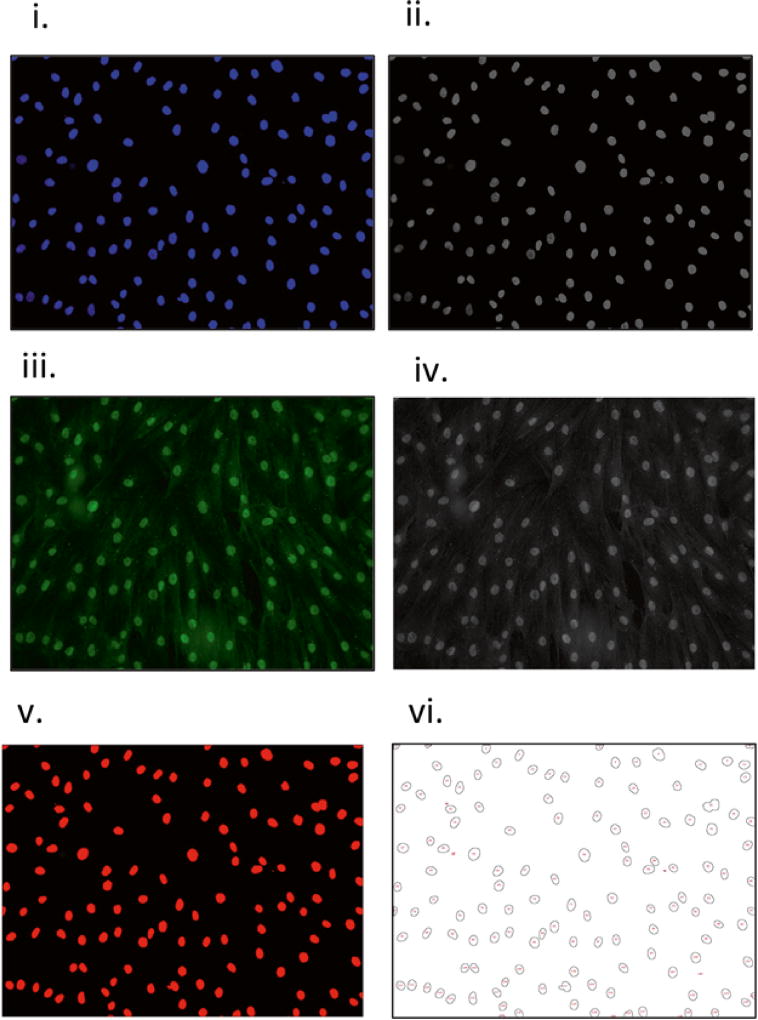
Using ImageJ, open the .tif files of the blue (DAPI-nuclear stained, Fig. 6i) and green (p65 stained, Fig. 6iii) channels for the first pair of images.
Using the menu labeled “Image,” select “Type” from the drop-down menu, and select 8 bit to convert the images from RGB color to 8-bit images, generating two new image files (Fig. 6ii, iv).
Select the nuclear 8-bit image (Fig. 6ii), and from the ImageJ menu, select “Image” and then “Adjust Threshold” from the drop-down menu, selecting “Default” for the thresholding algorithm, “Red,” and “dark background,” to generate a nuclear mask (Fig. 6v).
Select “Analyze” from the top ImageJ menu and “Set Measurements” from the drop-down menu, and select the options that you would like to have calculated. Select “mean gray value” and “limit to threshold” (see Note 15). Essential step: select “Redirect to,” and from the drop-down menu, select the file name corresponding to the 8-bit image of the p65 stain.
From the drop-down menu in ImageJ, select “Analyze,” then “Analyze Particles.” From the pop-up window, select “Show,” then “Outlines,” and select “Display Results.” Select “OK” (Fig. 5).
A result file is generated with multiple columns corresponding to the values chosen in Subheading 3.4, step 4. An image file named “Drawing of name of image file.tif ” with the numbered outlines of the nuclear masks (Fig. 6vi) will appear simultaneously with the file of values, labeled “Results” (see Note 16).
Save the result file, or directly copy and paste these values into Excel or your graphing software of choice.
Repeat steps 1–7 for all nuclear/p65 image pairs.
Graph the data or perform statistical analyses using Prism, Excel, or other software (Fig. 7).
Fig. 6.
Workflow of image analysis. An image of nuclear staining is opened (i) and converted to an 8-bit raw image (ii), as is done for the p65 stained image (iii–iv). Automatic thresholding of the 8-bit nuclear image will result in a thresholded image (v). Following particle analysis, a data file of numerical values and drawing with numbered nuclei is generated (vi), which permits visual correlation of numerical data with the corresponding individual cells
Fig. 5.
Quantitation of nuclear p65. For quantitation, two single-color images are obtained corresponding to nuclear and p65 stains (left, above). Using ImageJ, a nuclear mask is created using the image of the stained nuclei (dashed white oval) which is then applied to the image corresponding to p65 staining so that the mean nuclear pixel intensity is calculated for each cell in the image. Hundreds or thousands of individual cell nuclear intensities can be determined per 10× or 4× image, respectively
Fig. 7.
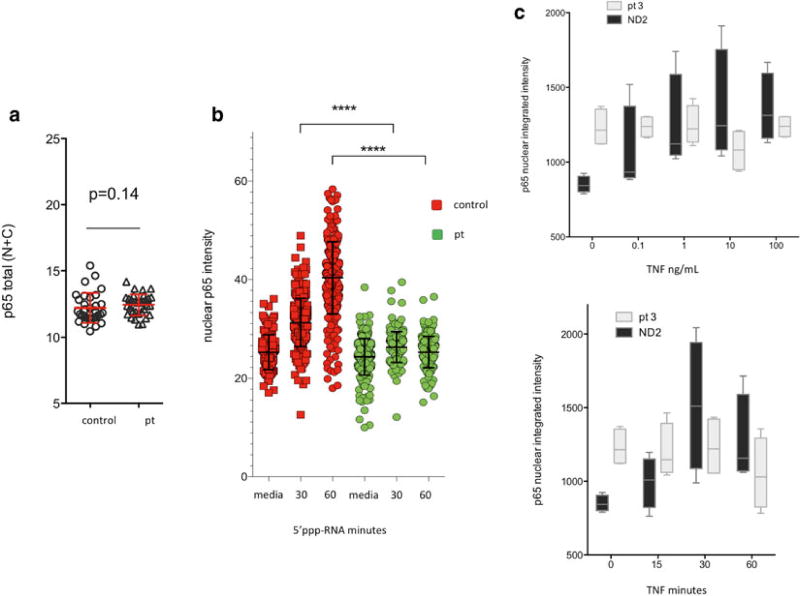
Visual representation of data. (a) Total p65 intensity per well is calculated for all samples analyzed to determine that differential staining of samples did not occur, and each data point corresponds to a well of a 96-well plate. N nucleus, C cytoplasm, pt patient samples. (b) Data can be graphed from pooling multiple images, with each individual data point corresponding to the mean fluorescence intensity of each individual cell nucleus. (c) Alternatively, the total nuclear integrated intensity per well can be plotted using n replicates (corresponding to n individual wells of a 96-well plate) per condition
Footnotes
This protocol describes the use of primary dermal fibroblasts; however, other adherent cells such as monocyte-derived macrophages or dendritic cells can also be used, which require specific medium and culture conditions to permit pure culture.
Nuclear p65 accumulation can be readily observed in human dermal fibroblasts treated with IL-1β (10 µg/mL), LPS (100 ng/mL), poly(I:C) (10 µg/mL), and 5′ppp-RNA (Invivogen, San Diego, CA, USA).
Alternatively, if your imaging system only allows two-color (red/green) detection, you may want to stain the nuclei with either a red or green nucleic acid stain. Syto 21 Molecular Probes (Carlsbad, CA, USA) stock is 1:10,000 and is a suitable green nuclear stain, and Syto 59 Molecular Probes (Carlsbad, CA, USA) is used at 1:1,000 and stains nuclei red. These are added simultaneously to the addition of the primary antibody.
Although developed primarily for live cell imaging, we use the IncuCyte ZOOM for convenient phase contrast and two-color fluorescent imaging of fixed cells. However, any fluorescence microscope can be used for this technique. If whole well or predetermined patterns of images are not automatically taken by an image acquisition device, we recommend having an investigator acquire images blinded to sample identity with a collaborator to perform image analysis in order to avoid inadvertent bias in image acquisition.
The biopsy site is chosen taking into consideration that scarring following wound healing is usually minimal but often leaves a residual area of hypopigmentation, which may be of cosmetic importance. Usually, we biopsy the medial (inner) aspect of the forearm, which is devoid of hair and has the skin that is under low tension.
We have found that one biopsy sample can be expanded to 20T75 tissue culture flasks within five passages to permit cryopreservation of 20 individual tubes. These can be stored in liquid nitrogen and thawed individually and used for experiments lasting several weeks before a new vial is thawed. This permits sequential experiments to be performed on cells that are of relatively equal passage number.
An important control is to stimulate some cells in the presence of inhibitors of canonical IKK activation or of p65 translocation. For that latter, MG-132 at 10 µM, added 1 h prior to stimulation, works well.
Just enough lidocaine to produce a small “blister” of fluid under the skin is sufficient. After about a minute, efficacy of anesthesia can be tested by jabbing the biopsy site with the scalpel tip. If done correctly, the only pain felt during the procedure should be from the first “jab” of the syringe into the skin.
The tissue sample can be kept in DMEM for up to 24 h at ambient temperature before the next step is performed. This potentially allows overnight shipment of sample from remote locations.
It is helpful to plate cells at sufficient density to allow rapid growth. If cells are plated very sparsely, they will take a very long time (months) to reach confluence, and culture may fail. If a tissue sample is particularly small or has decreased numbers of cells, centrifuge the sample (400 × g × 10 min) and resuspend in 1.5 mL of culture medium in one well of a six-well dish.
If cells are plated using a coverslip/microscopy slide method, empirically determine the number of cells to plate to obtain cell confluence 1–2 days after replating.
It is essential to stimulate all cells from one particular time point simultaneously. To achieve this, the use of a multichannel pipettor is essential. When adding the TNF containing the medium to the cells, it is preferable to add a larger volume of more dilute solution in order to minimize the effects of pipetting error.
Staining may be performed for longer, up to overnight, if convenient.
If using a conventional fluorescence microscope, add 20 µL antifade reagent with DAPI to each coverslip and place the coverslip onto a microscopy slide. Allow to dry overnight at room temperature, protected from light. Seal the edges of the glass coverslip with nail polish or other sealant and allow this to dry.
For some analyses, it may be useful to calculate the total nuclear intensity of p65 staining, rather than the mean value per nucleus, in which case “integrated intensity” should be selected.
It is useful to review each image alongside the corresponding data values. As a simple visual control, confirm that the numbered nuclei corresponding to the brightest p65 staining generated higher mean pixel intensity or integrated intensity values and that the nuclei with the dimmest p65 staining correspond to the lowest values. Depending on the quality of nuclear staining, small particles corresponding to debris may need to be excluded from the analysis by empirically modifying the default size range.
References
- 1.Hanson EP, Monaco-Shawver L, Solt LA, Madge LA, Banerjee PP, May MJ, Orange JS. Hypomorphic nuclear factor-kappaB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol. 2008;122(6):1169–1177.e1116. doi: 10.1016/j.jaci.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, Bonnet M, Puel A, Chable-Bessia C, Yamaoka S, Feinberg J, Dupuis-Girod S, Bodemer C, Livadiotti S, Novelli F, Rossi P, Fischer A, Israel A, Munnich A, Le Deist F, Casanova JL. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112(7):1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahtela J, Nousiainen HO, Stefanovic V, Tallila J, Viskari H, Karikoski R, Gentile M, Saloranta C, Varilo T, Salonen R, Kestila M. Mutant CHUK and severe fetal encasement malformation. N Engl J Med. 2010;363(17):1631–1637. doi: 10.1056/NEJMoa0911698. [DOI] [PubMed] [Google Scholar]
- 4.Pannicke U, Baumann B, Fuchs S, Henneke P, Rensing-Ehl A, Rizzi M, Janda A, Hese K, Schlesier M, Holzmann K, Borte S, Laux C, Rump EM, Rosenberg A, Zelinski T, Schrezenmeier H, Wirth T, Ehl S, Schroeder ML, Schwarz K. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl J Med. 2013;369(26):2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 6.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa] B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 7.Conte MI, Pescatore A, Paciolla M, Esposito E, Miano MG, Lioi MB, McAleer MA, Giardino G, Pignata C, Irvine AD, Scheuerle AE, Royer G, Hadj-Rabia S, Bodemer C, Bonnefont JP, Munnich A, Smahi A, Steffann J, Fusco F, Ursini MV. Insight into IKBKG/NEMO locus: report of new mutations and complex genomic rearrangements leading to incontinentia pigmenti disease. Hum Mutat. 2014;35(2):165–177. doi: 10.1002/humu.22483. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, Wallach D, Horwitz MS. Identification of a cell protein (FIP-3) as a modulator of NF-kappaB activity and as a target of an adenovirus inhibitor of tumor necrosis factor alpha-induced apoptosis. Proc Natl Acad Sci U S A. 1999;96(3):1042–1047. doi: 10.1073/pnas.96.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]