Abstract
Background
The methylotrophic yeast Pichia pastoris is a well-studied host organism for recombinant protein production, which is usually regulated either by a constitutive promoter (e.g. promoter of glyceraldehyde-3-phosphate dehydrogenase; PGAP) or an inducible promoter (e.g. promoter of alcohol oxidase 1; PAOX1). Both promoter systems have several advantages and disadvantages; with one of the main disadvantages being their lack of tunability. Various novel promoter systems, which are either inducible or de-repressed, allowing higher degrees of freedom, have been reported. Recently, bi-directional promoter systems in P. pastoris with two promoter systems regulating recombinant expression of one or more genes were developed. In this study, we introduce a novel bi-directional promoter system combining a modified catalase promoter system (PDC; derepressible and inducible) and the traditional PAOX1, allowing tunable recombinant protein production.
Results
We characterized a recombinant P. pastoris strain, carrying the novel bi-directional promoter system, during growth and production in three dynamic bioreactor cultivations. We cloned the model enzyme cellobiohydralase downstream of either promoter and applied different feeding strategies to determine the physiological boundaries of the strain. We succeeded in demonstrating tunability of recombinant protein production solely in response to the different feeding strategies and identified a mixed feed regime allowing highest productivity.
Conclusion
In this feasibility study, we present the first controlled bioreactor experiments with a recombinant P. pastoris strain carrying a novel bi-directional promotor combination of a catalase promoter variant (PDC) and the traditional PAOX1. We demonstrated that this bi-directional promoter system allows tunable recombinant protein expression only in response to the available C-sources. This bi-directional promoter system offers a high degree of freedom for bioprocess design and development, making bi-directional promoters in P. pastoris highly attractive for recombinant protein production.
Keywords: Pichia pastoris, Bi-directional promoter, Bioprocess development, Mixed feed, Tunable recombinant protein production
Background
The methylotrophic yeast Komagataella phaffii, also known as Pichia pastoris, is extensively used as host organism for recombinant protein production (e.g. [1–7]). The main advantages of P. pastoris are its fast growth, its ability to use the cheap substrate methanol as sole carbon source, its ability to perform typical eukaryotic post-translational modifications and the possibility of secreting the recombinant product [5, 6, 8–10]. Usually, recombinant protein production in P. pastoris is either regulated by a constitutive promoter, like the promoter of glyceraldehyde-3-phosphate dehydrogenase (PGAP), or an inducible promoter, like the promoter of alcohol oxidase 1 (PAOX1) (e.g. [7, 8, 11, 12]). Both of these prominent promoter systems are characterized by several advantages, but also disadvantages.
The strong, constitutive PGAP allows high product yields in rather short process times [13]. However, cell growth and recombinant protein production are directly linked causing a high metabolic burden for the cells, which might lead to the production of unwanted metabolites or even cell death [14].
On the other hand, biomass formation can be decoupled from recombinant protein production using the inducible PAOX1, which is tightly regulated and gives high expression levels [12, 13, 15, 16]. However, the safety aspect for industrial large-scale manufacturing processes with PAOX1-driven P. pastoris production strains needs particular attention, as the storage of large volumes of hazardous, flammable methanol is highly undesirable. Apart from the fact that methanol makes the fermentation process dangerous and environmentally non-friendly, methanol metabolism leads to great heat evolution and high oxygen consumption, which pose additional challenges for cultivations at large scales.
Furthermore, a common disadvantage of both promoter systems PGAP and PAOX1 is the lack of tunability. It would be highly advantageous to be able to adjust recombinant protein production to different growth conditions and environmental stress to reduce metabolic burden and thus the formation of unwanted metabolites. Besides, it would be highly advantageous to have an expression system in yeast where two recombinant genes can be controlled, regulated and tuned separately. Potential applications of such a system include the balanced co-expression of the individual peptide chains of dimeric proteins, or the consecutive expression of a chaperone followed by the tailored expression of the target product to boost the yield of correctly folded and active product (Fig. 1). Recently, we have used such a system for the recombinant production of the enzyme horseradish peroxidase [17]. However, such a tunable, bi-directional promoter system is not possible using a combination of the promoter systems PGAP and PAOX1 due to the aforementioned disadvantages. Thus, a lot of effort is ongoing to identify and investigate novel promoter systems, which are either inducible or de-repressed under limiting conditions [12, 18–26].
Fig. 1.
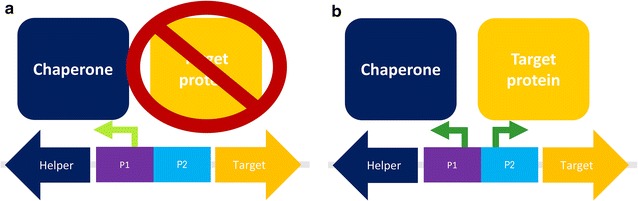
A bi-directional promoter system, where both promoters can be controlled and tuned separately allows a the consecutive and b the concomitant production of a chaperone and a target protein
In this study, we introduce a novel modified catalase promoter system (PDC), which is a 500 bp fragment of the DNA upstream of the peroxisomal catalase gene, active under both limiting conditions and by induction, allowing a high degree of freedom in its regulation and tunability. Furthermore, we present our results with respect to the characterization of a novel bi-directional promoter system, where we combined this novel PDC variant and PAOX1 in a bi-directional manner. In this feasibility study, we cloned two gene variants of the model enzyme cellobiohydrolase 2 (CBH2) downstream of either promoter to demonstrate the proof-of-concept that the expression level of this product can solely be tuned by induction conditions using the bi-directional promoter system. We used the same product downstream of the two promoters to exclude the possibility that different products are differently well expressed due to product-specific features, not only affecting productivity but also strain physiology. Thus, we ensured that the effects on productivity and physiology only resulted from induction conditions and the consequent regulation of the two promoters and not from product-specific features. In summary, our results nicely demonstrate tunability of a novel bi-directional promoter system solely in response to cultivation conditions, which widens the toolbox for P. pastoris. A potential future application of this system could be the subsequent production a chaperone and a target protein, as schematically shown in Fig. 1.
Methods
Host organism and model protein
A P. pastoris BSYBG11 strain harbouring a novel bi-directional promoter system, comprising the modified derepressible and inducible PDC and the methanol inducible PAOX1, was constructed by Bisy e.U. (Hofstaetten/Raab, Austria). BSYBG11 is a killer-plasmid-free next generation platform strain, based on the NRLY 11430 wildtype strain, where the AOX1 gene was deleted according to Sturmberger et al. [27]. The lignocellulolytic enzyme CBH2 derived from Trichoderma reesei [28] was used as a model protein in this study and cloned downstream of either promoter. Furthermore, the strain carried a Zeocin resistance gene, and the pre-pro signal sequence of the alpha-mating factor allowed product secretion.
Bioreactor cultivations
All bioreactor cultivations were carried out in a 5 L lab scale glass bioreactor (Infors, Switzerland). Monitoring and control of process parameters was done by the Lucullus process information management system (PIMS; Biospectra, Switzerland). pH was controlled at pH 5.0 and temperature at 30 °C. The dissolved oxygen concentration (dO2) was kept above 30% with a cascaded control of agitation and addition of pure oxygen.
Strain specific physiological parameters, such as substrate uptake rates (qs) and biomass yields (YX/S), are prerequisites for designing efficient fed-batch strategies. Thus, dynamic cultivations with shifts in qs and repeated substrate pulses were done to evaluate these strain specific physiological parameters, according to our previous studies (e.g. [29–32]). Based thereon, mixed feed cultivations were carried out to analyze tunability of recombinant protein production. A summary of the three dynamic cultivations performed in this study and their respective goals is shown in Table 1.
Table 1.
Dynamic cultivations performed with the recombinant P. pastoris strain carrying a novel bi-directional promoter system
| Experiment | Substrate | Feeding strategy based on qs [g g−1 h−1]a | Goals |
|---|---|---|---|
| FB1 | Glycerol | Batch on glycerol Fed-batch on glycerol with step-wise decrease of qs,gly 0.567–0.450–0.282–0.142–0.101–0.070–0.050 g g−1 h−1 (each step was done for 2 h) Methanol pulses: 0.5% (v/v) adaptation pulse at 30 °C—two 2% (v/v) pulses at 30 and 20 °C, respectively |
Determination of strain specific physiological parameters (qs,max,gly; qs,gly for maximum qp; qs,adapt,MeOH; qs,max,MeOH) Characterization of recombinant expression profile at different process parameters |
| FB2 | Glycerol and methanol | Batch on glycerol 0.5% (v/v) methanol adaptation pulse Constant MeOH feed at qs 0.02–0.025 g g−1 h−1 and concomitant glycerol feed with stepwise increases: 0.025-0.059-0.139-0.239 g g−1 h−1 (each step was done for 2 h) |
Find operating window for maximum qp |
| FB3 | Glycerol and methanol | Batch on glycerol Fed-batch at qs,gly = 0.212 g g−1 h−1 0.5% (v/v) methanol adaptation pulse followed by a fed-batch on methanol at qs = 0.034 g g−1 h−1 Mixed feed cultivation at qs,gly = 0.135 g g−1 h−1 and qs,MeOH = 0.035 g g−1 h−1 |
Demonstrate tunability of recombinant protein production |
aThe qs values were adjusted based on our findings during this study (vide infra)
Prior to all cultivations, precultures were prepared with frozen cryo-stocks in yeast nitrogen base medium (YNB) supplemented with Zeocin. The preculture was incubated at 30 °C and 230 rpm overnight. The batch phase for all cultivations was started by adding the preculture to the bioreactor [10% (v/v)]. After the batch, different feeding strategies were applied (Table 1). All cultivations were carried out in twofold basal salt medium (BSM) with a final glycerol concentration of 60 g L−1 in the batch medium. Samples were taken throughout the cultivations for offline analyses. In all the cultivations, the feed rate (FR) was calculated from biomass (cx) and substrate (cs) concentrations, volume of the bioreactor (VR) and specific substrate uptake rates (qs) and controlled using a feed forward strategy (Eq. 1).
| 1 |
Offline data analysis
Biomass concentration was determined by optical density measurements at 600 nm and dry cell weight measurement, as described before (e.g. [29–32]). Protein concentration was measured at 595 nm by the Bradford assay using the Sigma-Aldrich protein assay kit with bovine serum albumin as standard in the range of 0.2–1.2 mg mL−1. The specific productivity (qp) was determined from the total extracellular protein concentration measured in the cell-free cultivation broth and the respective dry cell weight. Product formation and electrophoretic purity was checked by SDS-PAGE.
Results and discussion
In this study, a recombinant P. pastoris strain harbouring a novel bi-directional promoter system with the two promoters PDC/PAOX1 was characterized and analyzed for tunable recombinant production of the model enzyme CBH2. The goal of this proof-of-concept study was to show that expression can be stirred solely by induction conditions using this novel bi-directional system, which is why we used the same model product to exclude product-dependent variations in productivity as well as product-dependent effects on physiology. We performed three dynamic bioreactor cultivations to characterize the recombinant P. pastoris strain (Table 1). In all of these cultivations we determined a maximum specific growth rate (µmax) on glycerol of 0.27 h−1, a qs,max,gly of 0.57 g g−1 h−1 and a biomass yield (YX/S) on glycerol of 0.47 g g−1. These values compare well with values we found for a MutS benchmark strain before [33], indicating that the introduced genetic construct had no negative impact on the physiology of the P. pastoris strain.
Fed-batch 1 (FB1)
In the first dynamic fed-batch, we determined strain specific physiological parameters and characterized the recombinant expression profile at different process parameters (Table 1). After complete consumption of glycerol in the batch phase, we stepwise decreased qs,gly in the subsequent fed-batch to find qs,gly where the PDC promoter was fully active. We determined the specific productivity (qp) as a measure for promoter activity. As shown in Table 2, the PDC promoter was actually never repressed, not even at high qs,gly. However, we found an optimum in qp at a qs,gly of 0.28 g g−1 h−1. At lower qs,gly, qp decreased again, as the cells came close to their maintenance metabolism, which was also visible in the specific yields (Table 2). Closing C-balances underlined the validity of the calculated physiological strain specific parameters.
Table 2.
Dynamic glycerol fed-batch phase to characterize the PDC in FB 1
| Steps | qs,gly (g g−1 h−1) | µ (h−1) | qp (mg g−1 h−1) | (Cmol Cmol−1) | YX/S (Cmol Cmol−1) | C-balance (−) |
|---|---|---|---|---|---|---|
| 1 | 0.567 | 0.318 | 0.307 | 0.28 | 0.67 | 0.95 |
| 2 | 0.450 | 0.251 | 0.362 | 0.34 | 0.67 | 1.01 |
| 3 | 0.282 | 0.132 | 0.492 | 0.37 | 0.56 | 0.92 |
| 4 | 0.142 | 0.068 | 0.349 | 0.44 | 0.51 | 0.95 |
| 5 | 0.101 | 0.056 | 0.124 | 0.46 | 0.47 | 0.93 |
| 6 | 0.070 | 0.039 | 0.121 | 0.51 | 0.43 | 0.94 |
| 7 | 0.050 | 0.023 | 0.073 | 0.58 | 0.37 | 0.95 |
After the dynamic glycerol phase in FB1, we added a 0.5% (v/v) methanol adaptation pulse, followed by two 2% (v/v) pulses at each 30 and 20 °C, respectively, to analyze the adaptation characteristics to methanol and determine the specific uptake rate of methanol (qs,MeOH) as well as qp at both temperatures (Table 3).
Table 3.
Methanol pulses at 30 and 20 °C, respectively, to characterize the PDC in FB1
| Phases | qs,MeOH (g g−1 h−1) | qP (mg g−1 h−1) |
|---|---|---|
| 0.5% adapt | 0.011 | 0.021 |
| 2% MeOHT=30 °C | 0.035 | 0.115 |
| 2% MeOHT=20 °C | 0.039 | 0.056 |
The adaptation time, which is the time taken for adaptation of the culture to the new substrate methanol [29, 30], of the strain carrying the bi-directional promoter system to methanol at 30 °C was only 3 h, which was much lower compared to a MutS benchmark strain, where we had found adaptation times of more than 6 h [29, 33]. This drastic reduction might be due to the concomitant presence of two promoters, which are inducible by methanol.
As shown in Table 3, the specific uptake rate of methanol was higher at 20 °C than at 30 °C. However, qp was higher at 30 °C, which is why we chose this temperature for the subsequent fed-batch phases on methanol.
Fed-batch 2 (FB2)
In FB2, we adapted one of our previous mixed feed strategies to allow fast physiological strain characterisation as well as bioprocess development in a mixed feed environment [34]. We constantly fed methanol at a qs = 0.022–0.025 g g−1 h−1, corresponding to around 75–80% of qs,max,MeOH at 30 °C (Table 3), and concomitantly fed glycerol, which we stepwise increased (Table 1). In the different phases, we determined specific rates and yields, to demonstrate tunability of the system and to find a good operating window allowing highest qp (Table 4). Again, closing C-balances underlined the validity of the calculated strain specific parameters.
Table 4.
Dynamics in FB2 to analyze tunability and to find an operating window for the bi-directional promoter system allowing highest qp
| Steps | qs,gly (g g−1 h−1) | qs,MeOH (g g−1 h−1) | µ (h−1) | qp (mg g−1 h−1) | (Cmol Cmol−1) | YX/S (Cmol Cmol−1) | C-balance (−) |
|---|---|---|---|---|---|---|---|
| 1 | 0.025 | 0.022 | 0.005 | 0.109 | 0.78 | 0.14 | 0.92 |
| 2 | 0.059 | 0.024 | 0.009 | 0.372 | 0.71 | 0.23 | 0.94 |
| 3 | 0.139 | 0.025 | 0.031 | 0.349 | 0.55 | 0.42 | 0.97 |
| 4 | 0.239 | 0.023 | 0.112 | 0.099 | 0.42 | 0.57 | 0.99 |
As shown in Table 4, qp could be tuned by adjusting different qs ratios. Interestingly, the highest qp was achieved in the presence of methanol at a rather low qs,gly. Compared to the qp at a similar qs,gly in FB1, which was 0.073 mg g−1 h−1 at qs,gly = 0.050 g g−1 h−1, we obtained a more than fivefold higher qp of 0.372 mg g−1 h−1 at qs,gly = 0.059 g g−1 h−1 in the presence of methanol. Furthermore, we found a 1.5-fold higher qp of 0.115 mg g−1 h−1 compared to the sole presence of methanol (Table 3). Increasing qs,gly in the presence of methanol had a negative impact on qp, which we ascribe to repression effects, as reported in similar studies before [34]. Offline analysis confirmed no accumulation of methanol in the mixed-feed phase.
Fed-batch 3 (FB3)
In the final cultivation (FB3), we demonstrated tunability of recombinant protein production by adjusting different feeding regimes. We analyzed productivity as well as the the strain specific, physiological parameters on glycerol, on methanol as well as in a mixed feed environment (Table 1). The results are summarized in Table 5.
Table 5.
Strain specific, physiological parameters determined in FB3
| Phase | qs,gly (g g−1 h−1) | qs,MeOH (g g−1 h−1) | µ (h−1) | qp (mg g−1 h−1) | (Cmol Cmol−1) | YX/S (Cmol Cmol−1) | C-balance (−) |
|---|---|---|---|---|---|---|---|
| Glycerol | 0.212 | – | 0.122 | 0.368 | 0.40 | 0.57 | 0.97 |
| Methanol | – | 0.034 | 0.003 | 0.097 | 0.83 | 0.15 | 0.98 |
| Mixed feed | 0.135 | 0.035 | 0.106 | 0.573 | 0.54 | 0.41 | 0.95 |
The qp in the glycerol fed-batch phase was comparable to the results we obtained in FB1 (Table 1). At a qs,gly = 0.212 g g−1 h−1 we calculated a qp = 0.368 mg g−1 h−1. In the subsequent fed-batch phase on methanol, where we adjusted a qs,MeOH = 0.034 g g−1 h−1, which was close to qs,max,MeOH, we calculated a qp of 0.097 mg g−1 h−1. When we concomitantly fed glycerol at a qs,gly = 0.135 g g−1 h−1, we were able to boost qp to 0.573 mg g−1 h−1, which was the highest value we obtained in all the experiments performed in this study. This value was higher than the qp we achieved in the mixed feed environment at qs,gly = 0.139 g g−1 h−1 in FB2, probably due to a higher qs,MeOH. Again, no methanol accumulation was identified during this cultivation. Closing C-balances underlined the validity of the calculated strain specific parameters. We analyzed the cell-free cultivation broth at different time points of the cultivation on SDS-PAGE gels. As shown in Fig. 2, the main protein fraction in the cell-free cultivation broth constituted for the recombinant product CHB2, which is why it was legitimate to use the total extracellular protein content to calculate productivities.
Fig. 2.
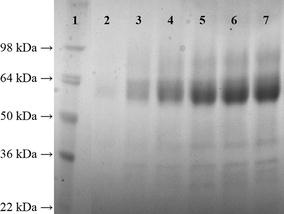
SDS-PAGE gel of cell-free cultivation broth taken at different time points during FB3. Lane 1, protein ladder; lane 2, sample after batch on glycerol; lanes 3–5, samples during fed-batch at qs,gly = 0.212 g g−1 h−1 (samples were taken every 2 h); lane 6, sample after methanol fed-batch; lane 7, sample after mixed feed phase. Size CBH2 = ca. 60 kDa
Conclusions
In this study we present the first controlled bioreactor experiments with a novel bi-directional promotor combination of a catalase promoter variant (PDC), which shows high activity in the presence of glycerol, but is also inducible by methanol, and the traditional PAOX1. By performing dynamic bioreactor cultivations, we physiologically characterized the recombinant strain and determined conditions allowing high productivity of the strain in only three experiments. Finally, we demonstrated that this bi-directional promoter system allows tunable recombinant protein expression solely in response to the available C-sources. This bi-directional promoter system offers a high degree of freedom for bioprocess design and development making bi-directional promoters in P. pastoris highly attractive for recombinant protein production.
Authors’ contributions
KM and VR conducted the experiments. CS generated the Pichia pastoris strain. OS planned and supervised the study and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors acknowledge the TU Wien University Library for financial support through its Open Access Funding Programme.
Competing interests
Bisy has an interest to commercialize bidirectional promoters.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by the Steirische Wirtschaftsförderungsgesellschaft and the program “Ideenreich”.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CBH2
cellobiohydrolase 2
- qP
strain specific productivity [mg g−1 h−1]
- qS
strain specific substrate uptake rate [g g−1 h−1]
- PDC
catalase promoter variant
- PAOX1
alcohol oxidase promoter
- PGAP
glyceraldehyde-3-phosphate dehydrogenase promoter
- PIMS
process information management system
- dO2
dissolved oxygen concentration [%]
- YX/S
yield of biomass per substrate [Cmol Cmol−1]
- YNB
yeast nitrogen base
- BSM
basal salt media
- FR
feed rate [L h−1]
- cS
concentration of substrate [g L−1]
- cX
biomass concentration [g L−1]
- VR
volume of the reactor [L]
yield of carbon dioxide per substrate [Cmol Cmol−1]
Footnotes
Vignesh Rajamanickam and Karl Metzger contributed equally to this work
Contributor Information
Vignesh Rajamanickam, Email: vignesh.rajamanickam@tuwien.ac.at.
Karl Metzger, Email: karlmetzger@me.com.
Christian Schmid, Email: christian.schmid@tugraz.at.
Oliver Spadiut, Phone: +43 1 58801 166473, Email: oliver.spadiut@tuwien.ac.at.
References
- 1.Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 2.Weinacker D, Rabert C, Zepeda AB, Figueroa CA, Pessoa A, Farias JG. Applications of recombinant Pichia pastoris in the healthcare industry. Braz J Microbiol. 2013;44:1043–1048. doi: 10.1590/S1517-83822013000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spohner SC, Muller H, Quitmann H, Czermak P. Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J Biotechnol. 2015;202:118–134. doi: 10.1016/j.jbiotec.2015.01.027. [DOI] [PubMed] [Google Scholar]
- 4.Cregg JM. Pichia protocols. 2nd ed. In: Cregg JM, editor. Methods in molecular biology. Totowa, NJ, US; 2007. p. 268.
- 5.Cregg JM, Cereghino JL, Shi JY, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 6.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 7.Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;13:329–332. doi: 10.1016/S0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]
- 8.Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- 9.Daly R, Hearn MT. Expression of the human activin type I and II receptor extracellular domains in Pichia pastoris. Protein Expr Purif. 2006;46:456–467. doi: 10.1016/j.pep.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Spadiut O, Herwig C. Dynamics in bioprocess development for Pichia pastoris. Bioengineered. 2014;5:401–404. doi: 10.4161/bioe.36152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang WH, Potter KJH, Plantz BA, Schlegel VL, Smith LA, Meagher MM. Pichia pastoris fermentation with mixed-feeds of glycerol and methanol: growth kinetics and production improvement. J Ind Microbiol Biotechnol. 2003;30:210–215. doi: 10.1007/s10295-003-0035-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhang AL, Luo JX, Zhang TY, Pan YW, Tan YH, Fu CY, Tu FZ. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol Biol Rep. 2009;36:1611–1619. doi: 10.1007/s11033-008-9359-4. [DOI] [PubMed] [Google Scholar]
- 13.Cos O, Ramon R, Montesinos JL, Valero F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Factories. 2006;5:17. doi: 10.1186/1475-2859-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nocon J, Steiger M, Mairinger T, Hohlweg J, Russmayer H, Hann S, Gasser B, Mattanovich D. Increasing pentose phosphate pathway flux enhances recombinant protein production in Pichia pastoris. Appl Microbiol Biotechnol. 2016;100:5955–5963. doi: 10.1007/s00253-016-7363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looser V, Bruhlmann B, Bumbak F, Stenger C, Costa M, Camattari A, Fotiadis D, Kovar K. Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv. 2015;33:1177–1193. doi: 10.1016/j.biotechadv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Mochizuki S, Hamato N, Hirose M, Miyano K, Ohtani W, Kameyama S, Kuwae S, Tokuyama T, Ohi H. Expression and characterization of recombinant human antithrombin III in Pichia pastoris. Protein Expr Purif. 2001;23:55–65. doi: 10.1006/prep.2001.1479. [DOI] [PubMed] [Google Scholar]
- 17.Krainer FW, Gerstmann MA, Darnhofer B, Birner-Gruenberger R, Glieder A. Biotechnological advances towards an enhanced peroxidase production in Pichia pastoris. J Biotechnol. 2016;233:181–189. doi: 10.1016/j.jbiotec.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Shen SG, Sulter G, Jeffries TW, Cregg JM. A strong nitrogen source-regulated promoter for controlled expression of foreign genes in the yeast Pichia pastoris. Gene. 1998;216:93–102. doi: 10.1016/S0378-1119(98)00315-1. [DOI] [PubMed] [Google Scholar]
- 19.Resina D, Serrano A, Valero F, Ferrer P. Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J Biotechnol. 2004;109:103–113. doi: 10.1016/j.jbiotec.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 20.Menendez J, Valdes I, Cabrera N. The ICL1 gene of Pichia pastoris, transcriptional regulation and use of its promoter. Yeast. 2003;20:1097–1108. doi: 10.1002/yea.1028. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Zhang Y, Zhang X, Yan C, Zhang Y, Xu X, Zhang W. Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci Rep. 2016;6:27352. doi: 10.1038/srep27352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol. 2015;32:9–17. doi: 10.1016/j.sbi.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Prielhofer R, Maurer M, Klein J, Wenger J, Kiziak C, Gasser B, Mattanovich D. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb Cell Factories. 2013;12:5. doi: 10.1186/1475-2859-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol. 2013;30:385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Vogl T, Ruth C, Pitzer J, Kickenweiz T, Glieder A. Synthetic core promoters for Pichia pastoris. ACS Synth Biol. 2014;3:188–191. doi: 10.1021/sb400091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogl T, Sturmberger L, Kickenweiz T, Wasmayer R, Schmid C, Hatzl AM, Gerstmann MA, Pitzer J, Wagner M, Thallinger GG, et al. A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth Biol. 2016;5:172–186. doi: 10.1021/acssynbio.5b00199. [DOI] [PubMed] [Google Scholar]
- 27.Sturmberger L, Chappell T, Geier M, Krainer F, Day KJ, Vide U, Trstenjak S, Schiefer A, Richardson T, Soriaga L, et al. Refined Pichia pastoris reference genome sequence. J Biotechnol. 2016;235:121–131. doi: 10.1016/j.jbiotec.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady SK, Sreelatha S, Feng Y, Chundawat SP, Lang MJ. Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps. Nat Commun. 2015;6:10149. doi: 10.1038/ncomms10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietzsch C, Spadiut O, Herwig C. A fast approach to determine a fed batch feeding profile for recombinant Pichia pastoris strains. Microb Cell Factories. 2011;10:85. doi: 10.1186/1475-2859-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietzsch C, Spadiut O, Herwig C. A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microb Cell Factories. 2011;10:14. doi: 10.1186/1475-2859-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spadiut O, Dietzsch C, Herwig C. Determination of a dynamic feeding strategy for recombinant Pichia pastoris strains. Methods Mol Biol. 2014;1152:185–194. doi: 10.1007/978-1-4939-0563-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spadiut O, Zalai D, Dietzsch C, Herwig C. Quantitative comparison of dynamic physiological feeding profiles for recombinant protein production with Pichia pastoris. Bioprocess Biosyst Eng. 2013 doi: 10.1007/s00449-013-1087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krainer FW, Gmeiner C, Neutsch L, Windwarder M, Pletzenauer R, Herwig C, Altmann F, Glieder A, Spadiut O. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris. Sci Rep. 2013;3:3279. doi: 10.1038/srep03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zalai D, Dietzsch C, Herwig C, Spadiut O. A dynamic fed batch strategy for a Pichia pastoris mixed feed system to increase process understanding. Biotechnol Prog. 2012 doi: 10.1002/btpr.1551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


