Abstract
Based on the increasing number of multi-drug resistant bacteria in periprosthetic infections, improvement of the antibacterial activity of commonly used biomaterials must be achieved. The broad-spectrum, high antimicrobial efficacy has made silver nanoparticles a promising new antibacterial agent. However, there is still a serious lack of knowledge concerning the impact of nanosilver on bone cells.
For this reason a study was conducted to evaluate the influence of silver nanoparticles on osteoclastogenesis of human peripheral blood mononuclear cells. Upon incubation with subtoxic concentrations of nanosilver the cells did not exhibit changes in osteoclast differentiation and podosomal structures. However, the osteoclasts were able to uptake the nanoparticles, accumulating them in endo-lysosomal compartments. Furthermore, nanosilver exposure led to an increase in oxidative stress and a decrease in clathrin-dependent endocytosis on the mRNA level.
In conclusion, our results indicate nanosilver-induced cell stress at higher concentrations. For this reason antibacterial benefits and possible health risks should be weighed in more detail in further studies.
1. Introduction
Bone tissue is a highly dynamic part of the endoskeleton comprised of a difficile interplay of bone formation and resorption. One key player in that sophisticated remodeling process are osteoblasts, which derive from mesenchymal stem cells and produce collagen as well as hydroxyapatite crystals, forming the extracellular bone matrix [1], [2]. The second key player are osteoclasts, which evolve from hematopoietic stem cells and are able to dissolve the organic as well as the inorganic compounds of the bone matrix by release of hydrogen ions and proteases, like cathepsin K [3], [4]. The activated, mature osteoclasts appear as multinucleated, polarized cells that exhibit adhesion structures called podosomes [5]. These podosomes are a prerequisite for sealing zone formation, marking the establishment of the ruffled border and therefore bone resorption [6], [7]. Only if balance is found between osteoblast and osteoclast activity, healthy bone structures and successful anchorage of implants can be achieved.
Bacterial colonization of implant surfaces is a common incidence in surgery and orthopedics and poses a devastating situation for the patient [8], [9], [10]. Treatment of periprosthetic infections proves to be difficult due to biofilm formation that protects the sessile bacteria against antimicrobial agents and the immune defense [11]. An already existing biofilm is highly stable and extremely difficult to eradicate, why bacterial adhesion and therefore biofilm formation has to be inhibited in the first place. Since the majority of implant-associated infections are caused by gram positive staphylococci [11], aminoglycosides like gentamicin have become a popular preventive additive for biomaterials [12]. However, in the last decades an increase of infections caused by multi-drug resistant organisms (MDRO) can be observed [11]. This circumstance requires the design of non-conventional antibiotics for infectionprophylaxis that allow broad spectrum efficacy, including MDRO. Silver nanoparticles (AgNPs) have been demonstrated to assure high antimicrobial efficacy while revealing low cytotoxicity [13], [14], [15], [16]. Due to the versatile mechanisms of bacterial inhibition, including disruption of bacterial membrane, interference with energy production and inhibition of replication, silver-resistance cannot easily be gained [17], [18], [19].
However, the potential use of silver nanoparticles for medical applications demands the thorough investigation of cellular interaction with the material. Therefore, a study was conducted to examine the interaction of pure AgNPs with human osteoclasts. Additionally, cellular uptake and intracellular distribution of nanosilver were studied.
2. Methods
2.1. Nanoparticles and nanoparticle characterization
The AgNPs were provided by ras materials GmbH (Regensburg, Germany). The nanoparticles are embedded in an aqueous stabilizing matrix composed of 4% polyoxyethylene glycerol trioleate (PGT) and 4% polyoxyethylene sorbitan monolaurate (Tween20), exhibiting a nominal silver content of 10 w/w%. Pure stabilizing matrix was included into the experiments as an internal control to distinguish between matrix-induced and nanoparticles-induced alterations. All stock solutions were prepared by dilution in sterile deionized water. The stocks were further diluted in cell medium to achieve the final concentrations for the experiments. All samples were kept in the dark to avoid light-induced reduction of silver. Medium without test substance supplement served as a negative control.
The hydrodynamic diameter and the zeta potential of 20 μg/g AgNPs diluted in cell medium or deionized water were determined by conducting Dynamic Light Scattering with a Malvern Zetasizer Nano Range (Malvern Instruments GmbH, Herrenberg, Germany) at 25 °C and an excitation of 633 nm. To define the agglomeration behavior of the AgNPs in the corresponding cell medium UV–vis spectrometry was applied. Therefore, 20 μg/g AgNPs diluted in cell medium or deionized water were transferred to a 1 cm cuvette and the UV–vis spectrum was measured at wavelengths between 350 and 700 nm with an 8453 UV–vis spectrophotometer (Hewlett-Packard, Palo Alto, California, USA).
2.2. Cell culture
Peripheral blood mononuclear cells (PBMCs) were isolated from human buffy coats of healthy anonymous donors. Buffy coats were provided by the blood donation center of the Justus-Liebig-University Giessen, Germany. Cells from the donors were not pooled. Lymphocytes and monocytes were separated from the blood by density gradient centrifugation based on Ficoll (LeucoSep, greiner bio-one, Frickenhausen, Germany). Subsequently, 10.5 × 105 PBMCs per cm2 were seeded into 24-well plates and incubated with DMEM high glucose including 10% fetal calf serum, 100 U/ml penicillin, 100 μg/g streptomycin and 25 ng/ml macrophage-colony stimulating factor (M-CSF) (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). To induce osteoclastogenesis, 25 ng/ml receptor activator of NFκB ligand (RANKL) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), 1 ng/ml transforming growth factor-β1 (TGF-β1) (Sigma-Aldrich) and small pieces of calf bone slices were given to the monocytes after 72 h. Additionally, the corresponding amount of AgNPs was given to the cells. The cells were incubated at 37 °C and 6% CO2 and the osteoclast medium, including the AgNPs, was changed twice per week.
2.3. Cell morphology and adhesion
After 8, 18 and 28 days changes in outer cell morphology were determined using a Leica microscope TYPE 090-135.002 (Leica Microsystems GmbH, Wetzlar, Germany) equipped with a Nikon Ds-Fi1 digital camera (Nikon, Duesseldorf, Germany). Concentrations above 3000 ng/g nanosilver revealed strong cytotoxicity indicated by cell death. Therefore, sublethal concentration of ≤3000 ng/g nanosilver were used in the following experiments.
Additionally, cell morphology was examined by fluorescence microscopy. Therefore, PBMCs were incubated with 1000 ng/g AgNPs for 8, 18 and 28 days. Subsequently, the samples were fixed with 4% paraformaldehyde in PBS, pH 7.3, for 10 min. Permeabilization was achieved by incubation with 0.1% triton X-100 in PBS for 5 min. F-actin was stained with 5 μg/ml phalloidin-TRITC (Sigma-Aldrich Chemie GmbH) for 40 min in the dark, followed by DNA staining with 2 μg/ml DAPI (Roth, Karlsruhe, Germany) for 15 min in the dark. Finally, the samples were mounted on glass coverslips and kept at 4 °C. Analysis was done using a confocal laser scanning microscope (TCS SP5, Leica Microsystems GmbH).
2.4. Time-of-flight secondary ion mass spectrometry
Cellular uptake of AgNPs was determined conducting Time of Flight-Secondary Ion Mass Spectrometry (ToF-SIMS). Therefore, PBMCs were seeded on silicon wavers (Silicon Materials, Kaufering, Germany) and incubated with 1000 ng/g AgNPs. After 28 days, the medium was discarded and the cell layer was fixed for 30 min at 4 °C with 2% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2–7.4) containing 2% glutardialdehyde and 0.02% picric acid. Samples were kept in the dark at 4 °C for further analysis with a TOF.SIMS 5-100 instrument (ION-TOF GmbH, Muenster, Germany). The instrument is equipped with a 25 keV Bi-cluster primary ion gun and a 10 keV C60 gun for sputtering. Further experimental details are given in [20]. Data analysis was done using the software “SurfaceLab 6.3” (ION-TOF GmbH).
2.5. Transmission electron microscopy
To define the destination of the nanoparticles inside the cell, PBMCs were incubated with 1000 ng/g AgNPs for 28 days. Subsequently, the cells were fixed for 30 min on ice with 2% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2–7.4) with 2% glutardialdehyde and 0.02% picric acid, followed by a 20 min fixation with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.2–7.4). The samples were dehydrated and embedded in epon, an epoxy resin. Ultrathin cuts were prepared and applied to collodion coated copper grids. Examination was done using a LEO 912 transmission electron microscope (Carl Zeiss AG, Oberkochen, Germany) equipped with a Slow Scan Dual Speed CCD camera TRS Sharpeye (Albert Troendle Prototypentwicklung, Moorenweis, Germany).
2.6. Real time reverse transcription polymerase chain reaction
Cells incubated with different concentrations of nanosilver were lyzed after 18 and 28 days, respectively. RNA was isolated according to the protocol of the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) and 1000 ng RNA was reversely transcribed using the cDNA QuantiTect Reverse Transcription Kit (Qiagen GmbH). Real time polymerase chain reaction was conducted applying the QuantiFast SYBR Green PCR Kit (Qiagen GmbH). All primers were purchased from Eurofins MWG Operon (Ebersberg, Germany) (Table 1). The results were finally normalized to the reference gene beta-2-microglobulin (β2MG).
Table 1.
Listing of primers used to examine changes in osteoclast gene expression.
| Gene | Role | Forward primer (5′–3′) | Reverse primer (3′–5′) |
|---|---|---|---|
| β2MG (NM_004048) | Reference | TCTCTCTTTCTGGCCTGGAG | CAACTTCAATGTCGGATGGA |
| CtsK (NM_000396.3) | Differentiation | TTCCGCAATCCCGATGGAATA | GCAAAGCTCACCACAGGTAG |
| PICALM (NM_001206946.1) | Endocytosis | ATTTTCTACCCCTAGTTCTTC | AGTAGCAGAGAAAGGATCTC |
| Nqo1 (NM_000903.2) | Oxidative stress | AGCTGGAAGCCGCAGACCTTG | TGAACACTCGCTCAAACCAGCC |
| CHOP (NM_001195053.1) | ER stress | ACCCTGCTTCTCTGGCTTGGC | GGAGAGTGAGGCCTCTGGGAG |
β2MG = beta-2-microglobulin, CtsK = cathepsin K, PICALM = phosphatidylinositol binding clathrin assembly protein, Nqo1 = NADPH quinone oxidoreductase 1, CHOP = C/EBP homologous protein.
2.7. Statistics
Statistical analysis was done using “SPSS Statistics 20.0” software (IBM Corporation, Armonk, NY, USA). Experiments were conducted as duplets and repeated five times. The treatment groups were compared to the negative control by a Mann–Whitney-U test. The results are expressed as means ± SD. p ≤ 0.05 was considered to be statistical significant.
3. Results
3.1. Nanoparticle characterization
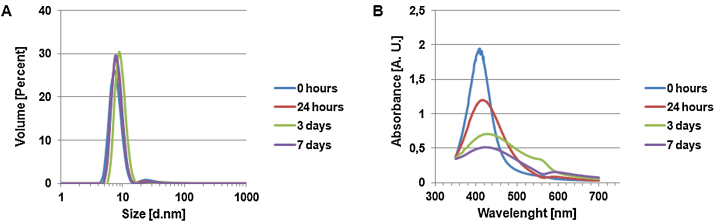
The hydrodynamic diameter of at least 95% of the nanoparticles diluted in osteoclast medium remained constantly between 5 and 10 nm, while a small amount displayed a size distribution of about 20–50 nm (Fig. 1A). The zeta potential, a term for the stability of a colloidal dispersion, was measured to be slightly negative (−6 mV), therefore indicating a tendency to nanoparticle agglomeration. These results correspond to the behavior of AgNPs diluted in deionized water [21].
Fig. 1.
Characterization of 20 μg/g AgNPs diluted in osteoclast medium. (A) AgNP size distribution was obtained by applying dynamic light scattering. (B) Nanoparticle agglomeration was examined by UV–vis spectrometry.
UV–vis analysis revealed a typical surface plasma resonance (SPR) peak at about 409 nm, slightly shifting to a higher wavelength with time. Due to particle agglomeration, a second peak is located at 560 nm shifting to about 590 nm at day 7. This was not observed for particles diluted in deionized water [21]. Additionally, a time-dependent decrease in absorbance as well as broadening of the SPR peak was detected, indicating attachment of medium components to the nanoparticle surface (Fig. 1B).
3.2. Nanoparticle-induced changes in cell morphology
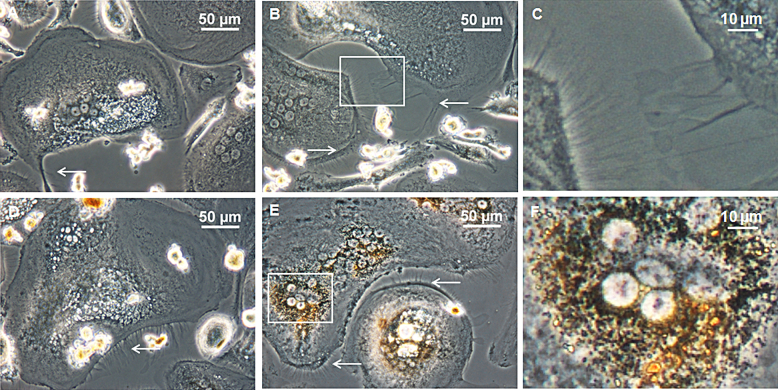
Upon incubation of the PBMCs with different concentrations of AgNPs no inhibition of preosteoclast fusion and osteoclast differentiation could be detected by light microscopy. At the early time point of 8 days, mainly two different cell types could be distinguished in the AgNP approach and the negative control: mononucleated, round-shaped cells and mononucleated, spindle-shaped cells (Data not shown). After 18 days, a third phenotype of round-shaped, multinucleated cells appeared, while the number of mononucleated cells decreased (Data not shown). After 28 days the cell culture was dominated by large, multinucleated cells of 100–250 μm. Few osteoclast-like giant cells with sizes exceeding 400 μm were detected (Fig. 2). The mature osteoclasts incubated with nanoparticles exhibit multiple filopodia and lamellipodia, while in the control the amount of those cytoplasmatic projections was reduced (Fig. 2). Additionally, orange coloring was observed in the peri-nuclear region of cells incubated with higher amounts of AgNPs, indicating nanoparticle uptake (Fig. 2F).
Fig. 2.
Morphological observation using inverted light microscopy. PBMCs were incubated with (B) 300 ng/g, (D) 1000 ng/g and (E) 3000 ng/g AgNPs for 28 days. (A) Cells without nanoparticle supplementation served as a negative control. (C) and (F) Magnified section of (B) and (E), respectively. White arrows indicate filopodia and lamellipodia.
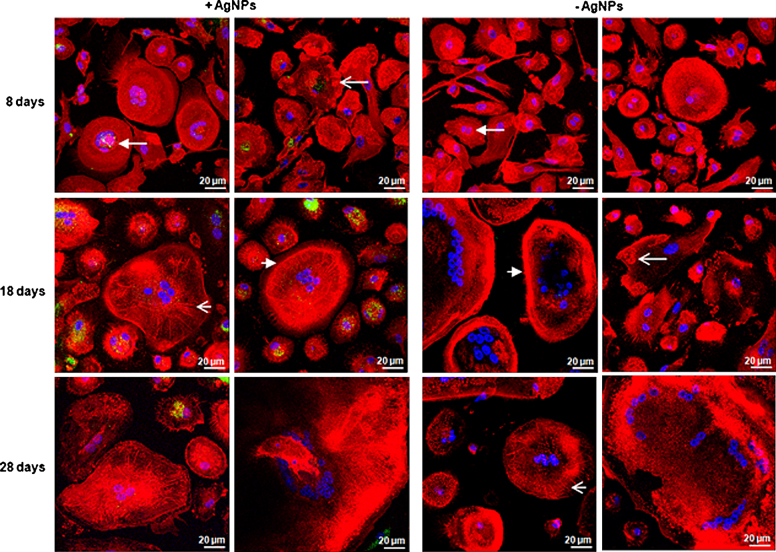
Fluorescence microscopy confirmed former observations. No inhibition in osteoclastogenesis could be detected following AgNP exposure. Contrary to the negative control, a larger number of filopodia/lamellipodia was detected, especially in the early time points. Three different podosome formations were found in all approaches: podosome clusters, podosome rings and podosome belts. Additionally, radial actin structures were visible in the AgNP approach and the negative control (Fig. 3). Based on the autofluorescence of AgNPs, the nanoparticles appeared as green dots in fluorescence microscopy. Already after 8 days AgNP uptake was visible. The nanoparticles accumulated peri-nuclear. However, the quantity of AgNPs differed in the various cell types. Especially, in the larger, multinucleated cells less AgNPs were visible (Fig. 3).
Fig. 3.
Cell morphology and nanoparticle uptake determined by fluorescence microscopy. PBMCs were incubated with 1000 ng/g AgNPs for 8, 18 or 28 days, while cells without AgNP supplementation served as a negative control. Osteoclasts showed formation of podosome clusters (open arrow), podosome rings (closed arrow), radial actin arrangements (short, open arrow) and podosome belts (short, closed arrow).
3.3. Cellular uptake of nanoparticles
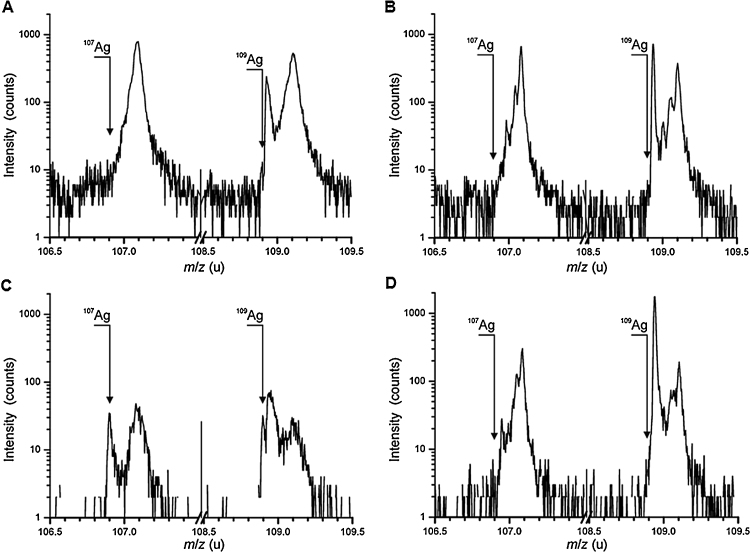
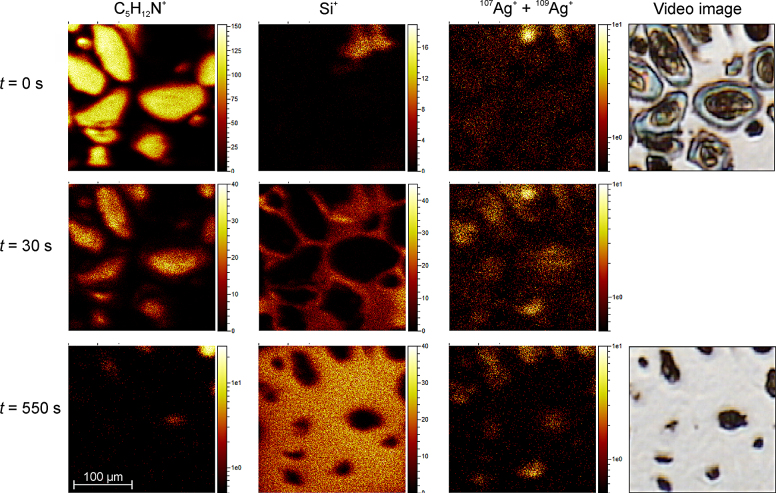
ToF-SIMS was used to perform mass spectrometric analysis of organic and inorganic components inside the cells incubated with 1000 ng/g AgNPs. Analysis revealed few amounts of silver on the outer cell membrane upon addition of AgNPs (Fig. 4A). With ongoing cell abrasion there is an increase in the signals of the silver isotopes 107Ag and 109Ag (Fig. 4C). In the control no silver could be detected associated with the membrane or located inside the cells (Fig. 4B and D). To depict the cells, the mass signal at m/z = 86.10 u (C5H12N+), a fragment of phosphatidylcholine, was chosen. Before abrasion (t = 0 s), a high C5H12N+ signal but only low silver count rates were detected directly on the cell surface. With ongoing cell abrasion (t = 30 s and t = 550 s), the signals for both silver isotopes increase. Additionally, the silicon signal is given to demonstrate removal of the cells. This silicon signal is increasing with decrease of the cell signal (Fig. 5).
Fig. 4.
Nanoparticle uptake determined by time of flight-secondary ion mass spectrometry. Osteoclasts were incubated with 1000 ng/g AgNPs for 28 days. Mass spectra of the cell surface (A) with AgNPs and (B) without AgNPs. Mass spectra of the cell interior. (C) with AgNPs and (D) without AgNPs.
Fig. 5.
Mass images obtained from time-of-flight secondary ion mass spectrometry. Osteoclasts were incubated with 1000 ng/g AgNPs for 28 days. The mass signals C5H12N+, Si+ and 107Ag+ + 109Ag+ are displayed in dependence of the sputter time. t = 0 s is a surface image and t = 550 shows the final situation after nearly complete removal of the cell. In the right column the starting and the final image of the internal video camera are depicted.
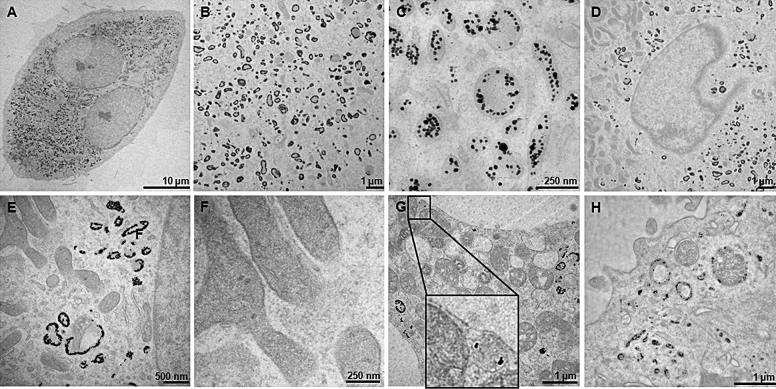
Transmission electron microscopy revealed cellular accumulation of AgNPs in larger quantities (Fig. 6A). The nanoparticles taken up by the osteoclasts are found inside endo-lysosomal structures, appearing as single nanoparticles or smaller clusters (Fig. 6B and C). Despite their small size, no nanoparticles could be found inside the nucleus (Fig. 6D). Additionally, no AgNPs were present inside organelles, like mitochondria or the endoplasmatic reticulum (ER) (Fig. 6E and F). According to ToF-SIMS, few nanoparticles were detected on the cell membrane, partly enclosed in vesicles (Fig. 6G). Furthermore, phagosome/pinosome-like structures could be observed inside the cells incubated with AgNPs (Fig. 6H).
Fig. 6.
Cellular distribution of nanoparticles examined by transmission electron microscopy. (A) Nanoparticle distribution inside osteoclasts incubated with 1000 ng/g AgNPs for 28 days. (B) AgNPs entrapped inside endo-lysosomal structures. (C) Higher magnification of (B). (D–F) Absence of particles inside organelles. (G and H) AgNPs detected inside vesicles and phagosome/pinosome-like structures.
3.4. Nanoparticle-induced changes in gene expression
Incubation with AgNPs during osteoclast differentiation left cathepsin K mRNA level unchanged for 18 days but led to a slight, not significant, decrease after 28 days. Additionally, a not significant increase in Nqo1, a marker for oxidative stress, could be observed for all tested concentrations. After 28 days Nqo1 levels were significantly elevated for the highest AgNP concentration. In contrast, no changes in ER stress, displayed by the marker CHOP, were detected at any time point. Furthermore, PICALM, a marker for clathrin-dependent endocytosis, was significantly decreased after 28 days upon incubation with 1000 ng/g and 3000 ng/g AgNPs (Fig. 7).
Fig. 7.
mRNA expression of specific osteoclast cell markers. Cells were incubated with different concentrations of AgNPs for 18 and 28 days. Results were compared to the negative control without AgNPs (broken line). *p ≤ 0.05.
No changes in gene expression were observed following incubation with the stabilizing matrix w/o AgNPs (data not shown).
4. Discussion
The increasing number of MDRO in biomaterial-associated infections necessitates the development of new antibacterial agents. In the present study we evaluated the cytocompatibility of silver nanoparticles toward osteoclasts. Albers et al. [22] demonstrated impairment of primary mouse osteoclast viability and differentiation at a concentration ≥128 μg/ml after 72 h. At sublethal concentrations stimulation of cell viability and differentiation capacity was observed. These findings could not be confirmed in our study.
Our results indicate no influence on osteoclastogenesis upon incubation of PBMCs with sublethal concentrations of AgNPs. The cells revealed no significantly changed mRNA levels of cathepsin K and displayed unaltered morphology as well as podosomal structures as described by Jurdic et al. [5]. Interestingly, additional radial actin structures could be observed in the mature osteoclasts of all approaches. Such actin arrangements have been described before by Takito et al. as zipper-like structures [23]. The author explains two different mechanisms of podosome belt formation in osteoclasts: On one hand, podosomal clusters become apparent after mononuclear cell fusion, which develop to actin rings and finally to a podosome belt. On the other hand, mononuclear cells with actin rings fuse to form large syncytia with zipper-like structures. These radial actin arrangements further break down leaving a podosome belt. The first mechanism has been described to form smaller osteoclasts, while the second mechanism can generate very large osteoclasts with more nuclei. Both mechanisms were observed in our cell culture approaches, which might explain the wide osteoclast size distribution.
A high number of filopodia/lamellipodia was visible on the osteoclast surface upon incubation with nanoparticles. Filopodia/lamellipodia presence in combination with podosomal structures is a known characteristic for osteoclasts seeded on nonosteoid materials [24]. Filopodia are thin membrane protrusions comprised of parallel bundles of filamentous actin, which are involved in a number of cellular processes, like cellular migration and adhesion [25]. Because less filopodia were seen on osteoclasts not exposed to AgNPs, additional filopodial function must be assumed. In macrophages involvement of filopodia in particle uptake has been repeatedly described [26], [27], [28]. Since nanoparticle accumulation inside osteoclasts could be confirmed in the present study, filopodial and lamellipodial involvement in particle uptake is plausible.
AgNPs uptaken by the cells end up in endo-lysosomal structures, but not in the cytosol or in organelles. However, mononucleated or small, multinucleated cells revealed a higher degree of AgNP accumulation than large, multinucleated osteoclasts. Mature osteoclasts possess a high transcytose activity that might lead to removal of uptaken nanoparticles [29]. This mechanism of AgNP removal by elevated transcytosis activity, could also explain the significantly reduced mRNA levels in clathrin-dependent endocytosis upon incubation with AgNPs. Corisdeo et al. demonstrated induction of cathepsin K mRNA expression by RANKL, which is endocytosed by a clathrin-dependent pathway [30], [31]. Therefore the reduced levels of PICALM might lead to the slight decrease in cathepsin K mRNA expression observed after incubation with AgNPs for 28 days.
In a previous study we have described the same endo-lysosomal accumulation pattern for AgNPs in mesenchymal stem cells and osteoblasts, finally leading to oxidative stress, ER stress and cell death [21]. Also in the present study nanoparticle supplementation led to induction of oxidative stress on the mRNA level. However, no nanoparticles could be detected inside mitochondria or the ER, which implies that silver ions released from the particles translocate into the organelles interfering with mitochondrial and ER function. Interestingly, ER stress, indicated by the CHOP marker, was not induced but down-regulated during AgNP exposure. These findings were unexpected and have to be investigated in further studies. Regarding long-time exposure, this time-dependent increase in oxidative stress combined with continuous cellular nanoparticle uptake might lead to reduction of the lethal concentration and to an increase in cellular toxicity over time. However, for medical application nanosilver will be firmly attached to the appropriate biomaterial preventing cellular nanoparticle uptake. Additionally, the in vivo system will ensure a steady flow of fluids reducing the local accumulation of silver nanoparticles and therefore possibly increasing the biocompatibility.
5. Conclusion
In conclusion, it can be stated that nanosilver exposure at sublethal concentrations does not induce alterations in osteoclastogenesis. However, in vitro testing led to accumulation of AgNPs and induction of the reactive oxygen species production. Therefore, further studies are needed to weigh the antibacterial benefit and the potential health risks of AgNPs.
Transparency document
Acknowledgements
The authors would like to thank Steffi Weghenkel, Ivonne Bergen and Iris Schütz for their technical assistance. Our special thanks go to ras materials GmbH, Regensburg, Germany for providing us with the nanosilver colloidal dispersion.
This work was financially supported by the Federal Ministry of Education and Research [03X0103D].
Footnotes
Available online 18 October 2014
References
- 1.Quarles L.D., Yohay D.A., Lever L.W., Caton R., Wenstrup R.J. Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J. Bone Miner. Res. 1992;7:683–692. doi: 10.1002/jbmr.5650070613. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Vaes G. Cellular biology and biochemical mechanism of bone resorption. A review of recent developments on the formation, activation, and mode of action of osteoclasts. Clin. Orthop. Relat. Res. 1988;231:239–271. [PubMed] [Google Scholar]
- 4.Yamashita D.S., Dodds R.A. Cathepsin K and the design of inhibitors of cathepsin K. Curr Pharm Des. 2000;6:1–24. doi: 10.2174/1381612003401569. [DOI] [PubMed] [Google Scholar]
- 5.Jurdic P., Saltel F., Chabadel A., Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur. J. Cell Biol. 2006;85:195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Väänänen H.K., Zhao H., Mulari M., Halleen J.M. The cell biology of osteoclast function. J. Cell. Sci. 2000;113:377–381. doi: 10.1242/jcs.113.3.377. [DOI] [PubMed] [Google Scholar]
- 7.Luxenburg C., Geblinger D., Klein E., Anderson K., Hanein D., Geiger B., Addadi L. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lentino R. Prosthetic joint infections: bane of orthopedists, challenge for infectious disease specialists. Clin. Infect. Dis. 2003;36:1157–1161. doi: 10.1086/374554. [DOI] [PubMed] [Google Scholar]
- 9.Jafari S.M., Coyle C., Mortazavi S.M., Sharkey P.F., Parvizi J. Revision hip arthroplasty: infection is the most common cause of failure. Clin. Orthop. Relat. Res. 2010;468:2046–2051. doi: 10.1007/s11999-010-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renaud A., Lavigne M., Vendittoli P.A. Periprosthetic joint infections at a teaching hospital in 1990–2007. Can. J. Surg. 2012;55:394–400. doi: 10.1503/cjs.033610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campoccia D., Montanaro L., Arciola C.R. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27:2331–2339. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y., Tai C.L., Hsieh P.H., Ueng S.W.N. Gentamicin in bone cement: a potentially more effective prophylactic measure of infection in joint arthroplasty. Bone Joint Res. 2013;2:220–226. doi: 10.1302/2046-3758.210.2000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho K.H., Park J.E., Osaka T., Park S.G. The study of antimicrobial activity and preservative effects of nanosilver ingredient. Electrochim. Acta. 2005;51:956–960. [Google Scholar]
- 14.Kim J.S., Kuk E., Yu K.N., Kim J.H., Park S.J., Lee H.J., Kim S.H., Park Y.K., Park Y.H., Hwang C.Y., Kim Y.K., Lee Y.S., Jeong D.H., Cho M.H. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 15.AshaRani P.V., Low Kah Mun G., Hande M.P., Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Gutierrez F., Olive P.L., Banuelos A., Orrantia E., Nino N., Sanchez E.M., Ruiz F., Bach H., Av-Gay Y. Synthesis, characterization, and evaluation of antimicrobial and cytotoxic effect of silver and titanium nanoparticles. Nanomedicine. 2010;6:681L 688. doi: 10.1016/j.nano.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q.L., Wu J., Chen G.Q., Cui F.Z., Kim T.N., Kim J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000;52:662–668. doi: 10.1002/1097-4636(20001215)52:4<662::aid-jbm10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Sondi I., Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004;275:177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Morones J.R. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 20.Kokesch-Himmelreich J., Schumacher M., Rohnke M., Gelinsky M., Janek J. ToF-SIMS analysis of osteoblast-like cells and their mineralized extracellular matrix on strontium enriched bone cements. Biointerphases. 2013;8:17. doi: 10.1186/1559-4106-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauksch L., Hartmann S., Rohnke M., Szalay G., Alt V., Schnettler R., Lips K.S. Biocompatibility of silver nanoparticles and silver ions in primary human mesenchymal stem cells and osteoblasts. Acta Biomater. 2014;10:439–449. doi: 10.1016/j.actbio.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 22.Albers C., Hofstetter W., Siebenrock K.A., Landmann R., Klenke F.M. In vitro cytotoxicity of silver nanoparticles on osteoblasts and osteoclasts at antibacterial concentrations. Nanotoxicology. 2011;7:30–36. doi: 10.3109/17435390.2011.626538. [DOI] [PubMed] [Google Scholar]
- 23.Takito J., Nakamura M., Yoda M., Tohmonda T., Uchikawa S., Horiuchi K., Toyama Y., Chiba K. The transient appearance of zipper-like actin superstructures during the fusion of osteoclasts. J. Cell. Sci. 2012;125:662–672. doi: 10.1242/jcs.090886. [DOI] [PubMed] [Google Scholar]
- 24.Saltel F., Destaing O., Bard F., Eichert D., Jurdic P. Apatite-mediated actin dynamics in resorbing osteoclasts. Mol. Biol. Cell. 2004;15:5231–5241. doi: 10.1091/mbc.E04-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 26.Kress H., Stelzer E.H., Holzer D., Buss F., Griffiths G., Rohrbach A. Filopodia act as phagocytic tentacles and pull with discrete steps and a load-dependent velocity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11633–11638. doi: 10.1073/pnas.0702449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Möller J., Lühmann T., Chabria M., Hall H., Vogel V. Macrophages lift off surface-bound bacteria using a filopodium-lamellipodium hook-and-shovel mechanism. Sci. Rep. 2013;3:2884. doi: 10.1038/srep02884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vonna L., Wiedemann A., Aepfelbacher M., Sackmann E. Micromechanics of filopodia mediated capture of pathogens by macrophages. Eur. Biophys. J. 2007;36:145–151. doi: 10.1007/s00249-006-0118-y. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H. Membrane trafficking in osteoblasts and osteoclasts: new avenues for understanding and treating skeletal diseases. Traffic. 2012;13:1307–1314. doi: 10.1111/j.1600-0854.2012.01395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narducci P., Bortul R., Bareggi R., Nicolin V. Clathrin-dependent endocytosis of membrane-bound RANKL in differentiated osteoclasts. Eur. J. Histochem. 2010;54:e6. doi: 10.4081/ejh.2010.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corisdeo S., Gyda M., Zaidi M., Moonga B.S., Troen B.R. New insights into the regulation of cathepsin K gene expression by osteoprotegerin ligand. Biochem. Biophys. Res. Commun. 2001;285:335–339. doi: 10.1006/bbrc.2001.5127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.