Abstract
Recently, clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 bacterial immunity system has opened a promising avenue to treat genetic diseases that affects the human hematopoietic stem cells (HSCs). Therefore, finding a highly efficient delivery method capable of modifying the genome in the hard-to-transfect HSCs, combined with the advanced CRISPR-Cas9 system, may meet the challenges for dissecting the hematologic diseases mechanisms and facilitate future clinical applications. Here, we developed an effective HSC-specified delivery microfluidic chip to disrupt the cell membrane transiently by inducing a rapid mechanical deformation that allowed for delivering biomaterials into cytoplasm from the surrounding matrix. Compared with the previous designs, the new nano-silicon-blade structure was specifically optimized for HSCs. Using the silicon substrate, the sharpness and rigidity of the nano-blade constriction was largely enhanced to improve the biomaterials delivery efficiency. We achieved highly efficient delivery results by transporting various macro-molecules into the HSCs. Moreover, the treated HSCs possess high viability and maintain inherent pluripotency after the delivery via the Nano-Blade Chip (NB-Chip). Subsequently, we disrupted p42 isoform in C/EBPα on the NB-Chip and induced HSCs into a myeloid proliferation behavior. In conclusion, the NB-Chip provides a harmless, rapid and high-throughput gene editing approach for the HSC study and therapeutics.
Introduction
For decades, hematopoietic stem cells (HSCs) have shown promising potentials in research and clinical applications. Particularly in recent years, the current genetic modification technologies, allowing precise manipulation of the biological functions, have created many new research possibilities in the HSC study and potential treatments of hematologic diseases.1–3 Numerous methods have been developed to dissect the mechanisms between differentiation and proliferation of the HSCs in vivo and in vitro, for example, animal models and genome engineering via retroviral vectors.4–6 Although these methods have greatly improved our understanding of the HSC mechanisms, the long-time costs and high expenses associated with generating new animal models and the unknown potential risks that accompany the use of the retroviral vectors have built serious obstructions towards the use of these technologies.7 The prevalent electroporation delivery method provides an impressive efficiency genome editing way, but the high voltage pulse inevitably decreases cell viability and may also alter the original properties of the HSCs.8,9 Consequently, an urgent need still exist to find a harmless, rapid, high-throughput method that also possesses high delivery efficiency with minimum external stimuli, to accelerate the research and therapeutic applications for the HSCs.
Micro and nano fluid platforms, which use precise fabrication techniques and the physical properties of flow and operate at the micron and nano scale, show strong potential in delivery applications.10,11 Recently, our lab developed a microfluidic chip that uses physical constrictions to deform and shear cells membrane for the macro-molecules delivery.12 With rapid mechanical deformation, disruptions that facilitate the passive diffusion of biomaterials from the surrounding matrix into the cytoplasm, will be created temporally on the membrane.13,14 As we effectively delivered Cas9 ribonucleoprotein (RNP) complex into human CD4+ T cells, this broad applicable delivery chip is then considered more suitable for the HSCs, when compared with the electroporation method, due to utilizing physical interaction forces without electronic stimuli.15 In order to satisfy the HSC delivery demands, polydimethylsiloxane (PDMS) was replaced by silicon as the chip substrate material. The silicon material allowed for the fabrication resolution to be increased to nanoscale which was necessary for achieving the nano-silicon-blade constriction while at the same time exhibiting a less deformable surface.16,17 The delivery parameters were optimized accordingly. Using the optimized delivery chip (the Nano-Blade Chip, NB-Chip), we achieved very impressive results when delivering macro-molecules or plasmids into the HSCs. Delivered HSCs also keep inherent pluripotency longer time when compared with HSCs treated with the electroporation method. We then delivered Cas9 protein complex with designed single-guide RNA (sgRNA) into the human HSCs on the NB-Chip to disrupt the CCAAT/enhancer-binding protein-α (C/EBPα/CEBPA) p42 isoform. This mutation target is known for inducing the acute myeloid leukaemia (AML) in which myeloid progenitor proliferation is uncontrollable but the differentiation ability is blocked.14 Biochemical and functional analysis proved that the NB-Chip effectively delivered Cas9 RNP complex into the human HSCs and disrupted the C/EBPα p42 isoform in vitro. Thus, as expected, the NB-Chip provides a harmless, rapid and more effective delivery method for future applications. (Fig. 1)
Fig. 1.
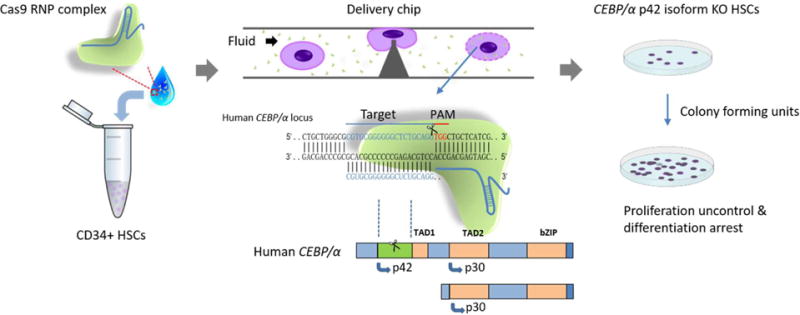
Experimental scheme of Cas9 ribonucleoprotein (RNP) complex delivery for genome editing in the HSCs via the NB-Chip. Transient deformation can disrupt cell membrane and create holes on surface that facilitate the diffusion of Cas9 RNP complex. The knock out, p42 isoform in C/EBPα, HSCs perform uncontrollable proliferation with arrested differentiation, a similar symptom to acute myeloid leukaemia (AML).
Materials and methods
Materials and Reagents
SU8-3010 photoresist was purchased from MicroChem. PDMS was purchased from Fisher Scientific. Tygon tubing was purchased from Saint-Gobain. Fetal bovine serum (FBS), trypsin, and penicillin-streptomycin were purchased from Fisher Scientific. StemSpanTMSFEM and StemSpanTMC110 were purchased from StemCells Technologies for culture and expansion of the HSCs. Dulbecco’s modified Eagle’s medium (DMEM) insulin, hydrocortisone, and phosphate-buffered saline (PBS) were purchased from Life Technologies. FITC-labeled, cascade blue dextran with different molecule weight (3 kD, 10 kD, 70 kD) and Cas9 Nuclease (3 μg/μL) were purchased from Thermofisher. The 20-bp target sequences of sgRNAs targeting C/EBPα were synthesized by Thermofisher. The sequences of the indicated sgRNAs were as follows:
sgEGFP-1, GGGCGAGGAGCTGTTCACCG;
sgEGFP-2, GAGCTGGACGGCGACGTAAA;
sgC/EBPα-1, CGTGCGGGGGGCTCTGCAGG;
sgC/EBPα-2: GCGCGTGCGGGGGGCTCTGC.
Chip design and fabrication
The NB-Chip pattern was designed with AutoCAD (Autodesk), and the microfabrication was performed in the Microelectronics Research Center at UT-Austin. Briefly, we utilized a 500-um thickness silicon wafer as substrate, then 450 nm of silicon oxide (SiO2) was deposited on the wafer by wet thermal oxidation (1050 °C). Then microchannels were patterned on the SiO2 film by standard photolithography using a NR9-500P negative photoresist (Futurrex Franklin, NJ, USA) and contact aligner (K.Suss MA6 mask aligner). Next, we transferred the channel pattern into SiO2 layer by reactive ion etch (RIE) in CF4 plasma (Plasmatherm BatchTop, 15 sccm CF4, 100 mTorr, 175W RF). Then 10-μm depth microchannels were etched into the silicon by Bosch based Deep Silicon Etch (DSE) on an Inductively Coupled Plasma Etcher (Plasma-Therm Versaline ICP) to form the nano-silicon-blade structures. The etching depth was monitored using a Dektak 150 Optical Profilometer. The PDMS microfluidic chips as control were fabricated using standard photolithography procedures.
Cell culture and colony-forming unit assays
Primary human CD34+ HSCs were purchased from StemCells Technologies and grown in StemSpanTMSFEM with 1% StemSpanTMC110, 0.5% L-glutamine and 0.5% penicillin-streptomycin in a humidified atmosphere of 5% CO2/95% air at 37°C. MDA-MB-231 cells were purchased from ATCC and grown in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were cultured in same environments during and after the delivery. For colony forming units assays (CFUs), CD34+ HSCs were plated in methylcellulose media (HSC-CFU complete with Epo, StemMACS) and total colonies were scored at 14 d following plating and classified according to their morphology.
Western blotting and flow cytometry
For western blotting after knockout, the HSCs and the MDA-MB-231 cells were allowed to recover in culture for 2 or 3 d after the delivery, respectively. The primary antibodies used were anti-C/EBPα (ab15048, Abcam), and anti-actin (A3853, Sigma-Aldrich). For flow cytometry analysis after RFP-expressed plasmid delivered into HSCs and sgEGFP-mediated knockout in MDA-MB-231 on the NB-Chip, cells were allowed to recover in culture for 3 d, followed by fluorescence analysis with a BD LSR Fortessa cell analyser, respectively.
Results
Delivery Mechanisms and Results
The delivery chip was designed with AutoCAD software and fabricated using photolithography and RIE technologies (More details in materials and methods). The structure of the NB-Chip is composed of cell-scattering zones near the chip inlet and outlet, and a deformation zone in the center of the chip containing several parallel nano-silicon-blade structures (Fig. 2A and S1A). As we know, the cell membrane will experience a rapid mechanical deformation when passing through the nano-blade structures, generating several transient membrane holes once the interaction force exceeds the phospholipid bilayer stress limitation.12,15,16 The interaction force, consisting of compressive and shear forces between the cell membrane and the constrictions on chip, is mainly determined by the size of the contact area. To achieve highly efficient delivery results, it is necessary to use harder materials and sharpen the constrictions to increase pressure on the phospholipid bilayer and create membrane holes large enough to enable macro-molecules passing through and diffusing into the cytoplasm. Delivered cells also need to be retained with significant viability after the cell membrane recovers from the transient disruptions. The delivery operation was also optimized including fluid rate, cell and target biomaterial concentrations and repeats time of the nano-blades. We utilized silicon as the substrate material instead of PDMS for two main reasons. First, the Young’s modulus of silicon is around 106 times higher than PDMS, which will provide a larger interaction pressure when the cell is passing through the nano-blade deformation zone. Second, the fabrication resolution on the nano-blade can be improved down to nanoscale along with the 10-μm depth blade structure which PDMS cannot achieve. Fluid distribution and the interaction force comparison between the optimized and previous design were simulated using COMSOL software. (Fig. 2B and Fig. S1B)
Fig. 2.
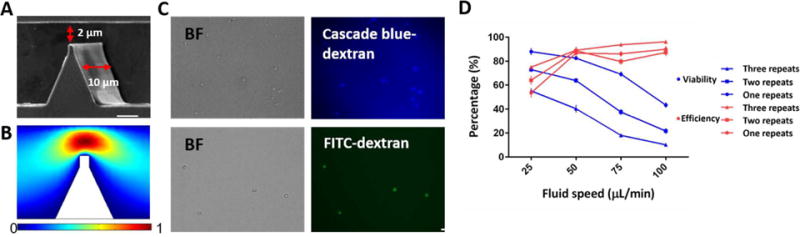
Chip design and efficiency test. (A) Scanning electron microscopy image of constrictions on chip are shown. The red arrow indicates the 2μm narrow deformable zones with 200 nm-radius spike constrictions and the 10 μm-depth channel. Scale bar, 2 μm. (B) Fluid velocity simulation at deformation zone by COMSOL software with 50 μL/min at the inlet. (C) Representative images of the delivery of two types of dextran into HSCs under the optimized conditions. Scale bar, 50 μm. (D) Delivery efficiency and cell viability after 1 d were calculated for different fluid rates and different repeat times of barriers in each channel (n = 5).
Next, we tested delivery efficiency and cell viability under different repeats of the nano-silicon-blade structure, cell concentrations and fluid rates. (Fig. 2C, D) Based on the cell membrane disrupting and recovering mechanism14, we considered that the number of the membrane holes has insignificant effects on the delivery results. Because during the additional 30 min incubation time after the delivery, target biomaterials will eventually diffuse into cytoplasm even if only a few membrane holes existed, under the influences of the concentration difference of the target delivery biomaterials between the surrounding matrix and the cytoplasm. To verify this phenomenon, we simulated the delivery process and analysed the force distributions using COMSOL. (Fig. 3A, B, Movie S1). As expected, the pressure on the nano-blade side is much higher than that on the flat side. We observed that utilizing the nano-blade constrictions only on one side will noticeably enhance the viability with only a slightly decrease of the delivery efficiency. (Fig. 4A) Considering all mentioned impact factors and using cascade blue 3 kD dextran, we optimized fluid rate to be 50 μL/min, cell concentration to be 1×l05/mL and one nanosilicon blade structure in one side per zone as final parameters. After optimization, we tested and compared the chip performance of the new design versus the previous version. Cascade blue 3 kD, 10 kD dextran and fluorescein isothiocyanate (FITC)-labeled 70 kD dextran were delivered into the HSCs via both chips and the delivery efficiency and cell viability were calculated. As expected, results showed that both chips possess similar cell viability, but the NB-Chip achieved noticeably higher delivery efficiency than the previous one (Fig. 4B). We achieved ~70% delivery efficiency with ~80% cell viability as our best results when delivering 70kD FITC labeled dextran into the HSCs. (Fig. S2) To test biologically functional macro-molecules on chip, we delivered plasmids, expressing RFP as a reporter, into the HSCs and tested the expression percentage. We obtained ~40% of RFP expressed HSCs and ~60% viability after culture for 2 d. (Fig. 4C) These data are consistent with the previous reports that plasmids have more toxicity when compared with the Cas9 RNP complex, which has a more straightforward approach to edit genes. Therefore, we chose to deliver Cas9 RNP complex instead of plasmids into the HSCs to edit the genome via our NB-Chip.
Fig. 3.
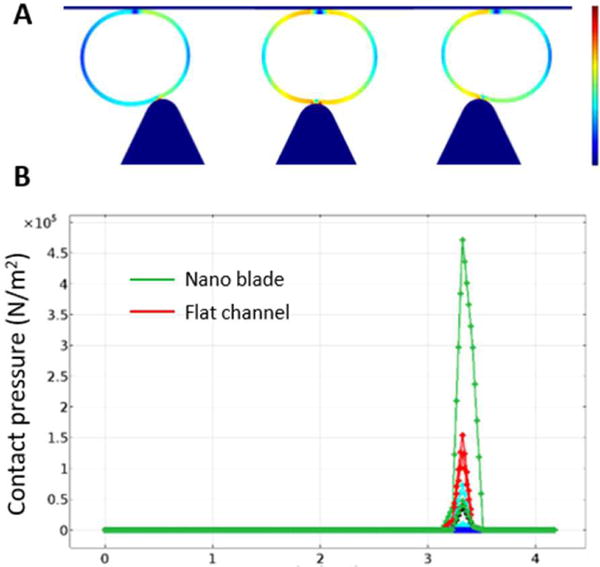
Contact pressure distribution analysis. (A) Contact pressure distribution when cell is passing a constriction only in one side. (B) The value of the highest pressure in each side implies that the nano-blade creates larger pressure than the flat channel side.
Fig. 4.
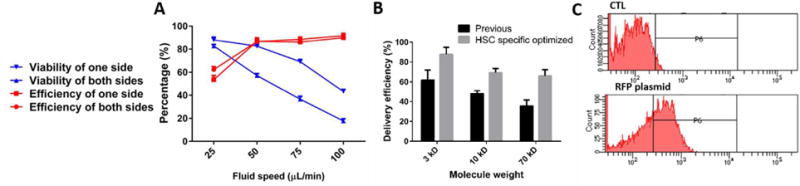
Delivery efficiency of the NB-Chip. (A) Delivery efficiency and cell viability after 1 d were calculated for at different fluid rates and constrictions in both sides and one side using 3 kD macro-molecules (n = 5). (B) Comparisons of delivery efficiency for different weight macro-molecules between optimized design and previous version (n = 5). (C) Histograms of RFP plasmid expression levels assessed by flow cytometry 2 d after treatment.
Functional assessment of the delivered HSCs
Preservation of the original biological functions after delivery plays a critical role in functional assessment of the delivery methods, especially in HSCs, which possesses a delicate balance between the differentiation and the proliferation in vivo.5,19 Compared with the retroviral approach, the electroporation method can avoid the risk of the viral integration into the HSCs genome.20,21 However, disrupting the membrane using high voltage pulses may produce unexpected dysfunctions.22 To assess the performance of our chip compared with the electroporation method, we first delivered the FITC-labeled 70 kD dextran into the HSCs to test the delivery efficiency.23 Both methods can achieve high efficiency after the delivery (~70%) (Fig. 5A).
Fig. 5.
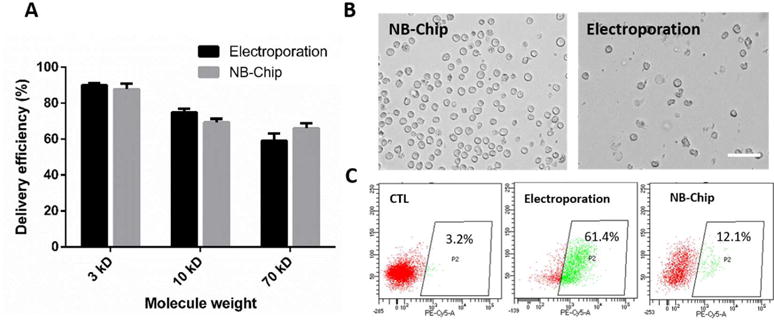
Delivery performance of the NB-Chip. (A) Comparison of the delivery efficiency for different weight of the macro-molecules between our chip and electroporation method. (B) Representative images of the HSC morphology after treatment by the NB-Chip and the electroporation method with Cas9 delivery only. Scale bar, 50 μm. (C) The surface marker CD38 expression level in HSCs were calculated after cells treatment with the deformation chip and electroporation method after 3 d by flow cytometry and the percentage of CD38+ cells is indicated.
Next, we measured the expression level of the surface marker CD38, which is associated with the differentiation24 in HSCs, after only delivering the Cas9 protein via both methods. Image results showed that the HSCs treated by the NB-Chip presenting better morphologies in comparison with the electroporation method, which indicates that the interaction force involved in the NB-Chip has a minor influence on the HSCs cell membrane and the cytoplasm than electrical stimuli (Fig. 5B). Interestingly, the HSCs treated by the NB-Chip reported lower CD38 expression level than the electroporation method after 2 d, which is more similar with the control group. (Fig. 5C) These results proved our hypothesis that treating the HSCs with high voltage pulses will inevitably induce some unexpected stimuli whereas disrupting the cell membrane by the transient deformation can mainly maintain the HSCs inherent pluripotency for longer time. Considering these advantages, we confirmed that the NB-Chip delivery strategy may provide a more capable platform for the HSCs gene editing in clinical applications.
Efficient gene disruption in human HSCs via the NB-Chip
Encouraged by the results obtained above, we assessed the feasibility and the efficiency of delivery with the Cas9 RNP complex. First, the MDA-MB-231 cells stably expressing enhanced green fluorescent protein (EGFP) were chosen to demonstrate the editing ability of our delivery chip.15 SgRNAs were designed to target the EGFP, and Cas9 protein was pre-complexed with the sgRNA to generate a ribonucleoprotein. Then we delivered the Cas9 RNP complex into MDA-MB-231 cells under a new set of specified conditions and analysed the results by flow cytometry after 3 d. As expected, the percentage of EGFP expressed cells decreased from ~100% to 18% which is consistent with previous reports (Fig. 6A). These results gave us confidence to deliver the Cas9 RNP complex into the hard-to-transfect and stimulus sensitive HSCs.
Fig. 6.
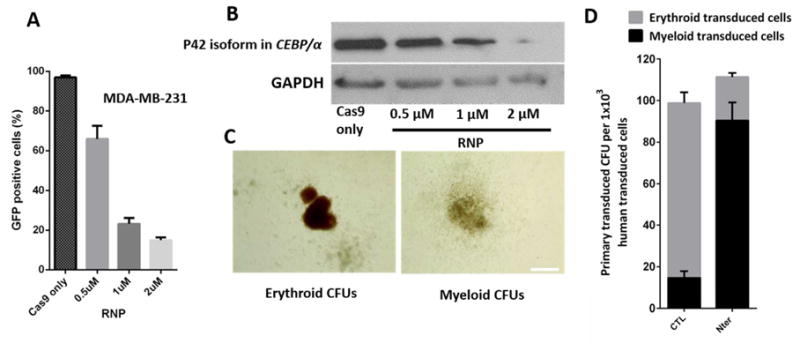
Evaluation of Cas9 RNP complex delivery results. (A) MDA-MB-231 cells stably expressing EGFP with different concentrations of Cas9 RNP targeting EGFP delivered through the deformation chip. The percentage of EGFP positive cells is shown after 3 d (n = 5). (B) Analysis of p42 isoform expression level under different concentrations of Cas9 RNP complex delivered Into HSCs. (C) Representative CFU experiments after two-week culture of normal HSCs and p42 isoform KO HSCs. Scale bar: 500 μm. (D) Analysis of the effect of the p42 Isoform functions on HSCs by CFU assay, 1×103 HSCs after transfection were plated on the CFU medium. The number of erythrold and myeloid colonies per cells are reported, (n = 5)
We chose to disrupt the C/EBPα in HSCs, which is a member of the CEBP family of bZIP transcription factors encoding two different translational isoforms of p42 and p30 (42 and 30 kD, respectively) with the same open reading frame, by using the alternative AUG codons.25 The two isoforms are expressed in a balanced way in HSCs but while p42 has the ability to block cell proliferation and induce differentiation of adipocytes and granulocytes, p30 has not.26 Mutations in acute myeloid leukaemia (AML) observed in ~9% of patients generally occurred in the C/EBPα, and specifically abolish translation of the full-length (p42) C/EBPα leading to an alteration between the two isoforms.16 It is meaningful to use the Cas9 RNP complex to target isoforms in genome engineering, and the ability to disrupt or correct the pathogenic genome sequence in HSCs could have broad applications to clinically reintroduce the genome engineered cells into patients. In our experiments, the designed Cas9 RNP complex was delivered by the NB-Chip and protein expression was analysed by western blot. We observed that the p42 expression level is correlated with the Cas9 RNP complex concentration from 0 to 2 μM. (Fig. 6B) To assess the function of the genome engineered HSCs, we performed colony forming units (CFU) both in WT and p42 disrupted HSCs. We analysed the number and the type of CFU colonies using an inverted fluorescent microscope after 14 d. As expected, the p42 isoform disruption group had a significant accumulation in the total number of the CFU colonies and the percentage of the myeloid colony type visibly increased when compared with WT group. (Fig. 6C and D) Thus, our data strongly suggests the NB-Chip can edit target genome isoform in a highly efficient level. Still, additional Cas9-mediated homology-directed repair (HDR) for C/EBPα intended to correct mutants and posterior reintroduction of the genome corrected cells into mouse models, to evaluate our chip clinical potential, will be studied later.
Conclusions
Highly efficient and effective delivery methods for the HSCs have great potentials in research and clinical applications.27 The NB-Chip uses the mechanical deformability of cells to generate transient membrane disruptions that facilitate biomaterials diffusion into the cytoplasm from the extracellular matrix. Generally, there are mainly three directions to improve the delivery efficiency on the deformation chip including increasing the deformation degree (narrowing the deformation zone), using faster fluid flow and enhancing the contact pressure. For HSCs, known as small and hard-to-transfect cells, which are lacking strong deformation ability due to the existence of large nuclei, narrowing the deformation zone will severely decrease viability after the delivery. Utilizing faster flow speed is not appropriate for increasing the delivery efficiency while keeping cell viability at same time. So, a potential way to increase the delivery efficiency while maintaining cell viability is to enhance a very localized pressure and create deliverable channels/pores during an extremely short time. The sharp constriction blade is a unique way of fulfilling such two purposes. Moreover, the cell membrane recovery mechanisms illustrate that cell membrane fixing will not occur immediately after the membrane disruption. To help cells recover from the membrane disruptions, we reduced the number of constriction repeats and left only one-side with nano-blades instead of both sides, the delivery results were mainly maintained but the cell viability was increased to an impressive level (~ 85%). We verified the deliver efficiency to be around the same level with that of the optimized electroporation strategies in dextran delivery, and established our advantages that the NB-Chip is capable of maintaining the pluripotency of the HSCs for longer time than current physical approaches. The NB-Chip should also broaden potential applications by the non-virus and unsevere stimulus method.
As Cas9 RNP complex is less toxic and produces a straightforward interaction with genes, combining the NB-Chip with the Cas9 RNP complex apparently promotes the gene engineering efficiency in the HSCs. In consequence, we disrupted the p42 isoform in C/EBPα using our chip by inducing the Cas9 RNP complex into the HSCs, although now we still do not have methods to observe which cells are edited, based on our biochemical analysis, we can still conclude that this platform disrupted p42 isoform in a highly efficient and accurate way. In conclusion, the effective, high-throughput and CRISPR/Cas9 based microfluidic delivery NB-Chip makes it possible to screen the functions of genetic mutants associated with hematologic diseases, such as AML, in a low-cost and rapid way. Still, we hope that the negligible stimuli advantage of the NB-Chip may provide more opportunities for the hematologic diseases therapies in the future.
Supplementary Material
Insight, innovation, integration.
Finding a highly efficient delivery method for the hard-to-transfect HSCs will significantly promote the research of genetic hematologic diseases and broaden its clinical potential. Now, the existing delivery methods cannot perfectly meet the demands of harmless, rapid and high efficiency for the HSC gene editing. Here we developed a HSC-specified delivery microfluidic chip with nano-silicon-blade constrictions to achieve highly efficient delivery results by disrupting the cell membrane via a transient deformation mechanically. Besides the high efficiency and viability advantages, the Nano-Blade Chip (NB-Chlp) also Induces minimum stimuli to maintain the HSC inherent pluripotency during and after the delivery, which may provide more opportunities for the hematologic diseases therapies in the future.
Acknowledgments
We are grateful for funding support from R01 CA180083, R56 AG049714, and R21 CA191179.
References
- 1.Adams GB, Scadden DT. Nature immunology. 2006;7:333–337. doi: 10.1038/ni1331. [DOI] [PubMed] [Google Scholar]
- 2.Copelan EA. New England Journal of Medicine. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 3.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q. Proceedings of the National Academy of Sciences. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack M, Wulffraat N, Leboulch Pa, Lim A, Osborne C, Pawliuk R, Morillon E. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 8.Talele S. Emerging Trends in Computing, Informatics, Systems Sciences, and Engineering. Springer; 2013. pp. 567–575. [Google Scholar]
- 9.Hoiles W, Krishnamurthy V, Cranfield CG, Cornell B. Biophysical journal. 2014;107:1339–1351. doi: 10.1016/j.bpj.2014.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Bao N, Paris L, Geahlen R, Lu C. Anal Chem. 2008;80:9840–9844. doi: 10.1021/ac801940w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Fei B, Geahlen RL, Lu C. Lab Chip. 2010;10:2673–9. doi: 10.1039/c0lc00131g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X, Liu Z, Chan Jo M, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L. Science advances. 2015;1:e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han MJ, Lytton-Jean A. Proceedings of the National Academy of Sciences. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak OS, Young YN, Marple GR, Veerapaneni S, Stone HA. Proceedings of the National Academy of Sciences. 2015;112:9822–9827. doi: 10.1073/pnas.1512152112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Liu Z, Ma Y, Zhang K, Qin L. Advanced Biosystems. 2017 doi: 10.1002/adbi.201600007. [DOI] [PubMed] [Google Scholar]
- 16.Nerlov C. Nature Reviews Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 17.Sukharev S, Betanzos M, Chiang CS, Guy HR. Nature. 2001;409:720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 18.Wiggins P, Phillips R. Proceedings of the National Academy of Sciences. 2004;101:4071–4076. doi: 10.1073/pnas.0307804101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Proceedings of the National Academy of Sciences. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng T, Zhan Y, Wang HY, Witting SR, Cornetta KG, Lu C, Control J. Release. 2010;144:91–100. doi: 10.1016/j.jconrel.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Zhan Y, Cao Z, Bao N, Li J, Wang J, Geng T, Lin H, Lu C, Control J. Release. 2012;160:570–576. doi: 10.1016/j.jconrel.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Human gene therapy. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 23.Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, Hsu JI, Hoegenauer KA, Rooney CM, Goodell MA. Cell reports. 2016;17:1453–1461. doi: 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenzie JL, Gan OI, Doedens M, Dick JE. Experimental hematology. 2007;35:1429–1436. doi: 10.1016/j.exphem.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Quintana-Bustamante O, Smith SLL, Griessinger E, Reyal Y, Vargaftig J, Lister TA, Fitzgibbon J, Bonnet D. Leukemia. 2012;26:1537–1546. doi: 10.1038/leu.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. New England Journal of Medicine. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 27.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S. Journal of biotechnology. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


